Abstract
Somatic cell mutagenesis is a powerful tool for characterizing receptor systems. We reported previously two complementation groups of mutant cell lines derived from CD14-transfected Chinese hamster ovary–K1 fibroblasts defective in responses to bacterial endotoxin. Both classes of mutants expressed a normal gene product for Toll-like receptor (TLR)4, and fully responded to stimulation by tumor necrosis factor (TNF)-α or interleukin (IL)-1β. We identified the lesion in one of the complementation groups in the gene for MD-2, a putative TLR4 coreceptor. The nonresponder phenotype of this mutant was reversed by transfection with MD-2. Cloning of MD-2 from the nonresponder cell line revealed a point mutation in a highly conserved region resulting in a C95Y amino acid exchange. Both forms of MD-2 colocalized with TLR4 on the cell surface after transfection, but only the wild-type cDNA reverted the lipopolysaccharide (LPS) nonresponder phenotype. Furthermore, soluble MD-2, but not soluble MD-2C95Y, functioned to enable LPS responses in cells that expressed TLR4. Thus, MD-2 is a required component of the LPS signaling complex and can function as a soluble receptor for cells that do not otherwise express it. We hypothesize that MD-2 conformationally affects the extracellular domain of TLR4, perhaps resulting in a change in affinity for LPS or functioning as a portion of the true ligand for TLR4.
Keywords: sepsis, signal transduction, Toll-like receptors, Gram-negative bacteria, lipopolysaccharide
Introduction
A crucial first line defense against infectious illnesses is the ability to sense the presence of invading microorganisms. Gram-negative bacterial sepsis is a common cause of septic shock and death 1 and may begin abruptly when pathogens evade mucosal or integumentary structures and invade the bloodstream. A major part of the systemic inflammatory reaction is the response to endotoxin (LPS), the main component of the outer membrane of Gram-negative bacteria 2. The conserved lipid A portion of LPS is the structural basis for the toxic activation of host cells 3. The innate immune response to LPS is vital to effectively combat an infection, as animals with impaired LPS recognition are hypersusceptible to invasive bacterial disease 4. However, in normal hosts, when the organism burden is very large, an excess of inflammatory mediators produced by LPS-activated cells may result in systemic inflammation and the sepsis syndrome, a life-threatening condition that is characterized by fever, hypotension, compromised cardiac performance, coagulopathy, and multiple end-organ failure 5.
The task of defining the composition of the mammalian cellular receptor complex responsible for the recognition of LPS is of major scientific interest. The monocyte differentiation antigen, CD14, has long been known to bind LPS as the first step in endotoxin-induced activation, but the subsequent pathway of signal transduction is less well established. CD14 is linked to the plasma membrane of phagocytic leukocytes by a glycosyl-phosphatidylinositol anchor, and can also function as a soluble receptor. The lack of transmembrane and cytoplasmic domains, as well as the unique CD14-independent pharmacology of lipid A analogues suggested that a separate signal transduction molecule was necessary for LPS-induced signals 6.
The response to microbial products occurs in lower organisms. Thus, advances in the understanding of nonmammalian biology have had a great impact upon our understanding endotoxin recognition. Drosophila Toll was initially identified for its role in development 7 8, but molecular cloning of Toll led to the realization that the protein was a member of the IL-1 receptor family. Subsequent observations established that Toll expression was involved in innate immunity 9. Mammalian orthologs of Toll 10 11, referred to as Toll-like receptors (TLRs), have been identified and characterized not only for their role in LPS recognition 12 13, but for their role in mediating immune responses to a variety of pathogenic organisms (for a review, see reference 14).
TLR4 has been defined as the main cellular signal transducer for LPS. The first clue that TLR4 was involved in LPS signal transduction came from the discovery that the LPS hyporesponsive mouse strains C3H/HeJ and C57BL/10ScCr have mutations in this gene 12 15 16. Cellular transfection studies demonstrating a gain of function in TLR4-transfected cells 13, as well as the phenotypic characterization of a mouse with a targeted deletion of TLR4 17 18, strengthened the hypothesis that TLR4 is an LPS signal transducer. Unlike previous studies with CD14 6, TLR4 expression was found also to confer the unique species-specific pharmacology of human and rodent cells to certain lipid A analogues, including the lipid A precursor lipid IVa and Rhodobacter sphaeroides lipid A 19 20. Thus, TLR4 not only is the primary receptor involved in sensitizing cells to the presence of LPS, but it also is responsible for the fine specificity of lipid A recognition.
Although TLR4 has been shown to be a critical component for cell activation by LPS, the fully functional LPS signaling complex appears to require additional components. Recently, a small, extracellular protein known as MD-2 was found to enhance sensitive responses to LPS in cell lines that were transfected with TLR4. Expression of TLR4 in MD-2–deficient cells failed to enable these cells to respond to LPS 21 although other reports suggested that MD-2 expression might not be absolutely essential 13 22. Neither a natural mutant in MD-2 nor an animal with a targeted mutation was available for study. We present here the first loss of function data, suggesting the nearly absolute importance of MD-2 for LPS signaling either as a cell-derived or blood-derived LPS receptor component.
We have reported previously two complementation groups of mutants in a CD14-transfected Chinese hamster ovary–K1 fibroblast (CHO) cell line that are defective in LPS signaling 23 but are fully responsive to the cytokines TNF-α and IL-1β. TLR4 gene expression is normal in both groups of cells. However, we identified a point mutation in the gene coding for MD-2 as responsible for the complete loss of function in one of these mutants. This mutant, nonfunctional MD-2 still colocalizes on the cell surface with TLR4. Thus, the LPS receptor complex appears to require CD14, TLR4, and MD-2 for efficient function. Furthermore, our analysis of the second complementation group suggests that the LPS receptor consists of at least one additional, and as yet unidentified, component.
Materials and Methods
Cells and Reagents.
Reagents were obtained from Sigma-Aldrich, unless otherwise indicated. PBS, Ham's F-12, DMEM, and trypsin-versene mixture were from BioWhittaker. Low endotoxin fetal bovine serum (FBS) was from Hyclone. Ciprofloxacin was a gift from Miles Pharmaceuticals. Hygromycin B was purchased from Calbiochem. Protein-free LPS derived from Escherichia coli strain K-235 was a gift from S. Vogel (Uniformed Services University of Health Sciences, Bethesda, MD). Synthetic lipid A (compound 506) was a gift from S. Kusumoto (Osaka University, Toyonaka, Japan). Human IL-1 was purchased from Genzyme. CHO cell lines were cultured in Ham's F-12 containing 10% FBS, 10 μg/ml ciprofloxacin, and 400 U of hygromycin per milliliter in 5% saturated CO2 atmosphere at 37°C. HEK 293 cells were cultured in DMEM containing 10% FBS and 10 μg/ml ciprofloxacin.
Cloning of TLR4 from CHO Cells.
The sequence of Chinese hamster TLR4, cloned from both a CHO/CD14 library and peritoneal macrophages, has been reported previously (GenBank/EMBL/DDBJ accession no. AF153676; reference 19). Total RNA was extracted from the cell lines 3E10 (wild-type), 7.7 (a member of complementation group A), and 7.19 (a member of complementation group B), respectively, and 1 μg of RNA was reverse transcribed using Superscript II reverse transcriptase according to the manufacturer's protocol (Life Technologies). Overlapping fragments of CHO-TLR4 from 3E10, 7.7, and 7.19 were generated using the following primer pairs: 5′-aggttgccactctcacttcc-3′/5′-atgtcaggcttggcagattcactt-3′, basepair (bp) 80–1,383; and 5′-ttcaaatggcaaaccttagcagtc-3′/5′-atgattctttgcctgagttggtga-3′, bp 1,238–2,992. Multiple fragments from three independent PCR reactions from all three cell lines were sequenced at the Boston Medical Center Core facility (Boston, MA) and in the laboratory of one of the authors (D.A. Schwartz).
Cloning of MD-2 from CHO Cells.
PCR was performed on a previously described cDNA library constructed with the ZAP Express cDNA Gigapack III Gold cloning kit (Stratagene) from polyadenylated mRNA derived from CHO/CD14 cells 24. Cross-species primers were generated based upon the homologous regions of the published sequences for human and mouse MD-2. An internal sequence of hamster MD-2 was determined by sequencing the PCR products. This sequence was the basis of the construction of a set of hamster-specific internal primers. A combination of plasmid-specific and hamster-specific MD-2 primers were used in a three-step nested PCR, using the CHO/CD14 cDNA library as a template to derive the full-length cDNA sequence of CHO MD-2 and the flanking untranslated regions. Based on these sequences, primers were constructed to amplify the complete coding region of the gene. For the cloning of full-length MD-2, total RNA was extracted from the cell lines 3E10 (wild-type), 7.7 (complementation group A), and 7.19 (complementation group B), respectively, and reversed transcribed as described above. One microliter of the resulting cDNA was used to amplify full-length MD-2 using the primer pair 5′-GTGGAAAGTGTTGGAGATA-3′ and 5′-TAAAAACATATATTCTTAATTTATT-3′. Sequences of the resulting products were confirmed by analyzing the products of two independent PCR reactions on multiple sequence runs (at least twice for each PCR product). Furthermore, the area of the C95Y mutation described in this manuscript has been amplified and sequenced separately using internal PCR primers from each of the three cell lines on at least four additional occasions. The sequence for MD-2 from the wild-type cell line 3E10 was in complete agreement with the sequence derived from the CHO cell cDNA library. The sequence data for hamster MD-2 are available from GenBank/EMBL/DDBJ under accession no. AF325501.
Expression Plasmids.
The cDNA of MD-2 was generated by PCR using site-specific primers and directionally cloned into XhoI and BamHI sites of the mammalian expression plasmid pEFBOS 25 to express the Flag epitope at the COOH-terminal end of the protein. The proper sequence, orientation, and frame were confirmed by sequencing. The expression plasmid for nontagged human TLR4 (hTOLL) in the vector pcDNA3 was a gift from C. Janeway and R. Medzhitov (Yale University, New Haven, CT). The pELAM-luc reporter plasmid that transcribes firefly luciferase from a nuclear factor (NF)-κB–dependent promoter has been described previously 13.
Site-directed Mutagenesis of Human MD-2.
The cDNA of human MD-2 was cloned into XhoI and BamHI sites of the mammalian expression plasmid pEFBOS as described previously. A mutant human MD-2C95Y was generated by site-directed mutagenesis using the primer pair 5′-GCAAAGAAGTTATTTACCGAGGATCTGATGACGATTAC-3′ and 5′-GTAATCGTCATCAGATCCTCGGTAAATAACTTCTTTGC-3′ and using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's protocol. Proper introduction of the point mutation was confirmed by DNA sequencing.
Immunoblotting for Activated c-Jun Kinase.
Cells were plated at a density of 5 × 10 5 cells per well in six-well dishes and incubated overnight. The following day, the cells were stimulated with LPS (100 ng/ml), TNF-α (30 ng/ml), or IL-1β (5 ng/ml) for 15 min, followed by lysis of the cell monolayer in 1× SDS sample buffer containing β-mercaptoethanol, sonication for 5 s, and boiling at 95°C for 5 min. Proteins were immunoblotted with an Ab specific for phospho-c-Jun NH2-terminal kinase (JNK; Promega) as described previously 22.
Transient Transfection and NF-κB Luciferase Reporter Assay.
Cells were plated at a density of 5 × 105 cells per well in six-well dishes and incubated overnight. The following day, the cells were transiently cotransfected with 1 μg of the indicated plasmid plus 1 μg of pELAM-luc using the Superfect transfection reagent (QIAGEN) as per the manufacturer's protocol (final vol 1.5 ml). After a 2-h incubation with the DNA, the cells were washed with PBS and incubated overnight. The next day, cells were stimulated for 5 h. Cell activation was determined by measuring luciferase activity of the total cellular lysate using an assay kit from Promega according to the manufacturer's instructions. Data are reported as the mean of triplicate determinations ± SD.
IL-6 Assay.
Cells were seeded at 2 × 104 cells per well in a 24-well dish and incubated at 37°C overnight. The following day, cells were washed twice with PBS and stimulated with the indicated concentrations of LPS or IL-1β for 7.5 h in a total volume of 250 μl F12 medium containing 2% FCS. Supernatants were analyzed for bioactive IL-6 in the B9 cell proliferation assay as described previously 19 26. To examine the ability of transfected MD-2 to complement the ability of the CHO/CD14 mutants to release IL-6, cells were seeded at 4 × 104 cells per well in a 24-well dish and transfected the next day with 0.2 μg of pcDNA3, the expression plasmid for wild-type or the mutant MD-2 plasmid using the effectene transfection reagent (QIAGEN), according to the manufacturer's protocol. After overnight incubation, cells were washed twice, stimulated for 7.5 h, and analyzed for bioactive IL-6.
Semiquantitative Reverse Transcriptase PCR of MD-2 mRNA.
To assess the levels of MD-2 transcript in the cell lines 3E10 and 7.19, total RNA was extracted and 5 μg of RNA was reverse transcribed as above. The resulting cDNA was set up in a master mix to a final volume of 1 μl cDNA per sample, and primers for amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 5′-GTCATCATCTCCGCCCCTTCTGC-3′ and 5′-GATGCCTGCTTCACCACCTTCTTG-3′) or full-length MD-2 were added, respectively. The products of the PCR reactions were removed from the PCR block after the indicated number of cycles, stored on ice until PCR was completed and resolved by electrophoresis on an ethidium bromide stained 2.5% agarose gel.
Flow Cytometry Analysis of Cell Surface Expressed MD-2.
Cells were plated at a density of 5 × 105 per well in six-well dishes and incubated overnight. The following day, the cells were transiently transfected with MD-2 expression plasmids coding for a Flag epitope tagged MD-2 and incubated for 24 h. Cells were harvested by incubating 20 min at 37°C with 1 mM Na2EDTA. Cells were washed with PBS containing 1% FBS and labeled with the anti-Flag mAb M2 (20 μg/ml in PBS per 1% FBS; Sigma-Aldrich) for 30 min on ice. Cells were then washed with PBS with 1% FBS and counterstained with FITC-conjugated sheep anti–mouse IgG (1:100 dilution; Sigma-Aldrich) for 30 min on ice, followed by washing in PBS with 1% FBS. The stained cells were analyzed for Flag epitope expression using a FACScan™ microfluorimeter with the CELLQuest™ software package (Becton Dickinson).
Immunoprecipitation.
HEK 293 cells were seeded at a density of 5 × 106 per 100-mm dish, cultured overnight, and transiently transfected with 8 μg of total DNA per condition. After 48 h, cells were washed once with ice-cold PBS and lysed in buffer consisting of 50 mM Tris, 1% NP40 (American Bioanalytical), 0.05% CHAPS, 1 mM PMSF, and 0.1% protease inhibitor cocktail (Sigma-Aldrich), adjusted to pH 7.5. After incubation on ice for 30 min, samples were cleared by centrifugation (10 min, 12,000× g) and supernatants were transferred to chilled fresh tubes containing 50 μl of anti-Flag M2 agarose in lysis buffer (1:2 packed gel). A 50-μl portion of 2× sample buffer containing 5% 2-mercaptoethanol was added to each sample. Samples were boiled for 3 min, spun down (30 s, 10,000 g), and subjected to SDS-PAGE (10% acrylamide) and Western blot analysis, using a horseradish peroxidase–conjugated anti-Flag mAb (2 μg/ml M2) and an horseradish peroxidase–conjugated anti-Myc mAb (0.2 μg/ml 9E10; Santa Cruz Biotechnology, Inc.).
Results
CHO cells lack the differentiation antigen, CD14, and do not respond to LPS, even though they express TLR4 19. After transfection with the cDNA for CD14, the phenotype of these cells with respect to endotoxin responses dramatically changes to resemble phagocytic leukocytes 27. LPS-responsive CHO/CD14 cells differ from cell lines of leukocytic origin because they are remarkably easy to manipulate genetically. Thus, CHO/CD14 are a practical cell line in which to screen for LPS nonresponder mutants. We selected for abnormalities in one of the key events in LPS-mediated responses, the activation of the transcription factor NF-κB 28 29. The CHO/CD14 cell lines employed in these investigations were stably transfected with an NF-κB–responsive reporter construct that was used to discriminate between LPS responder and nonresponder cells derived from methanesulfonic acid ethyl ester (a point mutagen)–treated stocks. The mutants that were derived were examined by complementation analysis and assigned to one of two groups, designated group A or B, each representing a mutation in a different gene 23.
To define the genetic lesion in each of the mutants, we focused on individual mutants from each of the known complementation groups: clone 7.7 (a member of the A complementation group) and clone 7.19 (a member of the B complementation group).
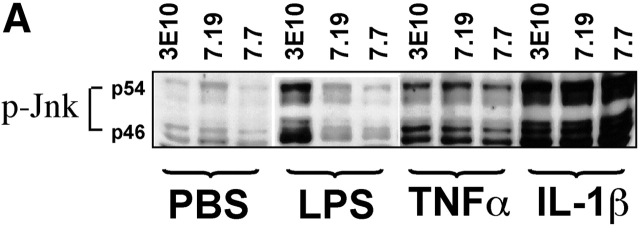
Wild-type CHO cells transfected with CD14 show a variety of responses to stimulation with LPS, including the translocation of the transcription factor NF-κB, the activation of certain mitogen-activated protein (MAP) kinases, and the release of inflammatory mediators such as arachidonic acid 27 and IL-6 19. To further characterize the defect in the mutant cell lines, we investigated the activation of MAP kinases as well as the secretion of IL-6 in response to stimulation with LPS. Neither of the mutant cell lines phosphorylated the JNK kinase, one of the MAP kinases known to be activated by LPS (Fig. 1 A), but showed normal phosphorylation of the JNK kinase in response to the cytokines TNF-α and IL-1β. Similarly, both mutants were impaired in their ability to secrete IL-6 in response to LPS, whereas they showed a normal response to stimulation with IL-1β (Fig. 1 B), indicating that the mutation has a broad effect on the capacity of these cells to show a physiological response to LPS. Neither of the mutants responded to concentrations as high as 5 μg LPS per milliliter as assessed by cotransfection with an NF-κB–dependent reporter plasmid and a control vector, whereas both cell lines responded to IL-1β (Fig. 2, lanes 1–3) and TNF-α 23. The IL-1 receptor cascade and TLR4 are thought to share many downstream signaling components in the NF-κB pathway, including MyD88 and TRAF6 30 31 32 33. The normal response of the mutant lines to IL-1β indicates that the mutations responsible for the LPS nonresponder phenotype affect gene products in the NF-κB pathway that are upstream of these signaling molecules, presumably receptor components.
Figure 1.
Mutant CHO-CD14 cells show defects in response to LPS stimulation, but not in response to the cytokines TNF-α and IL-1β. (A) Wild-type 3E10 cells and mutant cell lines 7.7 and 7.19 were stimulated for 15 min with LPS, TNF-α, or IL-1β. Cells were lysed and proteins were resolved by PAGE, electroblotted, and immunoblotted with an Ab specific for phospho-JNK. The blot shown is representative of three independent experiments. (B) Wild-type 3E10 and the mutant cell lines 7.7 and 7.19 were stimulated with LPS or IL-1β at the indicated concentrations for 7.5 h. Supernatants were assayed for bioactive IL-6. The results shown are mean values of triplicate determinations ±SD and are representative of three independent experiments. +, 3E10; ▴, 7.19; X, 7.7.
Figure 2.
Mouse MD-2 confers LPS signaling when expressed in the LPS nonresponder mutant 7.19, but not on the mutant 7.7. LPS nonresponder mutant cell lines 7.7 (A) and 7.19 (B) were transiently cotransfected with the control plasmid pcDNA3, plasmid coding for human TLR4 (hTOLL), or mouse MD-2, plus the pELAM-luc reporter plasmid as described in Materials and Methods. The next day, cells were stimulated with medium alone, 2.5 ng of IL-1β per milliliter, or 5 μg of LPS per milliliter for 5 h. Total cellular lysates were assayed for luciferase activity. The results shown are mean values of triplicate determinations ±SD and are representative of three independent experiments.
First we focused on the genes for TLR2 and TLR4 because these genes had been reported to be putative signaling receptors for LPS 12 32 34. Both genes were cloned and sequenced from wild-type CHO/CD14 cells. We found that wild-type CHO cells (as well as normal Chinese hamsters) do not express functional TLR2 35, thus indicating that TLR2 expression is not essential for responses to LPS. Subsequently, this conclusion was confirmed in the TLR2 knockout mouse 18. In contrast, CHO cells express a full-length and functional transcript for TLR4 19. Cloning of TLR4 from cDNA derived from the mutants 7.7 and 7.19 revealed that both complementation groups have a functional wild-type transcript for TLR4 (data not shown) that is identical to the previously reported sequence of hamster TLR4. Consistent with these data, transient transfection of the mutants 7.7 and 7.19 with the human gene for TLR4 failed to have an effect on the phenotype of these cells (Fig. 2, lanes 4–6).
Next, we sought to determine the role of MD-2 in the mutant cell lines because of the reports that MD-2 potentiated the activity of TLR4. Transfection with the gene for mouse MD-2 conferred responsiveness to one of the mutants, clone 7.19, a member of complementation group B. Transfection of mouse MD-2 into clone 7.7 had no effect. The ability of mouse MD-2 to impart full responsiveness to LPS to mutant 7.19 is shown in Fig. 3. Note how the MD-2 transfected mutant cell line responded similarly to a wild-type cell line at all concentrations of LPS that were tested.
Figure 3.
Transfection of the mutant 7.19 with mouse MD-2 confers normal sensitivity to LPS. The parental cell line 3E10 was cotransfected with plasmid pcDNA3 and the pELAM-luc reporter plasmid (black bars), as described in the legend to Fig. 2. Similarly, mutant strain 7.19 was cotransfected with mouse MD-2 and pELAM-luc reporter plasmid (hashed bars). Subsequently, cells were stimulated with medium alone, LPS, or IL-1β for 5 h, and cellular lysates were assayed for luciferase activity. The results shown are mean values of triplicate determinations ±SD and are representative of three independent experiments.
We cloned hamster MD-2 from all three cell lines by PCR using reverse-transcribed mRNA as template. Sequencing of the gene products from several independent amplification reactions revealed that the MD-2 transcript expressed by mutant 7.19 has a single-point mutation at position 284 of the gene (Fig. 4 A), leading to an amino acid exchange at position 95 of the protein from a cysteine to a tyrosine (MD-2C95Y). This C95Y mutation was not found in mutant 7.7 mRNA (data not shown). Fig. 4 B shows a partial sequence alignment of the gene for MD-2 from three different species, including the wild-type CHO cell sequence and the sequence of the mutant 7.19. As shown, the cysteine residue in position 95 is present in human, mouse, and hamster MD-2 in a highly conserved region of the gene. These data underscore both the importance of this residue and this conserved domain for proper MD-2 structure and function.
Figure 4.
The mutant 7.19 has a point mutation in the gene coding for MD-2. (A) MD-2 was sequenced from 3E10, 7.19 and mutant 7.7 (data not shown); a partial sequence of MD-2 from clone 3E10 and 7.19 is shown in this figure. Mutant 7.19 has a point mutation at position 284 of the coding region resulting in a conversion of the codon for cysteine to tyrosine (codon triplet marked by dashed line). The affected codon corresponds to amino acid position 95 of the protein. (B) Sequence alignment of bp 268–300 from human, mouse, wild-type CHO MD-2 from 3E10 cells, and MD-2 from the mutant 7.19, showing that the codon for cysteine at position 284 in the gene for MD-2, as well as most of the surrounding amino acids, are conserved in a variety of species.
MD-2 is thought to colocalize on the cell surface with TLR4 21. One possibility for the LPS nonresponder phenotype observed is that the mutant MD-2 failed to be transcribed, translated, and exported to the cell surface. We used semiquantitative reverse transcription PCR to determine if there were gross abnormalities in MD-2 mRNA expression in the LPS nonresponder CHO/CD14 mutant cell lines and found that transcript levels of the mutant species of MD-2 were comparable to the parental wild-type cell line (Fig. 5 A). To evaluate if the C95Y mutation prevented protein expression, we transfected both forms of MD-2 into 7.19 cells. After allowing cells sufficient time for protein expression to occur, surface expression of the Flag-tagged proteins was measured by flow microfluorimetry. Although the level of protein expression was somewhat lower with the mutant cDNA construct, both forms of MD-2 were well expressed on the surface of CHO 7.19 (Fig. 5 B). Wild-type MD-2 and mutant MD-2C95Y were expressed on the surface of 41 and 12.5% of the cells, respectively. Transfected cells were assayed simultaneously for LPS-inducible NF-κB activation using a luciferase reporter construct (Fig. 6 A). In contrast to the results found with protein expression, the mutant form of MD-2 failed to enable detectable translocation of NF-κB at a concentration of 1 μg of LPS per milliliter, a concentration that is ∼100,000-fold greater than ordinarily necessary to observe activation in wild-type CHO/CD14 cells, whereas the expression of the wild-type form of MD-2 enabled the mutant cell line to respond. The same effect was observed when transiently transfected cells were assayed for secretion of the cytokine IL-6 (Fig. 6 B). The expression of the wild-type form of MD-2 enabled a clear dose-dependent cytokine response to LPS, whereas MD-2C95Y did not. Thus, the C95Y mutation resulted in a loss of cell signaling capabilities that was strikingly out of proportion to the diminishment in the level of protein bound to the cell surface.
Figure 5.
The mutant MD-2C95Y is transcribed in normal quantities and expressed on the cell surface. (A) Semiquantitative PCR of MD-2 and GAPDH on reverse transcribed mRNA from 3E10 and 7.19 cells. (B) FACS® analysis showing surface expression of wild-type MD-2 and MD-2C95Y in transiently transfected 7.19 cells. Cells were transiently transfected with MD-2 expression plasmids coding for Flag epitope–tagged MD-2. After overnight incubation, cells were detached with 1 mM Na2EDTA, washed, and stained with anti-Flag mAb and FITC-conjugated sheep anti–mouse IgG secondary Ab as described in Materials and Methods. Controls were stained with the secondary Ab only.
Figure 6.
Transfection of the nonresponder mutant 7.19 with wild-type CHO MD-2 enables a response to LPS, whereas transfection with MD-2C95Y does not. (A) The mutant strain 7.19 was cotransfected with either wild-type hamster MD-2 or hamster MD-2C95Y plus the pELAM-luc reporter plasmid. The next day cells were stimulated with medium alone, IL-1β, or LPS at the indicated concentrations for 5 h. Total cellular lysates were assayed for luciferase activity. (B) The mutant strain 7.19 was cotransfected with pcDNA3 control plasmid, wild-type hamster MD-2, or hamster MD-2C95Y. The next day cells were stimulated with LPS or IL-1β at the indicated concentrations for 7.5 h and supernatants were assayed for bioactive IL-6. The results shown are mean values of triplicate determinations ±SD and are representative of two independent experiments.
To define the importance of the cysteine residue at position 95 for the function of MD-2 in the human immune response to LPS, a mutant human MD-2 with an identical mutation was generated by site-directed mutagenesis. Transient cotransfection of HEK 293 cells with human TLR4 (hTOLL) and the wild-type human MD-2 or mutant human MD-2C95Y demonstrated that only the wild-type MD-2 conferred a response to LPS on these cells (Fig. 7), underscoring the probable general importance of the cysteine residue at position 95 for cell signaling in all species of mammals.
Figure 7.
A point mutation at position 95 of the human MD-2 gene abolishes cell activation by LPS. HEK 293 cells were cotransfected with either wild-type human MD-2 plus human TLR4 (hTOLL) or human MD-2C95Y plus TLR4 and the pELAM-luc reporter plasmid. The next day cells were stimulated with medium alone, IL-1β, or LPS at the indicated concentrations for 5 h. Total cellular lysates were assayed for luciferase activity. The results shown are mean values of triplicate determinations ±SD and are representative of three independent experiments.
One possible explanation for the lack of LPS responsiveness in MD-2C95Y–transfected cells is that the receptor component, while it is expressed at the cell surface, does not actually colocalize with TLR4, as has been reported previously for the wild-type gene product 21. To examine the physical interaction of the mutant MD-2 with TLR4, immunoprecipitation analysis was performed on HEK 293 cells that were transiently transfected with Myc-tagged human TLR4 plus either Flag-tagged human MD-2 or human MD-2C95Y. Fig. 8 indicates that both wild-type and MD-2C95Y coimmunoprecipitated with TLR4, implying that the mechanism involved in the loss of MD-2 function is more than a simple loss of the physical interaction between MD-2 and TLR4.
Figure 8.
MD-2 and MD-2C95Y colocalize with TLR4. HEK 293 cells were transiently transfected with human MD-2Flag (lane 1), MD-2Flag plus human TLR4Myc (lane 2), or huMD-2C95Y Flag plus TLR4Myc (lane 3). After 48 h cells were harvested, lysed, and immunoprecipitated with anti-Flag agarose. Western blot analysis was performed using an anti-Flag mAb or an anti–c-Myc mAb. Depicted is one representative experiment out of two.
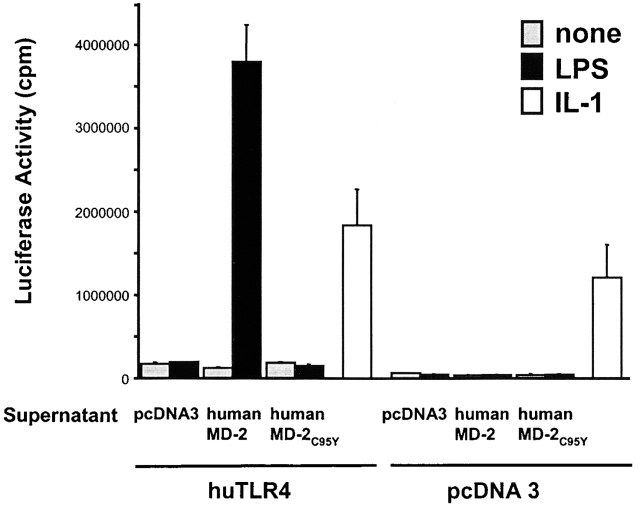
MD-2 is an extracellular protein with neither a predicted transmembrane domain nor linkage to the cell surface, although its mRNA does encode for a signal sequence. We hypothesized that MD-2 might be functional as a soluble receptor component and that expression of MD-2 transcript followed by translation and export to an extracellular site adjacent to TLR4 is not necessary for cells to respond to LPS. Rather, soluble MD-2 present in blood-derived products such as serum, or in tissue culture supernatants, might be sufficient to allow TLR4 to respond to the presence of LPS. To test this hypothesis, we transfected HEK 293 cells either with wild-type human MD-2 or mutant human MD-2C95Y and collected supernatants that were enriched in the secreted gene product. After allowing time for protein expression, these supernatants were then transferred to cell monolayers of HEK 293 cells that had been cotransfected with TLR4 plus an NF-κB–dependent reporter plasmid. Supernatants derived from cells that had been transfected with wild-type MD-2 conferred responsiveness on TLR4-expressing cells, whereas supernatants from cells expressing mutant MD-2C95Y did not (Fig. 9). These data suggest that MD-2 is not only secreted, but that the soluble receptor is functionally active. However, MD-2C95Y lacks the capability to function as a soluble receptor component.
Figure 9.
Human wild-type MD-2 but not MD-2C95Y–containing supernatants confer LPS responsiveness on TLR4-transfected cells. HEK 293 cells were transiently cotransfected with either human TLR4 (huTLR4) or empty vector (pcDNA3) plus the pELAM-luc reporter plasmid. The next day, supernatants from separate dishes of HEK 293 cells that had been transiently transfected overnight with either Flag-tagged human wild-type MD-2, human mutant MD-2C95Y, or pcDNA3 were transferred to TLR4- or pcDNA3-transfected cells. These supernatant-treated cells were then stimulated with LPS (100 ng/ml), IL-1β (2.5 ng/ml), or left untreated. After incubation for 5 h, the total cellular lysates were assayed for luciferase activity. The results shown are the mean values of triplicate determinations (±SD) and are representative of three nearly identical independent experiments.
Discussion
The ability of bacterial LPS to activate immune cells has been the subject of intense scrutiny because of its potential importance in understanding the etiology of the sepsis syndrome and other endotoxin-associated diseases. Even recently, activation models have been proposed based on the lipophilic characteristics of lipid A and the observation that CD14 transfers endotoxin into the lipid bilayer of animal cells very efficiently. These proposed models held that LPS signal transduction is initiated by nonspecific perturbations of lipid bilayer fluidity 36 37.
The observation that CD14 is a critical component of LPS responsiveness is now well established, but even this finding failed to resolve the dilemma of how LPS activates cells. While CD14 initially appeared to be a highly specific receptor for endotoxin, investigators quickly learned that the molecule interacted with a diverse repertoire of bacterial products 38 39 40. Furthermore, molecular genetic analysis of human and rodent CD14 demonstrated that this molecule lacked the ability to finely discriminate between nearly identical lipid A analogues with pharmacological properties that are completely different in different species of mammals 6, suggesting that CD14 lacked the specificity expected of a true LPS receptor. The identification of TLRs has radically changed this perspective concerning LPS-induced activation. In contrast to the uncertainty that previously surrounded our knowledge of the mechanism of cellular activation by LPS, the mechanism of how TLRs activate cells may be reasonably straightforward, involving CD14-mediated ligand–induced conformational changes in TLR4, receptor complex formation, and engagement of a subsequent signal transduction cascade.
However, this mechanism of signal transduction remains largely conjectural, and several important unanswered questions persist. Most importantly, despite molecular genetic observations suggesting direct binding of LPS to TLRs 19 20, clear-cut saturable ligand binding to TLRs has not been established by conventional techniques. This is not surprising in view of the difficulties that have been encountered in the past with LPS-binding assays due to the heterogeneity of the natural ligand, the difficulties in radiolabeling LPS to a highly specific activity, and the difficulties inherent in performing binding assays with a lipid. A potential difficulty associated with the development of ligand-binding assays for endotoxin–TLR4 interactions is the possibility that membrane intercalated endotoxin may bind to the transmembrane region of the signal transduction apparatus 41 in order to initiate signal transduction.
One of the most surprising aspects of TLR4 function appears to be the requirement of this receptor to engage LPS together with MD-2, a small, secreted protein. It has not been clear to what degree cells require MD-2 for LPS responses. Some investigators have observed LPS-induced activation in the absence of cotransfection with the gene for MD-2 13, although many laboratories have anecdotally reported problems repeating this observation. We propose that MD-2 is likely to be a biologically active serum constituent. The fact that supernatants derived from cells that have been transfected with MD-2 can confer responsiveness on cells transfected with only TLR4 (Fig. 9) supports the idea that MD-2 is secreted in a functionally active form and can influence cells that ordinarily do not express this gene product. Normal FBS apparently does not contain sufficient amounts of MD-2 to induce a strong response to LPS of cells transfected with TLR4 alone. We suggest that MD-2 needs to be present in abundance to have significant biological activity. Accordingly, the previous differences in LPS responses seen by different investigators who examined TLR4–transfected cells might, in fact, be due to differences in the source or the handling of the bovine serum.
While targeted mutant animals that lack MD-2 expression have not yet been generated, the loss of function data presented here support the conclusion that MD-2 is required for TLR4-mediated cell signaling to occur in response to LPS. The mutation in MD-2 identified from the nonresponder CHO/CD14 cell line is likely to be a critical clue to the mechanism by which MD-2 functions. Our data show that MD-2C95Y is apparently capable of binding to the cell surface and complexing with TLR4 so that the loss of simple binding of MD-2 to TLR4 does not explain the phenotype observed. One possibility is that MD-2 may influence ligand-induced TLR4 oligomerization or may be necessary for the assembly of an even larger multimeric receptor complex, and that MD-2C95Y is incapable of these important functions. Another consideration is that MD-2 might influence the affinity of TLR4 for LPS. An alternative consideration is the possibility that MD-2 binds LPS directly, as has recently been proposed 42; LPS–MD-2 complexes might conceivably function as the actual high affinity ligand for TLR4. Finally, LPS-bound MD-2 may even be the true ligand for TLR4, a role that is theoretically similar to the fly peptide spätzle. MD-2C95Y mutant may have lost its ability to bind LPS or may fail to make the proper conformational changes after being bound by endotoxin.
While the mechanism by which MD-2 supports TLR4 function is currently unknown, the importance of MD-2 in LPS responses is unambiguous, as demonstrated by the remarkable loss of function of the CHO/CD14 mutant cell lines and the lack of function of MD2C95Y as a soluble receptor. These data support the concept that the LPS receptor is a complex consisting of at least CD14, TLR4, and MD-2, but should not be seen as conclusive evidence that all of the components of the LPS receptor are currently known. Activation of TLR4 is thought to result in the initiation of signal transduction that is shared with the IL-1 receptor. Both MyD88 and TNF receptor–associated factor 6 knockout mice, for example, are defective in LPS- and IL-1β–induced activation 33 43. Our data demonstrate that the “A” complementation group of LPS nonresponder mutants, represented by clone 7.7, was normally responsive to stimulation by IL-1 β (Fig. 1 and Fig. 2). We hypothesize that in addition to CD14, TLR4, and MD-2, the complete LPS receptor is comprised of at least one additional gene product, represented by the mutation in the “A” complementation group. The hypothesized additional member of the LPS receptor may prove to be a homologous protein, such as a TLR, or a proximal molecule immediately adjacent to TLR4 in the LPS signal transduction pathway. The use of a cDNA library to expression clone the lesion in mutant 7.7 may identify this receptor component.
Acknowledgments
The authors would like to acknowledge Dr. Stefanie Vogel for her helpful discussions.
Andra Schromm is the recipient of DFG Schr 621/1-1 and is supported by SFB367, project B8; Philipp Henneke is the recipient of DFG He3127/1-1. This work was funded in part by National Institutes of Health grants RO1GM54060, AIPO150305, and AI32725 (to D.T. Golenbock, A. Schromm, P. Henneke, and B. Monks) and grants to D.A. Schwartz from the Department of Veteran Affairs (Merit Review), the National Institute of Environmental Health Sciences (ES07498 and ES09607), and the National Heart Lung and Blood Institute (HL62628 and HL64855). A. Yoshimura is supported by grant-in aid for Encouragement of Young Scientists from the Ministry of Education, Science, Sports, and Culture of Japan (12771332). E. Lien is supported by the Research Council of Norway and the Norwegian Cancer Society.
Footnotes
Abbreviations used in this paper: bp, basepair; CHO, Chinese hamster ovary–K1 fibroblast; FBS, fetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; JNK, c-Jun NH2-terminal kinase; MAP, mitogen-activated protein; NF, nuclear factor; TLR, Toll-like receptor.
References
- Sands K.E., Bates D.W., Lanken P.N., Graman P.S., Hibberd P.L., Kahn K.L., Parsonnet J., Panzer R., Orav E.J., Snydman D.R. Epidemiology of sepsis syndrome in 8 academic medical centers. Academic Medical Center Consortium Sepsis Project Working Group. JAMA. 1997;278:234–240. [PubMed] [Google Scholar]
- Raetz C.R., Ulevitch R.J., Wright S.D., Sibley C.H., Ding A., Nathan C.F. Gram-negative endotoxinan extraordinary lipid with profound effects on eukaryotic signal transduction. FASEB J. 1991;5:2652–2660. doi: 10.1096/fasebj.5.12.1916089. [DOI] [PubMed] [Google Scholar]
- Kotani S., Takada H., Tsujimoto M., Ogawa T., Takahashi I., Ikeda T., Otsuka K., Shimauchi H., Kasai N., Mashimo J. Synthetic lipid A with endotoxic and related biological activities comparable to those of a natural lipid A from an Escherichia coli re-mutant. Infect. Immun. 1985;49:225–237. doi: 10.1128/iai.49.1.225-237.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack R.S., Fan X., Bernheiden M., Rune G., Ehlers M., Weber A., Kirsch G., Mentel R., Furll B., Freudenberg M. Lipopolysaccharide-binding protein is required to combat a murine gram-negative bacterial infection. Nature. 1997;389:742–745. doi: 10.1038/39622. [DOI] [PubMed] [Google Scholar]
- Bone R.C. The pathogenesis of sepsis. Ann. Intern. Med. 1991;115:457–469. doi: 10.7326/0003-4819-115-6-457. [DOI] [PubMed] [Google Scholar]
- Delude R., Savedra R., Zhao H., Thieringer R., Yamamoto S., Fenton M., Golenbock D. CD14 enhances cellular responses to endotoxin without imparting ligand-specific recognition. Proc. Natl. Acad. Sci. USA. 1995;92:9288–9292. doi: 10.1073/pnas.92.20.9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K.V., Jurgens G., Nusslein-Volhard C. Establishment of dorsal-ventral polarity in the Drosophila embryogenetic studies on the role of the Toll gene product. Cell. 1985;42:779–789. doi: 10.1016/0092-8674(85)90274-0. [DOI] [PubMed] [Google Scholar]
- Schneider D.S., Hudson K.L., Lin T.Y., Anderson K.V. Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes Dev. 1991;5:797–807. doi: 10.1101/gad.5.5.797. [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Nicolas E., Michaut L., Reichhart J.M., Hoffmann J.A. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Medzhitov R., Preston-Hurlburt P., Janeway C.A., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Rock F.L., Hardiman G., Timans J.C., Kastelein R.A., Bazan J.F. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak A., He X., Smirnova I., Liu M.-Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr micemutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Chow J.C., Young D.W., Golenbock D.T., Christ W.J., Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- Means T.K., Golenbock D.T., Fenton M.J. The biology of Toll-like receptors. Cytokine Growth Factor Rev. 2000;11:219–232. doi: 10.1016/s1359-6101(00)00006-x. [DOI] [PubMed] [Google Scholar]
- Vogel S.N., Johnson D., Perera P.Y., Medvedev A., Lariviere L., Qureshi S.T., Malo D. Cutting edgefunctional characterization of the effect of the C3H/HeJ defect in mice that lack an Lpsn gene: in vivo evidence for a dominant negative mutation. J. Immunol. 1999;162:5666–5670. [PubMed] [Google Scholar]
- Qureshi S.T., Lariviere L., Leveque G., Clermont S., Moore K.J., Gros P., Malo D. Endotoxin-tolerant mice have mutations in toll-like receptor 4 (Tlr4) J. Exp. Med. 1999;189:615–625. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., Takeda Y., Takeda K., Akira S. Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharideevidence for TLR4 as the Lps gene product. J. Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., Takeda K., Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- Lien E., Means T.K., Heine H., Yoshimura A., Kusumoto S., Fukase K., Fenton M.J., Oikawa M., Qureshi N., Monks B. Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J. Clin. Invest. 2000;105:497–504. doi: 10.1172/JCI8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak A., Ricciardi-Castagnoli P., Citterio S., Beutler B. Physical contact between lipopolysaccharide and toll-like receptor 4 revealed by genetic complementation. Proc. Natl. Acad. Sci. USA. 2000;97:2163–2167. doi: 10.1073/pnas.040565397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu R., Akashi S., Ogata H., Nagai Y., Fukudome K., Miyake K., Kimoto M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Young D.W., Gusovsky F., Chow J.C. Cellular events mediated by lipopolysaccharide-stimulated toll-like receptor 4. MD-2 is required for activation of mitogen-activated protein kinases and Elk-1. J. Biol. Chem. 2000;275:20861–20866. doi: 10.1074/jbc.M002896200. [DOI] [PubMed] [Google Scholar]
- Delude R.L., Yoshimura A., Ingalls R.R., Golenbock D.T. Construction of a lipopolysaccharide reporter cell line and its use in identifying mutants defective in endotoxin, but not TNF-α, signal transduction. J. Immunol. 1998;161:3001–3009. [PubMed] [Google Scholar]
- Heine H., Delude R.L., Monks B.G., Espevik T., Golenbock D.T. Bacterial lipopolysaccharide induces expression of the stress response genes hop and H411. J. Biol. Chem. 1999;274:21049–21055. doi: 10.1074/jbc.274.30.21049. [DOI] [PubMed] [Google Scholar]
- Mizushima S., Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarden L.A., De Groot E.R., Schaap O.L., Lansdorp P.M. Production of hybridoma growth factor by human monocytes. Eur. J. Immunol. 1987;17:1411–1416. doi: 10.1002/eji.1830171004. [DOI] [PubMed] [Google Scholar]
- Golenbock D., Liu Y., Millham F., Freeman M., Zoeller R. Surface expression of human CD14 in Chinese hamster ovary fibroblasts imparts macrophage-like responsiveness to bacterial endotoxin. J. Biol. Chem. 1993;268:22055–22059. [PubMed] [Google Scholar]
- Shakhov A.N., Collart M.A., Vassalli P., Nedospasov S.A., Jongeneel C.V. κB-type enhancers are involved in lipopolysaccharide-mediated transcriptional activation of the tumor necrosis factor α gene in primary macrophages. J. Exp. Med. 1990;171:35–47. doi: 10.1084/jem.171.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H., Mitomo K., Watanabe T., Okamoto S., Yamamoto K. Involvement of a NF-κB-like transcription factor in the activation of the interleukin-6 gene by inflammatory lymphokines. Mol. Cell. Biol. 1990;10:561–568. doi: 10.1128/mcb.10.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Xiong J., Takeuchi M., Kurama T., Goeddel D.V. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- Medzhitov R., Preston-Hurlburt P., Kopp E., Stadlen A., Chen C., Ghosh S., Janeway C.A., Jr. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- Kirschning C.J., Wesche H., Ayres T.M., Rothe M. Human toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J. Exp. Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Adachi O., Ogawa T., Takeda K., Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- Yang R.B., Mark M.R., Gray A., Huang A., Xie M.H., Zhang M., Goddard A., Wood W.I., Gurney A.L., Godowski P.J. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature. 1998;395:284–288. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- Heine H., Kirschning C.J., Lien E., Monks B.G., Rothe M., Golenbock D.T. Cutting edgecells that carry a null allele for toll-like receptor 2 are capable of responding to endotoxin. J. Immunol. 1999;162:6971–6975. [PubMed] [Google Scholar]
- Wurfel M.M., Wright S.D. Lipopolysaccharide-binding protein and soluble CD14 transfer lipopolysaccharide to phospholipid bilayerspreferential interaction with particular classes of lipid. J. Immunol. 1997;158:3925–3934. [PubMed] [Google Scholar]
- Thieblemont N., Thieringer R., Wright S.D. Innate immune recognition of bacterial lipopolysaccharidedependence on interactions with membrane lipids and endocytic movement. Immunity. 1998;8:771–777. doi: 10.1016/s1074-7613(00)80582-8. [DOI] [PubMed] [Google Scholar]
- Kitchens R.L., Ulevitch R.J., Munford R.S. Lipopolysaccharide (LPS) partial structures inhibit responses to LPS in a human macrophage cell line without inhibiting LPS uptake by a CD14-mediated pathway. J. Exp. Med. 1992;176:485–494. doi: 10.1084/jem.176.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugin J., Heumann I.D., Tomasz A., Kravchenko V.V., Akamatsu Y., Nishijima M., Glauser M.P., Tobias P.S., Ulevitch R.J. CD14 is a pattern recognition receptor. Immunity. 1994;1:509–516. doi: 10.1016/1074-7613(94)90093-0. [DOI] [PubMed] [Google Scholar]
- Yu B., Hailman E., Wright S.D. Lipopolysaccharide binding protein and soluble CD14 catalyze exchange of phospholipids. J. Clin. Invest. 1997;99:315–324. doi: 10.1172/JCI119160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schromm A.B., Brandenburg K., Loppnow H., Moran A.P., Koch M.H., Rietschel E.T., Seydel U. Biological activities of lipopolysaccharides are determined by the shape of their lipid A portion. Eur. J. Biochem. 2000;267:2008–2013. doi: 10.1046/j.1432-1327.2000.01204.x. [DOI] [PubMed] [Google Scholar]
- Viriyakosol S., Kirkland T.N., Soldau K., Tobias P.S. MD-2 binds to bacterial lipopolysaccharide. J. Endotoxin Res. 2000;6:489–491. [PubMed] [Google Scholar]
- Lomaga M.A., Yeh W.C., Sarosi I., Duncan G.S., Furlonger C., Ho A., Morony S., Capparelli C., Van G., Kaufman S., van der Heiden A., Itie A. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]