Abstract
The isotype specificity of immunoglobulin (Ig) class switching is regulated by a cytokine which induces transcription of a specific switch (S) region, giving rise to so-called germline transcripts. Although previous studies have demonstrated that germline transcription of an S region is required for class switch recombination (CSR) of that particular S region, it has not been shown whether the level of S region transcription affects the efficiency of CSR. We addressed this question by using an artificial DNA construct containing a constitutively transcribed μ switch (Sμ) region and an α switch (Sα) region driven by a tetracycline-responsive promoter. The construct was introduced into a switch-inducible B lymphoma line and the quantitative correlation between Sα region transcription and class switching efficiency was evaluated. The level of Sα transcription was linearly correlated with CSR efficiency, reaching a plateau at saturation. On the other hand, we failed to obtain the evidence to support involvement of either RNA–DNA heteroduplex or trans germline transcripts in CSR. Taken together, it is likely that S region transcription and/or transcript processing in situ may be required for CSR. We propose that because of the unusual properties of S region DNA, transcription induces the DNA to transiently be single stranded, permitting secondary structure(s) to form. Such structures may be recognition targets of a putative class switch recombinase.
Keywords: secondary structures, recombinase, artificial constructs, tet inducible promoter, B lymphoma line
Introduction
Mature B cells expressing Ig on their surfaces become activated in peripheral lymphoid organs by encountering nominal antigens and help from T cells. Antigen-stimulated B cells actively proliferate in the germinal center and undergo two types of genetic alteration in their Ig gene loci. One is class switching which replaces the C region of the Ig H chain and changes isotypes from IgM to IgG, IgE or IgA, resulting in diversification of effector functions of the Ig recognizing a given antigen. The other alteration is somatic hypermutation (SHM) in the V regions of the Ig light and H chains, which generates Igs with diverse affinities to the antigen. Subsequently, B cells expressing high affinity Igs are selected by limited amounts of antigens, resulting in affinity maturation.
Class switch recombination (CSR) is accompanied by looping-out deletion of CH genes by recombination between μ switch (Sμ) region and one of downstream switch (S) regions located 5′ to each CH gene 1. S regions are composed of tandemly repetitive sequences 2 3 4 5. CSR can be divided into three steps: (a) selection of a target S region to be recombined with the Sμ region; (b) cleavage of that particular S and Sμ regions; (c) repair joining to generate a looped-out circular DNA and a deleted chromosomal IgH locus. The repair step of CSR has been well established to be mediated by the nonhomologous end joining (NHEJ) repair mechanism which is also involved in V(D)J recombination 6 7 8.
The target commitment of CSR to a particular S region is determined by transcription from an intronic (I) promoter located 5′ to each S region, which gives rise to synthesis of so-called germline transcripts 9 10. Mutations which abolish germline transcription severely affect CSR efficiency 11 12 13 14 15. As stimulation of B lymphocytes with different cytokines induces activation of specific I promoters, specificity of the CSR target S region appears to be determined by cytokines provided by T cells 16. The accessibility model proposes that such transcription of the S region enhances its accessibility to a putative cleaving enzyme 17. However, the exact molecular basis for accessibility or target selection is totally unknown. It has been speculated that germline transcripts per se may have some functional roles in CSR although they cannot encode any proteins. One hypothesis is the formation of transient heteroduplexes or R-loop with the S region, providing the specific structure for recognition by the cleaving enzyme 18 19 20. Splicing of germline transcripts has also been claimed to be required for CSR 21 22. Actual breakpoints of CSR have been clustered in S regions and its flanks whose sequences are spliced out from the germline transcripts 23 24. These results also suggest that transcription and cleavage steps might be coupled by an unknown mechanism.
Recently, activation-induced cytidine deaminase (AID) has been shown to play a critical role in a step between germline transcription and NHEJ repair, most likely in the cleavage process of CSR 25 26. All the isotypes other than IgM and IgD are absent in sera of AID−/− mice and hyper IgM syndrome type 2 patients who carry mutations in the AID gene. IgM+ spleen cells from AID−/− mice cannot accomplish CSR even after stimulation in vitro, although they generate germline transcripts and their NHEJ repair system is intact. AID has structural homology with an RNA editing enzyme APOBEC1 27 but its actual activity remains to be elucidated. AID is also involved in the SHM event because SHM is severely defective in AID−/− mice as well as hyper IgM syndrome type 2 patients 25 26. Although CSR and SHM occur independently, the two genetic events share AID as an essential molecule.
We have already shown that artificial DNA constructs containing two S regions, each transcribed by a constitutive promoter, can undergo CSR in a murine B lymphoma line CH12F3-2, which can switch efficiently from IgM to IgA by in vitro stimulation with cytokines and CD40 ligand (CD40L; reference 28). To investigate roles of germline transcription and its products in CSR, we generated a new DNA construct, in which one of the S region is regulated by a tetracycline (tet)-responsive promoter. Using CH12F3-2 transfectants with this construct, we showed that transcription of the S region is quantitatively associated with CSR efficiency. However, we could not obtain the evidence that germline transcripts per se are required for CSR. To explain these results, we propose that because of the unusual properties of S region DNA, transcription induces the DNA to transiently be single stranded, permitting secondary structure(s) to form. Such structures may be recognition targets of a putative class switch recombinase.
Materials and Methods
Constructs.
A tet-inducible substrate of CSR, pSCT(0,0) was constructed by modification of pSMC-24 29, which contains two transcription units driven by constitutively active human elongation factor-1α (EF1α) promoter and SRα promoter, respectively. SRα promoter was replaced with tet-inducible promoter taken from pTET-Splice (GIBCO BRL), without disturbing splicing regulatory elements. Into this plasmid designated as pSCT(0,0), FSμ-4, and FSα-2 fragments 30, which are segments of core Sμ and Sα region of mouse, were inserted to generate pSCT(μ,α).
To construct an expression vector, pCESS-V, TM-EGFP (enhanced green fluorescence protein) fragment 29 with a 151-bp spacer (SP) from the stuffer element of pEF-BOS 31 was inserted to modified pcDL·SRα 32 lacking splice acceptor sequence. Blasticidin S resistance marker (Bsr) from pSV2bsr (Kaken Seiyaku Co.) was further integrated to obtain pCESS-V. Two copies of FSα-2 fragment was inserted to pCESS-V to generate pCESS-V2Fα. pSCG(μ,2α), a switch substrate containing the constitutive promoter–driven S regions, was described previously 29.
CH12F3-2 Cell Transfectants and Their Analyses.
CH12F3-2 cell culture and flow cytometric analyses were performed as described 28 29. CSR induction of CH12F3-2 transfectants was done by stimulation with CD40L, IL-4, and hTGFβ1 as described 28. HTG204 cells are CH12F3-2 transfectants with SCG(μ,2α). FTZ14 cells are CH12F3-2 transfectants expressing a tet-responsive transactivator 25. CESS-V2Fa and SCT(μ,α) linearized with ScaI were introduced into HTG204 and FTZ14 cells, respectively, by electroporation. FTZ14 transfectants with SCT(μ,α) which showed a reasonable level of EGFP fluorescence (10–100 range of mean fluorescence intensity) without tet were selected. The integration status of SCT(μ,α) in the clones was examined by Southern blot analysis and transfectants with an intact single copy construct were used for further analyses.
Quantification of Pre- and Postswitch Transcripts.
Total cellular RNA isolation and reverse transcription (RT)-PCR for the pre- and postswitch transcripts (Pre-ST and Post-ST) were done as described previously 29, except for usage of TF primer (5′-CCA CGC TGT TTT GAC CTC CAT GAC A-3′) instead of SF for the amplification of Pre-ST II from SCT(μ,α). Quantification of bands after ethidium bromide staining was performed by densitometry (MediaGrabber™, v3.0; RasterOps Corporation). Northern blot analyses were performed by the standard procedure using HybondN+ membrane (Amersham Pharmacia Biotech). As probes, the SV40 ori fragment 30 and the BamHI/NotI fragment covering the EGFP segment from pSRa·TM·EGFP 29 were used to detect Pre-ST I, and Pre-ST II or Post-ST, respectively. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) fragment was prepared by PCR as an internal control.
Digestion-Circularization PCR.
Digestion-circulation (DC)-PCR analysis of SCT(μ,α) was carried out as described 33 with slight modifications. Genomic DNA was digested with BamHI and the endogenous c-myc gene locus served as control for the efficiency of digestion and ligation. The PCR primers and conditions used are as follows; primers DC-F (5′-GGA CCG GAT TGG ACT TCG CCT GTG A-3′) and DC-R (5′-CAC GCA AGG GCC ATA ACC CGT AAA GAG-3′), 40 cycles at 94°C for 30 s, 61°C for 30 s, and 72°C for 90 s; primers DC-myc-P1 (5′-CGG CAC ATG GAC TTG ATG TT-3′) and DC-myc-P2 (5′-TGA TGT TGG GTC AGT CGC AG-3′), 36 cycles at 94°C for 30 s, 53°C for 30 s, and 72°C for 90 s.
Genomic PCR and Determination of Breakpoints.
After CSR was induced at various concentrations of tet for 3 d, SCT(μ,α) transfectants #3 and #7 were expanded for another 3 d in a medium containing 500 ng/ml tet to prevent further CSR. We confirmed that percentages of CD8+ cells were unchanged (data not shown). Genomic DNAs were isolated from those cells by the standard procedure 34. PCR was done as described 30. A continuum of PCR products (3–5.5 kb) were amplified from genomic DNA using BI2F and L4 as primers. Clones of these fragments were made in pGEM-T Vector (Promega) and breakpoints of individual clones were determined using sequencing primers BOS-LV-1F (5′-CAG CCC CAG AGA CCA GAA GAT TG-3′) and CD8αI2R (5′-CGT CTC CCG GTC CAG GTC TCC CTC-3′).
Chromatin Immunoprecipitation Assay.
SCT(μ,α) transfectant #3 cells were cultured for 3 d with or without tet (500 ng/ml) and then an additional day with or without stimulation. Chromatin immunoprecipitation (ChIP) and quantitative PCR analyses were performed as described previously 35. Soluble chromatin prepared from 3 × 106 cells was used for each immunoprecipitation with 4 μg each of anti-acetylated H3 antibody (Upstate Biotechnology) or normal rabbit IgG (Santa Cruz Biotechnology, Inc.). Primer sequences used to amplify the tet or Iα promoter within chromatin immunoprecipitated DNAs are as follows. tetF, 5′-ATC GCC CTT CCC AAC AGT-3′; tetR, 5′-CTT TCT GGT TTT TCA GTT CCT CGA G-3′; IA-F, 5′-GAG GTG GAA CAG GAA GTG GGT GAG-3′; IA-R, 5′-TCA GTG TAC CAA TGA GCA GAG GAG-3′. CD19 and CD3ε promoter regions were used as control loci (unpublished data). All the PCR amplifications were performed in 30 cycles, except for 40 cycles in CD3ε primers, of 94°C for 15 s, 55°C for 30 s, 72°C for 1 min. Amplified bands were visualized by staining gels with SYBR Gold Nucleic Acid Gel Stain (Molecular Probes) and analyzing with a luminescent image analyzer (LAS-1000plus; FUJIFILM).
Bacterial RNaseHI Expression and Renaturation Gel Assay.
An Escherichia coli RNaseHI bicistronic expression vector with the tet-responsive promoter (GIBCO BRL) and internal ribosomal entry site (IRES)-EGFP segment (CLONTECH Laboratories, Inc.) was introduced to CH12F3-2 cells. G418 resistant cells with high EGFP expression without tet (Sigma-Aldrich) were further checked for the expression of RNaseHI as described previously 36 with some modifications. In brief, cellular extracts were electrophoresed for 16 h in a 12% polyacrylamide gel containing SDS. The running gel contained 107 counts per min polyrA-polydT (330 pmol of AMP). After renaturation for 10 h exchanging the renaturation buffer every 1.5 and 2 h, the gel was exposed overnight to X-ray film (Hyperfilm MP; Amersham Pharmacia Biotech).
Results
The Level of Cis Germline Transcription Is Positively Correlated with CSR Efficiency.
To examine whether germline transcription of the S region affects quantitatively the CSR efficiency, we constructed a new switch construct, SCT (μ,α), in which the downstream Sα region was driven by the tet promoter (Fig. 1). The construct was introduced into a CH12F3-2 transfectant, FTZ14, which expresses the tet responsible transactivator 25. Seven clones with integration of a single copy SCT(μ,α) were selected for tet-regulated EGFP expression. The CSR efficiency in the endogenous Ig and transgenic loci was estimated by surface expression of IgA and CD8α-EGFP fusion protein, respectively, 3 d after stimulation. All of the 7 clones were found to undergo CSR in the transgene locus only when tet was withdrawn from the medium (Fig. 2 A). By contrast, CSR efficiency in the endogenous Ig locus was not augmented by the removal of tet. The time courses of CSR in the endogenous Ig and transgene loci were similar (Fig. 2B Fig. c Fig. d).
Figure 1.
Structure of SCT(μ,α), CSR substrate with inducible germline transcripts. SCT(μ,α) contains the Sα region, transcription of which is regulated by tet promoter (pTET). The Sμ region is constitutively transcribed from the promoter of elongation factor 1α (pEF1α). CSR results in constitutive expression of the CD8α-EGFP fusion protein on surface, which can be monitored by FACS® analysis. Position of primers and BamHI sites is depicted on the map. Open, closed, and shaded triangles depict primers used for genomic or DC-PCR, RT-PCR, and ChIP assay, respectively. TM, transmembrane. pA, poly A. Neo, neomycin resistance gene with its own promoter. Rectangles, exons. Arrows, promoters.
Figure 2.

CSR in the switch substrate pSCT(μ,α) in CH12F3-2 carrying the tet responsible transactivator. (A) CSR efficiency in the presence or absence of tet. Seven FTZ14 transfectants with single copy integration of the switch construct SCT(μ,α) were assessed for CSR in SCT(μ,α) in the presence or absence of stimulation (stim) and/or germline transcription induction by tet removal. Cytokine stimulation was performed for 3 d. CD8α-EGFP and IgA expression was scored for CSR in transgene and endogenous loci, respectively. Black and white bars represent CD8α+EGFP+ and IgA+ cell frequencies by FACS® analyses, respectively. (B) FACS® profile of class switching in the endogenous and transgene loci of clones #3 and #7. CSR was assessed by surface expression of IgA and CD8α-EGFP fusion protein 156 h after tet removal and cytokine stimulation. Percentages indicate fractions of switched cells. Horizontal axis indicates EGFP expression while vertical axis indicates either CD8α or IgA staining. STIM, stimulation. (C and D) Time course of CSR in the endogenous (C) and transgenic (D) loci of clones #3 and #7 in the presence or absence of tet.
The clones #3 and #7 were further investigated for the quantitative relationship between Sα region transcription and CSR in detail. First, we quantitated the transgene Sα transcripts regulated by tet concentrations. The clones #3 and #7 were cultured for several days at various concentrations of tet. Total cellular RNA was isolated from these cells and the transgenic Sα transcripts (Pre-ST II) were measured by Northern blot analysis with an EGFP fragment as probe (Fig. 3 A). We confirmed that induction of the tet promoter is augmented synchronously in total population of cells (Fig. 3 B). The CSR efficiencies in the endogenous and transgenic loci, and the amount of transgenic Sα transcript expression were plotted versus the concentrations of tet (Fig. 3 C). The results showed that induction of Pre-ST II as well as appearance of CD8α-EGFP double-positive cells representing CSR in the transgene correlated inversely with the tet concentrations, indicating that CSR correlates with the transcription efficiency of the S region. The plot of the CSR efficiency versus the amount of Sα transcripts indicates that there may be at least two phases: (a) the initial proportional phase and (b) the plateau phase at high levels of transcription (Fig. 3 D). Saturation of the CSR efficiency with transcription of the Sα region indicates that another factor or process other than S region transcription is limiting under this condition.
Figure 3.
Positive correlation between germline transcription and CSR. (A) Quantitative analyses of preswitch RNA transcripts at various concentrations of tet. Expression of steady state Pre-ST I and II was examined by Northern analyses as described in Materials and Methods. Total cellular RNAs used were isolated from clones #3 and #7 cultured in the medium containing tet at indicated concentrations for 3 d. GAPDH mRNA was monitored to control the total amount of RNA. (B) FACS® analysis of clones #3 and #7 for expression of EGFP in the presence of various amounts of tet. Numbers and those in parentheses indicate tet concentrations and mean fluorescence intensity, respectively. Note that total population of cells gradually increases EGFP expression. (C) Germline transcription of the S region and CSR in clones #3 and #7 at various concentrations of tet. Steady state amounts of Pre-ST II in the presence of various amounts of tet were determined by normalization with internal GAPDH expression. Relative induction levels of Pre-ST II, IgA+, and CD8α+EGFP+ cells are shown by taking their maximal levels as 100. (D) Relationship between germline transcription and CSR of SCT(μ,α) in clones #3 and #7. Theoretical curve based on regression analysis was obtained by Microsoft EXCEL program.
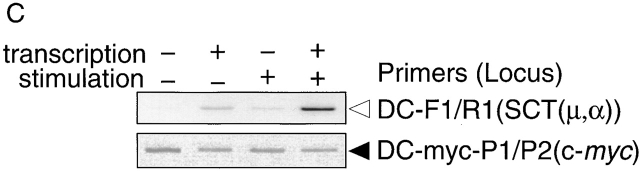
To further confirm that surface expression of the fusion protein (CD8α-EGFP) does represent CSR, we measured the appearance of Post-STs and recombination in the S region in clone #3. Post-ST expression increased in parallel with reduction of tet concentrations, which directly correlates with appearance of CD8+ cells (Fig. 4 A). Genomic PCR showed that increase in diffuse bands at the size range (3.0–5.5 kb) expected from CSR-mediated deletion between two S regions, indicating that DNA rearrangements clearly occurred in parallel with reductions of tet concentrations, which correlates with increase in CD8+ cell numbers (Fig. 4 B). DC-PCR further confirmed that S region recombination is dependent on Sα region transcription (Fig. 4 C). Recombination junction sequences were also determined by cloning genomic PCR products from 3.0- to 5.5-kb bands (Fig. 4 D). Their sequences clearly demonstrate that recombination took place between the Sμ and Sα regions and that occasional mutations were introduced in the proximity of the junctions as reported for the endogenous CSR junctions 23 24.
Figure 4.

Evidence for CSR in CD8α+EGFP+ cells. (A) Increase of Post-STs at various concentrations of tet. SCT(μ,α) transfectant clone #3 was stimulated to induce CSR for 3 d, keeping the concentration of tet as indicated before and after stimulation. Post-ST was determined by semiquantitative RT-PCR with five times serial dilutions of cDNA template. (B) Genomic PCR analyses. Genomic DNA from clone #3 was prepared as described in Materials and Methods, and PCR was performed with 0.5 μg of ScaI-digested DNA using BI2F and L4 primers. Arrowhead and bracket indicate unrearranged and rearranged bands, respectively. Note that rearranged bands are within the size range expected for recombination between Sμ and Sα regions. (C) DC-PCR analysis. Genomic DNA digested with BamHI was diluted and ligated to form circles followed by PCR amplification. The c-myc gene was used as control for ligation and PCR. (D) Nucleotide sequences surrounding 10 CSR junctions in SCT(μ,α) were determined after cloning of genomic PCR products isolated from 3.0 to 5.5-kb bands in B. Top, middle, and bottom rows indicate sequences of Sμ, determined junction, and Sα, respectively. Identical nucleotides are shown in uppercase letters. Underlined letters indicate mutations.
Relationship of Transcription with Histone Acetylation.
We have then examined whether increase in Sα transcription is correlated with the level of histone acetylation which has been shown to be a measure of chromatin opening of the locus 37. As shown in Fig. 5, even in the presence of high concentrations of tet, acetylation of histone H3 associated with the tet promoter region was already as high as those of the CD19 and Ig Iα promoters which are transcriptionally active in CH12F3-2 cells 28. Histone H3 acetylation in the tet promoter was not augmented even after either cytokine stimulation, transcription induction or both. As a negative control, histone H3 associated with the CD3 gene was not acetylated at all. The results are consistent with the idea that the chromatin structure of the Sα region in SCT(μ,α) may be in an open state before the transcription induction by tet removal.
Figure 5.
Acetylation states of H3 associated with tet promoter in SCT(μ,α) transfectant clone #3 before and after transcription induction. Soluble chromatins were prepared from SCT(μ,α) transfectant clone #3 cultured for 24 h in the presence or absence of S region transcription and switch stimulation. Antibodies against acetylated histone H3 (αAcH3) or rabbit IgG (Control) were used to isolate DNAs associated with the specific antigens. Fivefold serial dilutions of purified DNA were amplified by semiquantitative PCR with primers for tet, CD3ε, CD19, or Iα promoters. Total DNA before immunoprecipitation (IP) was used as an internal control of PCR. PCR cycles for CD3ε and the rest were 40 and 30, respectively.
CSR Is Not Affected by Expression of Germline Transcripts in Trans.
To examine whether the amount of S region transcripts is important for efficient CSR, we introduced an expression vector capable of overexpression of Sα germline transcripts (CESS-V2Fα) into HTG204 cells, transfectants of CH12F3-2 cells with SCG(μ,α) which contains the Sμ and Sα regions both constitutively transcribed (Fig. 6 A). CESS-V2Fα was designed to have the same structure as Sα transcripts of SCG(μ,2α) integrated in HTG204 cells except for an insertion of a 151-bp SP downstream of the EGFP gene (Fig. 6 A). This enables us to distinguish Sα transcripts of the switch substrate SCG(μ,2α) from those of CESS-V2Fα by the spacer size difference of RT-PCR products.
Figure 6.
Effect of overexpression of switch transcripts in trans on the CSR efficiency. (A) Germline transcript overexpression construct, CESS-V2Fα. The structure of Sα transcript driven by SRα promoter is the same as SCG(μ,2α), except for insertion of an 151-bp SP. Bsr stands for blasticidin resistance gene. TM, transmembrane. pA, poly A. CESS-V and SCG(μ,2α) (reference 29) are also shown for comparison. (B) CSR in the endogenous and transgene loci in five transfectants expressing germline transcripts in trans. The transfectants and control HTG204 cell were stimulated for 3 d with TGFβ, IL-4, and CD40L and surface marker expression after switching induction was quantitated by immunostaining and FACS®. The same analyses were carried out repeatedly and representative result is shown. CD8α+EGFP+ (2.49%) and IgA+ (20%) fractions of HTG204 after switch induction was taken as 100. (C) Expression levels of germline transcripts in each transfectant. Expression levels of cis (A) and trans (B) germline transcripts were estimated by competitive RT-PCR. Ratios of the respective germline transcripts to that of HTG204 and cis transcripts are shown below the gel photograph. The amount of each transcript was normalized by GAPDH signal.
Five HTG204 transfectants with CESS-V2Fα were compared for the switch efficiency with two control cell lines, parental HTG204, and its transfectant with CESS-V2Fα devoid of Sα (CESS-V). HTG204 switched 20 and 2.49% in the endogenous and transgenic loci, respectively, 3 d after stimulation. As shown in Fig. 6 B, the switch efficiencies of the endogenous and transgene loci were generally in parallel with each other as assessed by immunostaining and FACS® analysis. Comparative amounts of Sα germline transcripts from SCG(μ,α) and CESS-V2Fα were also semiquantitatively determined by RT-PCR (Fig. 6 C). HTG204 transfectants with CESS-V2Fα expressed variable amounts of Sα transcripts from CESS-V2Fα. By contrast, the amounts of Sα transcripts from SCG(μ,2α) were relatively stable among the transfectants. Nonetheless, the switching efficiencies of the five transfectants were similar to those of the control cells and had no correlation with the relative amounts of the two Sα transcripts, suggesting that trans germline transcripts may not affect the CSR efficiency.
CSR Is Not Affected by Bacterial RNaseHI Expression in CH12F3-2 Cells.
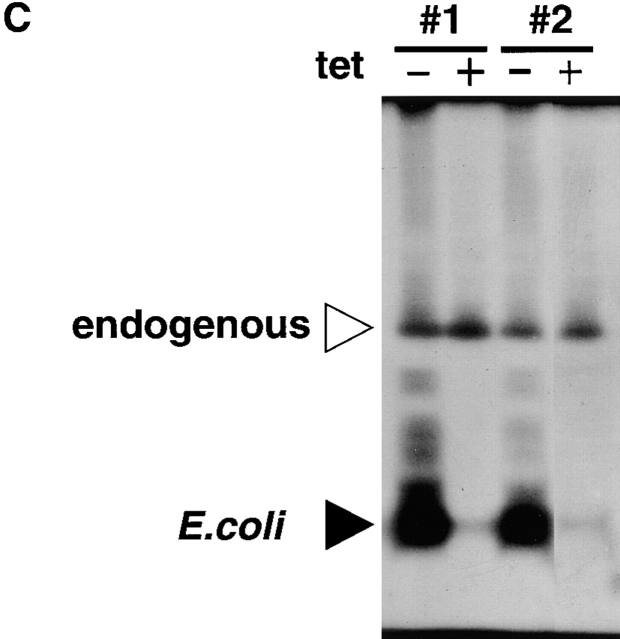
Several investigators have proposed a hypothesis that the nascent S region transcripts may form hybrid duplexes with the template DNA strand and that the hybrid or R-loop structure could be recognized by CSR recombinase 18 19 20. If this is the case, overexpression of E. coli RNaseHI may affect the efficiency of CSR in CH12F3-2 cells. A tet-inducible RNaseHI expression vector, RNaseHI-IRES-EGFP (Fig. 7 A), was introduced into CH12F3-2 cells and transfectants were selected by EGFP expression. The four transfectants showing EGFP expression only by tet removal were examined for the CSR efficiency in the presence or absence of tet (Fig. 7 B). All of them did not change the CSR efficiency with or without tet when stimulated for 1 or 3 d. Two of the four were confirmed for their expression of bacterial RNaseHI by the renaturation gel assay (Fig. 7 C). As E. coli RNaseHI has a small molecular size (18 kD), it can go through the nuclear pore without the nuclear localization signal 38. It is worth noting that CH12F3-2 cells express significant levels of endogenous RNaseH.
Figure 7.
Switching efficiency in CH12F3-2 cell lines is not affected by expression of E. coli RNaseHI. (A) E. coli RNaseHI-inducing vector. (B) Effects on CSR by RNaseHI expression in four transfectant clones. IgA expression on clones #1–4 and the parental line (FTZ14) which expresses only tet-repressible transactivator protein was assayed by FACS® 1 and 3 d after stimulation. Black and white bars indicate percentages of IgA+ cell with and without tet, respectively. RNase HI expression was induced by withdrawal of tet. (C) Renaturation gel assay of RNaseH activity in cellular extracts from RNaseHI transfected CH12F3-2 cells. Results from two transfectants (clone #1 and #2) are shown. The autoradiogram of the gel shows activity bands as black areas. Open and filled triangles indicate endogenous and bacterial RNaseH activities, respectively.
Discussion
We have examined the role of germline transcription of the S region and its products in CSR. Although S region transcription has been shown to be important for selecting a counterpart S region to be recombined with Sμ 16, it has not been examined whether the effect of S region transcription on CSR is all-or-none or quantitative. These results clearly indicate that the efficiency of germline transcription is linearly related to CSR efficiency over a broad range.
Germline transcription of an S region is believed to enhance accessibility of the S region 17. Accessibility of the locus to a putative recombinase is not clearly defined at the molecular level and may depend on several factors such as (a) chromatin structure, (b) target DNA structure, and (c) adapters required for recombinase binding to the locus. Opening of the chromatin structure may facilitate recognition of any DNA structure by CSR recombinase. We have shown that transcription does not change the histone acetylation level in the S region of SCT(μ,α), which is required for chromatin opening. As we have selected transfectants carrying randomly integrated transgenes whose S region promoters are active, it is likely that the S region in the transgene is already open before tet removal. In fact, RT-PCR analysis of SCT(μ,α) transfectants detected basal levels of Pre-ST II at a high concentration of tet (16 ng/ml) where CSR was hardly observed (data not shown).
Recombinase may not be able to bind a target unless the structure of the recognition target is appropriate. We have already shown that the primary sequence of the S region is not the recognition target of CSR recombinase 30. More recently, we have shown that class switch recombinase may recognize the secondary structure of the S region because a multiple cloning site sequence containing palindromes can replace the S region 29. Furthermore, CSR junctions are preferentially clustered around the neck region, namely the border between double-stranded and single-stranded region in the computer-generated secondary structure. Possible recognition structures may include RNA transcripts which can hybridize with template DNA 18 19 20. The experiments with induced overexpression of E. coli RNaseHI in CH12F3-2 cells failed to support involvement of such hybrids in CSR. In addition, constitutive expression of endogenous RNaseH in CH12F3-2 cells suggests that the stable levels of such hybrids may be low if present at all. We also showed that various levels of germline transcripts from the second transgene did not affect CSR of the initial switch construct. These results suggest that S region transcription per se may be critical to CSR. However, it is also possible that in situ processing of transcripts such as splicing may be important for CSR 21 22. Finally, CSR recombinase may require the RNA polymerase complex as adapter to bind the target DNA structure. These possibilities are not mutually exclusive with transcription-induced formation of the secondary structure in S regions.
Taking these results in toto, it is possible that transcription of the S region may be important for formation of the secondary structure in partially denatured S regions during transcription. If so, efficient transcription of S regions facilitates CSR by enhancing the formation of such secondary structures which probably depend on transient and partial denaturation of the S region during transcription. When transcription of the S region is limited by the ability of RNA polymerase to load onto the promoter, formation of secondary structure in the S region would be maximal at a certain point. Under these conditions, CSR recombinase could be limiting. It is therefore reasonable that an increase in transcription beyond a certain level does not yield more CSR products (Fig. 3 D).
The recent discovery of AID and characterizations of AID deficiency in mouse and man have clearly demonstrated that CSR and SHM share a common molecular mechanism 25 26. SHM as well as CSR has been shown to depend on transcription of the target DNA 39 40 41. Furthermore, increased transcription levels of a target construct were shown to induce higher mutation rates 42. Secondary structures in the variable region could be a recognition target by a putative mutator for SHM 43. In fact, formation of palindromic hairpin structures was predicted not only in S regions but also in V regions when they are single stranded 44. All these common features appear to support a scenario in which AID edits precursor mRNA for an endonuclease which recognizes secondary structures in single-stranded S and V region DNAs during efficient transcription 1. The search for target mRNA for a putative RNA editing activity of AID is a key to solve the molecular mechanism for both CSR and SHM.
Acknowledgments
We are grateful to Drs. K. Ikuta, S.K. Ye, Y. Sakakibara, A. Shimizu, and S. Takeda for their suggestions and critical reading of the manuscript. We thank Ms. K. Saito for preparation of the manuscript, Ms. T. Toyoshima, Y. Tabuchi, and K. Yurimoto for their technical assistance, and M. Muramatsu, S. Yoshikawa, X.C. Chen, Il-Mi Chung, T. Muto, T. Eto, and other members in Dr. Honjo's lab for sharing information and discussion.
This investigation was supported by Center of Excellence grant, and C.G. Lee was supported by the postdoctoral fellowship from Japanese Society for Promotion of Science.
Footnotes
Abbreviations used in this paper: AID, activation-induced cytidine deaminase; ChIP, chromatin immunoprecipitation; CSR, class switch recombination; DC, digestion-circularization; EGFP, enhanced green fluorescence protein; NHEJ, nonhomologous end joining; Post-ST, postswitch transcript; Pre-ST, preswitch transcript; RT, reverse transcription; S region, switch region; SHM, somatic hypermutation; SP, spacer; tet, tetracycline.
References
- Kinoshita K., Honjo T. Linking class switch recombination with somatic hypermutation. Nat. Rev. Mol. Cell Biol. 2001;2:493–503. doi: 10.1038/35080033. [DOI] [PubMed] [Google Scholar]
- Davis M.M., Kim S.K., Hood L.E. DNA sequences mediating class switching in alpha-immunoglobulins. Science. 1980;209:1360–1365. doi: 10.1126/science.6774415. [DOI] [PubMed] [Google Scholar]
- Dunnick W., Rabbitts T.H., Milstein C. An immunoglobulin deletion mutant with implications for the heavy-chain switch and RNA splicing. Nature. 1980;286:669–675. doi: 10.1038/286669a0. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Kawakami T., Takahashi N., Honjo T. Rearrangement of immunoglobulin γ1-chain gene and mechanism for heavy-chain class switch. Proc. Natl. Acad. Sci. USA. 1980;77:919–923. doi: 10.1073/pnas.77.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H., Maki R., Kurosawa Y., Roeder W., Tonegawa S. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature. 1980;286:676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- Casellas R., Nussenzweig A., Wuerffel R., Pelanda R., Reichlin A., Suh H., Qin X.F., Besmer E., Kenter A., Rajewsky K., Nussenzweig M.C. Ku80 is required for immunoglobulin isotype switching. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis J.P., Gu Y., Lansford R., Sonoda E., Ferrini R., Davidson L., Rajewsky K., Alt F.W. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J. Exp. Med. 1998;187:2081–2089. doi: 10.1084/jem.187.12.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolink A., Melchers F., Andersson J. The SCID but not the RAG-2 gene product is required for Sμ-Sε heavy chain class switching. Immunity. 1996;5:319–330. doi: 10.1016/s1074-7613(00)80258-7. [DOI] [PubMed] [Google Scholar]
- Stavnezer-Nordgren J., Sirlin S. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. EMBO J. 1986;5:95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos G.D., DePinho R.A., Zimmerman K.A., Lutzker S.G., Rosenberg N., Alt F.W. Secondary genomic rearrangement events in pre-B cellsVHDJH replacement by a LINE-1 sequence and directed class switching. EMBO J. 1986;5:3259–3266. doi: 10.1002/j.1460-2075.1986.tb04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Zou Y.R., Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- Jung S., Rajewsky K., Radbruch A. Shutdown of class switch recombination by deletion of a switch region control element. Science. 1993;259:984–987. doi: 10.1126/science.8438159. [DOI] [PubMed] [Google Scholar]
- Seidl K.J., Manis J.P., Bottaro A., Zhang J., Davidson L., Kisselgof A., Oettgen H., Alt F.W. Position-dependent inhibition of class-switch recombination by PGK-neor cassettes inserted into the immunoglobulin heavy chain constant region locus. Proc. Natl. Acad. Sci. USA. 1999;96:3000–3005. doi: 10.1073/pnas.96.6.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Gorham B., Li S.C., Bottaro A., Alt F.W., Rothman P. Replacement of germ-line ε promoter by gene targeting alters control of immunoglobulin heavy chain class switching. Proc. Natl. Acad. Sci. USA. 1993;90:3705–3709. doi: 10.1073/pnas.90.8.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Bottaro A., Li S., Stewart V., Alt F.W. A selective defect in IgG2b switching as a result of targeted mutation of the Iγ2b promoter and exon. EMBO J. 1993;12:3529–3537. doi: 10.1002/j.1460-2075.1993.tb06027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J. Immunoglobulin class switching. Curr. Opin. Immunol. 1996;8:199–205. doi: 10.1016/s0952-7915(96)80058-6. [DOI] [PubMed] [Google Scholar]
- Zhang T., Alt F.W., Honjo T. Regulation of class switch recombination of immunoglobulin heavy chain genes. In: Honjo T., Alt F.W., editors. Immunoglobulin Genes. 2nd ed. London Academic Press Ltd; London: 1995. [Google Scholar]
- Daniels G.A., Lieber M.R. RNA:DNA complex formation upon transcription of immunoglobulin switch regionsimplications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 1995;23:5006–5011. doi: 10.1093/nar/23.24.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaban M.E., Griffin J.A. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature. 1990;348:342–344. doi: 10.1038/348342a0. [DOI] [PubMed] [Google Scholar]
- Tian M., Alt F.W. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J. Biol. Chem. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- Hein K., Lorenz M.G., Siebenkotten G., Petry K., Christine R., Radbruch A. Processing of switch transcripts is required for targeting of antibody class switch recombination. J. Exp. Med. 1998;188:2369–2374. doi: 10.1084/jem.188.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M., Jung S., Radbruch A. Switch transcripts in immunoglobulin class switching. Science. 1995;267:1825–1828. doi: 10.1126/science.7892607. [DOI] [PubMed] [Google Scholar]
- Dunnick W., Hertz G.Z., Scappino L., Gritzmacher C. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.G., Kondo S., Honjo T. Frequent but biased class switch recombination in the Sμ flanking regions. Curr. Biol. 1998;8:227–230. doi: 10.1016/s0960-9822(98)70087-9. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Kondo S., Sugai M., Nazarea M., Imamura S., Honjo T. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int. Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- Tashiro J., Kinoshita K., Honjo T. Palindromic but not G-rich sequences are targets of class switch recombination. Int. Immunol. 2001;13:495–505. doi: 10.1093/intimm/13.4.495. [DOI] [PubMed] [Google Scholar]
- Kinoshita K., Tashiro J., Tomita S., Lee C.G., Honjo T. Target specificity of immunoglobulin class switch recombination is not determined by nucleotide sequences of S regions. Immunity. 1998;9:849–858. doi: 10.1016/s1074-7613(00)80650-0. [DOI] [PubMed] [Google Scholar]
- Mizushima S., Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe Y., Seiki M., Fujisawa J., Hoy P., Yokota K., Arai K., Yoshida M., Arai N. SRα promoteran efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol. Cell. Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.C., Paul W.E., Max E.E. Quantitation of immunoglobulin μ-γ1 heavy chain switch region recombination by a digestion-circularization polymerase chain reaction method. Proc. Natl. Acad. Sci. USA. 1992;89:6978–6982. doi: 10.1073/pnas.89.15.6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Firtsch E.F., Maniatis T. Molecular CloningA Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1989. [Google Scholar]
- Agata Y., Katakai T., Ye S.K., Sugai M., Gonda H., Honjo T., Ikuta K., Shimizu A. Histone acetylation determines the developmentally regulated accessibility for T cell receptor γ gene recombination. J. Exp. Med. 2001;193:873–880. doi: 10.1084/jem.193.7.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L.Y., Ma W.P., Crouch R.J. Ribonuclease H renaturation gel assay using a fluorescent-labeled substrate. Biotechniques. 1997;23:920–926. doi: 10.2144/97235rr01. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Acetylationa regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D. Nuclear protein import. Curr. Opin. Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- Fukita Y., Jacobs H., Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- Peters A., Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- Tumas-Brundage K., Manser T. The transcriptional promoter regulates hypermutation of the antibody heavy chain locus. J. Exp. Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachl J., Carlson C., Gray-Schopfer V., Dessing M., Olsson C. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J. Immunol. 2001;166:5051–5057. doi: 10.4049/jimmunol.166.8.5051. [DOI] [PubMed] [Google Scholar]
- Storb U., Klotz E.L., Hackett J., Jr., Kage K., Bozek G., Martin T.E. A hypermutable insert in an immunoglobulin transgene contains hotspots of somatic mutation and sequences predicting highly stable structures in the RNA transcript. J. Exp. Med. 1998;188:689–698. doi: 10.1084/jem.188.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolchanov N.A., Solovyov V.V., Rogozin I.B. Peculiarities of immunoglobulin gene structures as a basis for somatic mutation emergence. FEBS Lett. 1987;214:87–91. doi: 10.1016/0014-5793(87)80018-2. [DOI] [PubMed] [Google Scholar]