Abstract
The discovery of dendritic cell (DC)-specific intercellular adhesion molecule (ICAM)-3–grabbing nonintegrin (DC-SIGN) as a DC-specific ICAM-3 binding receptor that enhances HIV-1 infection of T cells in trans has indicated a potentially important role for adhesion molecules in AIDS pathogenesis. A related molecule called DC-SIGNR exhibits 77% amino acid sequence identity with DC-SIGN. The DC-SIGN and DC-SIGNR genes map within a 30-kb region on chromosome 19p13.2-3. Their strong homology and close physical location indicate a recent duplication of the original gene. Messenger RNA and protein expression patterns demonstrate that the DC-SIGN–related molecule is highly expressed on liver sinusoidal cells and in the lymph node but not on DCs, in contrast to DC-SIGN. Therefore, we suggest that a more appropriate name for the DC-SIGN–related molecule is L-SIGN, liver/lymph node–specific ICAM-3–grabbing nonintegrin. We show that in the liver, L-SIGN is expressed by sinusoidal endothelial cells. Functional studies indicate that L-SIGN behaves similarly to DC-SIGN in that it has a high affinity for ICAM-3, captures HIV-1 through gp120 binding, and enhances HIV-1 infection of T cells in trans. We propose that L-SIGN may play an important role in the interaction between liver sinusoidal endothelium and trafficking lymphocytes, as well as function in the pathogenesis of HIV-1.
Keywords: L-SIGN, adhesion receptor, chromosome 19p13.2-3, ICAM-3, HIV-1 gp120
Introduction
Dendritic cell (DC)-specific intercellular adhesion molecule (ICAM)-3–grabbing nonintegrin (DC-SIGN) has recently been identified as a DC-specific adhesion receptor that mediates the interaction between DCs and resting T cells through high affinity binding to ICAM-3, thereby facilitating the initiation of primary immune responses 1 2. DC-SIGN was shown to be identical to the previously reported type II membrane-associated C-type lectin 2 that binds HIV-1 envelope glycoprotein gp120 in a CD4-independent manner 3. The affinity of DC-SIGN exceeds that of CD4 for HIV-1 gp120 3, and upon capture of HIV-1, DC-SIGN does not appear to promote viral entry into the DC itself, but rather enhances infection of T cells in trans 1. DC-SIGN–associated HIV-1 remains infectious over a prolonged period of time, perhaps contributing to the infectious potential of the virus during its transport by DCs from the periphery to lymphoid organs.
A previous search by Yokoyama-Kobayashi et al. 4 for cDNA clones encoding type II membrane proteins resulted in the identification of a partial clone that was homologous, but not identical, to the cDNA encoding the molecule now known as DC-SIGN. The putative protein product contained a deletion of 28 amino acids in the cytoplasmic domain and was lacking the entire C-type lectin domain relative to the cDNA encoding DC-SIGN. More recently, Soilleux et al. 5 described the full-length cDNA sequence of the related gene, which they called DC-SIGNR. The genomic organization of DC-SIGN and DC-SIGNR was compared, indicating a high degree of similarity. Concomitant expression of the two genes in placenta, endometrium, and stimulated KG1 cells (a cell line that phenotypically resembles myeloid DCs) was observed, although the expression of DC-SIGNR was very low in both endometrium and stimulated KG1 cells 5.
While attempting to identify polymorphisms in the DC-SIGN gene, we also discovered the gene corresponding to the partial cDNA sequence described by Yokoyama-Kobayashi et al. 4. Tissue expression patterns of the DC-SIGNR gene indicated that it is expressed at considerably high levels in only two tissues, liver and lymph node, but not in monocyte-derived DCs. Therefore, we have called the molecule L-SIGN, liver/lymph node–specific ICAM-3–grabbing nonintegrin, which we believe more accurately depicts the function and expression pattern of this molecule than does DC-SIGNR. Here we refine the genomic organization of the SIGN gene complex, and also report the tissue distribution and functional characterization of the L-SIGN molecule.
Materials and Methods
Characterization of DC-SIGN and L-SIGN cDNA.
We have submitted the full DC-SIGN and L-SIGN cDNA sequences to GenBank/EMBL/DDBJ under accession nos. AF290886 and AF290887, respectively. The L-SIGN cDNA sequence represents a variant containing six repeats in exon 4. The 5′ and 3′ ends of the transcripts (except the 3′ end of the DC-SIGN mRNA) were determined by 5′ rapid amplification of cDNA ends (RACE; CLONTECH Laboratories, Inc.). The length of the 3′ end of the DC-SIGN mRNA was estimated based on Northern blot analysis data (transcript size) and reverse transcription (RT)-PCR data using forward primers specific for the 1.3-kb DC-SIGN cDNA sequence 3 and reverse primers specific for several GenBank/EMBL/DDBJ expressed sequence tags (ESTs) (e.g., AI472111 and AA454170), mapping downstream of the alleged 3′ end of DC-SIGN. A cDNA fragment containing the full coding sequence of L-SIGN (nucleotides [nt] 39–1184, GenBank/EMBL/DDBJ accession no. AF290887) was amplified from human placental mRNA (CLONTECH Laboratories, Inc.) and cloned into the expression vectors pcDNA3.1/V5-His/TOPO (pcDNA3-L-SIGN) and pCDM8 (pCDM8-L-SIGN).
Radiation Hybrid Mapping.
PCR-based radiation hybrid (RH) mapping with DC-SIGN– and L-SIGN–specific primers was performed using the Genebridge 4 RH panel (Research Genetics). The PCR results were submitted to the Gene Map server at the Sanger Center (http://www.sanger.ac.uk/Software/Rhserver). The chromosomal position of markers linked to the genes was determined searching the Genatlas database (http://web.citi2.fr/GENATLAS) and the genetic map of human chromosome 19 provided by the Marshfield Clinic (http://research. marshfieldclinic.org/genetics/).
Genotype Analysis of L-SIGN and DC-SIGN Exon4.
The repeat region in exon 4 was amplified with the following pairs of primers: L28, TGTCCAAGGTCCCCAGCTCCC, and L32, GAACTCACCAAATGCAGTCTTCAAATC, for L-SIGN; DL27, TGTCCAAGGTCCCCAGCTCC, and DI4R, CCCCGTGTTCTCATTTCACAG, for DC-SIGN. The cycle conditions were as follows: 94°C for 5 s and 68°C for 1 min. Alleles were distinguished by agarose gel electrophoresis and ethidium bromide staining.
Northern Blot Analysis.
Total RNA from cultured human immature DCs (see below) was isolated using Trizol (Life Technologies). 10 μg of the isolated RNA was electrophoresed on a 1% agarose gel, transferred to Hybond-XL (Amersham Pharmacia Biotech) as described 6, and used for Northern blot analysis along with two human multiple tissue Northern blots (CLONTECH Laboratories, Inc.). Three probes were subsequently hybridized to the blots: 1 an L-SIGN–specific probe (nt 100–183, GenBank/EMBL/DDBJ accession no. AF290887); 2 a probe recognizing both DC-SIGN and L-SIGN (nt 1–1233, GenBank/EMBL/DDBJ accession no. AF290886); and 3 an actin control probe (CLONTECH Laboratories, Inc.). Hybridization procedures were performed according to manufacturer specifications (CLONTECH Laboratories, Inc.).
Antibodies.
Anti–DC-SIGN mAbs AZN-D1 and AZN-D2 were described previously 2. mAb AZN-D3 was obtained by screening hybridoma supernatants of BALB/c mice immunized with THP-1–DC-SIGN cells 1 for the ability to stain both DC-SIGN and L-SIGN. Anti–DC-SIGN mAb AZN-D2 also cross-reacts with L-SIGN, as was initially determined by the staining of K562–L-SIGN cells (data not shown). Anti–L-SIGN rabbit antiserum was generated by immunization with two L-SIGN–specific peptides, PTTSGIRLFPRD and WNDNRCDVDNYW (Veritas, Inc. Laboratories).
Cells.
DCs were cultured from monocytes in the presence of 500 U/ml IL-4 and 800 U/ml GM-CSF (Schering-Plough; references 7 and 8). At day 7 the cells expressed high levels of MHC class I and II, αMβ2 (CD11b), αXβ2 (CD11c), DC-SIGN and ICAM-1, moderate levels of LFA-1 and CD86, and low levels of CD14, as measured by flow cytometry. Stable K562 transfectants expressing L-SIGN (K562–L-SIGN) were generated by cotransfection of K562 with the pCDM8–L-SIGN plasmid and the pGK-neo vector by electroporation 9. Stable K562–DC-SIGN transfectants were generated in a similar manner using pRc/CMV–DC-SIGN 2. THP-1–DC-SIGN cells were described previously 2. Stable THP-1–L-SIGN transfectants were generated by electroporation of THP-1 cells with pcDNA3–L-SIGN, selection for G418 resistance, and positive sorting for L-SIGN expression using mAb AZN-D3. All cell lines were maintained in RPMI 1640 supplemented with 10% fetal bovine serum in addition to specific cytokine or antibiotic requirements as indicated. K562 and THP-1 are monocytic cell lines. HEK293T are human embryonic kidney cells containing a single temperature-sensitive allele of SV-40 large T antigen. GHOST cells are HIV-indicator cells derived from human osteosarcoma cells 10. Hut/CC chemokine receptor (CCR)5 cells are the transformed human T cell line Hut78 stably transduced with CCR5.
Fluorescent Beads Adhesion Assay.
Carboxylate-modified TransFluorSpheres (488/645 nm, 1.0 μm; Molecular Probes) were coated with ICAM-3 as described previously for ICAM-1 11. Fluorescent beads were coated with M-tropic HIV-1MN envelope glycoprotein gp120 as follows: streptavidin-coated fluorescent beads were incubated with biotinylated F(ab′)2 fragment rabbit anti–sheep IgG (6 μg/ml; Jackson ImmunoResearch Laboratories) followed by an overnight incubation with sheep anti-gp120 antibody D7324 (Alto Bio Reagents, Ltd.) at 4°C. The beads were washed and incubated with 250 ng/ml purified HIV-1 gp120 (provided by Immunodiagnostics, Inc., through the National Institutes of Health AIDS Research and Reference Reagent Program) overnight at 4°C. The fluorescent beads adhesion assay was performed as described by Geijtenbeek et al. 11. In brief, cells were resuspended in adhesion buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM CaCl2, 2 mM MgCl2, 0.5% BSA) at a final concentration of 5 × 106 cells/ml. 50,000 cells were preincubated with mAb (20 μg/ml) for 10 min at room temperature. Ligand-coated fluorescent beads (20 beads/cell) were added and the suspension was incubated for 30 min at 37°C. Adhesion was determined by measuring the percentage of cells that bound fluorescent beads using flow cytometry on a FACScan™ (Becton Dickinson).
Detection of L-SIGN on Primary Human Liver Sinusoidal Endothelial Cells.
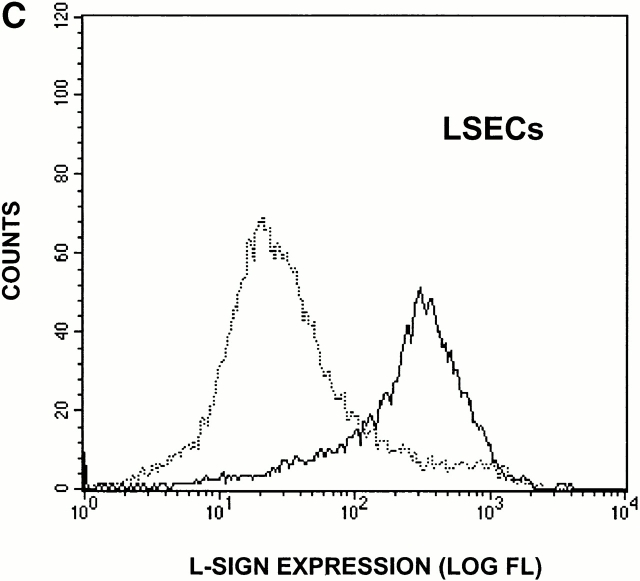
Liver tissue was obtained from a patient undergoing liver surgery after having received written consent. Isolation of primary human liver cells was performed as described previously 12. Cells were cultured on collagen type I–coated tissue culture plates in supplemented Williams E Medium 13. The day after isolation, liver cells were incubated with Texas red–labeled OVA (10 μg/ml; Molecular Probes) for 2 h and detached from the matrix by gentle trypsin treatment. Cells were stained with rabbit anti–L-SIGN antiserum followed by goat anti–rabbit Ig FITC (Dianova) and analyzed with a FACScan™ (Becton Dickinson) using CELLQuest™ software. OVA uptake was characteristic of liver sinusoidal endothelial cells (LSECs) only and not Kupffer cells, as verified by the costaining of OVA+ cells with an endothelial cell–specific marker, acetylated LDL, using confocal microscopy.
HIV-1 Infection Assays.
The infection assays were performed as described previously 1 2. Pseudotyped HIV-1 stocks were generated by calcium phosphate transfections of HEK293T cells with the proviral vector plasmid NL-Luc-E-R− containing a firefly luciferase reporter gene 14 and expression plasmids for either ADA or JRFL gp160 envelopes. Viral stocks were evaluated by limiting dilution on GHOST CXCR4/CCR5 and 293T-CD4-CCR5 cells. In HIV-1 cell capture assays, DC-SIGN or L-SIGN expressing THP-1 transfectants (250,000 cells) were preincubated with pseudotyped HIV-1 (multiplicity of infection ∼0.1 with regard to target cell concentration) in a total volume of 0.5 ml for 3 h to allow cellular adsorption of the virus. After 3 h incubation, cells were washed with 2 vol PBS and the THP-1 transfectants were cocultured with Hut/CCR5 targets (100,000 cells) in the presence of 10 μg/ml polybrene in 1 ml cell culture medium. Cell lysates were obtained after 3 d and analyzed for luciferase activity. In contrast, HIV-1 enhancement assays used suboptimal concentrations of virus (typically <0.05 multiplicity of infection) without a wash step. In brief, DC-SIGN or L-SIGN transfectants (50,000 cells) were incubated with identical virus concentrations (either pseudotyped HIV-1 or replication-competent M-tropic strain HIV-1JR-CSF), and after 2 h activated T cells (100,000 cells) were added. Cell lysates were obtained after several days and analyzed for either luciferase activity or p24 antigen levels. T cells were activated by culturing them in the presence of 10 U/ml IL-2 and 10 μg/ml PHA for 2 d.
Immunohistochemical Analysis.
Staining of the tissue cryosections was performed as described previously 2. 8-μm cryosections of the tissues were fixed in 100% acetone for 10 min, washed with PBS, and incubated with the first antibody (10 μg/ml) for 60 min at 37°C. After washing, the final staining was performed with the ABC-PO/ABC-AP Vectastain kit (Vector Laboratories) according to the manufacturer's protocol. Nuclear staining was performed with hematoxylin.
Results
Genomic Map of DC-SIGN and L-SIGN.
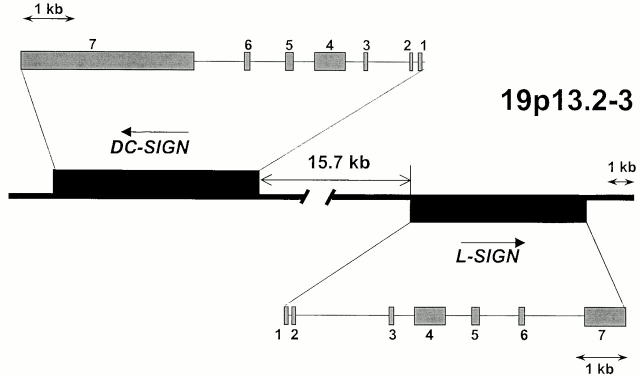
A fine map of the DC-SIGN/L-SIGN gene locus was determined using information from the human BAC clone CTD-2102F19 sequence, which is now available in GenBank/EMBL/DDBJ (accession no. AC008812; Fig. 1). DC-SIGN and L-SIGN are positioned in a head-to-head orientation 15.7 kb apart. RH mapping indicated that DC-SIGN and L-SIGN are located on chromosome 19p13.2-3, near the marker D19S912 (lod score values >11.1) with DC-SIGN positioned more telomeric. In agreement with the RH data, the D19S912 marker is found at a distance of ∼37 kb centromeric to L-SIGN on the BAC sequence.
Figure 1.
Schematic representation of the DC-SIGN/L-SIGN genetic map. Physical distances and gene orientation are based on the sequence provided from BAC clone CTD-2102F19 (GenBank/EMBL/DDBJ accession no. AC008812).
Soilleux et al. 5 reported a DC-SIGNR cDNA clone that contained eight exons with an additional 3′ intron spliced out of the 3′ untranslated region (UTR) compared with the cDNA clone described by Yokoyama-Kobayashi et al. 4. Our RT-PCR experiments on different tissues (liver, pancreas, lung, and placenta) showed that the splice variant described by Soilleux et al. is consistently present but only as a minor transcript, whereas the major transcript consists of seven exons (data not shown). Also, a transcript missing exons 2 and 6, as described by Yokoyama-Kobayashi et al. 4, was extremely rare in our hands.
Northern hybridization data (see below) indicated that DC-SIGN mRNA is 3 kb longer than that reported previously 3 5. We found that this difference is due to the presence of an additional 3-kb UTR in exon 7. Indeed, there was no canonical polyadenylation signal in either of the previously published DC-SIGN cDNA sequences, and a search of GenBank/EMBL/DDBJ sequences revealed short polyadenylated ESTs with putative poly(A) signal motifs mapping 3 kb downstream of the alleged 3′ end of DC-SIGN. RT-PCR experiments indicated that those ESTs correspond to DC-SIGN mRNA (data not shown). Based on these findings, we conclude that the full DC-SIGN transcript contains the additional 3 kb, resulting in a total of 4.3 kb.
Polymorphism in Exon 4 of L-SIGN.
Exon 4 of both DC-SIGN and L-SIGN contains repeats of 69 bp that encode repeating units of 23 amino acids. These repeats form a neck between the carbohydrate recognition domain and the transmembrane domain of the SIGN molecules. The L-SIGN cDNA clone isolated from placental mRNA contained the entire coding region of the gene, but only six full repeats were present in the sequence corresponding to exon 4, in contrast to seven full repeats identified in the cDNA reported by Soilleux et al. 5. This indicated that the repeat region of L-SIGN is polymorphic. Analysis of exon 4 in 350 Caucasian individuals showed the presence of seven alleles based on number of repeats (ranging from three to nine), the most common of which was the allele containing seven repeats (Table ). Strikingly, analysis of DC-SIGN exon 4 in 150 Caucasians did not reveal any variability.
Table 1.
Polymorphism of the Repeat Region in L-SIGN Exon 4
| No. of repeats | Allele frequency (percent) |
|---|---|
| 3 | 1 (0.3) |
| 4 | 25 (3.6) |
| 5 | 202 (28.9) |
| 6 | 86 (12.2) |
| 7 | 377 (53.9) |
| 8 | 2 (0.3) |
| 9 | 7 (1.0) |
Northern Blot Analysis of DC-SIGN and L-SIGN.
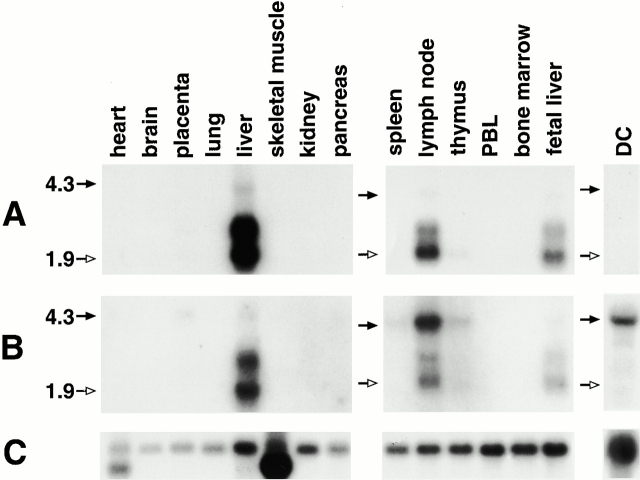
L-SIGN mRNA exhibits ∼90% similarity to DC-SIGN mRNA over the entire coding region, but there is only 53% similarity between exons 2 of the genes. Therefore, exon 2 sequence was used to generate a probe (84 nt) that was L-SIGN specific in Northern blot analysis. The probe hybridized to mRNA of ∼1.9, 2.6, and 4.2 kb in size in liver and lymph node, and a weak 1.9-kb band was detected in thymus (Fig. 2 A). The 1.9-kb band, which is prominent in lymph node and fetal liver, corresponds to the predicted size of L-SIGN. The upper bands (one of which, 2.6 kb, is substantial in adult liver) are likely to be alternative transcripts, but RACE and RT-PCR techniques have not indicated the presence of UTRs varying in length nor alternative splice variants. Therefore, we cannot exclude the possibility that a gene(s) with homology to L-SIGN and precisely the same expression pattern is present in humans, but a thorough search for such genes in the sequence databases has been unsuccessful. Finally, a polymorphism in the L-SIGN gene (e.g., exon 4 repeat expansion or alteration in the polyadenlation signal motif) could possibly explain the larger transcript size.
Figure 2.
Northern blot analysis of DC-SIGN and L-SIGN. Positions of the 4.3-kb (black arrows) and 1.9-kb (white arrows) sizes are marked on the left. (A) Hybridization with the L-SIGN–specific probe indicating expression of the gene in liver, lymph node, and weakly in thymus. (B) Hybridization with the probe recognizing both genes. 4.3-kb bands represent DC-SIGN mRNA. The light upper band (∼4.2 kb) evident in liver and lymph node using the L-SIGN–specific probe (Fig. 3 A) is distinct from DC-SIGN mRNA (4.3 kb) due to the specificity of the probe, intensity patterns, and slight differences in size. (C) Reprobing of the blots with the β-actin cDNA control probe.
Northern blots were reprobed with a 1.2-kb fragment containing the entire coding sequence of DC-SIGN, which recognizes both DC- and L-SIGN mRNA due to their high sequence similarity (Fig. 2 B). Once again, the bands representing L-SIGN transcripts were observed in liver, lymph node, and fetal liver. Additionally, a 4.3-kb transcript representing DC-SIGN was detected in monocyte-derived DCs and lymph node, and to a lesser extent, in placenta, spleen, thymus, and possibly liver.
L-SIGN mRNA was also detected in placenta and DCs using a more sensitive RT-PCR technique, in agreement with previously reported data 5, but the level of expression in these tissues is too low to be detected by Northern hybridization. The probe which recognizes both DC-SIGN and L-SIGN transcripts with nearly equal sensitivity clearly indicated differential tissue distribution of the two gene products: L-SIGN is primarily transcribed in liver and lymph node, whereas DC-SIGN is specifically expressed in DCs and tissues that accommodate DCs (Fig. 2; reference 1). Although both L-SIGN and DC-SIGN mRNAs are found in lymph node, it is likely that they are expressed by different cell types in this tissue. DCs, which are frequent in lymph node, are the source of DC-SIGN mRNA in this tissue 2, but L-SIGN mRNA is not detected by Northern blot analysis in DCs, peripheral blood lymphocytes, or spleen (Fig. 2). It is possible that L-SIGN expression may be inducible in certain leukocytes during specific stages of activation or, perhaps more likely, endothelial cells of the lymph node may constitutively express this receptor. Characterization of the mechanism involved in the differential tissue expression of these two highly homologous molecules will be of particular interest.
L-SIGN Is Expressed by Human LSECs and Not by DCs.
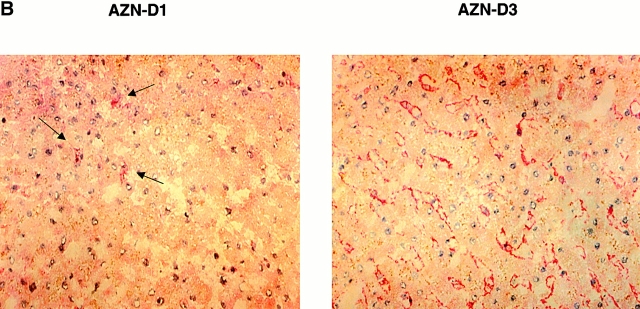
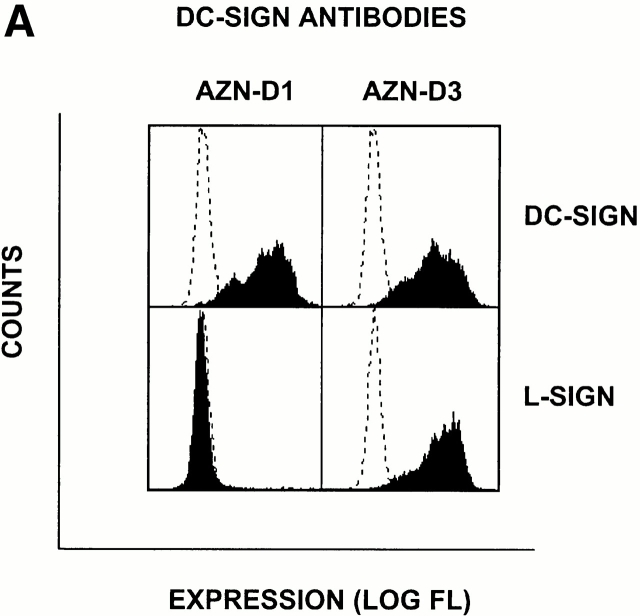
To identify the cells expressing L-SIGN molecules in vivo, we performed immunohistochemical analysis using a pair of anti–DC-SIGN mAbs, one of which, AZN-D3, crossreacted with L-SIGN, whereas another, AZN-D1, was DC-SIGN specific (Fig. 3 A). As expected from the Northern blot analysis, poor staining of liver tissue was observed using the DC-SIGN–specific mAb AZN-D1 (Fig. 3 B), and the rare cells detected with this antibody are probably DCs residing in liver. In contrast, the mAb AZN-D3 brightly stained cells lining the sinusoids of the liver (Fig. 3 B). mAbs against the endothelial cell–specific marker CD31 gave a similar staining pattern on serial liver sections (data not shown), suggesting that L-SIGN is expressed by LSECs. To support this idea, primary human LSECs were distinguished from the other hepatic cells by uptake of OVA, which is a unique characteristic of LSECs 15, and were tested for expression of L-SIGN directly. Staining of LSECs with polyclonal anti–L-SIGN antibodies indicated that L-SIGN is expressed exclusively by these cells in liver (Fig. 3 C).
Figure 3.
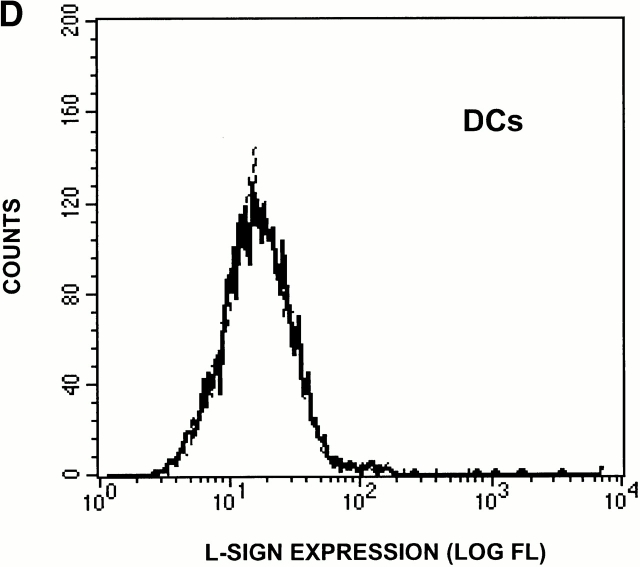
L-SIGN is expressed on LSECs and not on monocyte-derived DCs. (A) The antibody AZN-D1 is DC-SIGN specific, whereas AZN-D3 cross-reacts with L-SIGN. Stable DC-SIGN and L-SIGN K562 transfectants were stained with either AZN-D1 or AZN-D3. (B) Immunohistochemical analysis of DC-SIGN and L-SIGN expression in the human liver. Serial sections were stained with either AZN-D1 (DC-SIGN specific) or with AZN-D3 (detects both DC-SIGN and L-SIGN). AZN-D1 stains infrequent cells that may be DCs (arrows), whereas AZN-D3 stains cells lining sinusoids. (C) Expression of L-SIGN in liver is restricted to LSECs. 1 d after isolation, primary human liver cells were incubated with fluorochrome-labeled OVA. L-SIGN expression was determined by indirect immunofluorescence using an L-SIGN–specific polyclonal antibody. Cells that have taken up OVA (LSECs) and those that did not take up OVA (hepatocytes and other resident hepatic cells) are represented by solid and broken lines, respectively, by gating on the respective cell populations. 2 × 105 cells were analyzed. (D) L-SIGN is not expressed by monocyte-derived DCs. Immature DCs, cultured from monocytes in the presence of GM-CSF and IL-4, do not stain with anti–L-SIGN polyclonal antibody, as determined by FACScan™ analysis. Solid line indicates staining with anti–L-SIGN polyclonal serum, whereas broken line (hidden under solid lane) represents staining with rabbit preimmune serum.
Both AZN-D1 and AZN-D3 stained lymph node equally well (data not shown), but without sufficient definition to determine whether cellular staining patterns differed between the two antibodies. However, using L-SIGN–specific polyclonal antibodies, we found that L-SIGN is not expressed by monocyte-derived DCs (Fig. 3 D), which supports conclusions from the Northern blot analysis. Therefore, it is likely, that DC-SIGN and L-SIGN are expressed by different types of cells in the lymph node.
L-SIGN Binds ICAM-3 and HIV-1 gp120.
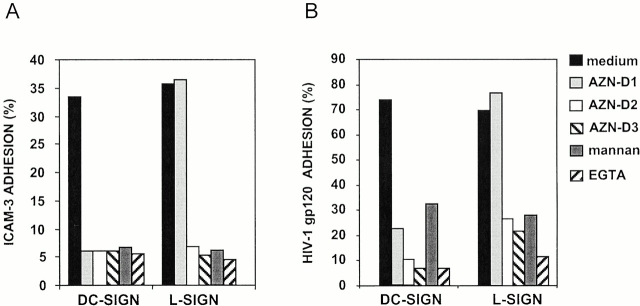
We predicted that L-SIGN and DC-SIGN would bind similar ligands given the nearly identical amino acid sequence of their extracellular domains. Both ICAM-3 and HIV-1MN gp120 have been shown to bind with high affinity to DC-SIGN in a Ca2+-dependent manner 1 2 3. Using a flow cytometry–based adhesion assay 11, K562 cells transfected with L-SIGN were shown to bind ICAM-3 with high affinity (Fig. 4 A). The L-SIGN–mediated binding was inhibited by the DC-SIGN/L-SIGN–specific mAbs AZN-D2 and AZN-D3, mannan, or EGTA, but not by the DC-SIGN–specific mAb AZN-D1, demonstrating that L-SIGN functions as a mannose-binding C-type lectin with a high affinity for ICAM-3. As predicted by the high homology to DC-SIGN, L-SIGN was also able to bind to HIV-1MN gp120 in a manner similar to that observed for DC-SIGN 1 (Fig. 4 B). Mock transfected cells did not bind either ICAM-3 or HIV-1MN gp120 (data not shown).
Figure 4.
L-SIGN binds ICAM-3 (A) and HIV-1 gp120 (B). Adhesion of ICAM-3 and gp120 to the K562–L-SIGN and K562–DC-SIGN cells was measured with the fluorescent bead adhesion assay (reference 11). The y-axis represents the percentage of cells binding ligand-coated fluorescent beads. The L-SIGN cross-reacting mAbs AZN-D2 (20 μg/ml) and AZN-D3 (20 μg/ml) inhibit the adhesion of ICAM-3 and gp120 to L-SIGN, in contrast to the DC-SIGN–specific mAb AZN-D1 (20 μg/ml). Adhesion of both ICAM-3 and gp120 to the K562 transfectants is also inhibited by either 20 μg/ml mannan or 5 mM EGTA. Adhesion of both ligands to mock transfectants was <5%. One representative experiment out of three is shown (SD < 5%).
L-SIGN Enhances HIV-1 Infection.
High affinity binding of L-SIGN to HIV-1 gp120 raised the possibility that, like DC-SIGN, L-SIGN might bind infectious HIV-1 and enhance infection of target cells in trans. To test the role of L-SIGN as a transreceptor in HIV-1 infection, THP-1 cells expressing either DC-SIGN or L-SIGN were pulsed with single round infectious HIV-luciferase pseudotyped with M-tropic HIV-1JRFL envelope glycoprotein, washed to remove unbound virus, and incubated with target cells permissive for HIV-1 infection. Infection was evaluated after 3 d. Both the L-SIGN– and DC-SIGN–transfected THP-1 cells captured infectious HIV-1 and transmitted the virus to target cells, while mock transfected THP-1 cells did not (Fig. 5 A).
Figure 5.
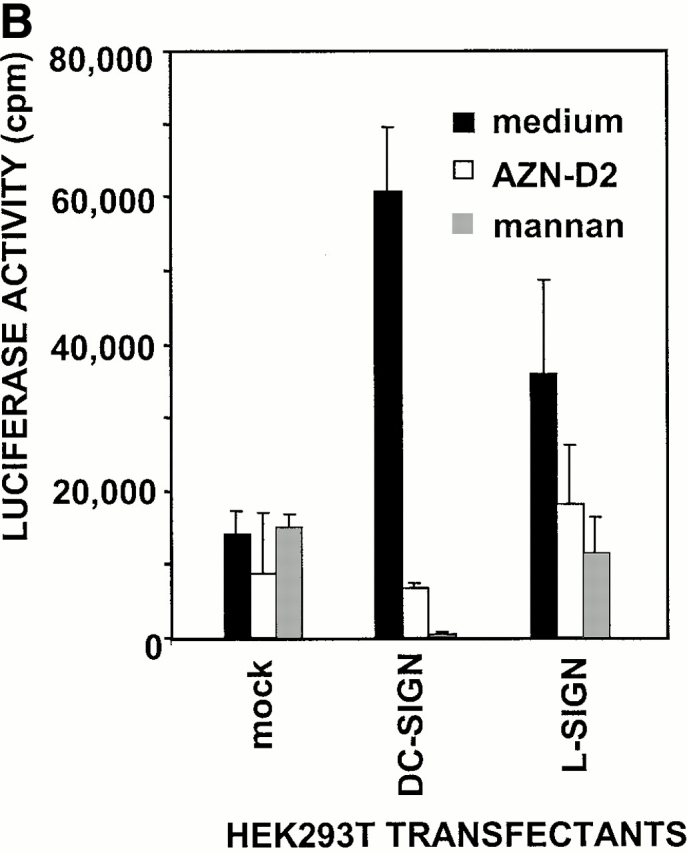
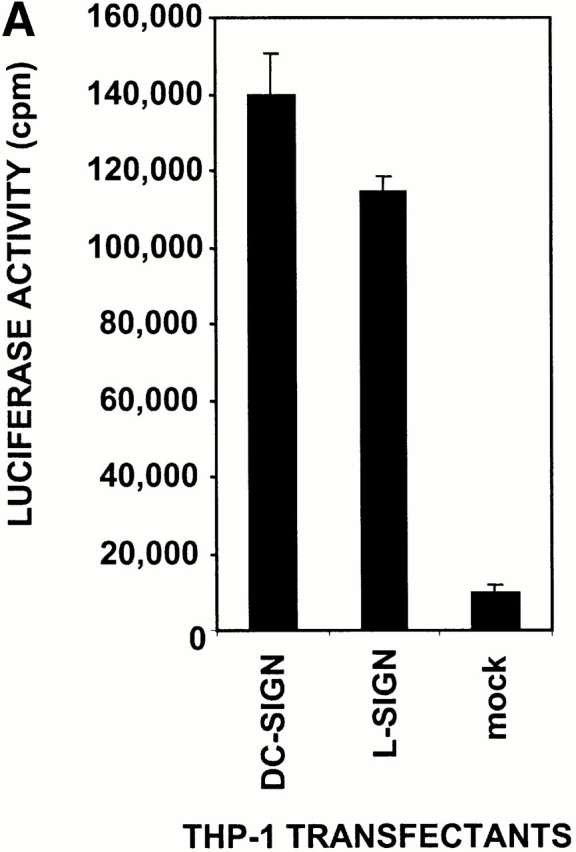
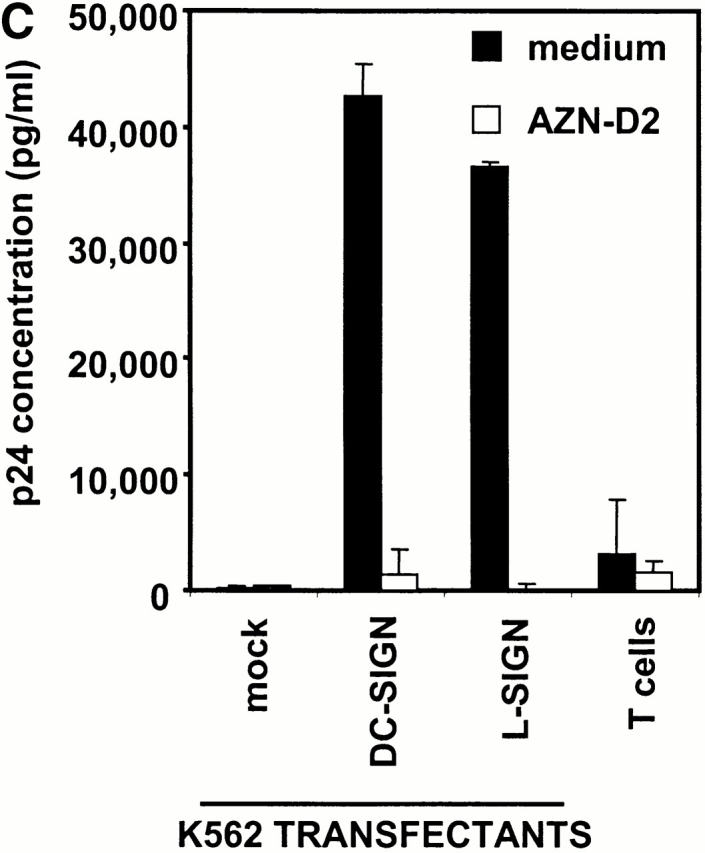
L-SIGN captures and enhances infection of T cells with HIV-1 in trans. (A) L-SIGN captures HIV-1 and transmits it to target cells. Stable DC-SIGN– or L-SIGN–expressing THP-1 transfectants were preincubated with HIV-luc/JRFL pseudovirions to allow capture of the virus. Cells were washed and THP-1 transfectants were cocultured with Hut/CCR5 target cells. Cell lysates were obtained after 3 d and analyzed for luciferase activity. For each of the coculture conditions employed, mock infected controls were uniformly <100 cps in activity. Each data set represents the mean of four separate wells of infected cells. One representative experiment out of two is shown. (B) L-SIGN enhances infection of T cells by pseudotyped HIV-1. HEK293T cells were transiently transfected with cDNA encoding DC-SIGN, L-SIGN, or empty vector. Control cells were preincubated with 20 μg/ml AZN-D2 or 20 μg/ml mannan. Low amounts of pseudotyped HIV-1ADA were added together with activated T cells as described previously (reference 1). Infectivity was determined after 2 d by measuring luciferase activity. One representative experiment of two performed is shown. Each experiment was done in triplicate wells. (C) L-SIGN enhances infection of T cells by replication competent HIV-1. Stable K562 transfectants of both L-SIGN and DC-SIGN were incubated with low virus concentrations of replication-competent M-tropic strain HIV-1JR-CSF (TCID50 100/ml). To determine the specificity, cells were preincubated with AZN-D2 (20 μg/ml). After 2 h, activated T cells were added as described previously (reference 2). Culture supernatants were collected at day 14 after K562-T cell coculture and HIV-1 production was measured using ELISA to determine p24 antigen levels. In control experiments, the same amount of virus was added directly to T cells. One representative experiment out of three is shown. Each data set represents the mean of three separate wells of infected cells.
Next we investigated whether L-SIGN would be able to capture a limiting concentration of HIV-1 and efficiently present the virus to the permissive cells promoting infection. HEK293T cells expressing DC-SIGN or L-SIGN, or mock transfected cells were incubated with low titers of HIV-luciferase pseudotyped with HIV-1ADA envelope glycoprotein. The unwashed cells were then cocultured with activated T cells. Minimal infection of target cells was observed from mock transfected HEK293T cells pulsed with HIV-1 (Fig. 5 B). However, HEK293T cells transfected with L-SIGN enhanced HIV-1 infection of T cells in trans, similar to DC-SIGN (Fig. 5 B). The DC-SIGN–mediated enhancement was inhibited with the cross-reactive AZN-D2 antibody, while partial inhibition was observed for L-SIGN, possibly because of some difference in the reactivity of this antibody to the two SIGN molecules that was evident under the conditions employed in this experiment. Mannan efficiently inhibited enhancement by both SIGN molecules.
Similar experiments to evaluate the ability of L-SIGN to enhance HIV-1 infection of T cells were performed using replication-competent virus. K562 cells transfected with L-SIGN, DC-SIGN, and empty vector were incubated with the M-tropic HIV-1JR-CSF strain at low virus concentrations for 2 h and subsequently cocultured with activated T cells (Fig. 5 C). No viral replication was observed using mock transfected K562 cells, while L-SIGN transfectants transmitted HIV-1 to target cells, resulting in viral replication with nearly the same efficiency as DC-SIGN transfectants. Almost complete inhibition of HIV-1 replication with the DC-SIGN/L-SIGN–specific antibody AZN-D2 indicated the specificity of these receptors to enhance HIV-1 infection. Thus, non-DC lineage cells expressing L-SIGN within liver and possibly in lymph node may also have the ability to capture and transmit HIV-1 to lymphocytes.
Discussion
The homologous human C-type lectins DC-SIGN and L-SIGN appear to be the products of a recent gene duplication. The corresponding proteins share the same domain organization and overlapping, if not completely identical, ligand specificity. The most diverse region of these molecules occurs in their cytoplasmic tails 5. It has been suggested that DC-SIGN–associated HIV-1 may be internalized, protecting it from degradation or inactivation 1. If so, the sequence variation in the cytoplasmic region of L-SIGN relative to DC-SIGN could affect the level of receptor internalization and viral uptake, perhaps explaining the consistent differences in efficiency of HIV-1 infection enhancement observed in our experiments between DC-SIGN and L-SIGN transfectants (Fig. 5).
Another obvious difference in SIGN genes is the repeat polymorphism in exon 4 of L-SIGN, which is conserved in DC-SIGN (Table ). The neck domain of L-SIGN may contain from three to nine repeats, while DC-SIGN always consists of seven repeats among the Caucasians tested. It is not clear whether the differences in exon 4 diversity of these genes is because of some distinction in the physical feature(s) of the genes or to selective processes acting on the genes differentially. The neck domain may be involved in oligomerization of the receptors 5 and variable numbers of repeats could potentially affect functional characteristics of the L-SIGN molecule, particularly in heterozygotes where heterooligomers might be present. However, our preliminary data indicated no difference between L-SIGN molecules containing six or seven repeats in ligand binding or in HIV-1 capture and enhancement experiments.
Although the SIGN genes have maintained sequence and functional similarity over their evolutionary history, regulatory elements determining their tissue distribution have evolved along unique paths. Northern blot analysis of mRNA expression clearly indicated expression of DC-SIGN in monocyte-derived DCs and in tissues where DCs reside, whereas expression of L-SIGN in DCs was undetectable (Fig. 2). Further, L-SIGN was not detected on monocyte-derived DCs using antibodies specific to L-SIGN (Fig. 3 C). Thus, it is most likely that unique cell types in the lymph node express one but not both SIGN molecules: L-SIGN could be expressed by endothelial cells, as it is in liver, whereas DC-SIGN is expressed by DCs in the T cell area of lymph node 2.
Liver sinusoids are specialized capillary vessels characterized by the presence of resident macrophages adhering to the endothelial lining. The LSEC–leukocyte interactions, which require expression of adhesion molecules on the cell surfaces, appear to constitute a central mechanism of peripheral immune surveillance in the liver 15. The mannose receptor as well as other costimulatory receptors such as MHC class II, CD80, and CD86, are known to be expressed on LSECs and to mediate the clearance of many potentially antigenic proteins from the circulation in a manner similar to DCs in lymphoid organs 15. L-SIGN may fit in this category of receptors on LSECs, as its tissue location and ligand-binding properties strongly implicate a physiologic role for this receptor in antigen clearance, as well as in LSEC–leukocyte adhesion. The high expression of ICAM-3 on apoptotic cells 16 may provide the means by which these cells are trapped by L-SIGN–expressing cells in the liver and subsequently cleared.
The mannose glycans present on gp120 appear to mediate HIV-1 adhesion to the SIGN molecules, although the contribution of the gp120 polypeptide backbone is not excluded 1. Several in vitro studies have shown that highly glycosylated HIV-1 gp120 is a strong ligand for a variety of mannose-binding lectins 17 18 19 20 21 22. Although the carbohydrate structures on the HIV envelope could be nonspecifically recognized by host lectins, the physiological consequences of such recognition will be specified by the functions of the binding molecule. The SIGN molecules are the first membrane-associated lectins identified to date that enhance HIV-1 infection. Interestingly, the expression of L-SIGN in liver sinusoids suggests that LSECs, which are in continual contact with passing leukocytes, can capture HIV-1 from the blood and promote transinfection of T cells. Moreover, prior studies have indicated that LSECs themselves may be susceptible to HIV-1 infection 23 24. Thus, it is possible that L-SIGN promotes infection of these cells, thereby establishing a reservoir for production of a new virus to pass on to T lymphocytes trafficking through the liver sinusoid.
Additional functional studies are necessary for understanding the normal physiologic role of L-SIGN and its possible role in HIV-1 pathogenesis. Its ability to enhance transinfection of T cells suggests that L-SIGN may contribute to HIV-1 susceptibility. Alternatively, if a physiologic function of L-SIGN involves antigen clearance, this receptor could play a protective role in clearance of the virus from the circulation. A clearer understanding of this receptor may provide insight into its potential use in novel therapy against HIV-1.
Acknowledgments
We thank Y. Liu, P. Karacki, and M. Chalaby for their technical contributions and the laboratory of J. Goudsmit for providing support. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-56000, and the Dutch Aids Foundation contract 5008.
Footnotes
A.A. Bashirova and T.B.H. Geijtenbeek contributed equally to this work.
Abbreviations used in this paper: CCR, CC chemokine receptor; DC, dendritic cell; DC-SIGN, DC-specific ICAM-3–grabbing nonintegrin; EST, expressed sequence tag; ICAM, intercellular adhesion molecule; L-SIGN, liver/lymph node–specific ICAM-3–grabbing nonintegrin; LSEC, liver sinusoidal endothelial cell; nt, nucleotides; RH, radiation hybrid; RT, reverse transcription; UTR, untranslated region.
References
- Geijtenbeek T.B., Kwon D.S., Torensma R., van Vliet S.J., van Duijnhoven G.C., Middel J., Cornelissen I.L., Nottet H.S., KewalRamani V.N., Littman D.R. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T.B., Torensma R., van Vliet S.J., van Duijnhoven G.C., Adema G.J., van Kooyk Y., Figdor C.G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- Curtis B.M., Scharnowske S., Watson A.J. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA. 1992;89:8356–8360. doi: 10.1073/pnas.89.17.8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama-Kobayashi M., Yamaguchi T., Sekine S., Kato S. Selection of cDNAs encoding putative type II membrane proteins on the cell surface from a human full-length cDNA bank. Gene. 1999;228:161–167. doi: 10.1016/s0378-1119(99)00004-9. [DOI] [PubMed] [Google Scholar]
- Soilleux E.J., Barten R., Trowsdale J. DC-SIGN, a related gene DC-SIGNR, and CD23 form a cluster on 19p13. J. Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- Chomczynski P. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal. Biochem. 1992;201:134–139. doi: 10.1016/0003-2697(92)90185-a. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α. J. Exp. Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani N., Gruner S., Brang D., Kampgen E., Lenz A., Trockenbacher B., Konwalinka G., Fritsch P.O., Steinman R.M., Schuler G. Proliferating dendritic cell progenitors in human blood. J. Exp. Med. 1994;180:83–93. doi: 10.1084/jem.180.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lub M., van Vliet S.J., Oomen S.P., Pieters R.A., Robinson M., Figdor C.G., van Kooyk Y. Cytoplasmic tails of β1, β2, and β7 integrins differentially regulate LFA-1 function in K562 cells. Mol. Biol. Cell. 1997;8:719–728. doi: 10.1091/mbc.8.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecilia D., KewalRamani V.N., O'Leary J., Volsky B., Nyambi P., Burda S., Xu S., Littman D.R., Zolla-Pazner S. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 1998;72:6988–6996. doi: 10.1128/jvi.72.9.6988-6996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijtenbeek T.B., van Kooyk Y., van Vliet S.J., Renes M.H., Raymakers R.A., Figdor C.G. High frequency of adhesion defects in B-lineage acute lymphoblastic leukemia. Blood. 1999;94:754–764. [PubMed] [Google Scholar]
- Hegenbarth S., Gerolami R., Protzer U., Tran P.L., Brechot C., Gerken G., Knolle P.A. Liver sinusoidal endothelial cells are not permissive for adenovirus type 5. Hum. Gene Ther. 2000;11:481–486. doi: 10.1089/10430340050015941. [DOI] [PubMed] [Google Scholar]
- Hild M., Weber O., Schaller H. Glucagon treatment interferes with an early step of duck hepatitis B virus infection. J. Virol. 1998;72:2600–2606. doi: 10.1128/jvi.72.4.2600-2606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor R.I., Chen B.K., Choe S., Landau N.R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- Knolle P.A., Gerken G. Local control of the immune response in the liver. Immunol. Rev. 2000;174:21–34. doi: 10.1034/j.1600-0528.2002.017408.x. [DOI] [PubMed] [Google Scholar]
- Moffatt O.D., Devitt A., Bell E.D., Simmons D.L., Gregory C.D. Macrophage recognition of ICAM-3 on apoptotic leukocytes. J. Immunol. 1999;162:6800–6810. [PubMed] [Google Scholar]
- Ezekowitz R.A., Kuhlman M., Groopman J.E., Byrn R.A. A human serum mannose-binding protein inhibits in vitro infection by the human immunodeficiency virus. J. Exp. Med. 1989;169:185–196. doi: 10.1084/jem.169.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattegno L., Ramdani A., Jouault T., Saffar L., Gluckman J.C. Lectin-carbohydrate interactions and infectivity of human immunodeficiency virus type 1 (HIV-1) AIDS Res. Hum. Retroviruses. 1992;8:27–37. doi: 10.1089/aid.1992.8.27. [DOI] [PubMed] [Google Scholar]
- Hammar L., Hirsch I., Machado A., De Mareuil J., Baillon J., Chermann J.C. Lectin effects on HIV-1 infectivity. Ann. NY Acad. Sci. 1994;724:166–169. doi: 10.1111/j.1749-6632.1994.tb38907.x. [DOI] [PubMed] [Google Scholar]
- Saifuddin M., Hart M.L., Gewurz H., Zhang Y., Spear G.T. Interaction of mannose-binding lectin with primary isolates of human immunodeficiency virus type 1. J. Gen. Virol. 2000;81:949–955. doi: 10.1099/0022-1317-81-4-949. [DOI] [PubMed] [Google Scholar]
- Ushijima H., Schroder H.C., Poznanovic S., Gasic M.J., Matthes E., Muller W.E. Inhibition of human immunodeficiency virus-1 infection by human conglutinin-like proteinin vitro studies. Jpn. J. Cancer Res. 1992;83:458–464. doi: 10.1111/j.1349-7006.1992.tb01950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milone M.C., Fitzgerald-Bocarsly P. The mannose receptor mediates induction of IFN-α in peripheral blood dendritic cells by enveloped RNA and DNA viruses. J. Immunol. 1998;161:2391–2399. [PubMed] [Google Scholar]
- Steffan A.M., Lafon M.E., Gendrault J.L., Schweitzer C., Royer C., Jaeck D., Arnaud J.P., Schmitt M.P., Aubertin A.M., Kirn A. Primary cultures of endothelial cells from the human liver sinusoid are permissive for human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA. 1992;89:1582–1586. doi: 10.1073/pnas.89.5.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housset C., Lamas E., Courgnaud V., Boucher O., Girard P.M., Marche C., Brechot C. Presence of HIV-1 in human parenchymal and non-parenchymal liver cells in vivo. J. Hepatol. 1993;19:252–258. doi: 10.1016/s0168-8278(05)80579-3. [DOI] [PubMed] [Google Scholar]