Bacteria remain a major cause of human disease and a formidable foe of the mammalian immune system. Bacter-ial entry into normally sterile compartments has long been recognized as a potential calamity for the mammalian host, in the most extreme circumstances resulting in circulatory collapse and multiorgan failure. In contrast to viruses, most bacteria are self-sufficient, producing their own lipid membranes, cell walls, nucleic acids, and proteins. Although many synthetic pathways used by bacteria and mammals are similar, others are subtly different and some are completely distinct. The mammalian immune system has evolved an array of mechanisms to recognize and respond to these distinct bacterial products upon infection. While it has been appreciated for over a century that the mammalian immune system uses receptors to specifically recognize bacterial products and initiate inflammatory defenses upon infection, the last decade has witnessed the molecular characterization of a number of remarkable pathways of immune defense against bacteria.
Recognition of signature bacterial products has generally been considered the domain of the innate immune system 1. Classically, innate immune receptors for bacterially derived compounds are encoded in the germline and do not require recombination for function. While it has been estimated that there may be roughly 100 distinct innate immune receptors (also known as pattern recognition receptors [PRRs]), this number is quite small when compared with the 1014–1018 distinct receptors that can be generated by the adaptive immune system 1. Two of the earliest identified receptors for bacterial products are the scavenger receptor (SR) and the mannose receptor (MR). These receptors function in phagocytosis and endosomal targeting and are found in macrophages, dendritic cells, and, in certain tissues, endothelial cells. MR is a 175-kD type I membrane glycoprotein from the calcium-dependent lectin family that specifically recognizes carbohydrates bearing large numbers of α-linked oligo-mannoses, a feature characteristic of microbial carbohydrates. MR uptake can lead to presentation in the context of MHC class II molecules and CD1 2 and can induce cytocidal and proinflammatory mechanisms. SRs consist of at least five classes of receptors with distinct structures and ligand binding properties. Class A SRs are homotrimeric membrane proteins that bind diverse ligands, including bacterial cell wall components from Gram-negative (LPS) or Gram-positive bacteria (lipotei-choic acid [LTA]) 3 4. As would be expected, SR-deficient mice are more susceptible to Listeria monocytogenes or Staphylococcus aureus infection and septic shock 5 6.
The recently identified family of mammalian toll-like receptors (TLRs) and the elucidation of some of their specificities has dramatically enhanced our understanding of the innate immune system 7. It is now clear that TLRs serve as recognition and signaling elements for bacterial substances as diverse as lipoproteins, peptidoglycan, LPS, LTA, and bacterial DNA. Thus, TLR-4 is the essential recognition and signaling element for LPS, while TLR-2 and TLR-6 play a role in recognition of bacterial lipoproteins 8. TLR-2 has been implicated in the recognition of bacterial peptidoglycan and lipoteichoic acids 9. Recently, the inflammatory and adjuvant effects of bacterial DNA were attributed to recognition of hypomethylated CpG sequences by the TLR-9 receptor 10.
Many decades ago, investigators recognized that bacterial culture supernatants harbored substances that readily induce inflammation by recruiting neutrophils to sites of bacterial infection and inducing their degranulation 11. Biochemical analyses revealed that the active components of this process were bacterial peptides, specifically small, hydrophobic peptides containing N-formyl methionine on the NH2 terminus 12. Even tripeptides, such as formyl-Met-Leu-Phe, could act as potent neutrophil and macrophage chemoattractants 13, and subsequently it was found that humans have two receptors for these bacterial products, now called the formyl peptide receptors (FPR1 and FPRL1). These receptors, like other receptors associated with chemotaxis, contain seven-transmembrane domains and are coupled to G proteins 14. Interestingly, although mice and humans both have versions of these receptors, the human receptor has ∼100-fold higher affinity for the f-Met peptides and is correspondingly more effective at directing inflammatory cells to sites of infection. Nevertheless, deletion of this receptor from mice results in enhanced susceptibility to some bacterial infections 15.
In contrast to innate immunity, adaptive immunity relies on recombined antigen receptors and their resulting greater diversity to recognize bacterial antigens. At the T cell level, receptor-mediated recognition was thought to be exclusive for peptides derived from the degradation of pathogen-derived proteins. However, this idea was turned on its head when Porcelli and Brenner 16 demonstrated that a subset of T lymphocytes recognize lipid antigens in the context of CD1, a non-MHC encoded transmembrane protein with great structural similarity to MHC class I molecules. Thus, lipids such as mycolic acid 17, derived from Mycobacterium tuberculosis, and lipoarabinomanan 18, derived from Mycobacterium leprae, are bound by CD1 and recognized by a range of T lymphocytes. Although it remains to be proven that these T cells play a role in protective immunity to mycobacterial infection, the concept that T lymphocyte specificity is exclusive for pathogen-derived peptides has been laid to rest.
The idea of selectively presenting signature bacterial molecules to T cells is not exclusive to CD1, however. Nearly a decade ago, the H2-M3 MHC class Ib molecule was found to present bacterially derived N-formyl methionine–containing peptides to CD8 T lymphocytes 19 20. This fascinating molecule had been discovered earlier as a restriction element for a very small population of peptides derived from the NH2 terminus of mitochondrially encoded proteins 21. The finding that N-formyl methionine was essential for high-affinity binding by H2-M3 suggested that this molecule might play a role in defense against bacterial infection 22, a good idea that was demonstrated to be correct in the setting of L. monocytogenes infection 19 20. Subsequent identifications of the antigenic L. monocytogenes–derived N-formyl peptides have enabled kinetic analyses of H2-M3–restricted T cell responses during bacterial infection 23 24 25. Two studies using highly quantitative methods to enumerate antigen-specific T cells have demonstrated that H2-M3–restricted T cell responses are substantial and occur ∼2 d earlier than CD8 T cell responses restricted by conventional MHC class Ia molecules 26 27. The large number of H2-M3–restricted T cells, achieving frequencies of 5–10% of CD8 T cells in some mouse strains, and their peak presence in vivo at the time of bacterial clearance suggest an important role for these T cells in the development of sterilizing immunity. H2-M3–restricted T cells are cytolytic and produce TNF and IFN-γ, cytokines associated with antilisterial immunity, further supporting the concept that H2-M3–restricted T cells participate in bacterial clearance 28. The most direct evidence for the role of H2-M3–restricted T cells in protective immunity comes from recent studies by Seaman et al. 29. Using mice that lack MHC class Ia molecules, they demonstrated that bacterial clearance depends almost exclusively on H2-M3–restricted CD8 T lymphocytes. That H2-M3–restricted T cells can provide protective immunity has also been demonstrated with the adoptive transfer of specific T cells 30.
How N-formyl methionine peptides are processed and presented by H2-M3 is unclear. Early studies by Kaufmann et al. 31 demonstrated that macrophages pulsed with heat-killed L. monocytogenes stimulate “MHC-unrestricted” (i.e., what were almost certainly H2-M–restricted) T cells, a finding that has been confirmed more recently with H2-M3–restricted CTL clones of defined peptide specificity 32. While presentation of mitochondrial peptides by H2-M3 appears to depend upon conventional transport from the cytosol into the endoplasmic reticulum (ER) by the transporter associated with antigen processing (TAP) 33, this does not appear to be absolutely necessary for the presentation of L. monocytogenes–derived peptides 32. For example, macrophages derived from TAP−/− mice are capable of presenting fMIGWII, an L. monocytogenes–derived peptide, to CD8 T cells 32.
Trafficking of newly synthesized H2-M3 molecules differs significantly from that of conventional MHC class Ia molecules. Using a specific mAb, Chiu et al. 34 found that while H2-M3 surface expression is very low in uninfected cells, exogenous administration of N-formyl methionine peptides dramatically increases surface expression. Remarkably, upregulation of most H2-M3 expression is TAP dependent, suggesting that exogenous peptides enter the cytosol and are transported into the ER by TAP. H2-M3 molecules appear to be sequestered in the ER for lack of N-formyl methionine–containing peptide ligands and are released for trafficking to the cell surface upon binding of bacterial peptides 34. This state of readiness, particularly for pathogen-derived peptides, distinguishes H2-M3 from other MHC class I molecules. In the setting of bacterial infection, therefore, the bulk of H2-M3 molecules on the cell surface will present pathogen-derived peptides, in contrast to MHC class Ia molecules, which overwhelmingly present peptides derived from the degradation of self-proteins (Fig. 1).
Figure 1.
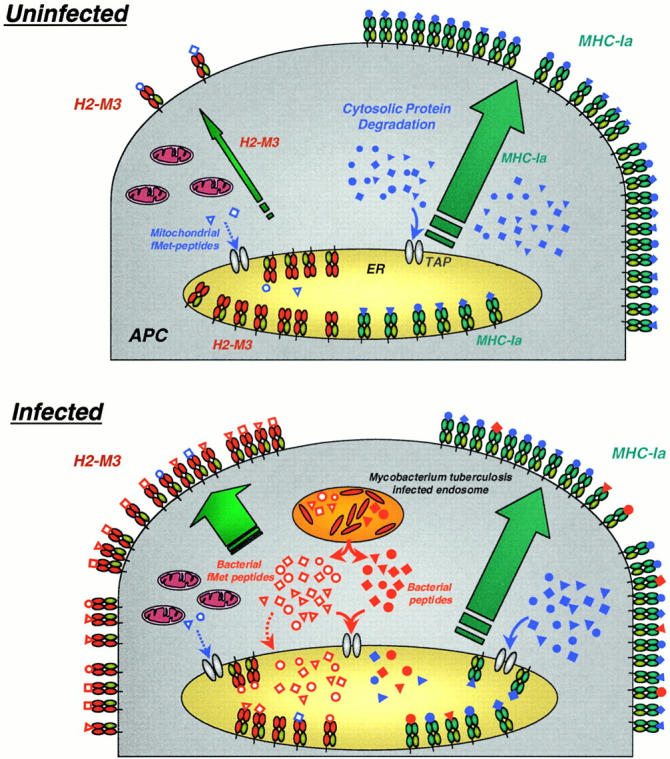
H2-M3 mobilization upon bacterial infection. Under normal circumstances (top panel), endogenous self-peptides (closed blue symbols) are generated in the cytosol and presented by classical MHC class Ia molecules. In contrast, empty class Ib molecules (H2-M3) are mostly retained in the ER, while a few mitochondrial formylated peptides are bound and presented by H2-M3 at the cell surface. After bacterial infection (bottom panel), empty ER-resident H2-M3 molecules are rapidly loaded with bacterially secreted formylated peptides (open red symbols) and form the majority of H2-M3 complexes on the cell surface. By contrast, peptides derived from bacterial antigens (closed red symbols) that are presented by MHC class Ia molecules are rare when compared with self-derived peptides.
Although it has been suspected that H2-M3 presents peptides from many classes of bacteria, until now the only pathogen for which H2-M3–mediated antigen presentation has been definitively demonstrated is L. monocytogenes. This has changed with the demonstration by Chun et al. 35 in this issue that N-formyl methionine peptides are presented by macrophages infected with M. tuberculosis. Using the recently sequenced M. tuberculosis genome database as a starting point, these investigators scanned for hydrophobic peptide sequences containing N-formyl methionine and otherwise conforming to the H2-M3 binding groove. The likelihood of successfully predicting which peptides are bound by H2-M3 was enhanced by knowledge of the H2-M3 crystal structure 36. After identifying a number of peptides, Chun et al. identified those that bound H2-M3 with the greatest affinity and further characterized these. Remarkably, T cell lines generated by peptide immunization of naive mice recognized and lysed M. tuberculosis–infected macrophages in an H2-M3–restricted fashion, indicating that mycobacterial peptides are presented on the macrophage surface. Importantly, mice infected with M. tuberculosis harbored formyl peptide–specific T lymphocytes that produced IFN-γ upon in vitro stimulation. This remarkable series of experiments demonstrates the existence of H2-M3–restricted T cells in the setting of M. tuberculosis infection, suggesting a role for H2-M3–restricted T cells in defense against one of the most important bacterial pathogens of humankind.
In contrast to L. monocytogenes, which enters the cytosol of infected cells by lysing the phagosome 37, M. tuberculosis resides in a vacuole during intracellular infection. Despite this important difference, N-formyl methionine peptides from both pathogens are presented by H2-M3. The finding that presentation of peptides by H2-M3 can be both TAP dependent and independent suggests that there may be several trafficking routes directing peptides into the H2-M3 groove. Thus, it is possible that M. tuberculosis–derived peptides access the cytosol and enter the ER in a TAP-dependent fashion. On the other hand, it is possible that peptides derived from M. tuberculosis follow the TAP-independent pathway demonstrated for peptides derived from heat-killed L. monocytogenes. An important difference between live mycobacteria and heat-killed L. monocytogenes is that the former use a clever mechanism to modify the vacuole, specifically by inhibiting acidification by excluding the proton ATPase, which may interfere with MHC class II antigen processing 38. Evading presentation by H2-M3 may be more difficult, perhaps in part due to their intracellular accumulation.
H2-M3, FPR, TLR, and CD1 have shown us that selective binding and recognition of bacterial molecules is a host defense strategy shared by the innate and adaptive arms of the mammalian immune system. Remarkably, in the setting of infection, CD1 molecules very likely bind the same glycolipid ligands as TLRs. Indeed, alterations in the lipid moiety of mycolic acids result in aberrant inflammatory responses that may, in part, reflect the roles of CD1 or TLRs in defense against M. tuberculosis 39. H2-M3 also shares the pool of bacterially secreted N-formyl methionine peptides with the chemotactic formyl peptide receptor. While it is unclear whether competition between innate and adaptive immune receptors for the same bacterial molecules occurs in vivo, it is conceivable that at limiting concentrations, receptors on highly prevalent cells such as neutrophils could deplete the local microenvironment of peptides for binding by less prevalent receptors. This kind of competition, therefore, potentially provides a mechanism for modulating the relative contributions of innate and adaptive inflammatory responses to bacterial infection. The convergent focus of different immune effector arms on the same pathogen-derived molecules underlines their importance as signals of infection but also suggests that the range of distinct bacterial molecules that can serve as such targets may be limited.
Although immunization with live, attenuated vaccines can be highly effective, the issue of potential adverse effects in immunocompromised individuals is always a bothersome concern. Therefore, the concept of immunizing and inducing protective immunity with nonliving components is tantalizing, but, in the case of intracellular bacterial infections such as M. tuberculosis, of unproven utility. Antigens bound by molecules such as CD1 and H2-M3 are particularly attractive as vaccine targets because they induce T cell–mediated immunity, but in a fashion that is not constrained by conventional MHC restriction. Therefore, the same bacterial molecules could potentially be used to immunize a population of individuals with diverse MHC haplotypes. It is even possible, since these molecules are detected by both the innate and adaptive immune systems, that these antigens could serve as their own adjuvants. In this context, the finding that a murine MHC class Ib molecule plays a role in the immune response to M. tuberculosis, arguably humanity's worst bacterial enemy, is exciting.
Chun et al. have taken advantage of two of the great advances of the last decade: the complete genome sequences of several important human pathogens and the increasing number of structurally defined, immune-related molecules. In the coming years, these invaluable databases will increasingly be recognized as indispensable tools for the study of antimicrobial immunity. To be sure, we have to remain clever as we investigate immune responses to bacterial pathogens, but it will certainly be a pleasure to replace months spent at the bench purifying antigenic peptides with a short trip on a laptop to the genome databases.
References
- Medzhitov R., Janeway C. Innate immunity. N. Engl. J. Med. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- Prigozy T.I., Sieling P.A., Clemens D., Stewart P.L., Behar D.M., Porcelli S.A., Brenner M.B., Modlin R.L., Kronenberg M. The mannose receptor delivers lipoglycan antigens to endosomes for presentation to T cells by CD1b molecules. Immunity. 1997;6:187–197. doi: 10.1016/s1074-7613(00)80425-2. [DOI] [PubMed] [Google Scholar]
- Hampton R., Golenbock D., Penman M., Krieger M., Raetz C. Recognition and plasma clearance of endotoxin by scavenger receptors. Nature. 1991;352:342–347. doi: 10.1038/352342a0. [DOI] [PubMed] [Google Scholar]
- Dunne D., Resnick D., Greenberg J., Krieger M., Joiner K. The type I macrophage scavenger receptor binds to Gram-positive bacteria and recognizes lipoteichoic acid. Proc. Natl. Acad. Sci. USA. 1994;91:1863–1867. doi: 10.1073/pnas.91.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Kurihara Y., Takeya M., Kamada N., Kataoka M., Jishage K., Ueda O., Sakaguchi H., Higashi T., Suzuki T. A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature. 1997;386:292–296. doi: 10.1038/386292a0. [DOI] [PubMed] [Google Scholar]
- Thomas C., Li Y., Kodama T., Suzuki H., Silverstein S., Khoury J.E. Protection from lethal Gram-positive infection by macrophage scavenger receptor–dependent phagocytosis. J. Exp. Med. 2000;191:147–156. doi: 10.1084/jem.191.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Janeway C. Innate immunity recognitionmechanisms and pathways. Immunol. Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- Ozinsky A., Underhill D., Fontenot J., Hajjar A., Smith K., Wilson C., Schroeder L., Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc. Natl. Acad. Sci. USA. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill D.M., Ozinsky A., Hajjar A.M., Stevens A., Wilson C.B., Bassetti M., Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:659–665. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Ward P., Lepow I., Newman L. Bacterial factors chemotactic for polymorphonuclear leukocytes. Am. J. Pathol. 1968;52:725–736. [PMC free article] [PubMed] [Google Scholar]
- Schiffmann E., Showell H., Corcoran B., Ward P., Smith E., Becker E. The isolation and partial characterization of neutrophil chemotactic factors from Escherichia coli . J. Immunol. 1975;114:1831–1837. [PubMed] [Google Scholar]
- Marasco W., Phan S., Krutzsch H., Showell H., Feltner D., Nairn R., Becker E., Ward P. Purification and identification of formyl-methionyl-leucyl-phenylalanine as the major peptide neutrophil chemotactic factor produced by Escherichia coli . J. Biol. Chem. 1984;259:5430–5439. [PubMed] [Google Scholar]
- Murphy P. The molecular biology of leukocyte chemoattractant receptors. Annu. Rev. Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- Gao J., Lee E., Murphy P. Impaired antibacterial host defense in mice lacking the N-formylpeptide receptor. J. Exp. Med. 1999;189:657–662. doi: 10.1084/jem.189.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcelli S.A., Morita C.T., Brenner M.B. CD1b restricts the response of human CD4-8- T lymphocytes to a microbial antigen. Nature. 1992;360:593–597. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- Beckman E.M., Porcelli S.A., Morita C.T., Behar S.M., Furlong S.T., Brenner M.B. Recognition of a lipid antigen by CD1-restricted alpha/beta T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- Sieling P.A., Chatterjee D., Porcelli S.A., Prigozy T.I., Mazzaccaro R.J., Soriano T., Bloom B.R., Brenner M.B., Kronenberg M., Brennan P.J. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- Pamer E.G., Wang C.R., Flaherty L., Lindahl K.F., Bevan M.J. H2-M3 presents a Listeria monocytogenes peptide to cytotoxic T lymphocytes. Cell. 1992;70:215–223. doi: 10.1016/0092-8674(92)90097-v. [DOI] [PubMed] [Google Scholar]
- Kurlander R.J., Shawar S.M., Brown M.L., Rich R.R. Specialized role for a murine class I-b MHC molecule in prokaryotic host defenses. Science. 1992;257:678–681. doi: 10.1126/science.1496381. [DOI] [PubMed] [Google Scholar]
- Loveland B.E., Wang C.R., Yonekawa H., Hermel E., Lindahl K.F. Maternally transmitted histocompatibility antigen of micea hydrophobic peptide of a mitochondrially encoded protein. Cell. 1990;60:971–980. doi: 10.1016/0092-8674(90)90345-f. [DOI] [PubMed] [Google Scholar]
- Shawar S.M., Cook R.G., Rodgers J.R., Rich R.R. Specialized functions of MHC class I molecules. I. An N-formyl peptide receptor is required for construction of the class I antigen Mta. J. Exp. Med. 1990;171:897–912. doi: 10.1084/jem.171.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz L.L., Dere B., Bevan M.J. Identification of an H2-M3-restricted Listeria epitopeimplications for antigen presentation by M3. Immunity. 1996;5:63–72. doi: 10.1016/s1074-7613(00)80310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulden P.H., Fischer P., Sherman N.E., Wang W., Engelhard V.H., Shabanowitz J., Hunt D.H., Pamer E.G. A Listeria monocytogenes pentapeptide is presented to cytolytic T lymphocytes by the H2-M3 MHC class Ib molecule. Immunity. 1996;5:73–79. doi: 10.1016/s1074-7613(00)80311-8. [DOI] [PubMed] [Google Scholar]
- Princiotta M.F., Lenz L.L., Bevan M.J., Staerz U.D. H2-M3 restricted presentation of a Listeria-derived leader peptide. J Exp Med. 1998;187:1711–1719. doi: 10.1084/jem.187.10.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerksiek K.M., Busch D.H., Pilip I.M., Allen S.E., Pamer E.G. H2-M3–restricted T cells in bacterial infectionrapid primary but diminished memory responses. J. Exp. Med. 1999;190:195–204. doi: 10.1084/jem.190.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M., Wang C., Forman J. MHC class Ib-restricted CTL provide protection against primary and secondary Listeria monocytogenes infection. J. Immunol. 2000;165:5192–5201. doi: 10.4049/jimmunol.165.9.5192. [DOI] [PubMed] [Google Scholar]
- Kerksiek K., Busch D., Pamer E. Variable immunodominance hierarchies for H2-M3 restricted peptides following bacterial infection. J. Immunol. 2001;166:1132–1140. doi: 10.4049/jimmunol.166.2.1132. [DOI] [PubMed] [Google Scholar]
- Seaman M., Perarnau B., Lindahl K.F., Lemonnier F., Forman J. Response to Listeria monocytogenes in mice lacking MHC class Ia molecules. J. Immunol. 1999;162:5429–5436. [PubMed] [Google Scholar]
- Rolph M., Kaufmann S. Partially TAP-dependent protection against Listeria monocytogenes by H2-M3–restricted CD8 T cells. J. Immunol. 2000;165:4575–4580. doi: 10.4049/jimmunol.165.8.4575. [DOI] [PubMed] [Google Scholar]
- Kaufmann S.H.E., Rodewald H.R., Hug E., Libero G.D. Cloned Listeria monocytogenes specific non-MHC-restricted Lyt-2+ T cells with cytolytic and protective activity. J. Immunol. 1988;140:3173–3179. [PubMed] [Google Scholar]
- Lenz L.L., Bevan M.J. H2-M3 restricted presentation of Listeria monocytogenes antigens. Immunol. Rev. 1996;151:107–121. doi: 10.1111/j.1600-065x.1996.tb00705.x. [DOI] [PubMed] [Google Scholar]
- Attaya M., Jameson S., Martinez C.K., Hermel E., Aldrich C., Forman J., Lindahl K.F., Bevan M.J., Monaco J.J. Ham-2 corrects the class I antigen-processing defect in RMA-S cells. Nature. 1992;355:647–649. doi: 10.1038/355647a0. [DOI] [PubMed] [Google Scholar]
- Chiu N.M., Chun T., Fay M., Mandal M., Wang C.R. The majority of H2-M3 is retained intracellularly in a peptide-receptive state and traffics to the cell surface in the presence of N-formylated peptides. J. Exp. Med. 1999;190:423–434. doi: 10.1084/jem.190.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T., Serbina N.V., Nolt D., Wang B., Chiu N.M., Flynn J.L., Wang C.-R. Induction of M3-restricted cytotoxic T lymphocyte responses by N-formylated peptides derived from Mycobacterium tuberculosis . J. Exp. Med. 2001;193:1213–1220. doi: 10.1084/jem.193.10.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.R., Castano A.R., Peterson P.A., Slaughter C., Lindahl K.F., Deisenhofer J. Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3. Cell. 1995;82:655–664. doi: 10.1016/0092-8674(95)90037-3. [DOI] [PubMed] [Google Scholar]
- Portnoy D.A., Chakraborty T., Goebel W., Cossart P. Molecular determinants of Listeria monocytogenes pathogenesis. Infect. Immun. 1992;60:1263–1267. doi: 10.1128/iai.60.4.1263-1267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill-Koszycki S., Schlesinger P., Chakraborty P., Haddix P., Collins H., Fok A., Allen R., Gluck S., Heuser J., Russell D. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- Glickman M.S., Cox J.S., Jacobs W. A novel mycolic acid cyclopropane synthetase is required for coding, persistence and virulence of Mycobacterium tuberculosis . Mol. Cell. 2000;5:717–727. doi: 10.1016/s1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]


