Abstract
High-affinity antibodies produced by memory B cells differ from antibodies produced in naive B cells in two respects. First, many of these antibodies show somatic hypermutation, and second, the repertoire of antibodies expressed in memory responses is highly selected. To determine whether somatic hypermutation is responsible for the shift in the antibody repertoire during affinity maturation, we analyzed the immunoglobulin lambda light chain (Igλ) repertoire expressed by naive and antigen-selected memory B cells in humans. We found that the Igλ repertoire differs between naive and memory B cells and that this shift in the repertoire does not occur in the absence of somatic hypermutation in patients lacking activation-induced cytidine deaminase (AID). Our work suggests that somatic hypermutation makes a significant contribution to shaping the antigen-selected antibody repertoire in humans.
Keywords: immunoglobulin repertoire, activation-induced cytidine deaminase, somatic hypermutation, memory B cell, affinity maturation
Introduction
The affinity of antibodies for their cognate antigens increases during immune responses 1. In depth analysis of hybridoma antibodies specific for influenza hemagglutinin or for small chemical haptens such as 2-phenyl-5-oxaz-olone or 4-hydroxy-3-nitrophenyl acetyl (NP) revealed that somatic hypermutation is one of the mechanisms that produce this increased affinity 2 3 4 5 6 7 8 9. For example, the VH186–2 + Igλ antibodies dominate the initial antibody response to NP, and mutation from TrpH33 to LeuH33 brings about a 10-fold increase in affinity of these VH186–2 + Igλ antibodies 8 10 11 12 13. Increased affinity is also accompanied by a shift in the antibody repertoire, and secondary high-affinity responses to NP are dominated by Igκ antibodies and not Igλ, suggesting that repertoire shifts contribute to affinity maturation 3 10 14 15. Little is known about this shift in the repertoire and how it relates to somatic hypermutation.
Activation-induced cytidine deaminase (AID) is a germinal center B cell–restricted molecule that carries cytidine deaminase activity and is required for switch recombination and somatic hypermutation in mice and humans 16 17 18. In the absence of AID, B cells are unable to undergo somatic hypermutation or produce secondary antibodies despite germinal center formation 17 18. Here we report on the Igλ antibody repertoire in humans deficient in AID. We find that AID is essential for the shift in repertoire between naive and antigen-selected memory B cells.
Materials and Methods
Patient Samples and Cell Preparation.
AID-deficient patients and AID mutations have been described 18. Patients P1, P13, P14, P17, and P18 were 10, 11, 4, 14, and 2 yr old, respectively at the time of blood donation, and they did not suffer from chronic infections. They were treated with intravenous Ig supplementation. Control donors C1, C2, C3, C4, C5, C6, and C7 were healthy and 32, 11, 35, 2, 28, 41, and 33 yr old, respectively when blood samples were obtained. Blood mononuclear cells were isolated on Ficoll gradients. Control CD19+ B cells were fractionated into naive CD19+IgM+CD27− and memory CD19+IgM+CD27+ B cells by cell sorting on FACS Vantage™. Due to absence of secondary isotypes in AID-deficient patients, AID B cells are all IgM+ and were therefore sorted into naive CD19+CD27− and memory CD19+CD27+ B cells without IgM staining. Antibodies used for staining were FITC–anti-CD19, PE–anti-CD27 (Immunotech/Beckman Coulter), and biotin-conjugated anti-IgM mAb (PharMingen), which was visualized with Streptavidin Red 670 (GIBCO BRL).
Reverse Transcription PCR, Cloning, and Sequencing.
Total RNA was extracted from 104–105 purified cells using TRIzol Reagent (GIBCO BRL) and reverse transcribed in a 10-μl reaction with Superscript II (GIBCO BRL). For reverse transcription (RT)-PCR reactions, 1 μl of cDNA was amplified for 30–35 cycles of 30 s at 94°C, 30 s at 58°C (VH1-Cμ) or at 55°C (Vλ-Cλ) and 30 s at 72°C with a final 10-min extension at 72°C using HotStarTaq™ DNA polymerase (QIAGEN) and the following primers: Vλ1−8 family consensus sense, 5′-GGG(G/A)TC(T/C)CTGA(C/T/G)CG(A/C/G)TTCTCTGG(C/G)TCC-3′; Vλ9 sense, 5′-ATCCCTGATCGCTTCTCAGTCTTG-3′; Vλ10 sense, 5′-GATCTCAGAGAGATTATCTGCATCC-3′; and Cλ antisense, 5′-CACAC(T/C)AGTGTGGCCTTGTTGGCTTG-3′. Sense FR1 VH1 and antisense Cμ primers were as described previously 19 20. RT-PCR products were run on 2% agarose gels, and PCR products were gel purified (Qiaquick™; QIAGEN) and cloned into TA vectors (Invitrogen). Double-stranded DNA sequences were obtained using antisense Cμ or Cλ primers and Dye Terminator Cycle Sequencing (PE Applied Biosystems). Sequences were analyzed using Ig BLAST®. When two or more identical sequences were found, they were counted as a single clone. Sequences were considered mutated when they displayed two or more nucleotide differences from their germline counterparts. Differences in gene distribution between naive and memory B cells were analyzed with chi-square tests (Cochran-Mantel-Haenszel test) adjusted by the Bonferroni method for multiple testing and they were considered significant when P values were equal to or less than 0.05.
Results and Discussion
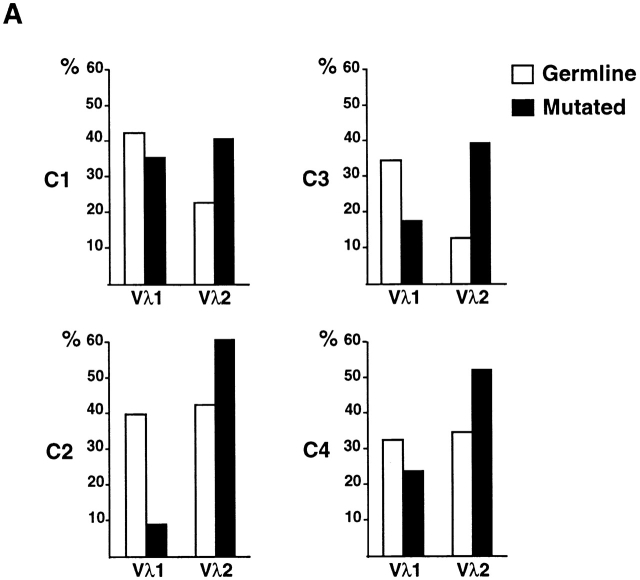
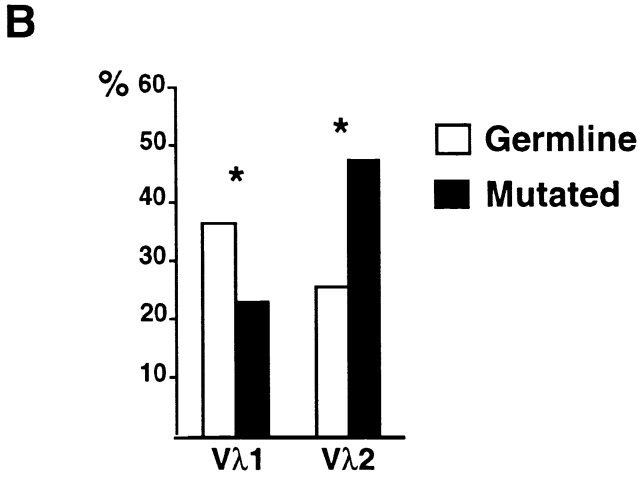
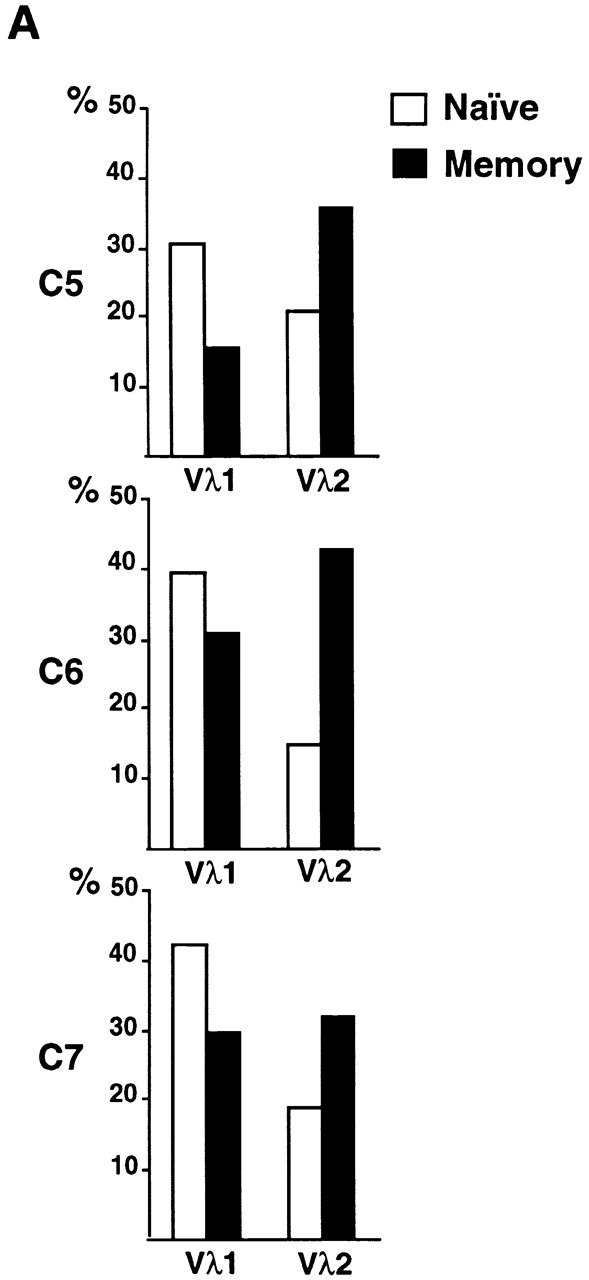
To determine whether there is a shift in repertoire between naive and antigen-selected B cell compartments in humans, we compared the unmutated germline Igλ sequences to mutated Igλ sequences obtained from CD19+ peripheral B cells from four control donors. In humans, Igλ light chains are found in 30–40% of all antibodies, and among the 10 Vλ gene families three (Vλ1, Vλ2 and Vλ3) represent >80–90% of all Vλ genes 21 22. We found that the distribution of Vλ1 and Vλ2, two of the most frequently used human Vλ families, differs between germline-encoded and mutated antibodies (Fig. 1 A; total of 239 individual sequences): Vλ1 is decreased and Vλ2 increased among mutated Igλs, and this difference is independent of the age of the donors (Fig. 1a and Fig. b).
Figure 1.
Igλ repertoire expressed in peripheral B cells from control donors. (A) Vλ1 and Vλ2 gene usage in germline-encoded (open bars) and mutated (solid bars) sequences from CD19+ peripheral B cells in four unrelated controls. 25, 15, 46, and 34 germline VλJλ sequences from donors C1, C2, C3, and C4 were compared with 33, 23, 33, and 29 mutated VλJλ sequences from the same individuals. The percent Vλ utilization is indicated on the y axis. (B) Combined total of Vλ1 and Vλ2 gene usage in germline and mutated VλJλ sequences from control donors. 121 germline-encoded and 118 mutated sequences were obtained from the four control donors. Asterisk (*) indicates statistically significant difference (Vλ1, P = 0.022; Vλ2, P = 0.0004).
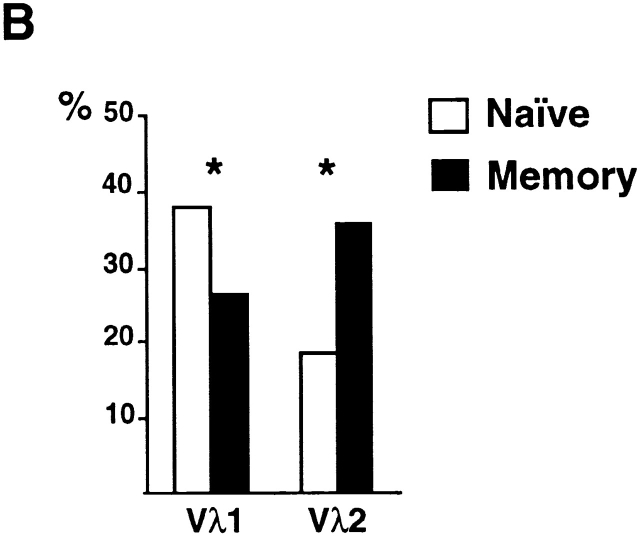
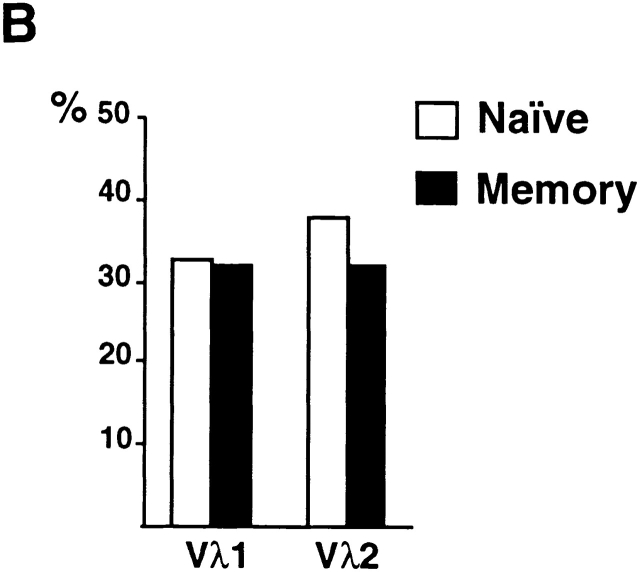
To further analyze the shift in Igλ repertoire between naive and memory B cells, we fractionated peripheral B cells using CD27 memory marker and isolated naive (CD19+IgM+CD27−) and memory (CD19+IgM+CD27+) B cells from control donors 23 24. The difference in Vλ distribution was also found when comparing naive and memory B cell compartments (Fig. 2; total of 262 sequences). Antibodies cloned from memory B cells were predominantly mutated and showed decreased Vλ1 and increased Vλ2 gene usage (Fig. 2a and Fig. b). We conclude that there is a shift in the Igλ repertoire between the naive and antigen-selected memory B cell compartments in humans.
Figure 2.
Igλ repertoire expressed in naive and memory B cells from control donors. (A) Vλ1 and Vλ2 gene usage in naive CD19+IgM+CD27− (open bars) and memory CD19+IgM+CD27+ (solid bars) B cells in three unrelated controls. 33, 33, and 52 VλJλ sequences from naive B cells from donors C5, C6, and C7, respectively were compared with 39, 42, and 63 VλJλ sequences from memory B cells from the same individuals. The percent Vλ utilization is indicated on the y axis. (B) Combined total Vλ1 and Vλ2 gene usage in CD19+ IgM+CD27− and CD19+IgM+ CD27+ B cells. Asterisk (*) indicates statistically significant difference (Vλ1, P = 0.035; and Vλ2, P = 0.0017).
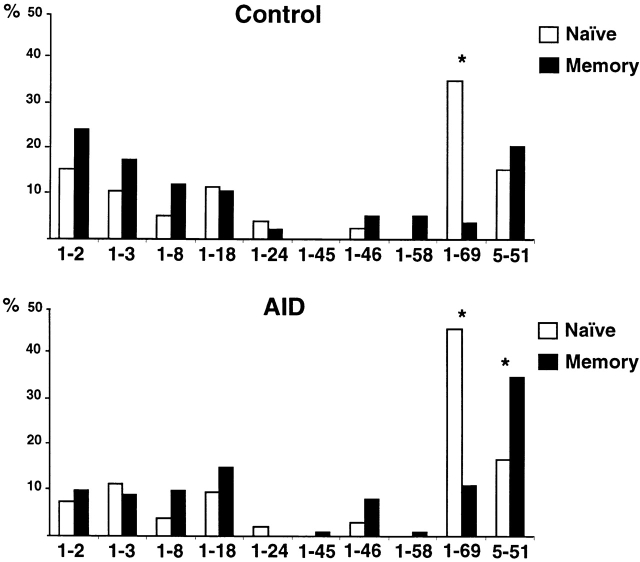
To determine whether the shift in Igλ repertoire between the naive and memory compartments is related to somatic hypermutation, we analyzed the Igλ genes expressed in naive and memory B cells from patients lacking activation-induced deaminase (AID) 17 18. AID has been shown to be essential for both hypermutation and switch recombination but does not appear to be necessary for normal B cell development in mice and humans 17 18. Patients with AID deficiency showed no secondary antibodies and no somatic mutation; nevertheless, these individuals displayed enlarged tonsils with germinal centers and showed normal numbers of CD19+CD27+ B cells 18. The CD27+IgM+ B cells found in AID-deficient patients resembled authentic CD27+ IgM + memory B cells in that they showed normal selection against VH1–69, a VH gene that is frequently found in B lymphoid chronic lymphocytic leukemias producing autoreactive antibodies (Fig. 3) 19 25 26. However, the antibodies expressed in antigen-selected memory B cells in five AID-deficient patients differed from the three controls in that they showed no mutations, and there was no shift in the Vλ repertoire between naive B cells and antigen-selected memory B cells (compare Fig. 2 and Fig. 4; total of 330 sequences). In particular, there was no increase in Vλ2 gene expression and no relative decrease in Vλ1 (Fig. 4). In addition, VH5–51 gene usage was favored in the memory CD27+ B cells from AID-deficient patients but not in normal controls (Fig. 3).
Figure 3.
VH1 and VH5 repertoire analysis of naive and memory B cells from control donors and AID-deficient patients. 77 control C5, C6, and C7 (top) and 105 AID (bottom) VH1 and VH5 sequences from naive B cells (open bars) are compared with 58 control and 112 AID sequences from memory B cells (solid bars). The percent VH1 and VH5 utilization is indicated on the y axis. Asterisk (*) indicates statistically significant difference (VH1–69, P < 0.0001 for both controls and AID-deficient patients; VH5–51, P = 0.03 for AID-deficient patients).
Figure 4.
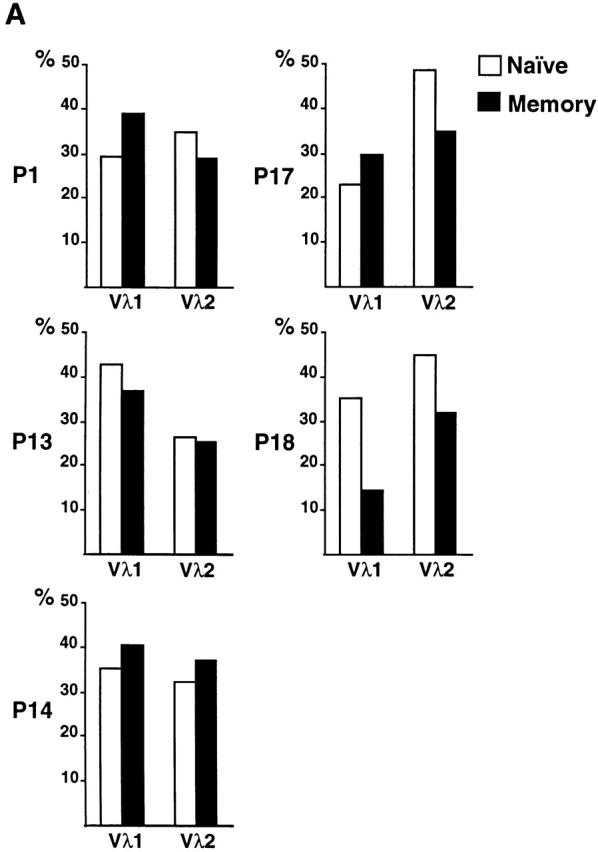
Vλ1 and Vλ2 gene usage in naive and memory B cells from AID-deficient patients. (A) Vλ1 and Vλ2 gene usage in naive CD19+CD27− (open bars) and memory CD19+CD27+ (solid bars) B cells in five unrelated AID-deficient patients. The patient numbers are as described in reference 18. 34, 30, 31, 35, and 31 VλJλ sequences from naive B cells from AID-deficient patients P1, P13, P14, P17, and P18, respectively were compared with 31, 35, 32, 37, and 34 VλJλ sequences from memory B cells from the same individuals. The percent Vλ utilization is indicated on the y axis. (B) Combined total Vλ1 and Vλ2 gene usage in CD19+CD27− and CD19+CD27+ B cells for AID-deficient patients.
Somatic hypermutation is known to increase antibody affinity during immune responses. However, the contribution of mutation to shaping the antibody repertoire has not been determined. We have found a global shift in the Igλ antibody repertoire between naive and memory B cells from normal donors. This shift in repertoire is associated with somatic hypermutation and is AID dependent. We conclude that AID and hypermutation make a significant contribution to shaping the antigen-selected memory B cell repertoire in humans.
Acknowledgments
We thank Dr. Mila Jankovic, Dr. Bernardo Reina San Martin, Dr. Eva Besmer, and members of the Nussenzweig lab for comments and discussions and Mrs. M. Forveille for excellent technical assistance.
This work was supported by grants from the National Institutes of Health to M.C. Nussenzweig and from Institut National de la Santé et de la Recherche Médicale. M.C. Nussenzweig is an investigator in the Howard Hughes Medical Institute.
Footnotes
M.C. Nussenzweig and A. Durandy contributed equally to this work.
References
- Siskind G.W., Eisen H.N. Effect of variation in antibody-hapten association constant upon the biologic activity of the antibody. J. Immunol. 1965;95:436–441. [PubMed] [Google Scholar]
- Weigert M.G., Cesari I.M., Yonkovich S.J., Cohn M. Variability in the lambda light chain sequences of mouse antibody. Nature. 1970;228:1045–1047. doi: 10.1038/2281045a0. [DOI] [PubMed] [Google Scholar]
- Griffiths G.M., Berek C., Kaartinen M., Milstein C. Somatic mutation and the maturation of immune response to 2-phenyl oxazolone. Nature. 1984;312:271–275. doi: 10.1038/312271a0. [DOI] [PubMed] [Google Scholar]
- Kaartinen M., Griffiths G.M., Hamlyn P.H., Markham A.F., Karjalainen K., Pelkonen J.L., Makela O., Milstein C. Anti-oxazolone hybridomas and the structure of the oxazolone idiotype. J. Immunol. 1983;130:937–945. [PubMed] [Google Scholar]
- Kaartinen M., Griffiths G.M., Markham A.F., Milstein C. mRNA sequences define an unusually restricted IgG response to 2-phenyloxazolone and its early diversification. Nature. 1983;304:320–324. doi: 10.1038/304320a0. [DOI] [PubMed] [Google Scholar]
- McKean D., Huppi K., Bell M., Staudt L., Gerhard W., Weigert M. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA. 1984;81:3180–3184. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablitzky F., Wildner G., Rajewsky K. Somatic mutation and clonal expansion of B cells in an antigen-driven immune response. EMBO J. 1985;4:345–350. doi: 10.1002/j.1460-2075.1985.tb03635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen D., Simon T., Sablitzky F., Rajewsky K., Cumano A. Antibody engineering for the analysis of affinity maturation of an anti-hapten response EMBO J. 7 1988. 1995 2001(published erratum at 8:2444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein C. Diversity and the genesis of high affinity antibodies. Biochem. Soc. Trans. 1987;15:779–787. doi: 10.1042/bst0150779. [DOI] [PubMed] [Google Scholar]
- Reth M., Hammerling G.J., Rajewsky K. Analysis of the repertoire of anti-NP antibodies in C57BL/6 mice by cell fusion. I. Characterization of antibody families in the primary and hyperimmune response. Eur. J. Immunol. 1978;8:393–400. doi: 10.1002/eji.1830080605. [DOI] [PubMed] [Google Scholar]
- Cumano A., Rajewsky K. Clonal recruitment and somatic mutation in the generation of immunological memory to the hapten NP. EMBO J. 1986;5:2459–2468. doi: 10.1002/j.1460-2075.1986.tb04522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French D.L., Laskov R., Scharff M.D. The role of somatic hypermutation in the generation of antibody diversity. Science. 1989;244:1152–1157. doi: 10.1126/science.2658060. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Akasako-Furukawa A., Shirai H., Nakamura H., Azuma T. Junctional amino acids determine the maturation pathway of an antibody. Immunity. 1999;11:329–338. doi: 10.1016/s1074-7613(00)80108-9. [DOI] [PubMed] [Google Scholar]
- Boersch-Supan M.E., Agarwal S., White-Scharf M.E., Imanishi-Kari T. Heavy chain variable region. Multiple gene segments encode anti-4-(hydroxy-3-nitro-phenyl)acetyl idiotypic antibodies. J. Exp. Med. 1985;161:1272–1292. doi: 10.1084/jem.161.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothwell A.L., Paskind M., Reth M., Imanishi-Kari T., Rajewsky K., Baltimore D. Heavy chain variable region contribution to the NPb family of antibodiessomatic mutation evident in a gamma 2a variable region. Cell. 1981;24:625–637. doi: 10.1016/0092-8674(81)90089-1. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Fais F., Ghiotto F., Hashimoto S., Sellars B., Valetto A., Allen S.L., Schulman P., Vinciguerra V.P., Rai K., Rassenti L.Z. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J. Clin. Invest. 1998;102:1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffre E., Papavasiliou F., Cohen P., de Bouteiller O., Bell D., Karasuyama H., Schiff C., Banchereau J., Liu Y.J., Nussenzweig M.C. Antigen receptor engagement turns off the V(D)J recombination machinery in human tonsil B cells. J. Exp. Med. 1998;188:765–772. doi: 10.1084/jem.188.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatovich O., Tomlinson I.M., Jones P.T., Winter G. The creation of diversity in the human immunoglobulin V(lambda) repertoire. J. Mol. Biol. 1997;268:69–77. doi: 10.1006/jmbi.1997.0956. [DOI] [PubMed] [Google Scholar]
- Farner N.L., Dorner T., Lipsky P.E. Molecular mechanisms and selection influence the generation of the human V lambda J lambda repertoire. J. Immunol. 1999;162:2137–2145. [PubMed] [Google Scholar]
- Klein U., Rajewsky K., Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genesCD27 as a general marker for somatically mutated (memory) B cells. J. Exp. Med. 1998;188:1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye S.G., Liu Y.J., Aversa G., Phillips J.H., de Vries J.E. Identification of functional human splenic memory B cells by expression of CD148 and CD27. J. Exp. Med. 1998;188:1691–1703. doi: 10.1084/jem.188.9.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sthoeger Z.M., Wakai M., Tse D.B., Vinciguerra V.P., Allen S.L., Budman D.R., Lichtman S.M., Schulman P., Weiselberg L.R., Chiorazzi N. Production of autoantibodies by CD5-expressing B lymphocytes from patients with chronic lymphocytic leukemia. J. Exp. Med. 1989;169:255–268. doi: 10.1084/jem.169.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borche L., Lim A., Binet J.L., Dighiero G. Evidence that chronic lymphocytic leukemia B lymphocytes are frequently committed to production of natural autoantibodies. Blood. 1990;76:562–569. [PubMed] [Google Scholar]