Abstract
To define the phenotype and T cell receptor (TCR) repertoire of CD1d-dependent T cells, we compared the populations of T cells that persisted in major histocompatibility complex (MHC)-deficient mice, which lack mainstream T cells, with those from MHC/CD1d doubly deficient mice, which lack both mainstream and CD1d-dependent T cells. Surprisingly, up to 80% of the CD1d-dependent T cells were stained by tetramers of CD1d/α-galactosylceramide, which specifically identify the previously described CD1d autoreactive Vα14-Jα18/Vβ8 natural killer (NK) T cells. Furthermore, zooming in on the CD1d-dependent non-Vα14 T cells, we found that, like Vα14 NK T cells, they mainly expressed recurrent, CD1d autoreactive TCR families and had a natural memory phenotype. Thus, CD1d-restricted T cells differ profoundly from MHC-peptide–specific T cells by their predominant use of autoreactive and semiinvariant, rather than naive and diverse, TCRs. They more closely resemble other lineages of innate lymphocytes such as B-1 B cells, γδ T cells, and NK cells, which express invariant or semiinvariant autoreactive receptors. Finally, we demonstrate that the MHC-restricted TCR repertoire is essentially non–cross-reactive to CD1d. Altogether, these findings imply that lipid recognition by CD1d-restricted T cells may have largely evolved as an innate rather than an adaptive arm of the mouse immune system.
Keywords: CD1, TCR, autoreactivity, T cell development, lipid antigens
Introduction
The MHC and the CD1 gene families diverged several hundred million years ago from the duplication of an ancestor locus 1. The CD1 locus of mammals includes up to five distinct isotypes encoding β2-microglobulin–associated molecules that, based on sequence homology, segregate into two groups. Group I contains CD1a, b, c, and e, and group II contains CD1d 2. In contrast to MHC, CD1 molecules are not polymorphic, and they present lipids rather than peptides to T cells 3 4. These two features have raised the prospect that CD1 might serve as the basis for population-wide vaccines directed against conserved microbial and tumor-specific glycolipids. However, the diversity of the CD1-restricted TCR repertoire is still unknown, as is the extent to which it might be able to generate effective primary and secondary adaptive immune responses against a broad range of antigens.
CD1-restricted T cells differ in several interesting ways from MHC-restricted T cells. For example, they include an inordinate proportion of double negative (DN) CD4−CD8− αβ T cells 3 5, a phenotype also associated with some unusual autoreactive, MHC-restricted T cells 6 7 8. They also include prominent T cell subsets expressing characteristics of innate rather than adaptive antigen recognition such as the CD1d autoreactive Vα14 NKT cells and the CD1c autoreactive Vδ1 T cells. Vα14 NKT cells express a TCR repertoire made of an invariant Vα14-Jα18 chain (Vα24-Jα18 in humans) mainly associated with the variable Vβ8 chains (Vβ11 in humans; reference 9) and uniformly recognize the glycolipid α-galactosylceramide (αGalCer) at picomolar concentrations 4. They also express several receptors associated with the NK lineage. Despite their limited TCR repertoire, Vα14 NKT cells constitute a disproportionate fraction of the mature T cell population of the adult spleen (2%), thymus (10%), bone marrow (20%), and liver (30%). Vδ1 γδ T cells constitute a major population of human intraepithelial intestinal lymphocytes and are considered to be part of the innate rather than adaptive immune system 10. These prominent Vα14 and Vδ1 CD1-restricted subsets seem to obey a logic that differs markedly from adaptive lymphocytes but closely resembles other “innate” lymphocytes, such as B-1 B cells, γδ T cells, and NK cells, which express invariant or semiinvariant antigen receptors and survey self-antigens or promiscuous (self plus foreign) antigens that are induced or exposed in conditions of infection, stress, or transformation 11 12 13 14 15.
However, there are indications that the CD1 system may have taken up a more recent evolutionary function of presenting a variety of lipid antigens to the adaptive immune system. Recent studies have shown that distinct mycobacterial lipid antigens could be recognized by in vitro–derived CD1-restricted T cell lines (for a review, see reference 3). In addition, patients recently infected with Mycobacterium tuberculosis exhibited detectable CD1c-restricted T cell responses to a semisynthetic isoprenoid glycolipid resembling natural mycobacterial antigens 16. Other studies have also suggested some diversity in both TCR repertoire and antigenic specificities 17 18 19 20 21 22 23.
The coexistence of innate and adaptive TCR recognition would be a unique and fascinating aspect of the CD1 system, likely to have important evolutionary implications for the protective value of lipid, as opposed to peptide, recognition. Therefore, we set out to define the respective sizes of the adaptive and innate T cell populations associated with CD1d, the only CD1 isotype expressed in mice, and the one shared by most, though perhaps not all 24, mammals thus far examined. To search for a diverse, adaptive population of CD1d-restricted T cells, we quantitatively examined the contribution of CD1d to the selection of CD4, CD8, and DN αβ T cells by comparing the size and composition of the residual T cell populations in mice deficient for MHC (which lack mainstream αβ T cells) versus mice doubly deficient for MHC and CD1d (which lack both the mainstream T cells and the CD1d-dependent T cells). By using fluorescent tetramers of CD1d1/αGalCer complexes to specifically identify Vα14 T cells, we were able to focus on the non–Vα14-based T cell population, determine its size and phenotype, and analyze its TCR repertoire. Further, we explored the possibility that classical MHC molecules might select T cells able to cross-react with CD1d that would contribute to the CD1d-restricted repertoire.
Our results indicate that the mouse CD1d-restricted TCR repertoire is limited in both size and diversity. It is dominated by cells bearing a few recurrent semiinvariant TCRs that exhibit the hallmarks of self-antigen–driven selection and the same “natural memory” NKT phenotype as Vα14 NKT cells. These surprising findings imply that recognition of lipid antigens by mouse CD1d-restricted T cells largely evolved in the context of innate rather than adaptive immunity.
Materials and Methods
Mice.
C57BL/6 mice were obtained from Taconic Farms. Mice deficient in KbDb or transporter associated with antigen presentation (TAP)-1 (referred to as MHC class I deficient), I-Ab (MHC class II deficient), CD1d1, or CD1d1/CD1d2 (CD1d deficient), and their crosses were used after 7–13 backcrosses to C57BL/6 25. All the results reported in this study were derived from comparative analysis of littermates expressing −/− versus +/− or +/+ genotypes. All mice were raised in a specific pathogen-free barrier environment at Princeton University, according to institutional animal care and use guidelines.
Flow Cytometry Studies and Monoclonal Antibodies.
To obtain liver or lung lymphocytes, the organs were first cut into small pieces and passed through a nylon mesh in PBS/1% BSA, then the cell suspension was centrifuged over a layer of Ficoll-Paque™ (Amersham Pharmacia Biotech) and the lymphocytes were collected at the interface between Ficoll and PBS-BSA. Fluorochrome or biotin-conjugated anti-pan TCR-β, Vβ8.1/8.2, Vβ7, Vβ2, Vβ6, Vα3.2, CD44, CD69, CD122, CD4, CD8, CD5, NK1.1, and heat stable antigen were obtained from BD PharMingen, as well as streptavidin-Cychrome, -allophycocyanin, or -PE. Anti-Vα8 monoclonal antibodies including KT 50, KT 65, and B21.14 26 27 were obtained from BD PharMingen and through Dr. B.J. Fowlkes (National Institutes of Health, Bethesda, MD). Samples were analyzed using a FACS Vantage™ (Becton Dickinson) equipped with argon and dye lasers, or a four-color FACSort™ equipped with argon and 635-nm diode lasers (Becton Dickinson), and CELLQuest™ software.
CD1d1/αGalCer Tetramers.
Recombinant soluble CD1d1 molecules with a biotinylation sequence were produced in a fly expression system, purified, and biotinylated as reported previously 28. Soluble biotinylated recombinant CD1d1 molecules were incubated for 18 h with a 2 μM stock solution of αGalCer in 0.5% Tween 20, and free αGalCer was removed by centrifugation dialysis in a Microcon YM-30 tube (Millipore). Tetramers were generated by mixing αGalCer-loaded monomers with fluorochrome-labeled streptavidin (streptavidin-PE, streptavidin-allophycocyanin, or streptavidin-Cychrome from BD PharMingen at 5:1) without further separation by gel filtration. Staining was performed by incubating cells on ice for 3 h with tetramers at a concentration equivalent to 3–5 μg/ml of CD1d1.
Killing Assay.
C57BL/6.CD1d−/− female mice were injected intraperitoneally with 3 × 106 spleen cells from male or female C57BL/6 or C57BL/6.CD1d−/− mice. 2 wk later, spleen cells were cultured with irradiated (3,000 rads) male C57BL/6 splenocytes for 5 d in culture medium supplemented with 10 U/ml IL-2 before assay against tritiated thymidine-labeled LPS blasts, as described (JAM test; reference 29).
T Cell Hybridoma Production and In Vitro Stimulation.
T cell hybridomas were obtained as described previously, by fusion with BW5147αβ2 of sorted NK1.1+ or NK1.1− MHC class II–deficient CD4 splenocytes that had been stimulated for 4–5 d with anti-CD3 and a mixture of IL-7 and IL-2 23 30. To detect CD1d autoreactivity, 5 × 104 hybridoma cells were mixed in a 96-well plate with 5 × 104 mouse CD1d transfectants (rat basophilic leukemia mast cells and/or mouse C57SV fibroblasts, as described) or with 5 × 105 cells of a 1:1 mixture of splenocytes and thymocytes, using as culture medium a 1:1 mixture of EHAA and RPMI 1640 (Biofluids) supplemented with 10% FCS, 5 × 10−5 M 2 ME, penicillin-streptomycin-gentamycin, and glutamine. Supernatants were harvested after 18–24 h to measure IL-2 release using CTLL cells as described. The IL-2 released by CD1d autoreactive hybrids in the presence of control untransfected cell lines or CD1d-deficient thymocytes/splenocytes was undetectable or at least five times lower than in the presence of CD1d-expressing cells.
TCR Gene Sequencing.
Reverse transcription PCR, primers, and methods were as described previously 23 31. TCR α and β chain nomenclature is according to the new World Health Organization-International Union of Immunological Societies (IUIS) nomenclature 32.
Results
The Size and Phenotype of the CD1d-dependent T Cell Population.
To improve our ability to identify CD1d-dependent T cells, we studied MHC-deficient mice, where the major “mainstream” population of MHC-restricted T cells is absent. We compared the size and phenotype of the T cell populations present in various lymphoid and nonlymphoid tissues of MHC only and MHC/CD1d doubly deficient mice. To minimize individual variations, we always compared littermates and usually cagemates derived from CD1d+/− intercrosses or crosses between CD1d−/− and CD1d+/− mice. In addition, all strains of mice deficient in MHC class II (I-Ab), MHC class I (KbDb or TAP), or CD1d were on the same C57BL/6 background.
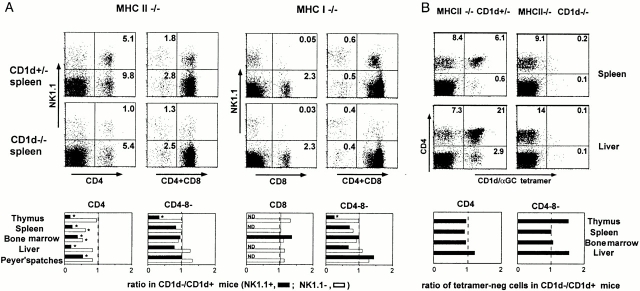
We studied NK1.1-positive and -negative αβ T cells separately, as NK1.1 is prominently expressed by some but not all CD1d-restricted T cells. Representative examples of FACS® dot plots displaying the CD4/CD8 versus NK1.1 profiles of TCR-α/β1 gated splenocytes are shown in Fig. 1 A for both the MHC class II–deficient and the MHC class I–deficient backgrounds. In addition, results from several experiments were compiled. The bar graphs in Fig. 1 A show the average values of the ratio of the cell number in the CD1d-deficient over that in the CD1d-sufficient background for several pairs of littermates (measured for 5–14 pairs of littermates per point) in the thymus, spleen, liver, bone marrow, and Peyer's patches.
Figure 1.
Residual αβ T cell subsets in MHC-versus MHC/CD1d–deficient mice. (A) NK1.1+ and NK1.1− subsets. FACS® dot plots represent the TCR-β1 gated splenocytes from MHC class II– and MHC class I–deficient mice stained with antibodies against NK1.1, CD4, and/or CD8. In the bar graphs, bars represent the average ratio of the numbers of each cell type in CD1d−/− mice versus CD1d+/− littermates (5–14 littermate pairs per data point). MHC class I−/− mice were either KbDb−/− or TAP−/−; results with KbDb−/− mice (shown in the FACS® dot plots) and TAP−/− mice (summarized in the bar graphs) were similar. Error bars are not shown for graphic clarity. ND, not detected (frequency <0.05%). *Statistically significant (P < 0.05). (B) CD1d/αGalCer+ and CD1d/αGalCer− subsets. The FACS® dot plots show the CD4 and CD1d/αGalCer tetramer profiles of TCR-β1 gated cells from MHC class II−/−CD1d+/− and MHC class II−/−CD1d−/− littermates, as indicated. The bar graphs represent the average ratio of the numbers of CD1d/αGalCer− cells in CD1d−/− mice versus CD1d+/− littermates (four to eight littermate pairs per data point).
As expected, in the MHC class II–deficient background, where the mainstream CD4 αβ T cell population is absent, a marked reduction of up to 70–80% of the CD4+NK1.1+ population, which is mainly composed of Vα14 T cells, was observed in the absence of CD1d. CD4+ NK1.1− cells in the spleen and the bone marrow of MHC class II–deficient mice also exhibited a 30–40% average decrease in the absence of CD1d, suggesting the presence of a CD1d-dependent subset among these cells, but in other tissues they were largely unaffected. Other populations in either the MHC class I– or MHC class II–deficient backgrounds were largely unaffected except for the CD4−CD8− DN cells, where a clear reduction was seen among the NK1.1+ cells of the thymus in CD1d-deficient mice. For the few residual CD8 cells in the MHC class I–deficient (TAP- or KbDb-deficient) backgrounds, which are very rare and mostly NK1.1−, no change in frequency was found in the absence of CD1d.
Thus, the CD1d-dependent population uncovered by this analysis is predominantly composed of cells which seem to qualitatively and quantitatively correspond to the Vα14 NK1.1+ T cells that were characterized previously 9 33 34. A population of NK1.1−CD4 T cells in the spleen and the bone marrow also seemed to be CD1d restricted, but their frequency was less than that of the canonical NK1.1+ Vα14 T cells in these tissues. We studied these cells in greater detail (see below). However, it should be noted that ∼50% of all CD4+TCR-α/β1 MHC class II–deficient splenocytes appeared to be CD1d independent. Their specificity and function remain to be determined. Finally, despite the fact that CD8 seems to be a coreceptor for CD1d 35 36 37, the study did not detect a sizable population of CD1d-dependent CD8 T cells.
CD1d/αGalCer+ T Cells Account for the Great Majority of CD1d-dependent T Cells.
In both mice and humans, αGalCer is uniformly and exclusively recognized by cells expressing the canonical mouse Vα14 or human Vα24 TCRs. Using fluorescent CD1d/αGalCer tetramer staining, we and others recently showed that a fraction of Vα14 T cells did not express NK1.1 and conversely, that a fraction of NK1.1+ T cells did not express the canonical Vα14 TCR 28 38 39. Thus, to refine our in vivo analysis of the CD1d-dependent T cell population, we reexamined the population of CD4 T cells in MHC class II–deficient mice, using CD1d/αGalCer tetramers to specifically determine the contribution of Vα14 T cells, independently of their NK1.1-positive or -negative cell surface phenotype. We stained cells from various tissues of CD1d-sufficient or -deficient, MHC class II–deficient mice with anti-CD4, anti–TCR-β, and tetramers. As expected, there were no CD4+ tetramer-positive cells in the TCR-β1 population of CD1d-deficient mice. However, the frequencies of CD4+ tetramer-negative cells were not detectably affected (Fig. 1 B). Thus, for example, the 6.1% of the total splenic αβ T cells that expressed the CD4+ tetramer-positive phenotype in MHC class II–deficient mice disappeared in the absence of CD1d (0.2%), whereas the 8.4% that were CD4+ tetramer-negative persisted (9.1%). A similar conclusion is reached after examination of the bar graphs in Fig. 1 B showing the average values of the ratio of the cell numbers in the CD1d-deficient over that in the CD1d-sufficient background for several pairs of littermates (measured for four to eight pairs of littermates per point) in the thymus, spleen, liver, and bone marrow.
Thus, after excluding the conventional MHC-restricted T cells by using MHC-deficient mice, and the canonical Vα14 T cells by staining with CD1d/αGalCer tetramers, our examination failed to find evidence for a sizable population of CD1d-dependent TCR-α/β1 T cells that did not use the Vα14 TCR. This result implies that the minor set of CD1d-dependent, NK1.1−CD4 T cells we had seen in the spleen and bone marrow is composed of Vα14 T cells and is in agreement with our recent finding that a subset of Vα14 T cells in wild-type C57BL/6 mice do not express NK1.1. These data, together with studies of other tissues such as the lung, gut, lymph nodes, and blood, where CD1d-dependent T cells are very rare (data not shown), indicate that the non-Vα14 CD1d-restricted T cells must be extremely rare.
The Non-Vα14, CD1d-dependent TCR Repertoire.
In apparent contradiction to the above in vivo findings, we and others have reported previously several examples of in vitro–derived CD4+ CD1d autoreactive hybridomas or clones that expressed non-Vα14 TCRs 17 22 23 30. To reexamine the frequency and the TCR repertoire of these cells, we undertook a large-scale, systematic study of T cell hybridomas derived from short-term TCR-β–activated CD4+ NK1.1-positive or -negative splenocytes from MHC class II–deficient mice. We measured the frequency of hybridomas expressing the canonical Vα14-Jα18 TCR α chain by PCR. We also tested for CD1d autoreactivity by measuring the IL-2 released after exposure to a CD1d-expressing thymic/splenic cell mixture or to various CD1d transfected cell lines versus CD1d-deficient thymic/splenic cells or untransfected cell lines. Table shows that out of >400 hybridomas examined, 82% of the hybrids derived from the NK1.1+ and 33% of those derived from the NK1.1− cells expressed the canonical Vα14 TCR. Whether originating from the NK1.1-positive or -negative cells, one third of these Vα14 hybrids expressed overt autoreactivity to CD1d. It is likely that the frequency of autoreactivity is substantially higher than one third because many hybridomas are intrinsically low IL-2 producers and the response to CD1d-expressing cells is only a fraction of their response to anti-CD3 stimulation (data not shown). Some other hybridomas did not express Vα14, yet were also CD1d autoreactive. Collectively, they represented 6.5 and 9.5% of the hybrids in the panels derived from the NK1.1-positive and -negative CD4 cells, respectively.
Table 1.
Frequency of CD4 Hybridomas Expressing CD1d-specific TCRs
| NK1.1-negative | NK1.1-positive | |||||
|---|---|---|---|---|---|---|
| Autoreactive | Nonautoreactive | Total | Autoreactive | Nonautoreactive | Total | |
| % | % | |||||
| Vα14-Jα18 | 10 | 23 | 33 | 29 | 53 | 82 |
| Vα3.2-Jα9 | 1.5 | 1.5 | 3 | 4 | 3 | 7 |
| Vα8 | 4 | ND | ND | 0.5 | ND | ND |
| Others | 4 | ND | ND | 2 | ND | ND |
Then we analyzed the TCR α and β chains of the non-Vα14 hybridomas that were CD1d autoreactive. While most used Vβ8 chains with diverse CDR3 junction sequences and lengths (data not shown), a recurrent usage of a few Vα chains was observed in a disproportionately large fraction.
A Vα3.2-Jα9 rearrangement was repeatedly found in several independent fusions. It was also expressed by the hybridoma 431.A11, obtained in a previously published fusion experiment (referred to as Vα3.2-Jα8 based on the previous TCR gene nomenclature). From the analysis of unrelated Vα3.2-Jα9 clones, represented in Table , a canonical, quasiinvariant amino acid junction appears to be recurrently used. Based on this canonical sequence, additional Vα3.2-Jα9+ clones could be identified by systematic reverse transcription PCR screening of the rest of the hybridomas that were not overtly CD1d autoreactive (∼55% of Vα3.2-Jα9+ hybridomas did not exhibit overt autoreactivity to CD1d, some of which may be low IL-2 producers that escaped detection by the in vitro assay). Interestingly, in contrast with the Vα14-Jα18 sequence, which is germline encoded, at least two or three nucleotide substitutions were almost systematically present in the Vα3.2-Jα9 junction, introducing, in 11/14 sequences, an arginine or a lysine in position 94 and a glycine in position 95. Together with the fixed amino acid length of the junctions, these substitutions are characteristic of an antigen-driven selection process. In addition, the requirement for nucleotide substitution most likely explains why the Vα3.2-Jα9/Vβ8 cells are less frequent than the Vα14-Jα18/Vβ8 cells. Interestingly, like Vα14 T cells, Vα3.2-Jα9/Vβ8 cells were found in both the NK1.1-negative and -positive compartments (3 and 7%, respectively; Table ), whether or not they were scored as overtly autoreactive to CD1d in the in vitro culture assay.
Table 2.
Nucleotide and Predicted Amino Acid Sequences of the Vα3-Jα9 Junctions of 14 T Cell Hybridomas Derived from CD4+TCR-α/β1 Splenocytes of MHC Class II−/− Mice
| Vα chain | Vβ chain | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Vα3 germline: | TGT | GCT | GTG | AGC | A | ||||||
| AGC | AAC | ATG | GGC | TAC | Ja9 germline | ||||||
| Cys | Ala | Val | 93 Lys | 94 Gly | Asn | Met | Gly | Tyr | |||
| 431.A11 | TGT | GCT | GTG | Aag | gGC | AAC | ATG | GGC | TAC | Vβ8.2Jβ2.4 | |
| S2M1 | Vβ8.2Jβ2.2 | ||||||||||
| F3F11.352 | Vβ8 | ||||||||||
| Lys | Gly | ||||||||||
| S2M2 | … | … | … | Aaa | gGC | … | … | … | … | Vβ8.1Jβ2.3 | |
| Arg | Gly | ||||||||||
| SPS3.3 | … | … | … | Aga | gGC | … | … | … | … | Vβ5 | |
| S2M15 | ND | ||||||||||
| Arg | Gly | ||||||||||
| DPS1.1 | … | … | … | Agg | gGC | … | … | … | … | Vβ8 | |
| 32A11.209 | Vβ8 | ||||||||||
| SPS5.2 | Vβ8 | ||||||||||
| Arg | Gly | ||||||||||
| 31F8.268 | … | … | … | cgg | gGC | … | … | … | … | Vβ8 | |
| Arg | Phe | ||||||||||
| F5G7.370 | … | … | … | cgg | ttc | … | … | … | … | Vβ8 | |
| Ser | Gly | ||||||||||
| F3B4.342 | … | … | … | AGC | gGC | … | … | … | ,,, | Vβ8 | |
| Ser | |||||||||||
| 31F1.264 | … | … | … | AGC | … | … | … | Vβ8 | |||
| SPS8.15 | ND | ||||||||||
Nucleotide additions are in bold, lowercase letters.
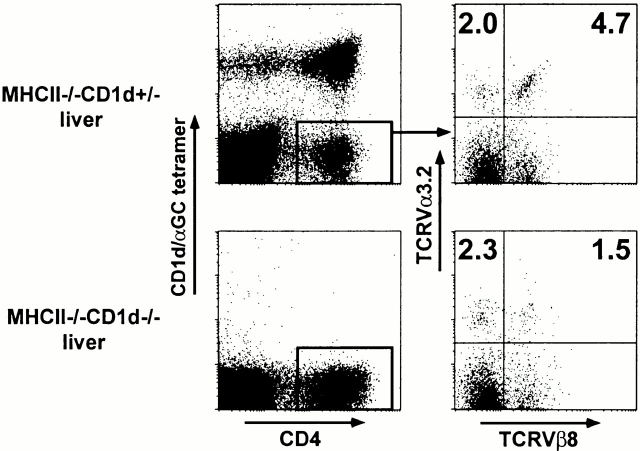
Because antibodies against both the Vα3.2 and Vβ8 gene products are available, we performed a flow cytometric analysis of these cells among MHC class II–deficient liver and spleen lymphocytes. By four-color analysis, gating on the residual CD4+ tetramer-negative lymphocytes, we measured an average frequency of 4.7% Vα3.2/Vβ8 cells in the liver and 2.7% in the spleen of a total of five to eight mice (Fig. 2, and Table ). The Vα3.2/non-Vβ8 cells represented only 2.4% on average of the cells in these tissues, indicating a bias towards Vβ8 expression (53–66%) among the Vα3.2 cells. In MHC class II/CD1d doubly deficient littermates, the frequency of Vα3.2/Vβ8+ cells decreased 2–3.6-fold, while the frequency of Vα3.2/non-Vβ8 cells was unchanged, confirming the CD1d restriction of the majority of the Vα3.2/Vβ8 population. Thus, we estimate that ∼2% of the total residual CD4 splenocytes in MHC class II–deficient mice express the CD1d autoreactive Vα3.2-Jα9/Vβ8 TCR. This is much less than the ∼40% of residual CD4 cells that are Vα14 T cells (Fig. 1 B) and is in good agreement with the hybridoma studies, supporting the notion that the hybridoma panels did not distort in major ways the TCR repertoire of MHC class II–deficient CD4 cells.
Figure 2.
CD1d-dependent Vα3.2/Vβ8 T cells. Liver lymphocytes from a pair of MHC class II–deficient and MHC class II/CD1d doubly deficient littermates were four-color stained with CD1d/αGalCer tetramers and anti-CD4, Vα3.2, and Vβ8. Right panels are gated from CD4+CD1d/αGalCer− cells as indicated.
Table 3.
Frequency of Vα3.2+/Vβ8+CD4+ T Cells in MHC Class II−/−CD1d+/− and MHC Class II−/−CD1d−/− Mice
| Vα3.2+/Vβ8+ | Vα3.2+Vβ8− | ||||
|---|---|---|---|---|---|
| Tissue | Mice | MHC class II−/−CD1d+/− | MHC class II−/−CD1d−/− | MHC class II−/−CD1d+/− | MHC class II−/−CD1d−/− |
| Spleen | 8 | 2.67 (1.01) | 1.43 (0.11) | 2.41 (0.18) | 2.13 (0.48) |
| Liver | 5 | 4.71 (2.13) | 1.31 (0.41) | 2.39 (0.42) | 2.84 (0.421) |
Data show percentage of CD1d/αGalCer tetramer-negative CD4+ T cells.
Another TCR α chain, Vα8, also in conjunction with Vβ8, was also disproportionately used by CD1d reactive hybridomas (Table ). Different Jα segments were used for each clone and nongermline elements were systematically introduced (Table ). Although the length of these junctions was usually restricted to 9–10 amino acids, there was no obvious conserved motif among them. In contrast, we made the surprising observation that none of our nine independent CD1d autoreactive Vα8 clones could be stained with any of the three available anti-Vα8 antibodies KT50, KT65, and B21.14 (data not shown), suggesting the use of a restricted subset of Vα8 genes. Indeed, further analysis of whole length Vα8 sequences showed that all the Vα8 genes belonged to the Vα8S3, Vα8S5, Vα8S8, and Vα8S15 subfamilies, which differ from other mouse Vα8S6, Vα8S7, and Vα8S9 genes by several amino acid changes in both the CDR1 and CDR2 regions 32. Thus, reactivity against CD1d seems to require a particular motif in a CDR loop of Vα8 (which cannot be deduced from the simple inspection of sequences [data not shown]) that is also the region targeted by the three anti-Vα8 monoclonal antibodies. CD1d autoreactive Vα8 cells accounted for 4% of hybridomas originating from NK1.1− and 0.5% of those originating from NK1.1+ CD4 cells (Table ). Their frequency may be underestimated because, like a fraction of Vα14 and Vα3.2 T cells, some of the CD1d-specific Vα8 cells may not exhibit overt autoreactivity to CD1d in our in vitro cytokine release assay and because there is no Vα-specific reagent to stain them in vivo. Conversely, the fact that these autoreactive receptors could not be found in a panel of control hybridomas derived from MHC class II/CD1d doubly deficient CD4 splenocytes emphasizes that they are authentic CD1d-restricted TCRs.
Table 4.
Nucleotide and Predicted Amino Acid Sequences of the Vα8-Jα Junctions of Nine CD1d Autoreactive T Cell Hybridomas Derived from CD4+TCR-α/β1 Splenocytes of MHC Class II−/− Mice
| Vα chain | Vβ chain | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vα8 germline: | TGT | GCT | TTG | AGT | GA | Ala | Ser | Leu | Gly | |||||
| T | GAA | CTG | GCC | AGT | TTG | GGG | Jα24 germline | |||||||
| 91 | 92 | 93 | 94 | |||||||||||
| Cys | Ala | Pro | Arg | Thr | ||||||||||
| TBA7 | TGT | GCT | ccc | cga | act | … | … | … | … | Vβ8.2Jβ2.6 | ||||
| Met | Thr | Thr | ||||||||||||
| S2M6 | … | … | atg | aca | act | … | … | … | … | Vβ8.2Jβ2.1 | ||||
| Gln | Gly | Gly | Arg | |||||||||||
| CC | TAC | CAG | GGA | GGC | AGA | Jα15 germline | ||||||||
| Asp | Gly | |||||||||||||
| SPS1.1 | … | … | … | … | GAt | ggg | … | … | … | … | ND | |||
| Arg | Gly | Ser | Ala | Leu | Gly | |||||||||
| TA | GAT | AGA | GGT | TCA | GCC | TTA | GGG | Jα18 germline | ||||||
| Gly | ||||||||||||||
| S2M9 | … | … | … | … | Ggg | … | … | … | … | … | … | Vβ8.2Jβ2.1 | ||
| CA | TCT | TCT | GGC | AGC | TGG | Jα22 germline | ||||||||
| Arg | Gly | |||||||||||||
| SAA8 | … | … | cct | cta | … | … | … | … | … | Vβ2Jβ2.1 | ||||
| Asn | Asn | Tyr | Ala | Gln | ||||||||||
| GA | AAT | AAC | TAT | GCC | CAG | Jα26 germline | ||||||||
| Gly | Arg | |||||||||||||
| SPS10.20 | … | … | … | Ggg | aGA | … | … | … | … | … | Vβ8 | |||
| Asn | Thr | Asn | Thr | Gly | ||||||||||
| T | AAC | ACC | AAT | ACA | GGC | Jα27 germline | ||||||||
| Asp | Leu | Pro | ||||||||||||
| P17 | … | … | … | … | GAt | ctc | ccC | … | … | … | … | … | Vβ14 | |
| Ser | Ser | Asn | Thr | Asn | ||||||||||
| TCT | TCC | AAT | ACC | AAC | Jα34 germline | |||||||||
| Ser | Pro | |||||||||||||
| SPS1.20 | … | … | Tcc | cCT | … | … | … | … | Vβ8 | |||||
| Ala | Asn | Tyr | Gly | Asn | Glu | |||||||||
| GCC | AAC | TAT | GGA | AAT | GAG | Jα48 germline | ||||||||
| Glu | Trp | Asp | ||||||||||||
| SPS1.18 | … | … | … | … | GAa | tgg | gAT | … | … | … | Vβ5 | |||
Nucleotide additions are in bold, lowercase letters.
Finally, some autoreactive hybridomas expressed neither a Vα3.2-Jα9 nor a Vα8 TCR α chain (Table ). Collectively, their frequency was less than the combined frequency of the Vα3.2-Jα9 and Vα8 cells (see Table ). It is possible that larger studies would also reveal recurrent TCR usage among these cells, as is suggested, for example, by the observation that 4/14 independent clones used Vα10, which was paired with Vβ11 in three cases.
Table 5.
TCR Gene Usage by Vα14/Vα3.2/Vα8− CD1d Autoreactive Hybridomas
| Hybridoma | NK1.1 origin | VαJα | Vβ |
|---|---|---|---|
| SPS3.10 | − | Vα10Jα5 | Vβ11 |
| SPS1.21 | − | Vα10Jα22 | ND |
| SPS8.23 | − | Vα10Jα26 | Vβ11 |
| S2M7 | − | Vα10Jα26 | Vβ11 |
| SPS8.1 | − | Vα15Jα21 | Vβ2 |
| P4 | + | Vα15Jα22 | Vβ8.2Jβ2.4 |
| SPS3.5 | − | Vα2Jα2 | Vβ5 |
| SBC12 | − | Vα3Jα23 | Vβ5.1Jβ2.4 |
| SPS9.1 | − | Vα4Jα48 | ND |
| 32E9.227 | + | Vα4 | Vβ8 |
| S2M16 | − | Vα5Jα33 | Vβ8.2Jβ1.4 |
| S2P16 | + | Vα12Jα18 | Vβ4 |
| DPS1.7 | + | Vα13Jα22 | Vβ9 |
| S2P23 | + | Vα18Jα23 | Vβ8 |
Table summarizes the estimated frequencies of all CD1d-restricted TCR families among CD4+ splenocytes in MHC class II–deficient mice, based on all our hybridomas and in vivo studies. Although a precise estimate cannot be provided for all the families, there is an overall excellent correlation between in vitro (hybridomas) and in vivo (mice) studies, strengthening the conclusion that the CD1d-restricted TCR repertoire is dominated by a few autoreactive TCR families.
Table 6.
Estimated Frequencies of CD1d-restricted TCRs in MHC Class II–deficient CD4+ Splenocytes
| TCR family | In vitro studies(hybridomas) | In vivo(mice) |
|---|---|---|
| % | % | |
| Vα14-Jα18 | 48 | 40 |
| Vα3.2-Jα9 | 4 | 2 |
| Vα8 | >4 | ND |
| Other | >4 | ND |
| Total | ND | 52 |
Non-Vα14, CD1d-restricted T Cells Have a Natural Memory Phenotype.
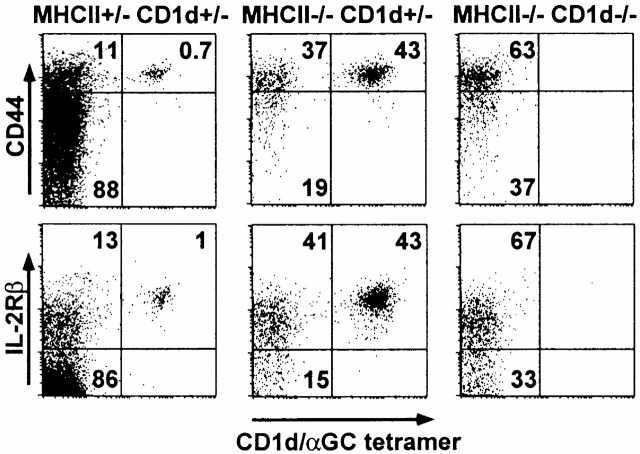
The above results suggested that a large fraction of CD4 cells expressing a CD1d-restricted TCR are in fact cells with overt or latent autoreactivity to CD1d. Therefore, like Vα14 T cells 9 and Vα3.2/Vβ8 T cells (data not shown), the other CD1d-dependent T cells should uniformly express a memory phenotype because of exposure to self. Fig. 3 shows that a majority of the CD4+CD1d/αGalCer− cells in MHC class II–deficient spleens did indeed express a memory CD44hiCD122hi phenotype. In MHC class II/CD1d doubly deficient littermates, the few tetramer-negative cells that exhibited a naive phenotype were slightly increased, further indicating the absence of a sizable component of naive CD1d-restricted non-Vα14 T cells. In support of this conclusion, direct staining of the CD1d-dependent Vα3.2/Vβ8 T cells in MHC class II–deficient mice showed a uniform memory phenotype (data not shown). Therefore, these results imply that most of the CD1d-dependent T cells express the characteristic phenotype of memory, antigen-experienced CD4 cells.
Figure 3.
Memory phenotype of CD1d-restricted T cells. Panels represent the FACS® dot plots of CD4 +TCR-β1 gated spleen cells of littermates of the indicated genotypes.
Taken together, our results strongly support the notion that nearly all of the non-Vα14 CD1d-restricted T cells are autoreactive, memory-type cells resembling Vα14 NK T cells. Their overall frequency is much lower than Vα14 NK T cells, most likely because their CD1d reactivity is not germline encoded, requiring several nucleotide insertions in the CDR3α region to generate the appropriate specificity, and therefore arising with lower probability.
Mainstream MHC-restricted CD8 Cells Do Not Cross-react onto CD1d.
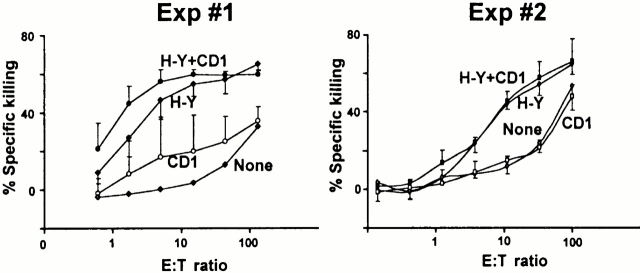
CD8 but not CD4 can function as a coreceptor for CD1d 18 35 40. To test the possibility that MHC molecules might select naive CD8 T cells capable of interacting with CD1d, we immunized C57BL/6.CD1d−/− female mice with C57BL/6.CD1d+/+ male spleen cells. This dual immunization with H-Y/Db and CD1d, both of which are highly expressed on dendritic cells and B cells, allowed us to compare the magnitude of the killer response against each of these antigens. H-Y/Db is a weak “minor” H antigen that is recognized by a population of CD8 cells whose frequency is very low, in the 10−5 range. In contrast, similar to allelic forms of MHC molecules, “CD1d” consists of a high density of various CD1d/glycolipid complexes to which the CD1d–deficient host is not tolerant. However, as shown in Fig. 4H-Y–specific killing was easily detected, whereas there was 10–50-fold lower CD1d-specific killing in several experiments. Furthermore, a significant fraction of this weak, inconsistent response might actually have been directed against CD1d peptides presented by MHC class I molecules, though, given the irregular response pattern, this possibility could not be directly tested. A similar result was obtained when CD1d/αGalCer was used as an antigen (data not shown). Therefore, the data support the notion that the MHC-restricted TCR repertoire is essentially unable to use CD1d as a restricting element, probably because of the divergent evolution of MHC and CD1.
Figure 4.
CD1d-deficient mice do not efficiently generate killer cells against CD1d. C57BL/6.CD1d−/− female mice were immunized with C57BL/6 male spleen cells and killing responses against LPS blasts from C57BL/6 males (H-Y + CD1), C57BL/6.CD1d−/− males (H-Y), C57BL/6 females (CD1), and C57BL/6.CD1d−/− females (none) were measured in vitro. Each graph represents separate experiments with three mice each.
Discussion
Before this study, the nature and diversity of the CD1-restricted T cells had not been systematically explored. However, this question has essential implications not only for the prospects of lipid vaccination but also, more generally, for our understanding of the evolution of lipid versus peptide antigen recognition by T cells.
Although there is evidence that CD1-restricted T cells may recognize diverse lipid antigens in a manner similar to MHC-restricted adaptive recognition of peptide antigens by mainstream αβ T cells 3 41, relatively few examples of CD1-restricted antigen-specific responses have been reported so far. On the other hand, and in contrast to MHC-restricted T cells, prominent subsets of CD1-restricted T cells expressing semiinvariant autoreactive TCRs have been identified, prompting the interesting suggestion that CD1 primarily evolved to survey self-lipid antigens in different intracellular compartments, only recently acquiring or being coopted for the ability to present microbial lipids to T cells 42.
We focused our study on CD1d, the only isotype expressed in the mouse. In addition to the conserved CD1d autoreactive population of mouse Vα14/human Vα24 NK T cells, the existence of other CD1d-restricted T cells, possibly associated with the adaptive rather than the innate arm of immunity, had been inferred from reports of non-Vα14 CD1d-restricted T cells 17 22 23 and from immunization experiments suggesting that CD1d-restricted CD8 T cell–mediated killing responses against peptide antigens could be elicited in vivo 18 37.
To estimate the size and diversity of the non-Vα14 CD1d-restricted TCR repertoire and to determine the frequency and phenotype of the corresponding T cells, we performed in vivo studies of MHC-deficient and MHC/CD1d doubly deficient mice using CD1d/αGalCer tetramers to identify the Vα14 T cells and zoom in on the non-Vα14–based CD1d-restricted population, combined with in vitro analysis of large panels of non–MHC-restricted hybridomas. The striking conclusion emerging from our studies was that, in sharp contrast with MHC class I– or class II–restricted T cells, the TCR repertoire of CD1d-restricted T cells is dominated by a restricted set of αβ TCRs. In addition to the Vα14 TCR, we found that some of the “diverse” TCRs previously identified in vitro were actually recurrently used, a finding that escaped recognition because of the limited size of the hybridoma panels examined. The most prominent of the non-Vα14 TCR families, Vα3.2-Jα9/Vβ8 and Vα8/Vβ8, were expressed by a sufficient number of independent clones that an in depth analysis was possible. This revealed that, unlike the canonical Vα14-Jα18, these TCRs were not germline encoded, a finding that is sufficient to explain why they would arise at a much lower frequency than the Vα14 TCRs during the random V-J recombination process. Particularly revealing was the study of the Vα3.2-Jα9 sequences, which disclosed that at least three nucleotide substitutions were required to generate a canonical motif, including arginine/lysine 94 and glycine 95. Despite their very low frequency (∼20 times less than Vα14 T cells), we were able to visualize the Vα3.2/Vβ8 cells in vivo by TCR-α and -β staining of the residual CD4+TCR-β1 cells in MHC class II–deficient mice, and demonstrate their dependence on CD1d by examining MHC class II/CD1 doubly deficient mice.
Interestingly, like the Vα14-Jα18 T cells themselves, the non-Vα14 CD1d-dependent T cell hybrids were often autoreactive, responding to CD1d-expressing thymocytes, splenocytes, and transfected CD1d+ lines. Like the Vα14-Jα18 sequences, the Vα3.2-Jα9 segments clearly exhibited signs of antigen-driven selection, suggesting in vivo selection by self-antigen. Unlike the Vα14 population, however, the non-Vα14 T cells do not respond to the surrogate antigen αGalCer 28 43. Their pattern of autoreactivity also differs from Vα14 T cells in that we previously showed they do not require endosomal targeting by the tyrosine-based endosomal targeting motif of CD1d to respond to their self-ligand 23. Altogether, these data indicate that, unlike the MHC-restricted TCR repertoire, the CD1d-restricted TCR repertoire is dominated by a limited range of specificities for distinct self-antigens rather than by a large set of specificities for a diverse universe of foreign antigens.
Our results do not exclude the existence of a functional diverse and naive “adaptive” CD1d-restricted TCR, but they suggest that it must be very limited in size compared with the autoreactive innate repertoire. An inherent limitation in our studies is that we could not directly visualize the CD1d-restricted T cells, but rather we inferred their frequencies from a comparison between MHC and MHC/CD1d doubly deficient mice. Thus, it is possible that a population of naive and diverse CD1d-restricted T cells might go undetected because it somehow requires the presence of MHC or MHC-restricted T cells. This is unlikely because, first, cell populations tend to expand rather than contract where space is available and second, the absolute numbers of Vα14-Jα18 and Vα3.2/Vβ8 CD1d-restricted T cells, which could be directly measured using flow cytometry, remained remarkably constant in the absence of MHC (data not shown). Another limitation of the study is that we could not ascribe a restriction element to up to 55% of the NK1.1− CD4 T cell hybridomas, because either they failed to exhibit CD1d autoreactivity or they did not express a canonical Vα14 or Vα3.2 TCR (Table ). It is possible that they include a proportion of CD1d-restricted T cells with diverse TCRs. However, this population would still be numerically inferior to the combined NK1.1+ and NK1.1− CD1d autoreactive T cells. In addition, it must include the residual MHC class II/CD1d–independent CD4 T cells found in vivo.
The phenotype of the CD1d-restricted T cells is consistent with the view that they predominantly express autoreactive TCRs. They do not exhibit a naive phenotype like the majority of MHC-restricted T cells but almost exclusively express high surface levels of several of the molecules associated with memory/effector cells, such as CD44 and IL-2Rβ. In addition, a significant fraction of them express NK receptors such as NK1.1, though the frequency of expression may depend on individual TCR families. Finally, they uniformly lack the expression of CD8, which was previously shown to induce negative selection of Vα14 autoreactive T cells, but rather express CD4 35 36. Thus, our study implies that (a) a majority of CD1d-dependent αβ T cells express an autoreactive TCR and a “natural memory” NKT phenotype, (b) a single germline encoded TCR family, Vα14-Jα18/Vβ8, Vβ7, Vβ2, is the main partner of CD1d, at least in the mouse, and (c) other natural examples of CD1d autoreactive αβ TCRs exist but they are less frequent, not germline encoded, and recurrent.
Although our results may seem to conflict with previous suggestions of a diverse CD1d-restricted TCR repertoire, a reanalysis of the published reports in the context of our data permits a resolution of some apparent contradictions. For example, it seems clear that other studies of T cell hybridomas and clones missed sighting the recurrent, antigen-driven non-Vα14 TCRs because the numbers of independent clones analyzed were small, inferior to five in each study. Conversely, in support of the view that CD1d-restricted T cells recognize a limited set of (mostly self) antigens, there are several indications that few exogenous antigens can be recognized in the context of CD1d. For example, mycobacterial lipoarabinomannan, an abundant mycobacterial product that can be recognized by human CD1b-restricted T cells 44 and also binds strongly to CD1d 28, failed to induce a CD1d-restricted T cell response in mice despite repeated immunizations 45. Furthermore, CD1d-deficient mice normally clear M. tuberculosis whether injected intravenously or inhaled 46 47. Likewise, immunization with a hydrophobic peptide carrying a CD1d binding motif (defined by screening of a peptide phage display library) elicited only a weak CD8 T cell response, even after strong priming by a simultaneous injection of different plasmids encoding CD1d, peptide, and B7. Attempts to derive in vitro lines or clones of these putative peptide-specific CD1d-restricted T cells were unsuccessful, preventing a full characterization of the responding T cells 18 37 40. Perhaps not surprisingly in light of our study, efforts by other investigators to obtain such cell lines specific for foreign peptides resulted instead in the generation of CD1d autoreactive clones expressing Vα14 and non-Vα14 TCRs 17. The inherently autoreactive nature of many non-Vα14 CD1d-restricted T cells is also highlighted by two independent reports that transgenic strains expressing non-Vα14 CD1d-restricted TCRs exhibited either overt autoreactivity and systemic lupus erythematosus in one case 48, or in the other case, the same memory/NKT phenotype as the Vα14 T cell subset 49. Finally, there is also good evidence from MHC-restricted TCR transgenic experiments that the DN NK1.1+ phenotype is often associated with autoreactivity 7 8.
Altogether, the results demonstrate a striking general bias of the naturally selected CD1d-restricted T cell population towards autoreactivity, explaining the natural memory phenotype and perhaps also the expression of inhibitory NK receptors to regulate it. This bias is contributed by a very restricted TCR α chain usage, which is often invariant, and a more promiscuous but still somewhat limited use of Vβ families, characteristically including Vβ8 with diverse VDJ junctions. Furthermore, the evolutionary divergence between CD1 and MHC molecules, which share only 30% identity, precludes the usage of MHC-restricted TCRs to take advantage of lipid presentation by CD1d because, as we showed, these TCRs do not cross-react onto CD1d molecules. Thus, we conclude that the population of αβ T cells available to recognize potential lipid antigens presented by CD1d is largely restricted to the natural memory/autoreactive NKT cell subset and its limited TCR repertoire. Our conclusion applies to the mouse CD1d system and cannot be extrapolated to other CD1 isotypes or to human CD1d. Further studies should clarify the extent to which these other CD1 isotypes may have taken up a more recent evolutionary function of presenting a variety of lipid antigens to the adaptive arm of the immune system.
There are striking resemblances between the CD1d-restricted T cell population, as defined in this study and B-1 B cells, marginal zone B cells, and γδ T cells. Like CD1d-restricted T cells, B-1 cells, marginal zone B cells, and γδ T cells are largely composed of antigen-driven autoreactive subsets expressing different types of semiinvariant germline-encoded antigen receptors with “supertypic” specificities for antigen families, and transgenic experiments have shown that, as for Vα14 T cells, the specificity of the antigen receptor drives a developmental program leading to acquisition of their peculiar differentiation and tissue distribution 12 50 51 52 53 54. Collectively, these studies delineate a family of lymphocytes expressing rearranged but germline-encoded self-antigen–reactive receptors that are present at relatively high frequencies and contribute to innate immunity. Their self-reactivity is somewhat difficult to comprehend within the context of the classical self/nonself discrimination model of tolerance, and in that regard, it is interesting that they usually prevent rather than aggravate chronic autoimmune conditions 15. It is increasingly envisioned that these innate cells may function as sentinels detecting modified self in conditions of tissue stress and damage 55 and act as regulators of the cytokine, chemokine, and costimulatory environment governing adaptive immune response. These innate lymphocytes are closer to NK cells, together with which they represent up to 20–50% of the lymphocyte population both in lymphoid organs and in tissues, than to classic adaptive B and T cells. Thus, they may not participate in memory formation to a diverse range of foreign antigens or be targets of conventional vaccine schemes.
Another important implication of this study is that there seem to exist systematic differences between MHC-driven and CD1d-driven T cell selection processes. What are the cause and the mechanisms underlying the peculiar development and differentiation of a majority of CD1d-restricted T cells and preventing the emergence of a naive, diverse CD1d-restricted population? One possibility is that only a few TCR-α genes have been evolutionarily selected to recognize CD1d, in comparison with those selected to recognize MHC products 56. In this scenario, given the lack of cross-reactivity of the MHC-restricted TCRs, the cells expressing these CD1d-specific genes would dominate the CD1d-restricted population, especially if they are autoreactive and strongly selected by chronic exposure to self-antigen. Another intriguing, nonexclusive possibility is that interaction with the cortical thymocyte, the CD1d-expressing cell type required for the selection of NKT cells 57, may not provide the same range of signals that specialized thymic epithelial cells provide for the selection of MHC-restricted low avidity T cells 58. In this scenario, only thymocytes expressing TCRs with high avidity for CD1d ligands would be selected. These T cells would achieve regulation of their autoreactive properties through the expression of inhibitory NK receptors. It is intriguing to speculate that such modes of selection might represent a primordial mechanism governing the selection and maintenance of the innate, preadaptive repertoire of T cells.
Acknowledgments
The authors would like to thank Bana Jabri and Polly Matzinger for their critical reading of the manuscript, members of Dr. Bendelac's laboratory for their discussions and support, Dr. Yasuhiko Koezuka, Kirin Brewery, Ltd., Gunma, Japan for providing αGalCer, and Vilma Zolyniene and the animal facility staff of Princeton University.
This work was supported by grants RO1AI38339 and CA87060 (to A. Bendelac) and AI62267 (to L. Teyton) from the National Institutes of Health and by a Leukemia and Lymphoma Society Fellowship (to K. Benlagha).
Footnotes
Abbreviations used in this paper: αGalCer, α-galactosylceramide; DN, double negative; TAP, transporter associated with antigen processing.
S.-H. Park and A. Weiss contributed equally to this work.
References
- Kasahara M., Nakaya J., Satta Y., Takahata N. Chromosomal duplication and the emergence of the adaptive immune system. Trends Genet. 1997;13:90–92. doi: 10.1016/s0168-9525(97)01065-2. [DOI] [PubMed] [Google Scholar]
- Calabi F., Jarvis J.M., Martin L.H., Milstein C. Two classes of CD1 genes. Eur. J. Immunol. 1989;19:285–292. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- Porcelli S.A., Modlin R.L. The CD1 systemantigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu. Rev. Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- Kawano T., Cui J., Koezuka Y., Toura I., Kaneko Y., Motoki K., Ueno H., Nakagawa R., Sato H., Kondo E. CD1d-restricted and TCR-mediated activation of Vα14 NK T cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- Porcelli S., Morita C.T., Brenner M.B. CD1β restricts the response of human CD4-8− T lymphocytes to a microbial antigen. Nature. 1992;360:593–597. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- Curnow S.J., Boyer C., Buferne M., Schmitt-Verhulst A.M. TCR-associated ζ-FceRIγ heterodimers on CD4−CD8−NK1.1+ T cells selected by specific class I MHC antigen. Immunity. 1995;3:427–438. doi: 10.1016/1074-7613(95)90172-8. [DOI] [PubMed] [Google Scholar]
- Legendre V., Boyer C., Guerder S., Arnold B., Hammerling G., Schmitt-Verhulst A.M. Selection of phenotypically distinct NK1.1+ T cells upon antigen expression in the thymus or in the liver. Eur. J. Immunol. 1999;29:2330–2343. doi: 10.1002/(SICI)1521-4141(199907)29:07<2330::AID-IMMU2330>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Schultz R.J., Parkes A., Mizoguchi E., Bhan A.K., Koyasu S. Development of CD4−CD8− αβTCR+ NK1.1+ T lymphocytes. Thymic selection by self antigen. J. Immunol. 1996;157:4379–4389. [PubMed] [Google Scholar]
- Bendelac A., Rivera M.N., Park S.-H., Roark J.H. Mouse CD1-specific NK+ T cells in development, specificity, and function. Annu. Rev. Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- Spada F.M., Grant E.P., Peters P.J., Sugita M., Melian A., Leslie D.S., Lee H.K., van Donselaar E., Hanson D.A., Krensky A.M. Self-recognition of CD1 by γ/δ T cellsimplications for innate immunity. J. Exp. Med. 2000;191:937–948. doi: 10.1084/jem.191.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway C.A., Jr. γδ T cellsresearch on the frontlines of defense. Res. Immunol. 1990;141:688–695. doi: 10.1016/0923-2494(90)90094-f. [DOI] [PubMed] [Google Scholar]
- Martin F., Kearney J.F. Positive selection from newly formed to marginal zone B cells depends on the rate of clonal production, CD19, and btk. Immunity. 2000;12:39–49. doi: 10.1016/s1074-7613(00)80157-0. [DOI] [PubMed] [Google Scholar]
- Havran W.L. A role for epithelial γδ T cells in tissue repair. Immunol. Res. 2000;21:63–69. doi: 10.1385/IR:21:2-3:63. [DOI] [PubMed] [Google Scholar]
- Diefenbach A., Raulet D.H. Natural killer cellsstress out, turn on, tune in. Curr. Biol. 1999;9:R851–R853. doi: 10.1016/s0960-9822(00)80044-5. [DOI] [PubMed] [Google Scholar]
- Benlagha K., Bendelac A. CD1d-restricted mouse Vα14 and human Vα24 T cellslymphocytes of innate immunity. Semin. Immunol. 2000;12:537–542. doi: 10.1006/smim.2000.0276. [DOI] [PubMed] [Google Scholar]
- Moody D.B., Ulrichs T., Muhlecker W., Young D.C., Gurcha S.S., Grant E., Rosat J.P., Brenner M.B., Costello C.E., Besra G.S., Porcelli S.A. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- Behar S.M., Podrebarac T.A., Roy C.J., Wang C.R., Brenner M.B. Diverse TCRs recognize murine CD1. J. Immunol. 1999;162:161–167. [PubMed] [Google Scholar]
- Lee D.J., Abeyratne A., Carson D.A., Corr M. Induction of an antigen-specific, CD1-restricted cytotoxic T lymphocyte response in vivo. J. Exp. Med. 1998;187:433–438. doi: 10.1084/jem.187.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamshiev A., Donda A., Prigozy T.I., Mori L., Chigorno V., Benedict C.A., Kappos L., Sonnino S., Kronenberg M., De Libero G. The αβ T cell response to self-glycolipids shows a novel mechanism of CD1β loading and a requirement for complex oligosaccharides. Immunity. 2000;13:255–264. doi: 10.1016/s1074-7613(00)00025-x. [DOI] [PubMed] [Google Scholar]
- Grant E.P., Degano M., Rosat J.P., Stenger S., Modlin R.L., Wilson I.A., Porcelli S.A., Brenner M.B. Molecular recognition of lipid antigens by T cell receptors. J. Exp. Med. 1999;189:195–205. doi: 10.1084/jem.189.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melian A., Watts G.F., Shamshiev A., De Libero G., Clatworthy A., Vincent M., Brenner M.B., Behar S., Niazi K., Modlin R.L. Molecular recognition of human CD1β antigen complexesevidence for a common pattern of interaction with αβ TCRs. J. Immunol. 2000;165:4494–4504. doi: 10.4049/jimmunol.165.8.4494. [DOI] [PubMed] [Google Scholar]
- Cardell S., Tangri S., Chan S., Kronenberg M., Benoist C., Mathis D. CD1-restricted CD4+ T cells in MHC class II–deficient mice. J. Exp. Med. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y.H., Jayawardena J., Weiss A., Lee D., Park S.-H., Dautry-Varsat A., Bendelac A. Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J. Exp. Med. 1999;189:103–110. doi: 10.1084/jem.189.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher C.C., Hiromatsu K., Naylor J.W., Brauer P.P., Brown K.A., Storey J.R., Behar S.M., Kawasaki E.S., Porcelli S.A., Brenner M.B., LeClair K.P. Conservation of a CD1 multigene family in the guinea pig. J. Immunol. 1999;163:5478–5488. [PubMed] [Google Scholar]
- Park S.-H., Guy-Grand D., Lemonnier F.A., Wang C.R., Bendelac A., Jabri B. Selection and expansion of CD8α/α(1) T cell receptor α/β(1) intestinal intraepithelial lymphocytes in the absence of both classical major histocompatibility complex class I and nonclassical CD1 molecules. J. Exp. Med. 1999;190:885–890. doi: 10.1084/jem.190.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonari K., Lovering E., Fairchild S., Spencer S. Two monoclonal antibodies specific for the T cell receptor Vα8. Eur. J. Immunol. 1989;19:1131–1135. doi: 10.1002/eji.1830190625. [DOI] [PubMed] [Google Scholar]
- Necker A., Rebai N., Matthes M., Jouvin-Marche E., Cazenave P.A., Swarnworawong P., Palmer E., MacDonald H.R., Malissen B. Monoclonal antibodies raised against engineered soluble mouse T cell receptors and specific for Vα8-, Vβ2-, or Vβ10-bearing T cells. Eur. J. Immunol. 1991;21:3035–3040. doi: 10.1002/eji.1830211220. [DOI] [PubMed] [Google Scholar]
- Benlagha K., Weiss A., Beavis A., Teyton L., Bendelac A. In vivo identification of glycolipid antigen–specific T cells using fluorescent CD1d tetramers. J. Exp. Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P. The JAM test. A simple assay for DNA fragmentation and cell death. J. Immunol. Methods. 1991;145:185–192. doi: 10.1016/0022-1759(91)90325-a. [DOI] [PubMed] [Google Scholar]
- Lantz O., Bendelac A. An invariant T cell receptor α chain is used by a unique subset of MHC class I–specific CD4+ and CD4−8− T cells in mice and humans. J. Exp. Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.L., Romero P., Widmann C., Kourilsky P., Maryanski J.L. T cell receptor genes in a series of class I major histocompatibility complex–restricted cytotoxic T lymphocyte clones specific for a Plasmodium berghei nonapeptideimplications for T cell allelic exclusion and antigen-specific repertoire. J. Exp. Med. 1991;174:1371–1383. doi: 10.1084/jem.174.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden B., Clark S.P., Kabelitz D., Mak T.W. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- Wang B., Chun T., Wang C.R. Comparative contribution of CD1 on the development of CD4+ and CD8+ T cell compartments. J. Immunol. 2000;164:739–745. doi: 10.4049/jimmunol.164.2.739. [DOI] [PubMed] [Google Scholar]
- Eberl G., Lees R., Smiley S.T., Taniguchi M., Grusby M.J., MacDonald H.R. Tissue-specific segregation of CD1d-dependent and CD1d-independent NK T cells. J. Immunol. 1999;162:6410–6419. [PubMed] [Google Scholar]
- Bendelac A., Killeen N., Littman D., Schwartz R.H. A subset of CD4+ thymocytes selected by MHC class I molecules. Science. 1994;263:1774–1778. doi: 10.1126/science.7907820. [DOI] [PubMed] [Google Scholar]
- Bendelac A., Hunziker R.D., Lantz O. Increased interleukin 4 and immunoglobulin E production in transgenic mice overexpressing NK+ T cells. J. Exp. Med. 1996;184:1285–1293. doi: 10.1084/jem.184.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangri S., Brossay L., Burdin N., Lee D.J., Corr M., Kronenberg M. Presentation of peptide antigens by mouse CD1 requires endosomal localization and protein antigen processing. Proc. Natl. Acad. Sci. USA. 1998;95:14314–14319. doi: 10.1073/pnas.95.24.14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda J.L., Naidenko O.V., Gapin L., Nakayama T., Taniguchi M., Wang C.R., Koezuka Y., Kronenberg M. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J. Exp. Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald H.R. CD1d-glycolipid tetramersa new tool to monitor natural killer T cells in health and disease. J. Exp. Med. 2000;192:F15–F20. doi: 10.1084/jem.192.5.f15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano A.R., Tangri S., Miller J.E.W., Holcombe H., Jackson M.R., Huse B., Kronenberg M., Peterson P.A. Peptide binding and presentation by mouse CD1. Science. 1995;269:223–226. doi: 10.1126/science.7542403. [DOI] [PubMed] [Google Scholar]
- Porcelli S.A., Brenner M.B. Antigen presentationmixing oil and water. Curr. Biol. 1997;7:508–511. doi: 10.1016/s0960-9822(06)00250-8. [DOI] [PubMed] [Google Scholar]
- Shinkai K., Locksley R.M. CD1, tuberculosis, and the evolution of major histocompatibility complex molecules. J. Exp. Med. 2000;191:907–914. doi: 10.1084/jem.191.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdin N., Brossay L., Koezuka Y., Smiley S.T., Grusby M.J., Gui M., Taniguchi M., Hayakawa K., Kronenberg M. Selective ability of mouse CD1 to present glycolipidsα galactosylceramide specifically stimulates Vα14+ NK T lymphocytes. J. Immunol. 1998;161:3271–3281. [PubMed] [Google Scholar]
- Sieling P.A., Ochoa M.T., Jullien D., Leslie D.S., Sabet S., Rosat J.P., Burdick A.E., Rea T.H., Brenner M.B., Porcelli S.A., Modlin R.L. Evidence for human CD4+ T cells in the CD1-restricted repertoirederivation of mycobacteria-reactive T cells from leprosy lesions. J. Immunol. 2000;164:4790–4796. doi: 10.4049/jimmunol.164.9.4790. [DOI] [PubMed] [Google Scholar]
- Burdin N., Brossay L., Kronenberg M. Immunization with α-galactosylceramide polarizes CD1-reactive NK T cells towards Th2 cytokine synthesis. Eur. J. Immunol. 1999;29:2014–2025. doi: 10.1002/(SICI)1521-4141(199906)29:06<2014::AID-IMMU2014>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Behar S.M., Dascher C.C., Grusby M.J., Wang C.R., Brenner M.B. Susceptibility of mice deficient in CD1D or TAP1 to infection with Mycobacterium tuberculosis . J. Exp. Med. 1999;189:1973–1980. doi: 10.1084/jem.189.12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza C.D., Cooper A.M., Frank A.A., Ehlers S., Turner J., Bendelac A., Orme I.M. A novel nonclassic β2-microglobulin-restricted mechanism influencing early lymphocyte accumulation and subsequent resistance to tuberculosis in the lung. Am. J. Respir. Cell. Mol. Biol. 2000;23:188–193. doi: 10.1165/ajrcmb.23.2.4063. [DOI] [PubMed] [Google Scholar]
- Zeng D., Dick M., Cheng L., Amano M., Dejbakhsh-Jones S., Huie P., Sibley R., Strober S. Subsets of transgenic T cells that recognize CD1 induce or prevent murine lupusrole of cytokines. J. Exp. Med. 1998;187:525–536. doi: 10.1084/jem.187.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skold M., Faizunnessa N.N., Wang C.R., Cardell S. CD1d-specific NK1.1+ T cells with a transgenic variant TCR. J. Immunol. 2000;165:168–174. doi: 10.4049/jimmunol.165.1.168. [DOI] [PubMed] [Google Scholar]
- Tatu C., Ye J., Arnold L.W., Clarke S.H. Selection at multiple checkpoints focuses V(H)12 B cell differentiation toward a single B-1 cell specificity. J. Exp. Med. 1999;190:903–914. doi: 10.1084/jem.190.7.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam K.P., Rajewsky K. B cell antigen receptor specificity and surface density together determine B-1 versus B-2 cell development. J. Exp. Med. 1999;190:471–477. doi: 10.1084/jem.190.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley M.J., Dal Porto J.M., Kawaguchi S., Cambier J.C., Nemazee D., Hardy R.R. A VH11Vκ9 B cell antigen receptor drives generation of CD5+ B cells both in vivo and in vitro. J. Immunol. 2000;164:4586–4593. doi: 10.4049/jimmunol.164.9.4586. [DOI] [PubMed] [Google Scholar]
- Hayakawa K., Asano M., Shinton S.A., Gui M., Allman D., Stewart C.L., Silver J., Hardy R.R. Positive selection of natural autoreactive B cells. Science. 1999;285:113–116. doi: 10.1126/science.285.5424.113. [DOI] [PubMed] [Google Scholar]
- Gerber D.J., Azuara V., Levraud J.P., Huang S.Y., Lembezat M.P., Pereira P. IL-4-producing γδ T cells that express a very restricted TCR repertoire are preferentially localized in liver and spleen. J. Immunol. 1999;163:3076–3082. [PubMed] [Google Scholar]
- Matzinger P. Tolerance, danger and the extended family. Annu. Rev. Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- Zerrahn J., Held W., Raulet D.H. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerrahn J., Volkmann A., Coles M.C., Held W., Lemonnier F.A., Raulet D.H. Class I MHC molecules on hematopoietic cells can support intrathymic positive selection of T cell receptor transgenic T cells. Proc. Natl. Acad. Sci. USA. 1999;96:11470–11475. doi: 10.1073/pnas.96.20.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]