Abstract
Using patient data from a unique single source outbreak of hepatitis B virus (HBV) infection, we have characterized the kinetics of acute HBV infection by monitoring viral turnover in the serum during the late incubation and clinical phases of the disease in humans. HBV replicates rapidly with minimally estimated doubling times ranging between 2.2 and 5.8 d (mean 3.7 ± 1.5 d). After a peak viral load in serum of nearly 1010 HBV DNA copies/ml is attained, clearance of HBV DNA follows a two or three phase decay pattern with an initial rapid decline characterized by mean half-life (t 1/2) of 3.7 ± 1.2 d, similar to the t 1/2 observed in the noncytolytic clearance of covalently closed circular DNA for other hepadnaviruses. The final phase of virion clearance occurs at a variable rate (t 1/2 of 4.8 to 284 d) and may relate to the rate of loss of infected hepatocytes. Free virus has a mean t 1/2 of at most 1.2 ± 0.6 d. We estimate a peak HBV production rate of at least 1013 virions/day and a maximum production rate of an infected hepatocyte of 200–1,000 virions/day, on average. At this peak rate of virion production we estimate that every possible single and most double mutations would be created each day.
Keywords: acute hepatitis B, kinetics, cytotoxic T lymphocytes, mutation, hepatocyte
Introduction
The kinetics of acute hepatitis B virus (HBV) infection have previously been studied in animal models but, until now, there has been limited quantitative data in humans with acute infection 1 2 and none based on the analytical sensitivity of PCR-based assays. Thus, the quantitation of the kinetics of self-limited acute infection has not been adequately studied. We have recently described a large, common source outbreak of acute hepatitis B, caused by transmission of a single HBV variant by “autohemotherapy”. An investigation to identify all cases, to characterize the epidemic, and to limit the transmission of the virus was begun as soon as the first patients were diagnosed. As a result of the early identification of patients infected with HBV in this outbreak, we could analyze data from several patients acutely infected from the same source before symptoms, and seroconversion to anti-HBe, i.e., during both the late incubation and clinical phases of the disease. In this paper, we report on a study of the dynamics of primary HBV infection in humans and estimate rates of viral production and clearance. From serial measurements of serum HBV DNA, we estimate viral doubling times (t d) and the time between infection and peak viral level. After the peak, viral levels decay in several phases which may correlate with underlying biological mechanisms encompassing decay of covalently closed circular (ccc)DNA, loss of infected hepatocytes, and clearance of free virus.
Materials and Methods
Patients and Data.
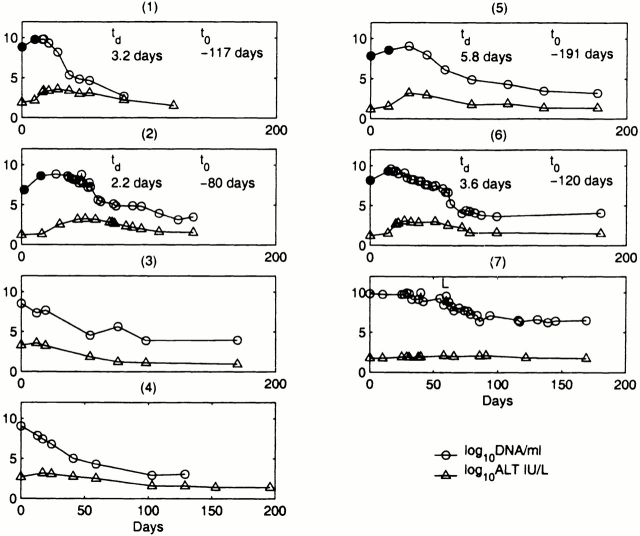
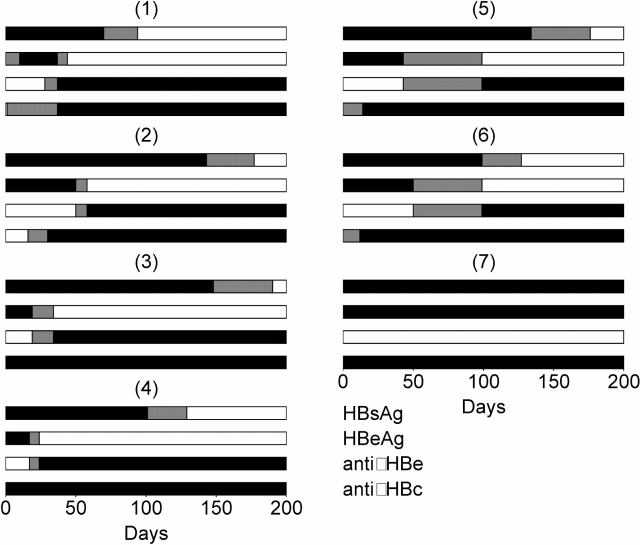
HBV DNA and alanine transaminase (ALT) levels in serum were obtained for seven patients with primary HBV infection (Fig. 1). ALT was measured by an in-house radioimmunoassay. Hepatitis B surface antigen (HBsAg), hepatitis B e antigen (HBeAg), anti-HBe antibody (anti-HBe), and antibody to hepatitis B core (anti-HBc) status (Fig. 2) were also determined by commercial enzyme immunoassay kits (Abbott Laboratories). Patient 7, who developed chronic hepatitis B infection, was identified in the incubation phase of infection and subsequently was commenced on the reverse transcriptase inhibitor lamivudine. We have reported previously that this outbreak of HBV was acquired from contamination of a multidose saline vial used in autohemotherapy 3. Sequencing of HBV DNA, from serum samples obtained at presentation, showed identical nucleotide sequence for the surface and core encoding regions in all of these patients. Thus, all patients were infected with an identical isolate of HBV and had a common source and mode of transmission.
Figure 1.
Serum log10 HBV DNA copies per ml and serum log10 ALT IU/liter values are displayed over the period of observation. In only one patient was there a serious comorbid condition: patient 7 who had cryptogenic organizing pneumonitis for which she was maintained on long-term steroid therapy before her autohemotherapy. Patient 7 failed to clear HBsAg, perhaps because of immunosuppresion, and was commenced on lamivudine 100 mg daily, which is denoted by “L” in the graph. Four patients were identified in the late incubation phase of acute infection when the initial serum HBV DNA was below the eventual peak. t d for serum HBV DNA were estimated by determining the rise in log10 viral level between the first and second measurements (•). The estimated t o before peak serum HBV DNA was determined using these t d.
Figure 2.
HBsAg, HBeAg, anti-HBe, and anti-HBc for the seven patients. Black denotes a positive assay, white a negative assay, and gray indicates time between a positive assay and a negative assay.
HBV DNA Quantification.
Serum samples for HBV DNA quantification were obtained prospectively and stored at −20°C. To reduce interassay variability, all samples derived from each patient were analyzed in the same assay, incorporating up to 41 samples. HBV DNA concentrations in serum were measured by the Amplicor HBV Monitor test (Roche Diagnostics Systems) which is a quantitative PCR based assay and the manufacturer's instructions were followed throughout. In brief, a 104-bp sequence of the precore/core region of HBV is amplified along with an internal standard (IS) to monitor the efficiency of the amplification reaction, one of the primers being biotinylated. Six HBV DNA standards (range 0–106 copies) are amplified at the same time. The amplicons are denatured and the biotinylated strand immobilized on a streptavidin-coated microwell plate. DNP-labeled oligonucleotide probes specific for HBV and the IS are then hybridized to the immobilized amplicons. After washing, the bound probe is quantitated using an ELISA procedure with anti-DNP–alkaline phosphatase conjugate and para-nitrophenylphosphate substrate. Absorbance of sample/HBV probe and sample/IS probe is measured at 405 nm. The HBV to IS signal ratio is calculated to correct for sample to sample differences in amplification efficiency. Finally, the ratios from the unknowns are compared with a standard curve generated from the HBV standards and the HBV concentrations of each sample are calculated. The assay quantitates virus between 4 × 102 and 4 × 107 particles per ml. Samples exceeding the upper limit were diluted in normal human serum and retested. Comparative quantitation of HBV DNA by this assay have been reported previously 4 5.
Results
Viral td and Time of Infection.
Patients 1, 2, 5, and 6 were identified in the incubation period when viral levels were below their eventual peak. For each of these patients, the t d of serum HBV DNA (t d) was estimated and ranged between 2.2 and 5.8 d with a mean of 3.7 ± 1.5 d. Because data was collected near the peak viral load, the true t d may have been underestimated.
The time of infection relative to the viral peak, t o, was estimated assuming that the infecting inoculum contained 103 HBV DNA particles (similar to that of a needle stick injury 6) and distributed throughout 3 liters of serum (Fig. 1). It seemed reasonable to assume that the amount of HBV-contaminated blood introduced into the multidose saline vial used to spread the infection was small because there was no obvious discoloration. This analysis yielded a mean time to infection before the viral peak of 127 ± 46 d. Analyzing the patient with the fastest t d, patient 2, where we may have caught the earlier stages of viral rise and hence obtained a more accurate estimate of the initial t d, the estimated time until peak virus level after infection was 80 d. This is consistent with the 60–110-d incubation period for the majority of HBV infections in humans 7 and the 8-wk time before viral peak for chimpanzees inoculated with a larger HBV dose 8. Furthermore, in patient 2, the t o was calculated to be around January 29, a date which coincides with her attendance for autohemotherapy on a weekly basis from January 12 to February 16. This provides some validation of our assumption that the infecting inoculum contained ∼103 HBV DNA particles, but unfortunately the inoculum was not available for examination and confirmation of the exact HBV content. However, we calculate that each 10-fold error in the initial concentration of HBV would change the estimate of the time from infection to the viral peak by 12.3 d. Because patient 2 could only have been infected while she attended for autohemotherapy, our estimate for her date of infection is at most off by 18 d, and thus the potential miscalculation of the dose is at most by a factor of 101.5.
Half-Lives of Viral Decay.
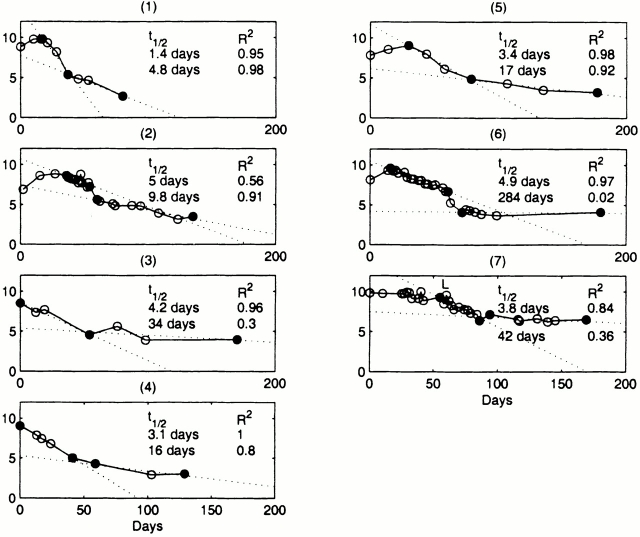
The decline in serum HBV DNA was multiphasic with an initial rapid decline over several days followed by a slower decline over several weeks. In patients 2 and 6, the transition between these phases was characterized by a rapid fall in viral DNA, which may represent a combination of both phases or a separate biological process. Using linear regression we estimated the rate of viral decay for each dominant phase (Fig. 3), and calculated the corresponding half-lives.
Figure 3.
Serum HBV DNA half-lives, linear regression lines, and R 2 statistics for the two dominant phases of viral decay for each patient. Endpoints for regression are noted with filled circles.
The initial decline in serum viremia was exponential as demonstrated by a straight-line fit to the data on a semilogarithmic plot with a mean R 2 statistic of 0.90 (excluding patient 7 who was on lamivudine therapy). The first phase decay of viral DNA had a mean half-life of 3.7 ± 1.2 d.
The final phase decay of viral DNA had a range of estimated half-lives that varied from 4.8 to 284 d, although we have limited confidence in the long half-life estimates. If we consider only those final phases where the regression lines have an R 2 statistic of 0.8 or better, then the range of half-life values was 4.8 to 17 d with a mean of 12 ± 6 d. This range is consistent with the half-life of infected hepatocytes as estimated using mathematical models in studies of patients chronically infected with HBV treated with lamivudine (half-lives of 10–100 d; reference 9) or adefovir (half-lives of 18 ± 7 d; reference 10).
Free Virus Half-Life.
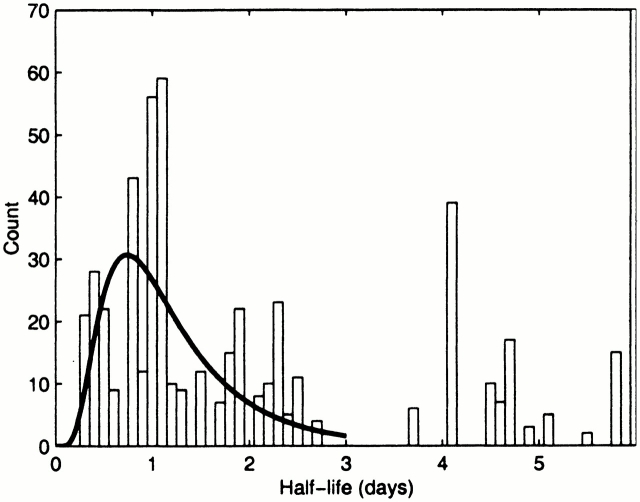
Based on drug perturbation experiments 9 10 as well as estimates of the half-life of other viruses, the half-life of HBV in serum is expected to be short and clearance of HBV should be the fastest kinetic process that we observe. We thus estimated the half-life of free virus by examining the rate of change of HBV DNA levels between individual data points. We excluded from the calculations patients 3, 4, and 5 because the time between data points was relatively large and would tend to mask the faster half-lives. A histogram of the half-lives in the range 0–6 d is shown in Fig. 4.
Figure 4.
Histogram of half-lives in the range 0–6 d for patients 1, 2, 6, and 7, obtained by determining the slopes between consecutive data points. Each slope was counted several times depending on the magnitude of the viral change. For example, if a half-life of 1 d was measured between time points 10 d apart, it was counted 10 times as many as a half-life of 1 d measured between time points 1 d apart.
The histogram in Fig. 4 displays two clusters of half-lives, the shortest between 0.3 and 2.7 d, and the second between 3.7 and 5.8 d. The second cluster is confirmation of the first phase half-life of 3.7 ± 1.2 d discussed above. Under the assumption that the shortest half-lives seen in the data correspond to the decay of free virus 9 10, we estimated the mean virion half-life by fitting a lognormal distribution to the data in the first cluster (Fig. 4). This gave a mean half-life of free virus of 1.2 ± 0.6 d. The median of this set of half-lives was 1.1 d. This is consistent with the values estimated in treatment studies for chronic HBV showing a free virus half-life of 1.1 d (for adefovir 10) and 1 d (for lamivudine 9). As the smallest time between data points was 1 d, there is an inherent bias towards underestimating these decay rates by our method. Consequently, the value derived here provides an underestimate of the decay rate and therefore an overestimate of the half-life. The same caveat can be placed on the literature estimates, where viral levels were measured every 7 d in one study 10, and on days 0, 2, 7, and weekly thereafter in the other 9.
Peak Viral Levels and Rate of Virion Production.
For the five patients where the first serum HBV DNA measurement was at or before the peak level, the maximum log10 viral level was determined. The mean maximum log10 viral level was 9.5 ± 0.5. The three largest log10 values were 9.8, 9.6, and 10.
A possible explanation for the uniform maximum serum HBV DNA levels seen, and indeed the stable high level for the chronically infected patient 7, is that at the peak of infection nearly all hepatocytes are infected. This would be in agreement with animal studies in which immunohistochemical analysis of liver specimens using antibody directed against the HBV core antigen has shown a >95% infection of hepatocytes in acutely infected woodchucks 11.
If in acute infection most hepatocytes become infected, then we can estimate the daily production of virus by an infected cell. When the peak viral load is reached the amount of virus in the serum is neither increasing nor decreasing so that the infection process is in steady state where the rate of viral production equals the rate of virion clearance. If there are I infected cells, each producing an average of p virions per day, then the rate of virion production is pI. If the peak viral burden is V and free virions are cleared at rate c per virion, then the total rate of clearance is cV. The estimated average half-life of free virions of 1.2 d implies that c = 0.58 d−1. HBV is clearly present in the 3 liters of serum (3 × 103 ml) of the prototypical 70-kg person. Its presence in the rest of body water is not well characterized. Thus, we obtain as a minimal estimate that in a person with a peak value of 1010 HBV DNA/ml, V = 3 × 1013 virions. If virus is present in all 15 liters of body water at the same concentration as serum, then V = 15 × 1013 virions. Thus, the daily clearance (and daily production at steady state) cV is between 1.7 × 1013 and 8.7 × 1013 virions. If there are 1011 hepatocytes in a typical adult liver 12 and 85% are infected, then to attain a peak viral load that is at steady state each infected hepatocyte would need to produce 200–1,000 virions per day. Virions are produced via transcription of cccDNA within the nucleus of infected hepatocytes forming RNA pregenomes that are subsequently reverse transcribed to form the virion HBV DNA 13. If there are 10–50 cccDNA per infected hepatocyte 14, we deduce that each cccDNA gives rise to a daily production of 4–100 new extracellular hepatitis B virions. This deduction does not take into account possible subpopulations of human cccDNA with potentially differing transcriptional activity, as has been reported for duck HBV (DHBV) DNA 15.
Basic Reproductive Number.
The basic reproductive number, R 0, is an estimate of the number of secondary infections that arise from one infected cell over the course of its life span at the beginning of infection when cells susceptible to infection (target cells) are not depleted. If R 0 is >1, then the infection grows, whereas if R 0 is <1 then the infection dies out. From a standard model of viral infection that has been applied to simian immunodeficiency virus (SIV 16) and HIV 17 infection in which it is a assumed that there is a delay of t days from the infection of a cell to the production of progeny virions, one finds that R 0 = (1 + r 0/δ) exp (r 0t), where r 0 is the initial growth rate of the virus and δ is the loss rate of an infected cell by death or other mechanisms. Using the estimates of the viral t d given in Fig. 1 we can estimate r 0, as by definition t d = ln 2/r 0 = 0.693/r 0. If we further assume that the final phase of viral decay reflects the loss of infected cells, then from the half-lives reported in Fig. 3 we can estimate δ, as by definition half-life = 0.693/δ. For patient 6 the estimate of the final phase half-life is not reliable, but for patients 1, 2, and 5 the R 2 statistic suggests that the estimates shown in Fig. 3 are valid. Computing the R 0 for these patients assuming a 1-d delay between infection and viral production, we find R 0 = 3.1, 7.5, and 4.4, for patients 1, 2, and 5, respectively, with a mean of 5.0. These estimates are most likely underestimates of R 0, as the number of target cells may have been reduced by the time the patients were first identified and our estimate of r 0 may be an underestimate of the true initial viral growth rate. Patient 2, the patient with the lowest initial viral load and hence the patient for whom the estimate of initial viral growth may be most accurate, has R 0 = 7.5. These estimates suggest that early in acute infection each HBV-infected cell will infect five or more other cells.
Discussion
For patients 1, 2, and 6 (Fig. 1), the initiation of clinical symptoms of acute hepatitis B infection coincided with the peak or the initial decline in serum HBV DNA levels. At this stage, symptoms were mild and consisted only of nonspecific lethargy, malaise, myalgia, and headache. By the time jaundice set in, 2–6 wk after the peak in serum HBV DNA, symptoms were deteriorating and viral levels were substantially reduced to ∼105–107 copies/ml.
Interestingly, a faster viral t d appeared to correlate with the severity of acute infection. Hence, patient 2 with the shortest t d became considerably jaundiced with a serum bilirubin peaking at 847 mmol/ml (normal range 5–17 mmol/ml) and experienced a decline in hepatic synthetic function with a serum albumin of 27 g/liter (normal range 35–50 g/liter) and a prolonged prothrombin time of 24.7 s (normal range <16 s). This was in contrast to patients 1 and 6, with t d of 3.2 and 3.6 d, respectively, who retained normal hepatic synthetic function though they did become jaundiced with serum bilirubin as high as 429 mmol/ml and 98 mmol/ml, respectively. Patient 5 with the longest t d of 5.8 d had a relatively mild illness, neither developing jaundice nor impairment of hepatic synthetic function. However, due to the limited sample size, these observations have no statistical power.
In this study we are able to characterize the kinetics of HBV clearance using a quantitative, sensitive assay for HBV DNA. The reason for the rapid first phase (3.7 ± 1.2 d) and subsequent phases of viral clearance are, however, unclear from our data. The resolution of acute hepatitis B involves a complex interplay of innate and adaptive immune responses and cellular and humoral immunity. In animal models, it has been suggested that the mechanism underlying initial viral clearance is the noncytopathic removal or inhibition, possibly by cytokines, of cccDNA replenishment 8. In our study, the mean half-life for the first phase of viral clearance is consistent with values obtained in animal models for the half-life of cccDNA 18 19. This suggests that the mechanism underlying the first phase of viral decay may indeed be related to clearance of cccDNA; however, we cannot exclude the possibility of inhibition of downstream steps in the viral life cycle, such as viral RNA, but proof of this requires direct experimental evidence. We have not made measurements of intrahepatic replicative intermediates of HBV DNA, nor of intrahepatic cellular immune responses. Thus, the sequential role of innate immunity, cytokine-mediated downregulation of HBV DNA, antigen-specific immune responses, and antibody neutralization can only be speculated on; however, quantitative analysis hints at their importance. The nonspecific symptoms experienced by patients as virus titre starts to fall are similar to those experienced by patients treated with IFN-α, suggesting a role for cytokine-mediated noncytolytic downregulation of the infection. However, proof would require accurate, repeated measurements of cytokines and replicative intermediates of hepatitis B viral DNA within the liver of acutely infected patients, and intrahepatic cytokine concentrations, but these studies, in patients with acute resolving hepatitis B, are ethically inadmissible.
The cccDNA is the transcriptional template for HBV replication 13 14 20. The loss of replicative templates thus would be expected to decrease the ability of infected cells to produce hepatitis B virions, and hence serum viral DNA should decrease accordingly. Thus, the administration of a reverse transcriptase inhibitor would abrogate the replenishment of the cccDNA pool, resulting in a decline of serum HBV DNA as cccDNA decays or cells harboring HBV cccDNA die. Patient 7, who remained negative for anti-HBe antibodies, was started on the reverse transcriptase inhibitor lamivudine. In support of the argument that the first phase of viral clearance is due to noncytopathic removal or inhibition of cccDNA replenishment, lamivude treatment resulted in a rate of decline of serum HBV DNA similar to the rate of viral decline observed during the first phase in patients not treated with lamivudine.
HBV is thought to be a noncytopathic virus and that cytotoxic T lymphocytes mediate clearance of HBV-infected cells through a cytolytic process 21 22. Serum ALT is a surrogate marker of cytolysis of hepatocytes 23 24. Consistent with the proposed noncytopathic nature of HBV, we observed for patients 2 and 6 that until the peak viral load was reached they remained asymptomatic with normal serum ALT (Fig. 1). For patients 1, 2, 3, 4, and 6, serum ALT peaked at 14 ± 6 d after the peak in serum HBV DNA level. By the time the peak serum ALT was achieved there was at least a one log, that is 90%, fall in the serum HBV DNA level. Therefore, our data suggests that the initial clearance of HBV is a process commenced before the main cytolytic loss of infected hepatocytes. This is in agreement with recent evidence from Guidotti et al. 8 that indicates in the chimpanzee 90% of acutely HBV-infected hepatocytes are cleared of infection by noncytolytic processes.
We have recently studied cellular immune responses in five HLA-A2–positive patients identified in this outbreak, with patients 1 to 5 in the study by Webster et al. 25 corresponding to patients 1, 4, 6, 2, and 7, respectively, of this study. Quantification of HBV-specific CD8+ cells, in blood, using HLA class I tetramers, suggest that adaptive immune mechanisms are present during the incubation phase, at least 4 wk before symptoms 25. Our results have also suggested that the pattern of reduction in HBV replication is not directly proportional to tissue injury during acute hepatitis B in man 25. As virus-specific immune responses and significant reductions in viral replication are seen during the incubation phase, it is likely that some immunological events central to viral control occur before symptomatic disease. Thus, overall our data are in general agreement that noncytolytic processes may contribute to the initial control of the virus but mechanisms involving cytolysis are also involved at this early stage. It is of note that in each of the patients that we studied, the initial fall from peak viral load was associated with some elevation of ALT. However, given that the peak ALT occurred 14 ± 6 d after the peak in serum HBV DNA level, the cytolytic mechanisms of viral clearance may take longer to initiate than noncytolytic mechanisms and may play a greater role once there has been a substantial fall in the serum viral load. After the initial decline in serum HBV levels, there appears to be a slower final phase of viral clearance characterized in a subset of patients by a half-life of 12 ± 6 d, that coincides with previous estimates of the rate of removal by the immune system of infected hepatocytes 9 10. The fact that serum aminotransferases are raised during this phase suggests that the latter phase may be related to death of hepatocytes by apoptosis and hepatocellular necrosis.
Antibodies to pre-S1 appear early in acute hepatitis B and may be important in elimination of viremia 26. We have not examined whether one of the phases of viral clearance (perhaps by the inhibition of HBV attachment to a hepatocyte receptor or the regulation of intrahepatic DNA levels and viral assembly) could be due to pre-S1 antibody. The pre-S1 protein (large HBsAg protein) is an essential component of the envelope of HBV virions, but is also found, although in far smaller amounts, on HBV tubular filaments (but not on 20-nm circular particles 27). We have no measure of the relative proportions of mature virions to filaments in plasma during the phases of acute hepatitis B, nor of antigen antibody complexing of anti–pre-S1 to expressed pre-S1, which if present could potentially limit the usefulness of anti–pre-S1 measurements. However, recently a good correlation was observed between pre-S1 antigen measured by enzyme-linked immunoassay, and HBV DNA measured by the Roche Amplicor assay 28. Thus, we believe that our measurements of viral kinetics are valid, but careful quantitation of pre-S1 antigen, antibody, and intracellular HBV DNA in animal models could shed further light on mechanisms of viral clearance in future studies.
Using a unique cohort of primary HBV-infected patients, we have characterized the early kinetics of HBV infection, during the late incubation phase and early clinical period of acute hepatitis B. We have shown that HBV can double as rapidly as every 2.2 d leading to the creation of viral loads that peak near 1010 HBV DNA copies (or particles)/ml at times as early as 80 d after infection. Our analysis suggests that even though extremely high serum viral loads are attained, if the vast majority of hepatocytes become productively infected then each cell need only produce 200–1,000 virions per day at the peak of infection. However, as noncytolytic processes diminish cccDNA, we would predict the virion production rate to drop dramatically. Although the virion production rate in infected humans is not known, Lin et al. 29 have shown that in DHBV infection the total DNA load in the liver is ∼500 copies per cell, which although it includes the DHBV cccDNA, is in the range that we predict for humans during acute infection.
Variability in HBV t d was observed among patients, even though the patients were infected from the same source. More rapid daily replication of HBV appeared to be associated with an increase in severity of acute hepatitis, suggesting that host factors play an important role in determining both HBV dynamics and the severity of subsequent liver disease.
From the estimated HBV t d and the half-life estimates of productively infected cells obtained from an examination of the final phase of viral decay, we were able to obtain the first estimates of the R 0 for HBV. This number indicates how many cells are infected by the progeny virions produced by an infected cell early in acute infection, i.e., before target cells are depleted. For the three patients for whom we could do this analysis, R 0 ranged between 3.1 and 7.4 with a mean of 5. In HIV infection, R 0 has been estimated as 19.3 17. Any intervention that reduces R 0 below 1 results in fewer than one secondary infected cell arising from each initial infected cell. Thus, the size of R 0 provides an estimate of how difficult it should be to extinguish infection via therapy, vaccination, or host response. The most accurate estimates of the R 0 are when target cells are unlimited. It is therefore likely that the estimates here are a small underestimate of the true R 0, as target cells are likely to be depleted during the course of acute hepatitis B infection. However, in keeping with other studies, such as those for acute HIV infection, it is unlikely that this underestimate will substantially increase the value of R 0 17. The significantly smaller R 0 that we report for HBV compared with that of HIV is in agreement with the observation that the vast majority of immunocompetent adults that are infected with HBV never develop chronic infection 30 and the efficacy of HBV vaccination 31.
If R 0 can be reduced below 1, say by vaccination, then infection should die out. However, bile duct cells and extrahepatic compartments may be a reservoir for hepadnaviruses that may harbor virus for long periods, although, at present, it would appear that in humans, the major burden of HBV replication occurs in the hepatic compartment 29.
Each virion undergoes reverse transcription before its expulsion from an infected cell. As HBV reverse transcriptase contains no error correction mechanism, this is an inherently error-prone process. The in vivo HIV error rate for reverse transcription is estimated as 3.4 × 10−5 base substitutions per site per replication cycle 32. If we assume conservatively that the error rate for HBV to be lower at say 10−6 base substitutions per site per reverse transcription then we can estimate the likely number of virions that contain mutations from wild-type. With 3,200 bases in the HBV genome, the average number of base changes is 0.0032 per reverse transcription and hence per virion. According to the Poisson distribution, we thus expect ∼0.32% of new virions to have a single mutation. Our calculations show that >1013 virions are produced daily and observed in the blood at peak infection. Therefore, we would expect at least 3.2 × 1010 point mutations to be produced per day, vastly more than the possible number of different single base changes in the HBV genome (Table ). Hence, we would expect all possible single base changes to be produced per day at peak infection. The calculations in Table show that this is also true for the majority of double mutations. This has important consequences for the development of mutations that escape the immune response, and also for mutations that provide resistance to reverse transcriptase inhibitors.
Table 1.
Rate of Generation of HBV Mutants at Peak Infection
| Base changes | Fraction | No. created per day | No. of possible mutants | Fraction of all possible mutants created per day |
|---|---|---|---|---|
| 0 | 0.9968 | 9.97 × 1012 | − | − |
| 1 | 0.0032 | 3.2 × 1010 | 9.6 × 103 | 1 |
| 2 | 5 × 10−6 | 5 × 107 | 4.6 × 107 | 0.66 |
| 3 | 5.4 × 10−9 | 5.4 × 104 | 1.5 × 1011 | 3.6 × 10−7 |
Further characterization of the kinetics of acute HBV infection should lead to better understanding of the biological processes underlying the virological, immunological, and liver pathological aspects of the resulting hepatitis.
Acknowledgments
We thank Dr. F. Mattes for assistance with the serological data and Dr. G. Colucci, at Roche (Basel, Switzerland) for providing the Amplicor Monitor test kits.
Portions of this work were done under the auspices of the U.S. Department of Energy. This work was funded in part by National Institutes of Health grant RR06555 and the Peter Samuel Research Fund.
Footnotes
Abbreviations used in this paper: ALT, alanine transaminase; ccc, covalently closed circular; DHBV, duck HBV; HBV, hepatitis B virus; HBsAg, hepatitis B surface antigen; IS, internal standard.
References
- Fong T.L., Akriviadis E.A., Govindarajan S., Valinluck B., Redeker A.G. Serum hepatitis B viral DNA in acute viral hepatitis B. Ann. Intern. Med. 1989;110:936–937. doi: 10.7326/0003-4819-110-11-936. [DOI] [PubMed] [Google Scholar]
- Fong T.L., Di Bisceglie A.M., Biswas R., Waggoner J.G., Wilson L., Claggett J., Hoofnagle J.H. High levels of viral replication during acute hepatitis B infection predict progression to chronicity. J. Med. Virol. 1994;43:155–158. doi: 10.1002/jmv.1890430210. [DOI] [PubMed] [Google Scholar]
- Webster G.J., Hallett R., Whalley S.A., Meltzer M., Balogun K., Brown D., Farrington C.P., Sharma S., Hamilton G., Farrow S.C. The molecular epidemiology of a large outbreak of hepatitis B linked to ‘autohaemotherapy’. Lancet. 2000;356:379–384. doi: 10.1016/S0140-6736(00)02529-0. [DOI] [PubMed] [Google Scholar]
- Pawlotsky J.M., Bastle A., Hezode C., Lonjon I., Darthuy F., Remine J., Dhumeaux D. Routine detection and quantification of hepatitis B virus DNA in clinical laboratories. J. Immunol. Methods. 2000;85:11–21. doi: 10.1016/s0166-0934(99)00149-4. [DOI] [PubMed] [Google Scholar]
- Noborg U., Gusdal A., Pisa E.K., Hedrum A., Lindh M. Automated quantitative analysis of hepatitis B virus DNA by using the cobra Amplicor HBV monitor test. J. Clin. Microbiol. 1999;37:2793–2797. doi: 10.1128/jcm.37.9.2793-2797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilbert A.R., Miller D.S., Scougall C.A., Turnbull H., Burrell C.J. Kinetics of duck hepatitis B virus infection following low dose virus inoculationone virus DNA genome is infectious in neonatal ducks. Virology. 1996;226:338–345. doi: 10.1006/viro.1996.0661. [DOI] [PubMed] [Google Scholar]
- McIntyre N. Clinical presentation of acute viral hepatitis. Br. Med. Bull. 1990;46:533–547. doi: 10.1093/oxfordjournals.bmb.a072414. [DOI] [PubMed] [Google Scholar]
- Guidotti L.G., Rochford R., Chung J., Shapiro M., Purcell R., Chisari F.V. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- Nowak M.A., Bonhoeffer S., Hill A.M., Boehme R., Thomas H.C., McDade H. Viral dynamics in hepatitis B virus infection. Proc. Natl. Acad. Sci. USA. 1996;93:4398–4402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiang M., Rooney J.F., Toole J.J., Gibbs C.S. Biphasic clearance kinetics of hepatitis B virus from patients during adefovir dipivoxil therapy. Hepatology. 1999;29:1863–1869. doi: 10.1002/hep.510290626. [DOI] [PubMed] [Google Scholar]
- Guo J.T., Zhou H., Liu C., Aldrich C., Saputelli J., Whitaker T., Barrasa M.I., Mason W.S., Seeger C. Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infections. J. Virol. 2000;74:1495–1505. doi: 10.1128/jvi.74.3.1495-1505.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock S.D., Dooley J. Diseases of the Liver and Biliary System 10th ed 1996. Blackwell Science Ltd; London, England: pp. 714 pp [Google Scholar]
- Mason W.S., Halpern M.S., England J.M., Seal G., Egan J., Coates L., Aldrich C., Summers J. Experimental transmission of duck hepatitis B virus. Virology. 1983;131:375–384. doi: 10.1016/0042-6822(83)90505-6. [DOI] [PubMed] [Google Scholar]
- Tuttleman J.S., Pourcel C., Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- Newbold J.E., Xin H., Tenca M., Sherman G., Dean J., Bowden S., Locarnini S. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J. Virol. 1995;69:3350–3357. doi: 10.1128/jvi.69.6.3350-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M.A., Lloyd A.L., Vasquez G.M., Wiltrout T.A., Wahl L.M., Bischofberger N., Williams J., Kinter A., Fauci A.S., Hirsch V.M., Lifson J.D. Viral dynamics of primary viremia and antiretroviral therapy in simian immunodeficiency virus infection. J. Virol. 1997;71:7518–7525. doi: 10.1128/jvi.71.10.7518-7525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little S.J., McLean A.R., Spina C.A., Richman D.D., Havlir D.V. Viral dynamics of acute HIV-1 infection. J. Exp. Med. 1999;190:841–850. doi: 10.1084/jem.190.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civitico G.M., Locarnini S.A. The half-life of duck hepatitis B virus supercoiled DNA in congenitally infected primary hepatocyte cultures. Virology. 1994;203:81–89. doi: 10.1006/viro.1994.1457. [DOI] [PubMed] [Google Scholar]
- Delaney W.E., Miller T.G., Isom H.C. Use of the hepatitis B virus recombinant baculovirus-HepG2 system to study the effects of (-)-beta-2′,3′-dideoxy-3′-thiacytidine on replication of hepatitis B virus and accumulation of covalently closed circular DNA. Antimicrob. Agents Chemother. 1999;43:2017–2026. doi: 10.1128/aac.43.8.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J., Mason W.S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Mondelli M., Vergani G.M., Alberti A., Vergani D., Portmann B., Eddleston A.L., Williams R. Specificity of T lymphocyte cytotoxicity to autologous hepatocytes in chronic hepatitis B virus infectionevidence that T cells are directed against HBV core antigen expressed on hepatocytes. J. Immunol. 1982;129:2773–2778. [PubMed] [Google Scholar]
- Chisari F.V., Ferrari C. Hepatitis B virus immunopathogenesis. Annu. Rev. Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- Feutren G., Lacour B., Bach J.F. Immune lysis of hepatocytes in cultureaccurate detection by aspartate aminotransferase release measurement. J. Immunol. Methods. 1984;75:85–94. doi: 10.1016/0022-1759(84)90227-8. [DOI] [PubMed] [Google Scholar]
- Kew M.C. Serum aminotransferase concentration as evidence of hepatocellular damage. Lancet. 2000;355:591–592. doi: 10.1016/S0140-6736(99)00219-6. [DOI] [PubMed] [Google Scholar]
- Webster G.J.M., Reignat S., Maini M.K., Whalley S.A., Ogg G.S., King A., Brown D., Amlot P.L., Williams R., Vergani D. Incubation phase of acute hepatitis B in mandynamic of cellular immune mechanisms. Hepatology. 2000;32:1117–1124. doi: 10.1053/jhep.2000.19324. [DOI] [PubMed] [Google Scholar]
- Gerken G., Manns M., Gerlich W.H., Hess G., Meyer zum Buschenfelde K.-H. Pre-S encoded surface proteins in relation to the major viral surface antigen in acute hepatitis B virus infection. Gastroenterology. 1987;92:1864–1868. doi: 10.1016/0016-5085(87)90617-2. [DOI] [PubMed] [Google Scholar]
- Heermann K.H., Goldmann U., Schwartz W., Seyffarth T., Baumgarten H., Gerlich W. Large surface proteins of hepatitis B virus containing the pre-S sequence. J. Virol. 1984;52:396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffello-Le Guillou D., Duclos-Vallee J.C., Eberle F., Capel F., Petif M.A. Evaluation of an enzyme linked immunosorbent assay for detection and quantification of hepatitis B virus preS1 envelope antigen in serum samplescomparison with two commercial assays for measuring hepatitis B virus DNA. Viral Hepatitis. 2000;7:387–392. doi: 10.1046/j.1365-2893.2000.00248.x. [DOI] [PubMed] [Google Scholar]
- Lin E., Luscombe C., Wang Y.Y., Shaw T., Locarnini S. The guanine nucleoside analog penciclovir is active against chronic duck hepatitis B infection in vivo. Antimicrob. Agents Chemother. 1996;40:413–418. doi: 10.1128/aac.40.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymans K.C. Risks of chronicity following acute hepatitis B virus infectiona review. Clin. Infect. Dis. 1995;20:992–1000. doi: 10.1093/clinids/20.4.992. [DOI] [PubMed] [Google Scholar]
- Szmuness W., Stevens C.E., Zang E.A., Harley E.J., Kellner A. A controlled clinical trial of the efficacy of the hepatitis B vaccine (Heptavax B)a final report. Hepatology. 1981;1:377–385. doi: 10.1002/hep.1840010502. [DOI] [PubMed] [Google Scholar]
- Mansky L.M., Temin H.M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]