Abstract
TWEAK, a new member of the tumor necrosis factor (TNF) family, induces cell death in some tumor cell lines, but its physiological functions are largely unknown. In this study, we investigated the expression and function of TWEAK in human peripheral blood mononuclear cells (PBMCs) by using newly generated anti–human TWEAK mAbs. Although freshly isolated PBMCs expressed no detectable level of TWEAK on their surfaces, a remarkable TWEAK expression was rapidly observed on monocytes upon stimulation with interferon (IFN)-γ but not with IFN-α or lipopolysaccharide. Cytotoxic activity of IFN-γ–stimulated monocytes against human squamous carcinoma cell line HSC3 was inhibited partially by anti-TWEAK mAb alone and almost completely by combination with anti-TRAIL (TNF-related apoptosis-inducing ligand) mAb. These results revealed a novel pathway of monocyte cytotoxicity against tumor cells that is mediated by TWEAK and potentiated by IFN-γ.
Keywords: TWEAK, TRAIL, IFN-γ, monocyte, cytotoxicity
Introduction
Some members of the TNF family, such as TNF-α, Fas ligand (FasL), and TNF-related apoptosis-inducing ligand (TRAIL) induce apoptotic cell death in a variety of tumor cells and nontransformed cells 1 2 3. These molecules have been characterized as playing critical roles in regulating immune responses and in the pathogenesis of various diseases 1 2 3. TWEAK is a recently identified member of the TNF family that exhibited cytotoxic activity against some tumor cell lines in vitro 4 5 6. It has also been reported that TWEAK activated nuclear factor (NF)-κB in some tumor cells 4 and induced proliferation of endothelial cells and angiogenesis 7. TWEAK mRNA has been found in various tissues and cell lines 4. However, its expression at the protein level and physiological roles remain largely unknown. It has been reported that TWEAK bound to DR3/TRAMP/LARD/APO-3/WSL1 5, which is a member of the TNFR family containing cytoplasmic death domain (DD) homologous to that in TNFR-I, Fas, TRAIL-R1, and TRAIL-R2 8. Like TNFR-I, DR3 could induce both apoptosis and NF-κB activation by recruiting TNFR-I–associated DD protein (TRADD), TNFR-associated factor (TRAF) 2, Fas-associated DD protein (FADD), and caspase-8 8. However, some conflicting results have been reported as to the TWEAK–DR3 interaction. For example, a study by Chicheportiche et al. 4 failed to demonstrate the binding of TWEAK to DR3. In addition, it has been reported that TWEAK induced apoptosis in KYM-1 cells lacking DR3 mRNA expression 6. These reports suggest the existence of unidentified TWEAK receptor distinct from DR3.
It is well known that monocytes play important roles in innate immunity and inflammatory responses via production of various cytokines and chemical mediators. In addition, previous studies indicated that monocytes kill some tumor cells directly and that IFNs are an effective inducer of monocyte cytotoxicity 9 10. TNF-α and nitric oxide (NO) have been considered to be the major mediators for IFN-stimulated monocyte cytotoxicity 9 11. However, there are conflicting data as to the involvement of TNF-α and NO, especially in the human system. For example, it has been reported that both IFN-α and IFN-γ failed to induce NO release and TNF-α production by human monocytes 12 13. Recently, Griffith et al. 13 have shown a critical contribution of TRAIL to the IFN-α– or IFN-γ–activated monocyte cytotoxicity against TRAIL-sensitive tumor cells. In that study, they demonstrated that similar levels of TRAIL expression could be induced on monocytes upon stimulation with IFN-α or IFN-γ. However, in general, IFN-γ is a much more potent inducer of monocyte cytotoxicity than IFN-α 10. This suggests a possible existence of additional death-inducing molecule(s) on monocytes that can be specifically inducible upon IFN-γ stimulation.
In this study, by using newly generated neutralizing mAbs against human (h)TWEAK, we examined the expression and function of TWEAK in human PBMCs. TWEAK was not found on freshly isolated PBMCs but was preferentially inducible on monocytes upon IFN-γ stimulation. We demonstrated that TWEAK is at least partly responsible for the IFN-γ–stimulated monocyte cytotoxicity against TWEAK-sensitive tumor cells. These results suggested possible roles for TWEAK in antitumor activity and inflammatory responses mediated by IFN-γ–stimulated monocytes.
Materials and Methods
Cells.
Mouse B lymphoma 2PK-3, T lymphoma L5178Y, myeloma P3U1 (P3X63Ag8U.1), and human colon adenocarcinoma HT-29 were obtained from American Type Culture Collection and cultured in RPMI 1640 containing 10% FCS, 100 μg/ml streptomycin and penicillin, and 2 mM glutamine (culture medium). Human oral squamous cell carcinoma HSC3 and gastric adenocarcinoma KATO-III were obtained from Japan Cancer Research Bank and maintained in the culture medium. 2PK-3–derived transfectants, including hTWEAK/2PK-3, hTRAIL/2PK-3 14, and hFasL/2PK-3 15, were also maintained in the culture medium.
Reagents.
Human IFN-γ, GM-CSF, TNF-α, IL-4, IL-15, and anti–TNF-α mAb (mAb1) were purchased from PharMingen. Human IL-12 was purchased from R & D Systems. Human IL-2, IFN-α, and IL-18 were provided by Shionogi, Toray, and Drs. H. Tsutsui, H. Okamura, and K. Nakanishi (Hyogo College of Medicine, Nishinomiya, Japan), respectively. PHA and LPS were purchased from Wako Pure Chemical, Ltd. Anti-CD3 mAb was purified from the supernatant of the hybridoma (OKT-3) purchased from American Type Culture Collection. A neutralizing anti–human TRAIL mAb (RIK-2, mouse IgG1/κ) was prepared as described previously 14. Control mouse IgG3 mAb (J606) and mouse IgM mAb (C48-6) were purchased from PharMingen.
Construction and Preparation of Soluble CD8–TWEAK Fusion Protein.
hTWEAK cDNA was prepared by reverse transcription PCR amplification of total RNA from human PBMCs with an oligonucleotide primer corresponding to the first six codons as the 5′ primer and that corresponding to the last six codons as the 3′ primer, according to the published sequence 4. The PCR product of 750 bp was subcloned into pCR3.1 TA cloning vector (Invitrogen), and the nucleotide sequence was confirmed using an automated sequencer (Applied Biosystems) and a fluoresceinated dye terminator cycle sequencing method. The cDNA encoding extracellular region of hTWEAK (amino acids 93–249) was amplified by PCR from the pCR3.1 plasmid carrying hTWEAK cDNA using AGAAGTGCACCTAAAGGC as the 5′ primer and GAGAAGGTCCAAGTGACT as the 3′ primer. EcoRV and NotI sites were introduced into the 5′ and 3′ primers, respectively. After EcoRV and NotI digestion, the PCR product of 470 bp was subcloned into HindIII- and NotI-digested pBluescript II SK (+), together with the 700-bp HindIII–EcoRV fragment encoding the extracellular region of human CD8α, which was derived from pGEM3 carrying human CD8α cDNA 16 (provided by H. Nakauchi, Tsukuba University, Tsukuba, Japan). This results in in-frame fusion of the human CD8α extracellular region to TWEAK (CD8–TWEAK). After confirmation of nucleotide sequence, the 1.2-kb XhoI–NotI fragment containing the fusion construct was transferred into XhoI–NotI sites of pMKITNeo (provided by Dr. K. Maruyama, Tokyo Medical and Dental University, Tokyo, Japan). COS7 cells were transfected with CD8–TWEAK/pMKITNeo by the standard DEAE dextran method. After 72-h culture, the supernatant was collected, and the concentration of CD8–TWEAK soluble fusion protein was evaluated by sandwich ELISA using two anti-CD8α mAbs (RPA-T8 and biotinylated OKT8) as described previously 16. Affinity-purified CD8–FasL 16 was used as standard.
Preparation of hTWEAK Transfectants.
hTWEAK/pCR3.1 vector was transfected into 2PK-3 cells (350 V, 800 μF) with a Gene Pulser (Bio-Rad Laboratories). After selection with 1 mg/ml G418 and cloning by limiting dilution, a stable transfectant, designated as hTWEAK/2PK-3, was selected by cytotoxic assay as described below. In a similar way, L5178Y cells stably expressing hTWEAK (hTWEAK/L5178Y) were prepared.
Generation of Anti-hTWEAK mAbs.
6-wk-old female BALB/c mice (Clear Japan) were immunized by intraperitoneal injection of hTWEAK/2PK-3 (107 cells) four times at 10-d intervals. 3 d after the final immunization, the splenocytes were fused with P3U1 mouse myeloma cells as described previously 14. After HAT selection, the mAbs that inhibited cytotoxic activity of CD8–TWEAK against HSC3 cells were screened. Two mAbs (CARL-1 and CARL-2) were identified by their inhibitory effects and cloned by limiting dilution. CARL-1 (mouse IgG3/κ) and CARL-2 (mouse IgM/κ) were purified from culture supernatants with a protein G column (Amersham Pharmacia Biotech) and a protein L column (CLONTECH Laboratories, Inc.), respectively.
51Cr-Release Assay.
51Cr-labeled target cells (104) and effector cells were mixed in U-bottomed wells of a 96-well microtiter plate at the indicated E/T ratios. After 8 or 18 h of incubation, cell-free supernatants were collected, and radioactivity was measured in a gamma counter. The percentage of specific 51Cr release was calculated as described before 14. RIK-2, CARL-1, and/or anti–TNF-α mAbs were added to a final concentration of 10 μg/ml at the start of cytotoxic assay.
Cell Viability Assay.
Tumor cells (5 × 103) were cultured with or without the indicated doses of CD8–TWEAK and/or IFN-γ (20 ng/ml) for 36 h in flat-bottomed wells of a 96-well microtiter plate. The cell viability was then determined by WST assay using a Cell Counting kit (Wako Pure Chemical, Ltd.) according to the manufacturer's instructions.
Flow Cytometric Analysis.
2PK-3 and hTWEAK/2PK-3 cells (106 cells) were incubated with 0.5 μg of biotinylated mAb for 1 h at 4°C, followed by PE-labeled avidin (PharMingen). After washing with PBS, the cells were analyzed on a FACSCalibur™ (Becton Dickinson), and the data were processed by using the CELLQuest™ program (Becton Dickinson). In some experiments, PBMCs or purified monocytes were stained with 0.5 μg of biotinylated mAbs, followed by PE-labeled avidin and Cy-Chrome–labeled anti-CD3 mAb, FITC-labeled anti-CD19, anti-CD56, or anti-CD14 mAb (PharMingen).
Cell Preparation and Activation.
PBMCs were prepared from healthy volunteers by Ficoll-Hypaque (Sigma-Aldrich) centrifugation. PBMCs (106 cells/ml) were cultured at 37°C for 24 or 48 h on 24-well plates precoated with anti-CD3 mAb (10 μg/ml) or in the presence of PHA (20 μg/ml), IFN-α (200 U/ml), IFN-γ (100 ng/ml), IL-2 (500 U/ml), IL-4 (50 ng/ml), IL-10 (50 ng/ml), IL-12 (20 ng/ml), IL-15 (150 ng/ml), or IL-18 (500 ng/ml). For preparation of monocytes, PBMCs were cultured on tissue culture dishes precoated with FCS for 1 h at 37°C. After removal of nonadherent cells, adherent cells were detached by suspending in ice-cold PBS containing 0.5% EDTA and then resuspended in the culture medium. Monocytes were further purified using a monocyte negative isolation kit (Dynal) according to the manufacturer's instructions. Purity of monocytes used was >93% CD14+ as estimated by flow cytometry.
Results
Characterization of Recombinant Soluble hTWEAK and hTWEAK Transfectants.
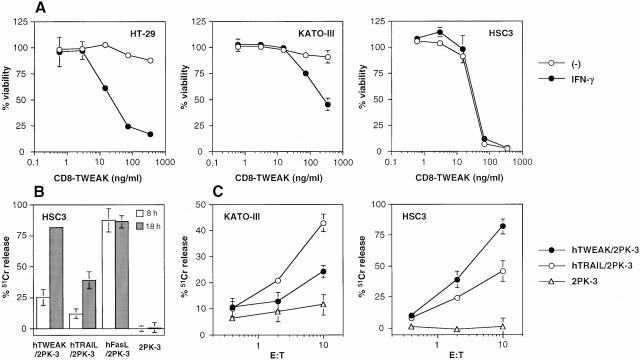
To characterize the functional properties of hTWEAK, we generated a soluble CD8–TWEAK fusion protein and stable cDNA transfectants expressing full-length hTWEAK. First, we constructed a CD8–TWEAK fusion protein composed of the extracellular domain of hTWEAK (amino acids 93–249) and human CD8α, as we previously generated a functional soluble form of FasL 16. We examined the cytotoxic activity of CD8–TWEAK against some tumor cell lines in the presence or absence of IFN-γ. As previously reported by others 4, colon adenocarcinoma HT-29 cells were highly sensitive to TWEAK-induced cell death only in the presence of IFN-γ (Fig. 1 A). Gastric adenocarcinoma KATO-III cells were also weakly sensitive to TWEAK only in the presence of IFN-γ. In contrast, oral squamous carcinoma HSC3 cells were highly sensitive to TWEAK even in the absence of IFN-γ (Fig. 1 A).
Figure 1.
Cytotoxic activity of TWEAK against tumor cell lines. (A) Cytotoxic activity of CD8–TWEAK. Target tumor cells (5 × 103) were cultured with the indicated concentrations of CD8–TWEAK in the presence (•) or absence (○) of IFN-γ (20 ng/ml). After 36-h culture, viability was estimated by the WST assay. Data represent mean ± SD of triplicate samples. Similar results were obtained in three independent experiments. (B) Cytotoxic activity of hTWEAK, hTRAIL, and hFasL transfectants against HSC3 cells. Cytotoxic activity of the indicated transfectants was tested against HSC3 cells in 8- (white bars) or 18-h (gray bars) 51Cr-release assay at an E/T ratio of 10. Data represent mean ± SD of triplicate samples. Similar results were obtained in three independent experiments. (C) Cytotoxic activity of hTWEAK or hTRAIL transfectants against KATO-III and HSC3 cells. Cytotoxic activity of hTWEAK/2PK-3 (•), hTRAIL/2PK-3 (○), or 2PK-3 (▵) was tested against KATO-III cells in the presence of IFN-γ (20 ng/ml) or HSC3 cells in 18-h 51Cr-release assay at the indicated E/T ratios. Data represent mean ± SD of triplicate samples. Similar results were obtained in three independent experiments.
We next generated stable transfectants expressing full-length TWEAK cDNA. Mouse B lymphoma 2PK-3 cells, which were totally resistant to CD8–TWEAK-induced cytotoxicity (data not shown), were transfected with hTWEAK/pCR3.1 by electroporation to make hTWEAK/2PK-3 cells. To verify the expression of TWEAK in hTWEAK/2PK-3 cells, we tested the cytotoxic activity of the transfectant against HSC3 target cells in 8- or 18-h 51Cr-release assay, along with the parental 2PK-3 cells, human FasL transfectant (hFasL/2PK-3), and hTRAIL transfectant (hTRAIL/2PK-3). As represented in Fig. 1 B, all of these transfectants lysed HSC3 target cells significantly, whereas no cytotoxicity was observed with the parental 2PK-3 cells. In contrast to a rapid time course of FasL-induced cytotoxicity, which reached a peak at 8 h, a more prolonged period of 18 h was required to substantially detect TWEAK- or TRAIL-induced cytotoxicity. Thus, we used 18-h 51Cr-release assay to evaluate the TRAIL- or TWEAK-mediated cytotoxicity in the following experiments. As represented in Fig. 1 C, both KATO-III cells and HSC3 cells were killed by TWEAK and TRAIL transfectants in an E/T ratio–dependent manner, whereas KATO-III cells were much less sensitive to TWEAK transfectant than HSC3. Similarly, hTWEAK/L5178Y cells, but not L5178Y cells, exhibited cytotoxic activity against these target cells (data not shown).
We and others have reported that some members of the TNF family, such as TNF-α and FasL, undergo processing by certain proteinases, resulting in the release of functional soluble forms from the cell surface 15 17. Likewise, we observed that the culture supernatants of the hTWEAK/2PK-3 and hTWEAK/L5178Y cells, but not those of parental cell lines, exhibited substantial cytotoxicity against HSC3 and KATO-III cells (data not shown). This suggested that TWEAK could be also released as a functional soluble form.
Characterization of Anti-hTWEAK mAbs.
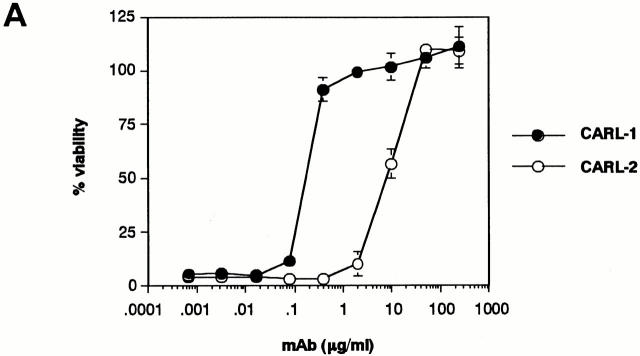
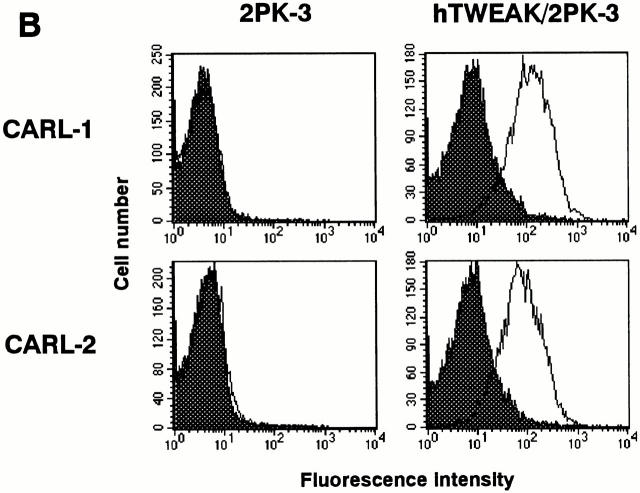
To characterize the expression and function of hTWEAK, we generated two mAbs that specifically bind to hTWEAK and block its cytotoxic activity. Hybridomas were prepared from splenocytes from mice immunized with the hTWEAK/2PK-3 cells. Two hybridomas producing CARL-1 and CARL-2 mAbs (mouse IgG3/κ and IgM/κ, respectively) were selected by their ability to block CD8–TWEAK cytotoxicity. As represented in Fig. 2 A, both CARL-1 and CARL-2 mAbs neutralized CD8–TWEAK cytotoxicity against HSC3 cells in a dose-dependent manner; CARL-1 was more effective than CARL-2. Both CARL-1 and CARL-2 reacted with hTWEAK/2PK-3, but not with 2PK-3, as estimated by cell surface staining (Fig. 2 B). These mAbs also stained hTWEAK/L5178Y but not L5178Y, hFasL/2PK-3, or hTRAIL/2PK-3 (data not shown), indicating the specific binding of CARL-1 and CARL-2 to hTWEAK.
Figure 2.
Characterization of anti-hTWEAK mAbs. (A) Inhibition of TWEAK cytotoxicity. Cytotoxic activity of CD8–TWEAK (100 ng/ml) was tested against HSC3 cells in the presence of serially diluted anti-hTWEAK mAbs, CARL-1 (•) or CARL-2 (○), by the WST assay. Data represent mean ± SD of triplicate samples. Similar results were obtained in three independent experiments. (B) Cell surface staining of hTWEAK transfectant. 2PK-3 and hTWEAK/2PK-3 cells were stained with biotinylated CARL-1 or CARL-2, followed by PE-labeled avidin (white histograms). Dark histograms indicate background staining with biotinylated control mAb plus PE-labeled avidin.
Expression of TWEAK on IFN-γ-stimulated Monocytes.
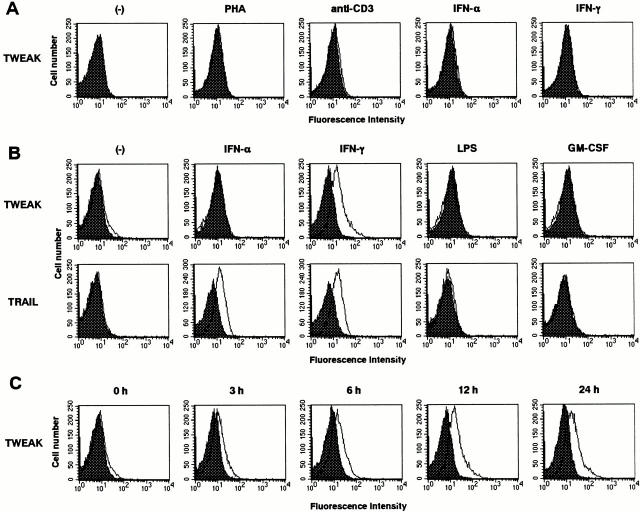
Although the expression of TWEAK at the mRNA level has been demonstrated in various tissues and cells 4, little is known about its expression and regulation at the protein level. Thus, we first examined the expression of TWEAK on human PBMCs by staining with the anti-hTWEAK mAb CARL-1. No significant level of cell surface TWEAK expression was observed on CD3+CD56− T cells, CD3−CD56+ NK cells, CD3−CD19+ B cells, or CD14+ monocytes in freshly isolated PBMCs (data not shown). We previously demonstrated that the expression of other death-inducing ligands of the TNF family such as FasL and TRAIL could be induced on peripheral blood (PB) T cells upon stimulation with certain mitogens or cytokines 15 18. However, even after the stimulation with PHA, immobilized anti-CD3 mAb, IFN-α, IFN-γ (Fig. 3 A), IL-2, IL-4, IL-12, IL-15, or IL-18 (data not shown) for 24–48 h, PB T cells did not express detectable level of TWEAK on their surfaces. Furthermore, in contrast to the FasL and TRAIL that were inducible on NK cells upon IL-2 stimulation 19 20, no detectable TWEAK expression was found on IL-2–activated NK cells (data not shown).
Figure 3.
Expression of TWEAK on human PB T cells and monocytes. (A) Expression of TWEAK on human PB T cells. PBMCs were cultured for 24 h on plates precoated with anti-CD3 mAb (10 μg/ml) or in the presence of PHA (20 μg/ml), IFN-α (200 U/ml), or IFN-γ (100 ng/ml). The cells were then stained with biotinylated CARL-1, followed by PE-labeled avidin and FITC-labeled anti-CD3 mAb. White histograms indicate the staining with CARL-1 on electronically gated CD3+ T cells. Dark histograms indicate the background staining with biotinylated control mAb and PE-labeled avidin. Similar results were obtained with PBMCs from three individuals. (B) Expression of TWEAK and TRAIL on human PB monocytes. Purified monocytes were stimulated with IFN-α (200 U/ml), IFN-γ (100 ng/ml), LPS (20 μg/ml), or GM-CSF (100 ng/ml) for 12 h and then stained with biotinylated CARL-1 and PE–avidin (white histograms). Dark histograms indicate the background staining with biotinylated control mAb and PE–avidin. Similar results were obtained with monocytes from three individuals. (C) Time course of TWEAK expression on monocytes after stimulation with IFN-γ (100 ng/ml). After the indicated time periods, the cells were stained with biotinylated CARL-1 and PE–avidin (white histograms). Dark histograms indicate the background staining with biotinylated control mAb and PE–avidin. Similar results were obtained with monocytes from three individuals.
It is well known that LPS strongly induces TNF-α production by monocytes. Furthermore, a recent study by Griffith et al. 13 showed that TRAIL could be induced on IFN-α– or IFN-γ–stimulated monocytes. Thus, we next examined whether surface TWEAK expression could also be induced on PB monocytes by LPS, IFN-α, IFN-γ, or GM-CSF. Purified monocytes were cultured with IFN-α (200 U/ml), IFN-γ (100 ng/ml), LPS (20 μg/ml), or GM-CSF (100 ng/ml) for 12 h, and the expression of TWEAK and TRAIL was then examined by cell surface staining with CARL-1. As represented in Fig. 3 B, a remarkable TWEAK expression could be observed on IFN-γ–stimulated monocytes. Interestingly, in contrast to the TRAIL expression that could be equivalently induced by both IFN-α and IFN-γ, TWEAK could be induced by IFN-γ but not IFN-α. Neither GM-CSF nor LPS was effective in inducing TWEAK and TRAIL expression. These results indicated a unique expression of TWEAK on IFN-γ–stimulated monocytes. We also examined the kinetics of TWEAK expression after stimulation with IFN-γ. As represented in Fig. 3 C, TWEAK appeared at 3 h and reached a peak at 12 h. A similar kinetics of TRAIL expression was observed (data not shown).
Involvement of TWEAK in IFN-γ–stimulated Monocyte Cytotoxicity.
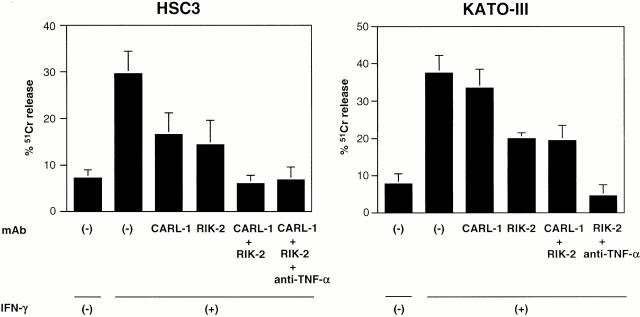
It has been reported that monocytes exerted cytotoxicity against some kinds of tumor cells, especially after the stimulation with IFN-γ 10. We then examined whether TWEAK is involved in the IFN-γ–stimulated monocyte cytotoxicity against tumor cells. We used highly TWEAK- and TRAIL-sensitive HSC3 cells and highly TRAIL- and weakly TWEAK-sensitive KATO-III cells as target cells. Neutralizing anti-TWEAK mAb (CARL-1) and neutralizing anti-TRAIL mAb (RIK-2) were used to assess the contribution of TWEAK and TRAIL to the cytotoxicity, respectively.
As shown in Fig. 4, IFN-γ–stimulated monocytes exhibited substantial cytotoxicity against HSC3 cells that was partially inhibited by either CARL-1 or RIK-2 alone and almost completely abrogated by the combination of both mAbs. This indicated that TWEAK and TRAIL expressed on IFN-γ–stimulated monocytes mostly account for the cytotoxic activity of IFN-γ–stimulated monocytes against HSC3 cells. In contrast, while IFN-γ–stimulated monocytes also efficiently lysed KATO-III cells, CARL-1 alone hardly inhibited this cytotoxicity. Even in combination with RIK-2 that alone partially inhibited this cytotoxicity, the addition of CARL-1 did not affect the cytotoxicity against KATO-III cells, indicating little contribution of TWEAK. As KATO-III cells were highly sensitive to TNF-α–induced cell death (data not shown), we tested neutralizing anti–TNF-α mAb in combination with RIK-2 and found that the addition of both mAbs mostly abrogated the IFN-γ–stimulated monocyte cytotoxicity against KATO-III cells (Fig. 4). This indicated that TRAIL and TNF-α mostly account for the IFN-γ–stimulated monocyte cytotoxicity against KATO-III cells. These results suggested that relative contributions of TWEAK, TRAIL, and TNF-α appear to be determined by the susceptibility of individual target cells to each death-inducing ligand.
Figure 4.
Contribution of TWEAK, TRAIL, and TNF-α to IFN-γ–stimulated monocyte cytotoxicity. Purified monocytes were stimulated with IFN-γ (100 ng/ml) for 12 h. Cytotoxic activity was then tested against HSC3 and IFN-γ–treated KATO-III cells by 18-h 51Cr-release assay at an E/T ratio of 50. Data represent mean ± SD of triplicate samples. Similar results were obtained in three independent experiments.
Discussion
In this study, we investigated the expression and function of TWEAK in human PBMCs by using newly established neutralizing mAbs against hTWEAK. No detectable level of TWEAK expression was found on each subpopulation of T cells, B cells, NK cells, and monocytes in freshly isolated PBMCs. However, a remarkable level of TWEAK expression could be induced on monocytes upon IFN-γ stimulation. IFN-γ–stimulated monocytes lysed HSC3 target cells by using both TWEAK and TRAIL. These results are the first indication that TWEAK is involved in monocyte cytotoxicity and that IFN-γ is a potent inducer of TWEAK expression on monocytes.
As represented in Fig. 3, monocytes expressed a substantial level of TWEAK upon IFN-γ stimulation. In contrast, T cells expressed no detectable level of TWEAK even after various stimulation including IFN-γ. We also observed that stimulation of NK cells with IFN-γ did not induce TWEAK expression (data not shown). Thus, so far, TWEAK appears to be preferentially expressed on monocytes. Molecular mechanisms for the restricted induction of TWEAK in monocytes remain obscure. This could not be explained by differential expression of IFN-γR on these cells, as IFN-γ upregulated MHC class I expression on T cells and NK cells as well as on monocytes (data not shown). Further study is now underway to explore the mechanisms for monocyte-restricted induction of TWEAK by IFN-γ.
A recent study using DR3–Ig fusion protein demonstrated the constitutive expression of DR3 ligand (TWEAK?) on murine CD4+ T cell clones, which was responsible for their cytotoxicity against mouse macrophages 21. These results seem to be inconsistent with our finding indicating the preferential expression of TWEAK on monocytes. Moreover, we observed that human monocytes were resistant to TWEAK-induced cell death in the presence or absence of IFN-γ (data not shown). These conflicting results might reflect possible differences between human and mouse systems. However, it should be noted that the definition of TWEAK as the DR3 ligand still remains controversial. In our preliminary study, we could not demonstrate the binding of hDR3–Ig fusion protein to our hTWEAK transfectants (our unpublished data). Furthermore, we could not detect DR3 expression in HSC3 cells or IFN-γ–treated HT-29 and KATO-III cells at the protein or mRNA levels, despite their susceptibility to TWEAK-induced cell death (our unpublished data). In this respect, it is possible that another DR3 ligand distinct from TWEAK was expressed on the murine T cell clones, which might mediate the cytotoxicity against murine macrophages. Of course, we also cannot rule out the possible induction of TWEAK expression in lymphocytes in certain culture conditions. Further studies are now underway to determine the TWEAK expression on various subpopulations of leukocytes, including PMNs and dendritic cells, after stimulation with cytokines and inflammatory mediators.
A previous study demonstrated the release of soluble TWEAK into the supernatant of 293-EBNA cells transiently transfected with full-length TWEAK cDNA 4. Consistently, we also observed soluble TWEAK in the supernatant of our hTWEAK transfectants, as assessed by cytotoxicity against HSC3 cells and sandwich ELISA using CARL-1 and CARL-2 mAbs (data not shown). Processing of membrane-bound TNF-α leads to the release of soluble form with substantial loss of biological activity against TNFR-II 22. Similarly, naturally processed soluble form of FasL mostly lacked cytotoxic activity and is speculated to act as an antagonist against membrane-bound FasL 23. In our preliminary study, we observed that the naturally processed soluble TWEAK had as potent cytotoxic activity (ED50 = 32 ng/ml) as CD8–TWEAK fusion protein (ED50 = 37 ng/ml) against HSC3 cells. This suggests possible function of TWEAK as a diffusible cytokine produced by IFN-γ–stimulated monocytes. Further biochemical analysis will be needed to clarify whether the naturally processed soluble TWEAK has biological activities similar to those of membrane-bound TWEAK.
It has been reported that monocytes killed tumor cells via TNF-α and TRAIL 9 13. Here, we demonstrated an additional pathway of monocyte cytotoxicity via TWEAK. Thus, TWEAK, TRAIL, and TNF-α might be largely responsible for mediating human monocyte cytotoxicity against various tumor cells. The relative contribution of each effector molecule appears to be determined by the activational state of monocytes and the sensitivity of individual tumor targets to each death-inducing ligand. A previous study in the murine system reported a critical role of monocytes in tumor rejection in vivo that was evoked by endogenous IFN-γ 24. Indeed, infiltration of monocytes has been frequently observed in various tumors 25. Furthermore, in a human clinical trial, adoptive transfer of IFN-γ–stimulated monocytes exerted some antitumor effects in patients 26. Direct cytotoxicity by IFN-γ–stimulated monocytes via death-inducing ligands might be involved in the beneficial effects.
In addition to the ability to induce cell death in tumor cells, TWEAK stimulates endothelial cells to proliferate in vitro and induces angiogenesis in vivo 7. We also observed that CD8–TWEAK induced intercellular adhesion molecule (ICAM)-1 upregulation and IL-8 secretion in human endothelial cells (our unpublished data). Therefore, TWEAK produced by IFN-γ–stimulated monocytes may play important roles not only as an antitumor effector mechanism but also as a proinflammatory cytokine. The anti-TWEAK mAbs generated in this study will be useful for further investigating the pathophysiological functions of TWEAK.
Acknowledgments
We thank Drs. H. Akiba, K. Takeda, and H. Nakano for helpful suggestions.
This work was supported by grants from the Princess Takamatsu Cancer Research Fund (99-23110), the Science and Technology Agency, the Ministry of Education, Science, Sports and Culture of Japan, and the Japanese Ministry of Health. M. Nakayama is a Research Fellow of the Japan Society for the Promotion of Science.
References
- Ashkenazi A., Dixit V.M. Death receptorssignaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- Golstein P. Cell deathTRAIL and its receptors. Curr. Biol. 1997;7:R750–753. doi: 10.1016/s0960-9822(06)90000-1. [DOI] [PubMed] [Google Scholar]
- Chicheportiche Y., Bourdon P.R., Xu H., Hsu Y.M., Scott H., Hession C., Garcia I., Browning J.L. TWEAK, a new secreted ligand in the tumor necrosis factor family that weakly induces apoptosis. J. Biol. Chem. 1997;272:32401–32410. doi: 10.1074/jbc.272.51.32401. [DOI] [PubMed] [Google Scholar]
- Marsters S.A., Sheridan J.P., Pitti R.M., Brush J., Goddard A., Ashkenazi A. Identification of a ligand for the death-domain-containing receptor Apo3. Curr. Biol. 1998;8:525–528. doi: 10.1016/s0960-9822(98)70204-0. [DOI] [PubMed] [Google Scholar]
- Schneider P., Schwenzer R., Haas E., Muhlenbeck F., Schubert G., Scheurich P., Tschopp J., Wajant H. TWEAK can induce cell death via endogenous TNF and TNF receptor 1. Eur. J. Immunol. 1999;29:1785–1792. doi: 10.1002/(SICI)1521-4141(199906)29:06<1785::AID-IMMU1785>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lynch C.N., Wang Y.C., Lund J.K., Chen Y.W., Leal J.A., Wiley S.R. TWEAK induces angiogenesis and proliferation of endothelial cells. J. Biol. Chem. 1999;274:8455–8459. doi: 10.1074/jbc.274.13.8455. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan A.M., O'Rourke K., Yu G.L., Lyons R.H., Garg M., Duan D.R., Xing L., Gentz R., Ni J., Dixit V.M. Signal transduction by DR3, a death domain-containing receptor related to TNFR-1 and CD95. Science. 1996;274:990–992. doi: 10.1126/science.274.5289.990. [DOI] [PubMed] [Google Scholar]
- Feinman R., Henriksen-DeStefano D., Tsujimoto M., Vilcek J. Tumor necrosis factor is an important mediator of tumor cell killing by human monocytes. J. Immunol. 1987;138:635–640. [PubMed] [Google Scholar]
- Le J., Prensky W., Yip Y.K., Chang Z., Hoffman T., Stevenson H.C., Balazs I., Sadlik J.R., Vilcek J. Activation of human monocyte cytotoxicity by natural and recombinant immune interferon. J. Immunol. 1983;131:2821–2826. [PubMed] [Google Scholar]
- MacMicking J., Xie Q.W., Nathan C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- Weinberg J.B., Misukonis M.A., Shami P.J., Mason S.N., Sauls D.L., Dittman W.A., Wood E.R., Smith G.K., McDonald B., Bachus K.E. Human mononuclear phagocyte inducible nitric oxide synthase (iNOS)analysis of iNOS mRNA, iNOS protein, biopterin, and nitric oxide production by blood monocytes and peritoneal macrophages. Blood. 1995;86:1184–1195. [PubMed] [Google Scholar]
- Griffith T.S., Wiley S.R., Kubin M.Z., Sedger L.M., Maliszewski C.R., Fanger N.A. Monocyte-mediated tumoricidal activity via the tumor necrosis factor–related cytokine, TRAIL. J. Exp. Med. 1999;189:1343–1354. doi: 10.1084/jem.189.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N., Yamaguchi N., Nakayama M., Kawasaki A., Akiba H., Okumura K., Yagita H. Involvement of TNF-related apoptosis-inducing ligand in human CD4+ T cell-mediated cytotoxicity. J. Immunol. 1999;162:2639–2647. [PubMed] [Google Scholar]
- Kayagaki N., Kawasaki A., Ebata T., Ohmoto H., Ikeda S., Inoue S., Yoshino K., Okumura K., Yagita H. Metalloproteinase-mediated release of human Fas ligand. J. Exp. Med. 1995;182:1777–1783. doi: 10.1084/jem.182.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N., Yamaguchi N., Nagao F., Matsuo S., Maeda H., Okumura K., Yagita H. Polymorphism of murine Fas ligand that affects the biological activity. Proc. Natl. Acad. Sci. USA. 1997;94:3914–3919. doi: 10.1073/pnas.94.8.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing A.J., Beckett P., Christodoulou M., Churchill M., Clements J., Davidson A.H., Drummond A.H., Galloway W.A., Gilbert R., Gordon J.L. Processing of tumour necrosis factor-α precursor by metalloproteinases. Nature. 1994;370:555–557. doi: 10.1038/370555a0. [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Yamaguchi N., Nakayama M., Eto H., Okumura K., Yagita H. Type I interferons (IFNs) regulate tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) expression on human T cellsa novel mechanism for the antitumor effects of type I IFNs. J. Exp. Med. 1999;189:1451–1460. doi: 10.1084/jem.189.9.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N., Yamaguchi N., Nakayama M., Takeda K., Akiba H., Tsutsui H., Okamura H., Nakanishi K., Okumura K., Yagita H. Expression and function of TNF-related apoptosis-inducing ligand on murine activated NK cells. J. Immunol. 1999;163:1906–1913. [PubMed] [Google Scholar]
- Tanaka M., Suda T., Haze K., Nakamura N., Sato K., Kimura F., Motoyoshi K., Mizuki M., Tagawa S., Ohga S. Fas ligand in human serum. Nat. Med. 1996;2:317–322. doi: 10.1038/nm0396-317. [DOI] [PubMed] [Google Scholar]
- Kaplan M.J., Ray D., Mo R.R., Yung R.L., Richardson B.C. TRAIL (Apo2 ligand) and TWEAK (Apo3 ligand) mediate CD4+ T cell killing of antigen-presenting macrophages. J. Immunol. 2000;164:2897–2904. doi: 10.4049/jimmunol.164.6.2897. [DOI] [PubMed] [Google Scholar]
- Grell M., Douni E., Wajant H., Lohden M., Clauss M., Maxeiner B., Georgopoulos S., Lesslauer W., Kollias G., Pfizenmaier K. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell. 1995;83:793–802. doi: 10.1016/0092-8674(95)90192-2. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Itai T., Adachi M., Nagata S. Downregulation of Fas ligand by shedding. Nat. Med. 1998;4:31–36. doi: 10.1038/nm0198-031. [DOI] [PubMed] [Google Scholar]
- Yoshida R., Yoneda Y., Kuriyama M., Kubota T. IFN-γ- and cell-to-cell contact-dependent cytotoxicity of allograft-induced macrophages against syngeneic tumor cells and cell linesan application of allografting to cancer treatment. J. Immunol. 1999;163:148–154. [PubMed] [Google Scholar]
- McBride W.H. Phenotype and functions of intratumoral macrophages. Biochim. Biophys. Acta. 1986;865:27–41. doi: 10.1016/0304-419x(86)90011-9. [DOI] [PubMed] [Google Scholar]
- Lesimple T., Moisan A., Toujas L. Autologous human macrophages and anti-tumour cell therapy. Res. Immunol. 1998;149:663–671. doi: 10.1016/s0923-2494(99)80036-4. [DOI] [PubMed] [Google Scholar]