Abstract
Angioimmunoblastic lymphadenopathy with dysproteinemia (AILD) is a peculiar T cell lymphoma, as expanding B cell clones are often present besides the malignant T cell clones. In addition, large numbers of Epstein-Barr virus (EBV)-infected B cells are frequently observed. To analyze the differentiation status and clonal composition of EBV-harboring B cells in AILD, single EBV-infected cells were micromanipulated from lymph nodes of six patients with frequent EBV+ cells and their rearranged immunoglobulin (Ig) genes analyzed. Most EBV-infected B cells carried mutated Ig genes, indicating that in AILD, EBV preferentially resides in memory and/or germinal center B cells. EBV+ B cell clones observed in all six cases ranged from small polyclonal to large monoclonal expansions and often showed ongoing somatic hypermutation while EBV− B cells showed little tendency for clonal expansion. Surprisingly, many members of expanding B cell clones had acquired destructive mutations in originally functional V gene rearrangements and showed an unfavorable high load of replacement mutations in the framework regions, indicating that they accumulated mutations over repeated rounds of mutation and division while not being selected through their antigen receptor. This sustained selection-free accumulation of somatic mutations is unique to AILD. Moreover, the survival and clonal expansion of “forbidden” (i.e., Ig-deficient) B cells has not been observed before in vivo and thus represents a novel type of viral latency in the B cell compartment. It is likely the interplay between the microenvironment in AILD lymph nodes and the viral transformation that leads to the survival and clonal expansion of Ig-less B cells.
Keywords: B lymphocytes, somatic hypermutation, EBV, immunoglobulin genes, single cell PCR
Introduction
Angioimmunoblastic lymphadenopathy with dysproteinemia (AILD) represents one of the most frequent T cell lymphomas in the Western world and usually takes an aggressive course. Affected patients suffer from generalized lymph node swelling, fever, rash, and predisposition to infections. Histologically, affected lymph nodes show an effaced structure, usually with disappearance of follicles and germinal centers (GCs) but presence of a diffuse network of follicular dendritic cells (FDCs), proliferation of venules and an infiltrate of polymorphous atypical T cells and also B immunoblasts 1.
In most cases of AILD, clonal expansion of T cells can be detected using Southern blot analysis or PCR strategies 2 3 4 5. In a proportion of cases also clonal expansion of B cells was demonstrated, and different expanded B cell clones were observed in sequential biopsies 6 7 8. Furthermore, diffuse large B cell lymphomas occasionally develop in AILD patients 9 10 11.
Epstein-Barr virus (EBV), a human herpes virus infecting >90% of adults, has been detected by in situ hybridization in B cells of 80–95% of AILD-involved lymph nodes and in some cases also in T cells 8 12 13. Whereas in healthy EBV carriers, EBV resides in B cells at a frequency of ∼1 in 105–106 14 15, in lymph nodes of AILD patients 1 in 10 to 1 in 500 B cells may be EBV-infected (12 13; see below).
EBV is also associated with a variety of other lymphomas. More than 95% of endemic Burkitt's lymphoma and ∼40% of classical Hodgkin's lymphoma (HL) show EBV infection of the tumor cells 16. Furthermore, during states of immunosuppression as in HIV-infected patients or in patients after organ transplantation, EBV+ high grade B cell lymphomas are observed at increased frequency 17 18. The lymphomas differ in the pattern of EBV latent gene expression. Only the EBV nuclear antigen (EBNA)1 is expressed in Burkitt's lymphoma, Hodgkin–Reed/Sternberg (HRS) cells of HL show in addition expression of the latent membrane proteins (LMPs) 1 and 2a, and lymphomas in immunodeficient states show expression of all nine latent proteins 16. In healthy carriers, EBV is found in a latent form in resting memory B cells in the peripheral blood, and the virus-infected cells lack expression of most, if not all, latent proteins 19. These cells are tightly controlled by virus-specific T cells 20 21.
The role of EBV and the origin and state of maturation of EBV-infected B cells in AILD are unknown. Here, we analyzed the Ig gene rearrangements of single EBV-infected cells micromanipulated from frozen tissue sections of six cases of AILD with a relatively high content of EBV-infected cells in order to define their clonal composition and stage of differentiation. In addition, from one of the six cases and three further cases with no or few EBV+ cells, single EBV− B cells were analyzed for comparison.
Materials and Methods
Tissues and Clinical Data.
Frozen biopsies taken for diagnostic purposes were from the Departments of Pathology of the Universities of Kiel and Henri Mondor. The six cases used for analysis of EBV-infected B cells were selected from 20 cases for their relatively high content of EBER+ cells (Fig. 1). Clinical data for the patients are summarized in Table . All cases were examined by at least two experienced hematopathologists.
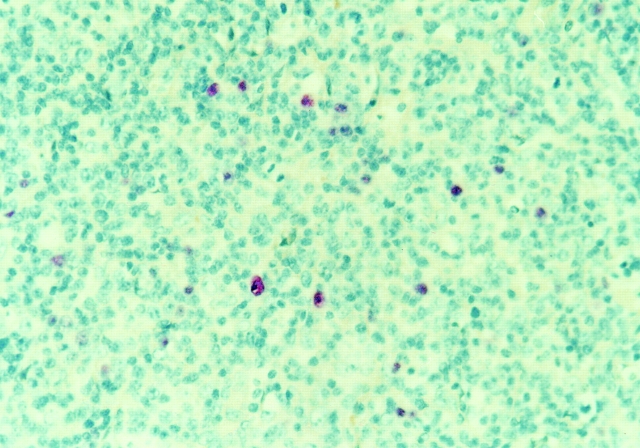
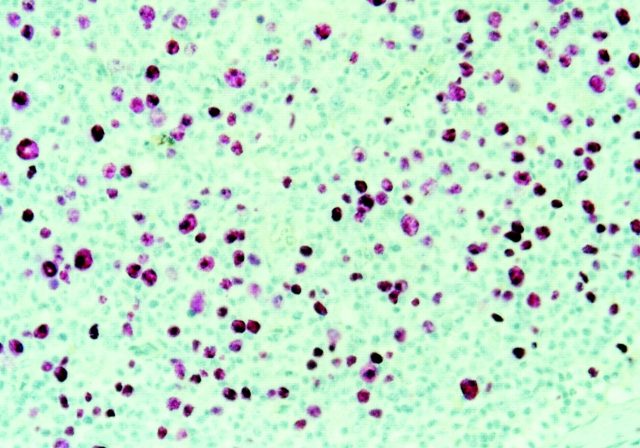
Figure 1.
EBER in situ hybridization for two cases of AILD. EBER in situ hybridization was performed on paraffin sections as described in Materials and Methods.
Table 1.
Description of Nine Cases of AILD
| Case | Sex | Age | Lymph nodebiopsy site | PercentageCD20+ cells | PercentageEBER+ B cells | EBNA1 | LMP1 | LMP2a | EBNA2 | BZLF1 | TCR-γlength distribution |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 72 | Cervical | 20 | 0.1–1 | + | − | − | − | − | 1 clone, polyclonal background |
| 2 | F | 81 | Cervical | 10 | 1–10 | ND | + | − | − | − | 1 clone, polyclonal background |
| 3 | F | 77 | Cervical | 20 | 0.1–1 | − | ND | − | ND | ND | NE |
| 4 | M | 86 | Axillary | 5 | 1–10 | + | + | + | − | − | monoclonal |
| 5 | M | 37 | Inguinal | 20 | >50 | − | + | + | − | − | monoclonal |
| 6 | F | 70 | Supraclavicular | 25 | 1–10 | + | NE | + | − | − | NE |
| 7 | M | 58 | Inguinal | 25 | 0 | ND | ND | ND | ND | ND | polyclonal |
| 8 | F | 43 | Axillary | 20 | <0.01 | ND | ND | ND | ND | ND | polyclonal |
| 9 | F | 73 | Axillary | 15 | <0.1 | ND | ND | ND | ND | ND | monoclonal |
CD20, EBNA1, LMP1, EBNA2, and BZLF1 expression were determined by immunohistochemistry; LMP2a expression by RT-PCR.
NE, not evaluable.
Immunostaining, EBER In Situ Hybridization, and Double Staining for EBER and CD20.
Immunostaining was performed on 5–8-μm thick paraffin sections using monoclonal antibodies against CD20, LMP1, BZLF1, and EBNA2 (all from Dako). For antigen retrieval, deparaffinated and rehydrated sections were subjected to 2 min of high pressure cooking in citrate buffer, pH 6. Specifically bound primary antibodies were detected using biotinylated secondary antibodies (Dako) and avidin-coupled alkaline phosphatase (Dako). Fast Red or BCIP/NBT (Dako) served as substrate for alkaline phosphatase.
Frozen tissue sections were used for Ki67 (polyclonal; Dako), CD20 and Ki67/CD20 double stainings. For the double stainings, horseradish peroxidase–coupled goat anti–rabbit and alkaline phosphatase–coupled goat anti–mouse antibodies were used with diaminobenzidine and Fast Red as substrates.
EBER in situ hybridization for detection of EBV-infected cells was carried out on paraffin and frozen sections using digoxigenin-labeled EBER1 and EBER2 probes as described previously 22. Plasmids for generation of probes were provided by G. Niedobitek, Department of Pathology, Friedrich-Alexander University, Erlangen, Germany. Double staining for CD20 and EBER was performed on frozen tissue sections. Before in situ hybridization, sections were incubated with peroxidase-coupled anti-CD20 antibody (CD20-EPOS; Dako) and visualized with Diaminobenzidine (Sigma-Aldrich) as chromogen. In situ hybridization was continued as described previously.
Fragment Length Analysis.
DNA was extracted from eight to ten 10-μm sections of frozen tissues using a silica column system (QiaAmp; Qiagen). TCR-γ rearrangements were amplified in two separate reactions with the consensus primers V1–8 and V9–11 and the same set of three Fam2-labeled J primers in both reactions 23. PCR products were analyzed with an automatic sequencer (ABI377; Applied Biosystems) and the GeneScan software.
Detection of Human Herpes Virus 8 in DNA from Whole Tissue Sections.
30–60 ng of DNA from frozen tissues (see Fragment Length Analysis) (equivalent to 0.6–1.2 × 104 cells) were used in a seminested PCR to amplify a 230-bp fragment of the ORF26 of human herpes virus (HHV)8. Both PCRs were performed in 1× PCR buffer (Boehringer) with 2 mM dNTPs (Amersham Pharmacia Biotech), 0.125 μM for each primer and 2.5 and 2.0 mM MgCl2 for the first and second round, respectively. In the first round HHV8US1 and HHV8LS1 were used, in the second round HHV8US1 together with HHV8LS2 (HHV8US1: 5′-GAAAGGATTCCACCATTGTGCTCG-3′; HHV8LS1: 5′-ATGGACAGATCGTCAAGCACTCGC-3′; HHV8LS2: 5′-CCGTGTTGTCTACGTCCAGACG-3′). After an initial cycle with 5 min at 95°C, 4 min at 65°C when the enzyme was added, and 1 min at 72°C, 39 cycles with 30 s at 95°C, 30 s at 61°C and 45 s at 72°C were performed in the first round of PCR. Cycle conditions for the second round of PCR were the same as for the first round, except that the initial denaturation at 95°C was reduced to 2 min. With this seminested PCR, HHV8 DNA could be detected in 60 pg of DNA (equivalent to ∼12 cells) extracted from a HHV8-positive Kaposi sarcoma.
Micromanipulation and Single Cell PCR.
Single EBER+, CD20+, EBER−/CD20+, Ki67+, and Ki67+/CD20+ cells were micromanipulated with a hydraulic micromanipulator from 5-μm sections of frozen tissues covered with buffer and transferred to 20 μl PCR buffer in PCR tubes as described previously 24 25. During all micromanipulation procedures aliquots of buffer covering the sections were taken as negative controls, usually four buffer aliquots for each 10 micromanipulated cells. After 2 h of proteinase K digestion, rearranged IgH, Igκ, and Igλ genes and a fragment of the EBNA1 gene were amplified by seminested PCR as described previously 25 26 27 28. In the first round of PCR, family-specific primers for the framework region I of the IgH, Igκ, and Igλ V gene segments were used together with sets of primers for the corresponding J segments and EBNA1-specific primers. For cases 4–6, in addition, family-specific DH primers binding 100–200-bp 5′ of the DH segments were used together with the IgH primer set. In second round PCRs the same family-specific V primers (and for cases 4–6 also the family-specific DH primers) were used in separate PCRs for each V (or DH) gene family together with nested J primers. PCR products were gel-purified and directly sequenced.
For case 5, 20 EBER-ISH+ cells were also analyzed for HHV8. In the first round of PCR, conditions were as described for the V gene rearrangement and EBNA1 PCR. The IgH- and EBNA1-specific primers were used together with HHV8US1 and HHV8LS1, each of the latter at 0.05 μM. In a separate second round for HHV8, HHV8US1 and HHV8LS2 primers were used for 40 cycles (for PCR conditions see Detection of HHV8 in DNA Extracted from Whole Tissue Sections). 10 single cells from a HHV8-positive cell line (CRO AP2; reference 29) served as positive control, and HHV8 was detected in 8 of 10 of these cells.
Reverse Transcription PCR.
RNA was isolated from 10–25 mg of frozen tissues (∼2–5 × 106 cells, high pure tissue RNA isolation kit; Boehringer). One-tenth of the RNAs were used for cDNA synthesis with random hexamers and AMV reverse transcriptase (1st strand cDNA synthesis kit for reverse transcription (RT)-PCR; Boehringer). The isolation of amplifiable RNA was tested with primers for β-actin (β-actinUS: 5′-CACCTTCTACAATGAGCTGCGTG-3′ and β-actinLSa: 5′-GGCGTACAGGGATAGCACAGC-3′; a 170-bp product from cDNA and a 610-bp product from genomic DNA), each 0.125 μM, in 1× PCR buffer (Boehringer) with 2 mM dNTPs and 2 mM MgCl2 and 1/100 of the cDNA as template in a PCR with an initial cycle of 2 min at 95°C, 4 min at 65°C, and 1 min at 72°C, followed by 24 cycles with 30 s at 95°C, 30 s at 61°C, and 45 s at 72°C. PCR products from all cases were sequenced to exclude the amplification of processed β-actin pseudogenes.
1/10 of the cDNAs served as templates for detection of LMP2a transcripts in a seminested PCR. In the first round LMP2aF1 and LMP2aRa were used, each 0.125 μM, in 1× PCR buffer (Boehringer) with 2 mM dNTPs and 2 mM MgCl2. Cycle conditions were 2 min at 95°C, 4 min at 65°C, and 1 min at 72oC followed by 39 cycles with 30 s 95°C, 30 s 61°C, and 45 s 72°C. 1 μl aliquots of the first rounds were used as templates in second rounds with the same PCR conditions as in the first round, except the usage of LMP2aF1 together with LMP2aRi and an annealing temperature of 57°C (LMP2aF1: 5′-TCTCAACACATATACGAAGAAGCG-3′; LMP2aRa: 5′-GCGGTCACAACGGTACTAACTG-3′; LMP2aRi: 5′-AACTGAGGCCGTGAAACACGAG-3′; a 123-bp product from cDNA and a >5,000-bp product from genomic DNA).
Results
Immunohistology, EBER In Situ Hybridization, RT-PCR, and TCR-γ Fragment Length Analysis.
All selected cases displayed the typical histological and immunohistological picture of AILD, i.e., proliferation of (partly atypical) CD3+ T cells, admixed with scattered CD20+ B immunoblasts and hyperplastic FDC networks. By EBER in situ hybridization, which detects all virus-harboring B cells, moderate to high amounts of EBV-positive cells were observed in six cases from which EBV-infected cells were micromanipulated (Fig. 1), while in three other cases no or few EBV-infected cells were detected (Table ). Immunohistology for BZLF1 (whose expression indicates that cells switched from latent to lytic infection 30 31) and EBNA2 (typical for cells expressing all latent EBV proteins) was negative in all cases analyzed. Scattered LMP1+ cells were observed in cases 2 and 4. In case 5, LMP1-positive cells accounted for about one- third of the EBER-positive cells, partly displayed the morphology of large blasts and were predominantly arranged in a nodular fashion throughout the lymph node. As LMP2a immunostainings could not be evaluated because of unspecific staining, RT-PCR of RNA isolated from whole tissue sections of cases 1–6 was used to detect LMP2a expression. Cases 4, 5, and 6 tested positive (Table ).
In a fraction of AILD cases, another human herpes virus, HHV8, associated with B cell malignancies and with transforming potential in animal models, has been detected 32 33 34. In a sensitive seminested PCR with DNA from cases 1–6, HHV8 DNA was detected only in case 5 (data not shown). However, when single EBV-infected cells from this same case were analyzed, no coinfection of EBV-positive cells with HHV8 was observed (see Materials and Methods). Therefore, the features of the EBV-infected B cells analyzed in this study and described below are not influenced by a coinfection of the cells with HHV8.
To analyze the cases for the presence of clonal T cell populations, a PCR amplification of TCR-γ gene rearrangements from DNA extracted from whole tissue sections and subsequent determination of length and intensity distributions of the fluorescent-labeled PCR products was performed 23 35. This approach is suitable to detect clonal expansions among TCR-γδ– as well as TCR-αβ–expressing T cells, since also the latter usually carry TCR-γ gene rearrangements. In all four of the six EBV-rich cases informative for TCR-γ rearrangements (cases 1, 2, 4, and 5; in cases 3 and 6 signals were too low for evaluation; Table ), the results were indicative of a single T cell expansion in each patient, thus supporting the diagnosis of T cell lymphoma at the molecular level. Among the three cases used for analysis of EBV-negative B cells only in case 9 a dominant T cell clone was detected.
Clonal Expansion among EBER+ and CD20+ Cells.
Between 50 and 115 single EBER+ cells were micromanipulated from frozen tissue sections of six EBV-rich cases (Table ). All cells were subjected to seminested PCR for rearrangements at the IgH, Igκ, and Igλ loci and in most instances also tested for EBV-infection in an EBNA1-specific PCR. From 31 to 58% of the micromanipulated cells at least one Ig gene rearrangement was obtained, and between 24 and 125 Ig rearrangements were amplified per case. Comparison of the Ig gene sequences identified clonal expansions among the EBV+ cells in all six cases (Table ). However, the number of clones and the extent of clonal expansion differed markedly between cases. In patients 1–3, 60–90% of the EBER+ cells could be assigned to several clones of varying sizes (case 1: 10 clones with 2–7 members; case 2: seven clones with 2–18 members; and case 3: six clones composed of 2–4 cells). In case 4, a dominant clone encompassing nearly half of the EBER+ cells was identified. In cases 5 and 6, all EBER+ cells belonged to a single clone.
Table 2.
PCR and Sequence Analysis of Rearranged Ig Genes of Single EBER+ and CD20+ Cells of AILD
| Case | Cells | Cells positive/cells analyzed | Cells with mutated rearrangements/cells informative | Cells assignedto clones | Uniquecells | Numberof clones | Cellsper clone | Clones withintraclonal diversity |
|---|---|---|---|---|---|---|---|---|
| % | % | |||||||
| EBV+ | ||||||||
| 1 | EBER+ | 59/103 | 56/56 (100) | 37 (63) | 22 | 10 | 2–7 | 3 |
| 2 | EBER+ | 53/115 | 50/50 (100) | 47 (89) | 6 | 7 | 2–18 | 6 |
| 3 | EBER+ | 26/56 | 13/23 (57) | 15 (58) | 11 | 6 | 2–4 | 2 |
| 4 | EBER+ | 48/101 | 44/44 (100) | 26 (54) | 22 | 3 | 2×2, 22 | 1 |
| 5 | EBER+ | 21/70 | 21/21 (100) | 21 (100) | 0 | 1 | 21 | 1 |
| 6 | EBER+ | 27/50 | 25/25 (100) | 27 (100) | 0 | 1 | 27 | 1 |
| EBV+/− | ||||||||
| 5 | CD20+ | 14/40 EBNA+ | 13/14 (93) | 12 (86) | 2 | 1 | 12 | 1 |
| 3/40 EBNA− | 3/3 (100) | 0 | 3 | 0 | 0 | 0 | ||
| 6 | CD20+ | 4/56 EBNA+ | 3/3 (100) | 4 (100) | 0 | 1 | 4 | 1 |
| 17/56 EBNA− | 13/15 (87) | 14 (82) | 3 | 1 | 14 | 1 | ||
| EBV− | ||||||||
| 2 | EBER−/CD20+ | 24/49 | 13/42 (31) | 4 | 20 | 2 | 2×2 | 0 |
| 7 | Ki67+ | 18/106 | 11/17 (65) | 11 (61) | 7 | 1 | 11 | 1 |
| 8 | Ki67+ | 26/140 | 5/19 (26) | 0 | 26 | 0 | 0 | 0 |
| 9 | CD20+ | 88/300 | 60/81 (74) | 2 | 86 | 1 | 2 | 0 |
PCR for DH/JH rearrangements was performed for cases 4, 5, and 6. For the large clone of case 4 a clonal DH6–25/JH2 rearrangement was coamplified with the clonal VH or Vκ rearrangement from 8 of 22 cells, for case 5 a clonal DH3–9/JH6 rearrangement was coamplified with the clonal VH rearrangement from 6 of 20 cells analyzed and for case 6 a clonal DH3–9/JH5 rearrangement was coamplified with the clonal VH rearrangement from 5 of 20 cells analyzed. All sequences have been deposited at the EMBL database under AJ347089-703 and AJ344348-350.
In all micromanipulation experiments aliquots of buffer covering the sections were aspirated as negative controls (usually four buffer aliquots for each 10 cells). For case 1, from 1 of 42 buffer controls a VH rearrangement also amplified from a cell belonging to a clone, and from another buffer aliquot three unique VH rearrangements were amplified. For case 4, a unique VH rearrangement was amplified from one of 41 buffer controls and from two of 43 buffer controls for case 6 the clonal VH rearrangement was amplified. For case 9 the same VH rearrangement was amplified from three buffer controls. This rearrangement was unrelated to any other rearrangement amplified in this study.
To analyze whether the large clonal expansions of cases 5 and 6 were restricted to EBV-positive cells and/or accounted for most B cells in the tissue, for both cases also CD20+ cells were micromanipulated and tested for infection with EBV in an EBNA1-specific PCR (Table ). In case 5, 14 of 17 CD20+ cells were positive for EBNA1. Thus, in this case ∼80% of the CD20+ B cells are EBV-infected. Furthermore, nearly all EBV-positive cells of this case (33 of 35 cells taking EBER+ and CD20+/EBNA1+ cells together) belonged to one clone. The three EBNA1 PCR-negative cells carried unique rearrangements. In contrast to case 5, in case 6 only a small fraction of the CD20+ cells was EBV-infected (4 of 21 Ig-PCR positive cells were also positive in the EBNA1-PCR). The four EBV-positive cells belonged to the same clone identified among the EBER+ cells. Surprisingly, also 14 of 17 CD20+ and EBNA1 PCR-negative cells could be assigned to this clone. Since the EBNA1-specific PCR works very efficiently and since 9 of the EBNA1-PCR-negative cells define a distinct subclone in the genealogical tree of this clone (Fig. 2), we conclude that in this case, the B cell population consisted largely of a single dominant clone and only about a quarter of the cells in this clone are EBV-infected, perhaps due to a single infectious event in the history of the clone.
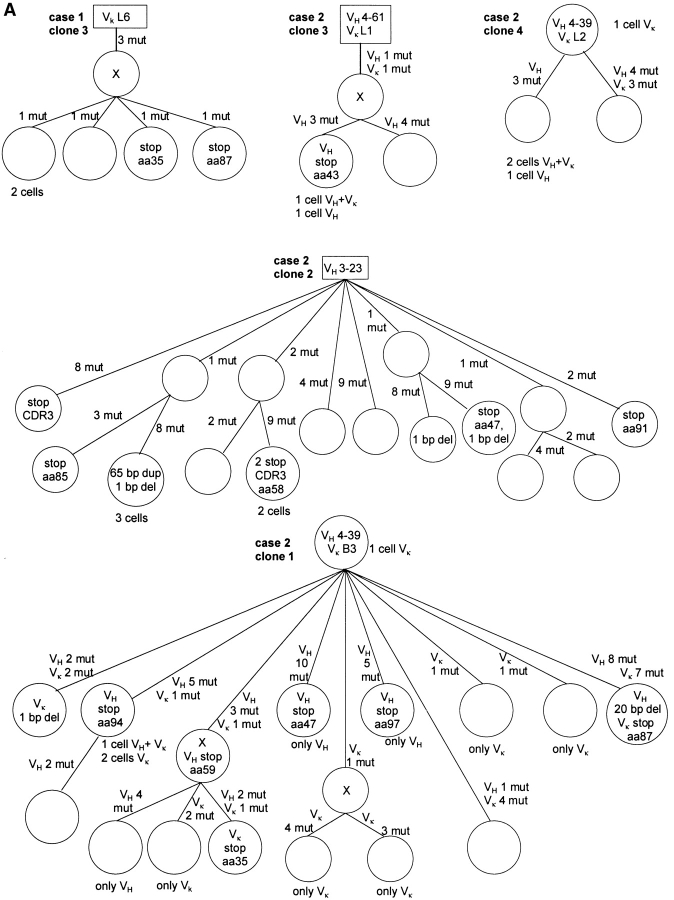
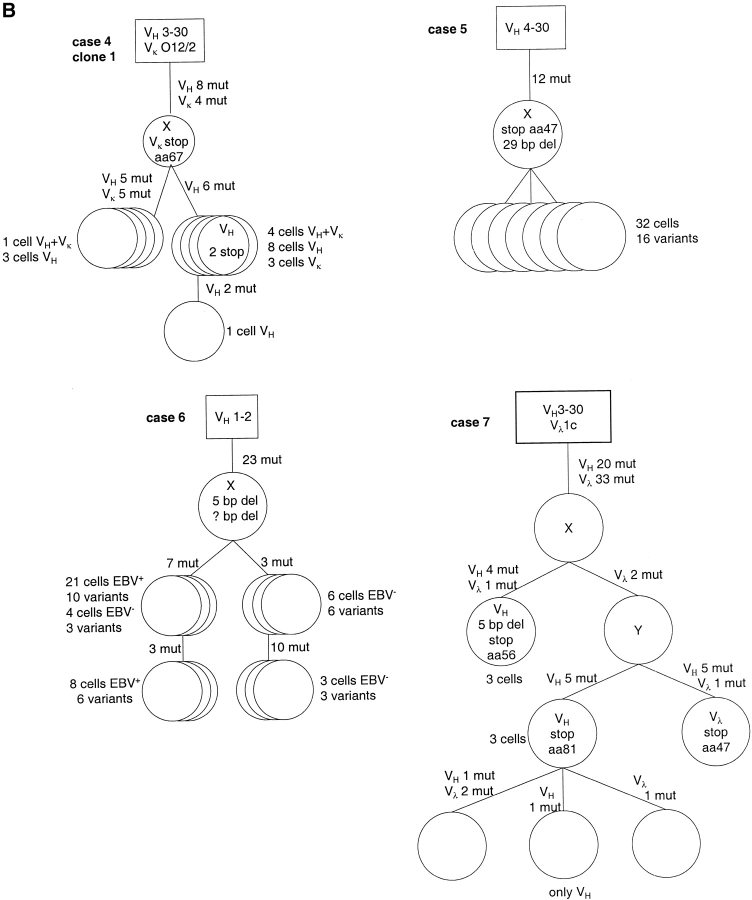
Figure 2.
Genealogical trees for examples of clonal expansions of EBV-infected B cells and an EBV-negative clone (case 7) with ongoing somatic hypermutation from six cases of AILD. Genealogical trees are based on alignments obtained with the GeneWorks software 2.5 (Intelligenetics) and only shown for originally potentially functional rearrangements. Presumed precursors are boxed. Assumed intermediates are included in the genealogical trees and marked as X. Inactivating mutations are shown within the circles (for rearrangements which acquired sequentially inactivating mutations only the initially inactivating ones are shown). For clone 2 of case 2, several pairs of rearrangements each with one shared mutation were observed, but together were partly incompatible regarding the construction of a genealogical tree. Therefore, a fraction of these shared mutations was likely due to independent mutation events. For three of such mutations a single mutation event was assumed (no mutation hot spot in the RGYW motif and no other mutations at the same position). In cases 1, 2, and 3 circles mostly represent single cells. If identical rearrangements were obtained from several cells, this is indicated below the circles. In cases 4, 5, and 6 the circles usually represent several cells with ongoing somatic hypermutation. Numbers of cells and variants identified are given beside the circles. In case 6 the EBV− cells were micromanipulated using a CD20 staining and subsequently tested in an EBNA1-PCR. The 21 EBV+ cells of one subclone are composed of 19 EBER-ISH+ cells and two cells tested positive for EBNA1, the eight EBV+ cells of a second subclone are composed of six EBER-ISH+ cells and two EBNA1+ cells. In case 7, one cell could not be fit in the genealogical tree, because VH and Vλ rearrangements would belong to different branches. These rearrangements may represent cellular contaminations during the micromanipulation procedure. aa, amino acid; del, deletion; mut, mutation.
Some CD20+/EBER− B cells were also micromanipulated from case 2 to analyze the clonal composition and differentiation status of EBV-negative B cells. Among the 42 rearrangements amplified from 24 EBER− and EBNA1 PCR-negative samples (two to three cells were analyzed together in each PCR) none was clonally related to those amplified from EBER+ cells. Only two pairs of clonally related cells were identified.
In addition, for a comprehensive comparison of EBV+ and EBV− B cells in AILD, from three further cases with no or few EBV+ cells, single proliferating (Ki67+) cells, B cells (CD20+) and proliferating B cells (Ki67+/CD20+) were micromanipulated (Ki67 staining was chosen because the analysis of these cases was originally focused on proliferating B cells in EBV− cases of AILD). In case 8, no clonally related B cells were identified and in case 9, only 2 of 71 informative cells were clonally related. A single EBV− B cell clone encompassing nearly 2/3 of the proliferating B cells was identified in case 7. As in this case no T cell clone was detected, this B cell clone may represent a B cell tumor that developed in the background of AILD as described previously 9 10 11 .
Taken together, in each of the six EBV-rich cases clonal B cell expansions were detected among the EBV-infected cells, ranging from polyclonal populations with several small clones to monoclonal expansions dominating the B cell population in the tissue. One case with a large monoclonal expansion included EBV+ as well as EBV− B cells. With the exception of one large clone in an EBV− case, EBV− B cells showed little tendency for clonal expansion.
Somatic Mutation in Rearranged Ig Genes of EBV-infected B Cells.
Rearranged Ig genes of EBV-infected B cells were evaluated for the presence of somatic mutations, considering members of clones and EBV-harboring cells not assigned to clones (unique cells) separately. Analysis of the sequences amplified from unique EBV-infected cells revealed that 55 of 58 informative cells from cases 1–5 carried mutated V genes. The average mutation frequency of 5.4% for VH rearrangements (n = 29; range 0.4–11.9%) is within the range typical for memory B cells 36.
Among the EBV-infected cells belonging to expanded clones, three types of clonal expansions were identified based on the presence and pattern of somatic mutation (two clones are not considered, see legend to Table ): (i) only three of the 26 clones carried unmutated V region genes. These three clones were all identified in case 3 and consisted of only 2–4 members. (ii) Nine clones identified in cases 1, 2, and 4 had mutated V genes with an average mutation frequency of 7% for VH rearrangements (n = 8, range 3.5–11.6%) but did not show intraclonal diversity. Most of these nine clones were small and defined by only two members. (iii) In 14 clones with mutated V genes intraclonal diversity was observed (Table and Fig. 2). Such clones were identified in all six cases. The extent of intraclonal diversity varied considerably between the clones, ranging from three sequence variants among 17 sequences (clone 1 of case 4) to 12 variants among 15 sequences (clone 2 of case 2). Also the numbers of shared mutations differed markedly between clones. For the dominant clones of cases 4–6 several shared mutations were identified, whereas for clones 1 and 2 of case 2 no mutations common to all clone members were observed (Fig. 2). The average mutation frequency of VH rearrangements for clones with ongoing mutation from cases 1–4 was 2.8% (identical mutated rearrangements were counted only once; n = 35, range 0–7.2%), and for the two clones of cases 5 and 6 the least mutated members showed 10 and 17% mutation frequency, respectively.
The clone detected among EBV− cells in case 7 showed also intraclonal diversity besides 20 and 33 shared mutations in its VH and Vλ rearrangements, respectively (Fig. 2).
Taken together, the vast majority of EBV-infected B cells carried somatically mutated V region genes, and many members of EBV+ B cell clones showed ongoing hypermutation, indicating that the virus preferentially resides in GC and perhaps also memory B cells.
Frequent Crippling Somatic Mutations in Clonally Expanding EBV-infected B Cells.
Surprisingly, the analysis of the mutation pattern disclosed a large number of crippling mutations in originally potentially functional V gene rearrangements. These destructive mutations were either nonsense mutations or duplications or deletions resulting in loss of the correct reading frame. Most destructive mutations were found in clones showing ongoing somatic mutation (Table ). For each of the large clones of cases 4–6, inactivating mutations were found in the in-frame V gene rearrangements (Fig. 2). In cases 5 and 6, amplification of DHJH rearrangements from the second IgH alleles confirmed that the inactivated VHDHJH rearrangements were indeed the originally functional ones (see legend to Table ). The 11 cells belonging to the EBV− clone of case 7 carried various different mutations rendering either the originally potentially functional VH or the functional Vλ rearrangement nonfunctional, while none of the shared mutations was crippling (Fig. 2).
Table 3.
Somatic Mutation Pattern of Ig Gene Rearrangements from EBER+ Cells of AILD
| Deletions and duplications | ||||||
|---|---|---|---|---|---|---|
| Rearrangements | No. ofrearrangements analyzed | Frequency of nonsensemutations | Percentage ofall mutations | Total(No.) | Destructive(No.) | Replacement/silent mutations in framework regions |
| % | ||||||
| Potentially functional | ||||||
| Unique cells (n = 39) | 55 | 0.2 (1/515) | 0.6 | 3 | 2 | 1.3 (143/110) |
| Clones without ongoing mutation (n = 8) | 12 | 0.0 (0/143) | 0.7 | 1 | 0 | 1.1 (36/33) |
| Clones with ongoing mutation (n = 12) | 78 | 6.5(16/247) 7.7 (19/247) | 6.4 | 17 | 11 | 2.7 (70/26) |
| Memory B cells | − | 0.0 | 0.2 | − | − | 1.0–1.6 |
| Nonfunctional | ||||||
| This study | 151 | 5.2 (27/522) | 3.5 | 19 | 14 | 2.5 (190/75) |
| Literature | − | 4.8 | 6.0 | 20 | 20 | 3.0 |
For clones only Ig gene rearrangements amplified at least two times are considered.
Nonsense mutations were found in originally potentially functional rearrangements of clones with ongoing mutation with an average frequency of 7.7% of all mutations (or 6.5% if, to estimate the frequency of destructive nonsense mutations, in V genes with several nonsense mutations only one is counted). This contrasts with the 0% (0 in 143 mutations) and 0.2% (1 in 515 mutations) nonsense mutations found in clones without ongoing mutation and unique cells, respectively (Table ). For mutations that a priori could not be subject to selection (i.e., mutations in out-of-frame rearrangements and mutations that occurred in already clonally inactivated rearrangements), a frequency of 5.2% nonsense mutations was observed, which is close to the calculated theoretical value of 4.7% assuming random mutagenesis without selection 37. Thus, the originally functional V genes of clones with ongoing mutations accumulated nonsense mutations with a frequency somewhat higher than in the case of nonproductive, nonselected rearrangements.
In normal GC B cells, deletions or duplications account for ∼6% of somatic mutation events, as has been calculated from their frequency in mutated out-of-frame rearrangements 38. In this study, the frequency of deletions/duplications in nonproductive rearrangements was somewhat lower (3.5%). In functional V region genes most deletions/duplications result in loss of the correct reading frame or cripple a V gene rearrangement because a large part of the rearrangement is lost or duplicated. Such deletions/duplications are therefore stringently counterselected, so that expressed V genes of post GC B cells only rarely carry such mutations (usually 3- or 6-bp deletions/duplications in the complementary determining regions). In line with this, deletions/duplications were only rarely observed in the present study in in-frame rearrangements of clones without ongoing mutation and in unique rearrangements, with frequencies of 0.7 and 0.6%, respectively (Table ). However, in originally functional rearrangements of clones with ongoing mutation, deletions and duplications were found with a frequency of 6.4%, and many of those were destructive, further indicating that these rearrangements acquired somatic mutations without selection for functionality.
In addition to the counterselection of destructive mutations, also the ratio of replacement (R) to silent (S) mutations in FRs of functional Ig gene rearrangements is indicative for selective pressure on a cell to express a functional antigen receptor. In cells selected for expression of a functional B cell receptor, R mutations in FRs are usually counterselected. Thus, for selected memory B cells a R/S ratio of 1.0 to 1.6 is usually observed, contrasting with the ratio of ∼ 3.0 for V gene rearrangements that acquire somatic mutations without selection, like out-of-frame rearrangements (Table ). The out-of-frame rearrangements analyzed in this study showed an average R/S value of 2.5. For the in-frame rearrangements of clones without ongoing mutation and the unique EBER+ cells, we observed R/S ratios of 1.1 and 1.3, respectively, indicative of stringent selection for expression of a functional antigen receptor, like in normal B cells. In contrast, the R/S ratio for clones with intraclonal diversity is 2.7, and thus similar to that of rearrangements that acquire mutations without selection for functionality.
Taken together, each of the three features of somatic hypermutation analyzed (the frequency of nonsense mutations, the frequency of deletions/duplications, and the R/S ratios) indicate that expanding EBV-infected B cells in AILD acquire somatic mutations without selection for expression of a functional antigen receptor.
Considering the obvious crippling Ig mutations, the frequencies of Ig-less B cells among all B cells can be calculated. In case 2, ∼1% of all B cells lost the capacity to express a functional antigen receptor, whereas in cases 5, 6, and 7 the fraction of Ig-less B cells amounts to 71, 86, and 56% of all B cells, respectively. For cases 1, 2, and 4 fractions of 4, 0, and 43% of Ig-less B cells among the EBER+ B cells were calculated.
Discussion
Clonal Expansion of EBV-infected B Cells in AILD.
In this study, clonal B cell expansions were detected among the EBV-infected cells in each of the six cases of AILD-type T cell lymphoma analyzed. The number of clones and their sizes differed markedly between cases, ranging from small oligoclonal to large monoclonal expansions encompassing nearly all B cells in the infiltrated tissue. In cases 2, 8, and 9, where large numbers of EBV−, CD20+ and Ki67+, proliferating B cells were analyzed, no large clones were detected among these cells (for case 6 see below). Only in case 7, where no T cell clone was detected, nearly all EBV− B cells belonged to a single clone, likely representing a tumor clone. Hence it appears that most large B cell clones in EBV-positive cases of AILD represent expanding EBV-infected B cells, pointing to a role of EBV in these expansions.
An exceptional situation was observed in case 6, where a large B cell clone was identified which was only partially EBV-infected (Fig. 2). The finding that EBV is restricted to a particular branch in the somatic mutation-based genealogical tree of this clone may indicate that EBV infection happened in a member of an already expanding B cell clone. Alternatively, EBV might have been present already in the founder cell of the clone but was later lost from a member of the clone that then established the EBV-negative branch in the genealogical tree. Such a loss of EBV has been described for some cases of Burkitt's lymphoma 39.
The observed spectrum of expansions among EBV-infected cells resembles the spectrum found in patients with posttransplantation lymphoproliferative disease 17 40 41. Like in posttransplantation patients, also in AILD, an initial polyclonal expansion of EBV-transformed B cells in a setting of insufficient control of these cells by cytotoxic T cells 42 may lead to the outgrowth of a dominant clone which perhaps acquired genetic changes conferring a growth advantage and occasionally may give rise to a malignant B cell clone. Indeed, in AILD patients EBV+ immunoblastic B cell lymphomas have been repeatedly observed 9 10 11. However, whereas EBV+ lymphomas in posttransplantation lymphomas are characterized by expression of the full spectrum of latent EBV proteins 43, the expanded cells detected in AILD do not express EBNA2.
EBV Is Largely Restricted to Memory and GC-like B Cells in AILD.
Although there are also many naive B cells present in the lymph node biopsies of AILD patients, as indicated from the analysis of EBV-negative cells in cases 2, 7, 8, and 9, the vast majority of EBV-infected B cells carried mutated Ig V genes and are thus memory or GC-like B cells. The somatic mutation pattern of rearranged V genes of the unique EBV+ cells indicates that they are mostly memory B cells selected for expression of a functional antigen receptor: with three exceptions no inactivating mutations were observed and the R/S ratio of 1.3 for the FRs is within the range typical for memory B cells (1.0–1.6) and considerably lower than the value of GC B cells (1.8; Table ). Also the EBV-positive B cells belonging to small clones without ongoing mutation likely represent selected memory B cells, as no inactivating mutations were observed and the R/S ratio is 1.1 (Table ). This propensity of EBV to reside and persist in long-lived memory B cells is observed also during primary infection with EBV in infectious mononucleosis, and in peripheral blood of healthy virus carriers 44 45.
About half of the clones showed ongoing somatic mutation in the course of clonal expansion (Table ). Since somatic hypermutation is regarded as a GC B cell–specific process 46, these clones of EBV-infected B cells resemble expanding GC B cells. Notably, some clones are composed of members without common mutations (including clones with unmutated members), indicating that in these cases unmutated (naive) B cells were originally EBV-infected and then induced to proliferate and differentiate to GC-like cells. For clones with shared mutations and intraclonal diversity (like clone 3 of case 1 and clone 1 of case 4; Fig. 2), it remains unclear whether naive, GC, or memory B cells were originally infected by EBV.
Thus, in AILD patients EBV is mainly found in two types of B cells: cells resembling memory B cells and showing relatively little tendency for clonal expansion and cells resembling GC B cells which are characterized by clonal expansion and ongoing somatic mutation.
Survival and Clonal Expansion of “Forbidden” B Cells in AILD.
In the clones with ongoing somatic mutation, the frequencies of nonsense mutations and deletions/duplications and the R/S ratios of originally potentially functional rearrangements were in the same range as is typical for nonfunctional out-of-frame rearrangements (Table ), indicating that EBV-infected B cells with ongoing somatic hypermutation are not selected for expression of a functional antigen receptor and that for most if not all cells, selection-free accumulation of mutations happened from the beginning of the mutation activity on through many rounds of proliferation and mutation.
Such a development of receptor-deficient B cells has not been observed before in normal or malignant human B cells in vivo, and is also distinct from the generation of “crippled” HRS cells in classical HL, as discussed below. Usually, B cells are stringently selected for expression of a functional surface receptor throughout their life and cells lacking an appropriate B cell receptor are efficiently and quickly eliminated 47. Even most transformed B cells in non-HLs still appear to depend on antigen receptor expression 48. Moreover, a sustained somatic hypermutation activity uncoupled from control of the mutating cells by selection is also unprecedented in normal and malignant B cells in vivo. These observations show that EBV infection in the setting of AILD can have a dramatic influence on the differentiation of B cells, allowing survival and clonal expansion of forbidden B cells.
How could EBV be involved in these processes? In transgenic mice expressing the LMP2a gene already early in B cell development, B lineage cells lacking a functional antigen receptor are generated and survive in the periphery, suggesting that LMP2a can mimic antigen receptor signaling 49. Since LMP2a expression was observed in the three cases with the highest content of EBER+ cells (Table ), it may be speculated, that the survival of cells with inactivated antigen receptor could at least partly be due to LMP2a expression. Moreover, LMP1, which simulates CD40 signaling and thus functions as a survival signal for B cells 50, was also found to be expressed by a fraction of the EBV-harboring B cells and may thus synergize with LMP2a in the rescue of Ig-deficient B cells.
It is likely that also the microenvironment in the AILD-affected lymph nodes plays a role in the development of the receptor-less B cells, because similar cells have not been seen in other instances where EBV-infected B cells are found and proliferate (see below). Whereas GCs are only rarely found in AILD, large aggregates of FDCs are usually present 7, and, together with the numerous T cells, may provide a microenvironment inducing or maintaining somatic hypermutation in some cells, similar to their proposed role in normal GCs. Moreover, AILD differs from most other B cell non-HLs by the production of a variety of cytokines in the tissue. Besides IL-6 and TNF-α also lymphotoxin is produced 51 52. Since the latter two cytokines are known to induce B cell growth 53 54, it may well be that the stimulation of EBV-infected B cells by these cytokines is involved in rescuing the crippled B cells from apoptosis and supporting their clonal expansion.
With respect to a potential role of the microenvironment in the survival of Ig-deficient B cells in AILD, it is interesting to compare AILD with infectious mononucleosis. In both diseases, EBV is largely restricted to memory and GC-like B cells, and expanded clones of virus-infected B cells are present 45. Moreover, IL-6, TNF-α, and lymphotoxin are also detectable in tonsils of patients affected by infectious mononucleosis 51. However, no crippled B cells were found among EBV-harboring cells in infectious mononucleosis, and no ongoing mutation was observed among members of expanded B cell clones 45. Perhaps, the lack of an extensive FDC network and a dominant population of T helper cells in infectious mononucleosis prevents the survival of crippled B cells in this disease. Moreover, whereas a state of immunodeficiency is observed in AILD 42, there is an intense immune response taking place against virus-harboring B cells in infectious mononucleosis, thereby selecting for B cells that downregulate immunogenic EBV proteins that might be essential for the survival of crippled B cells. Collectively, these factors likely prevent the survival of receptor-less B cells in infectious mononucleosis and also in healthy virus carriers.
Although the strong association between expanding mutating B cell clones with crippled V genes and the presence of EBV in the cells points to a decisive role of the virus in the rescue of the cells, it appears that in AILD occasionally also EBV− crippled B cells can survive and expand. This is shown here for a subset of the cells belonging to the large B cell clone in case 6 and the clone in case 7. In both instances these clones account for the vast majority of B cells in the tissue, and thus likely represent B cell tumors that acquired transforming events other than EBV. For the clone of case 7 the mutation pattern may indicate that the clone was initially dependent on a B cell receptor, as none of the many shared mutations (20 in VH and 33 in Vλ) is obviously crippling. A transforming event rendering the clone independent of a functional receptor likely occurred later in the course of its clonal expansion, now allowing the persistence of clone members with destructive mutations. The observation of ongoing mutation in this EBV− clone is in line with the idea that factors in the microenvironment of AILD tissues play an important role in triggering somatic hypermutation in expanding B cell clones (see above). The single dominant clones of crippled B cells found in cases 4 and 5 may also have acquired additional transforming events besides EBV infection and perhaps represent B cell tumors developing in the setting of AILD. Such events might render the expansion of B cell clones independent from EBV (like in case 6?).
EBV-infected B Cells in Lymphoid Malignancies: a Virus–Host Cell Relationship with Many Faces.
The three classical types of B cell lymphomas associated with EBV-infection, Burkitt's lymphoma, classical HL, and posttransplantation lymphomas, each show a distinct and characteristic pattern of EBV latent gene expression 16. In Burkitt's lymphoma, only the EBNA1 protein is expressed, which is needed for replication of the viral genome and hence maintenance of the virus in proliferating cells (latency I). The EBV-infected HRS cells of HL show, besides EBNA1, also LMP1 and 2a expression (latency II). In EBV-positive B cell lymphomas of immunocompromised patients all nine EBV-encoded latent proteins are expressed (latency III). The expression pattern of EBV-encoded latent genes in AILD is most similar to the situation in HL, since absence of EBNA2 expression and detection of LMP1-positive cells and LMP2a transcripts suggests a latency II profile. However, it appears that only a fraction of the EBV-carrying cells express LMP1. Whether this is also true for LMP2a expression, could not be determined because of the lack of a reliable anti-LMP2a antibody.
Crippling mutations have also been observed in the HRS cells of classical HL in ∼25% of the cases (both EBV+ and EBV−; references 22, 24, 25, 27, 28, 55 56 57 58). However, there are fundamental differences between the two situations. The mutation pattern of the V genes in HRS cells indicates that these cells were initially selected for antigen receptor expression, then acquired unfavorable V gene mutations (e.g., a nonsense mutation or a replacement mutation resulting in reduced affinity to the immunizing antigen) and were finally rescued from apoptosis by some transforming event, like EBV-infection in the EBV-positive cases of HL 25. Since ongoing mutation is not observed in the HRS cells in classical HL, it appears that the somatic hypermutation machinery is switched off when the tumor precursors entered the preapoptotic state. In contrast, in AILD, a population of EBV-infected B cells is driven into proliferation, activates the hypermutation process, and acquires somatic mutations without selection right from the beginning. Latency II was also described for very rare EBV-infected B cells with a GC phenotype from human tonsils 19. It is presently unclear whether these cells undergo somatic hypermutation, and if so, whether they would tolerate crippling mutations. Babcock et al. speculated that these cells will develop along the normal differentiation pathway into Ig+ memory B cells 19.
Taken together, in AILD two types of EBV-infected B cells can be discriminated. A subset of the cells likely represents virus-carrying memory B cells and thus resembles the cells seen in healthy virus carriers and during infectious mononucleosis. These cells show little tendency for clonal expansion. A considerable fraction of EBV-infected B cells is driven into massive proliferation and these cells show ongoing somatic mutation during clonal expansion without any obvious selection for functionality of the B cell receptor. Such a survival and clonal expansion of Ig-deficient B cells represents a novel type of viral latency in the B cell compartment. Although the factors defining this particular form of EBV persistence are far from clear, one may speculate that it may be the interplay between the particular microenvironment in AILD and the viral transformation of the cells that allows the survival of populations of forbidden B cells. In some cases, these EBV+ B cell clones may develop into B cell lymphomas.
Acknowledgments
We thank Christiane Gerhardt, Tanja Schaffer, and Ekaterini Hadzoglou for excellent technical assistance, Julia Kurth for comments on the manuscript, and Drs. B. Fabiani and A.-C. Baglin for providing tissue samples from patients.
This work was supported by the Deutsche Krebshilfe, Dr. Mildred Scheel Stiftung, and the Deutsche Forschungsgemeinschaft (SFB 502). R. Küppers is supported by the Heisenberg Programme of the Deutsche Forschungsgemeinschaft.
Footnotes
A. Bräuninger, T. Spieker, and K. Willenbrock contributed equally to this paper.
Abbreviations used in this paper: AILD, angioimmunoblastic lymphadenopathy with dysproteinemia; EBNA, EBV nuclear antigen; EBV, Epstein-Barr virus; FDC, follicular dendritic cell; GC, germinal center; HHV, human herpes virus; HL, Hodgkin's lymphoma; HRS, Hodgkin–Reed/Sternberg; LMP, latent membrane protein; RT, reverse transcription.
References
- Knecht H., Schwarze E.W., Lennert K. Histological, immunohistological and autopsy findings in lymphogranulomatosis X (including angio-immunoblastic lymphadenopathy) Virchows Arch. A Pathol. Anat. Histopathol. 1985;406:105–124. doi: 10.1007/BF00710561. [DOI] [PubMed] [Google Scholar]
- Griesser H., Feller A., Lennert K., Minden M., Mak T.W. Rearrangement of the β chain of the T cell antigen receptor and immunoglobulin genes in lymphoproliferative disorders. J. Clin. Invest. 1986;78:1179–1184. doi: 10.1172/JCI112700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor N.T., Crick J.A., Wainscoat J.S., Gatter K.C., Stein H., Falini B., Mason D.Y. Evidence for monoclonal T lymphocyte proliferation in angioimmunoblastic lymphadenopathy. J. Clin. Pathol. 1986;39:1229–1232. doi: 10.1136/jcp.39.11.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss L.M., Strickler J.G., Dorfman R.F., Horning S.J., Warnke R.A., Sklar J. Clonal T-cell populations in angioimmunoblastic lymphadenopathy and angioimmunoblastic lymphadenopathy-like lymphoma. Am. J. Pathol. 1986;122:392–397. [PMC free article] [PubMed] [Google Scholar]
- Theodorou I., Bigorgne C., Delfau M.H., Lahet C., Cochet G., Vidaud M., Raphael M., Gaulard P., Farcet J.P. VJ rearrangements of the TCR γ locus in peripheral T-cell lymphomasanalysis by polymerase chain reaction and denaturing gradient gel electrophoresis. J. Pathol. 1996;178:303–310. doi: 10.1002/(SICI)1096-9896(199603)178:3<303::AID-PATH475>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Lipford E.H., Smith H.R., Pittaluga S., Jaffe E.S., Steinberg A.D., Cossman J. Clonality of angioimmunoblastic lymphadenopathy and implications for its evolution to malignant lymphoma. J. Clin. Invest. 1987;79:637–642. doi: 10.1172/JCI112860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A.C., Griesser H., Schilling C.V., Wacker H.H., Dallenbach F., Bartels H., Kuse R., Mak T.W., Lennert K. Clonal gene rearrangement patterns correlate with immunophenotype and clinical parameters in patients with angioimmunoblastic lymphadenopathy. Am. J. Pathol. 1988;133:549–556. [PMC free article] [PubMed] [Google Scholar]
- Smith J.L., Hodges E., Quin C.T., McCarthy K.P., Wright D.H. Frequent T and B cell oligoclones in histologically and immunophenotypically characterized angioimmunoblastic lymphadenopathy. Am. J. Pathol. 2000;156:661–669. doi: 10.1016/S0002-9440(10)64770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abruzzo L.V., Schmidt K., Weiss L.M., Jaffe E.S., Medeiros L.J., Sander C.A., Raffeld M. B-cell lymphoma after angioimmunoblastic lymphadenopathya case with oligoclonal gene rearrangements associated with Epstein-Barr virus. Blood. 1993;82:241–246. [PubMed] [Google Scholar]
- Knecht H., Martius F., Bachmann E., Hoffman T., Zimmermann D.R., Rothenberger S., Sandvej K., Wegmann W., Hurwitz N., Odermatt B.F. A deletion mutant of the LMP1 oncogene of Epstein-Barr virus is associated with evolution of angioimmunoblastic lymphadenopathy into B immunoblastic lymphoma. Leukemia. 1995;9:458–465. [PubMed] [Google Scholar]
- Matsue K., Itoh M., Tsukuda K., Kokubo T., Hirose Y. Development of Epstein-Barr virus-associated B cell lymphoma after intensive treatment of patients with angioimmunoblastic lymphadenopathy with dysproteinemia. Int. J. Hematol. 1998;67:319–329. doi: 10.1016/s0925-5710(98)00005-x. [DOI] [PubMed] [Google Scholar]
- Anagnostopoulos I., Hummel M., Finn T., Tiemann M., Korbjuhn P., Dimmler C., Gatter K., Dallenbach F., Parwaresch M.R., Stein H. Heterogeneous Epstein-Barr virus infection patterns in peripheral T-cell lymphoma of angioimmunoblastic lymphadenopathy type. Blood. 1992;80:1804–1812. [PubMed] [Google Scholar]
- Weiss L.M., Jaffe E.S., Liu X.F., Chen Y.Y., Shibata D., Medeiros L.J. Detection and localization of Epstein-Barr viral genomes in angioimmunoblastic lymphadenopathy and angioimmunoblastic lymphadenopathy-like lymphoma. Blood. 1992;79:1789–1795. [PubMed] [Google Scholar]
- Wagner H.J., Bein G., Bitsch A., Kirchner H. Detection and quantification of latently infected B lymphocytes in Epstein-Barr virus-seropositive, healthy individuals by polymerase chain reaction. J. Clin. Microbiol. 1992;30:2826–2829. doi: 10.1128/jcm.30.11.2826-2829.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita E.M., Yang B., Lam K.M., Crawford D.H., Thorley-Lawson D.A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- Rickinson A.B., Kieff E. Epstein-Barr Virus. In: Fields B.N., Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott-Raven Publishers; Philadelphia: 1996. pp. 2397–2446. [Google Scholar]
- Knowles D.M., Cesarman E., Chadburn A., Frizzera G., Chen J., Rose E.A., Michler R.E. Correlative morphologic and molecular genetic analysis demonstrates three distinct categories of posttransplantation lymphoproliferative disorders. Blood. 1995;85:552–565. [PubMed] [Google Scholar]
- Biggar R.J., Rabkin C.S. The epidemiology of AIDS-related neoplasms. Hematol. Oncol. Clin. North Am. 1996;10:997–1010. doi: 10.1016/s0889-8588(05)70380-4. [DOI] [PubMed] [Google Scholar]
- Babcock G.J., Hochberg D., Thorley-Lawson D.A. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity. 2000;13:497–506. doi: 10.1016/s1074-7613(00)00049-2. [DOI] [PubMed] [Google Scholar]
- Callan M.F., Tan L., Annels N., Ogg G.S., Wilson J.D., O'Callaghan C.A., Steven N., McMichael A.J., Rickinson A.B. Direct visualization of antigen-specific CD8+ T cells during the primary immune response to Epstein-Barr virus In vivo. J. Exp. Med. 1998;187:1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz C., Bickham K.L., Subklewe M., Tsang M.L., Chahroudi A., Kurilla M.G., Zhang D., O'Donnell M., Steinman R.M. Human CD4+ T lymphocytes consistently respond to the latent Epstein-Barr virus nuclear antigen EBNA1. J. Exp. Med. 2000;191:1649–1660. doi: 10.1084/jem.191.10.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieker T., Kurth J., Küppers R., Rajewsky K., Bräuninger A., Hansmann M.-L. Molecular single cell analysis of the clonal relationship of small Epstein-Barr virus infected cells and Epstein-Barr virus harboring Hodgkin and Reed/Sternberg cells in Hodgkin's disease. Blood. 2000;96:3133–3138. [PubMed] [Google Scholar]
- Kneba M., Bolz I., Linke B., Bertram J., Rothaupt D., Hiddemann W. Characterization of clone-specific rearrangement T-cell receptor γ-chain genes in lymphomas and leukemias by the polymerase chain reaction and DNA sequencing. Blood. 1994;84:574–581. [PubMed] [Google Scholar]
- Küppers R., Rajewsky K., Zhao M., Simons G., Laumann R., Fischer R., Hansmann M.L. Hodgkin diseaseHodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc. Natl. Acad. Sci. USA. 1994;91:10962–10966. doi: 10.1073/pnas.91.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzler H., Küppers R., Hansmann M.L., Rajewsky K. Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J. Exp. Med. 1996;184:1495–1505. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuninger A., Küppers R., Spieker T., Siebert R., Strickler J.G., Schlegelberger B., Rajewsky K., Hansmann M.L. Molecular analysis of single B cells from T-cell-rich B-cell lymphoma shows the derivation of the tumor cells from mutating germinal center B cells and exemplifies means by which immunoglobulin genes are modified in germinal center B cells. Blood. 1999;93:2679–2687. [PubMed] [Google Scholar]
- Bräuninger A., Hansmann M.L., Strickler J.G., Dummer R., Burg G., Rajewsky K., Küppers R. Identification of common germinal-center B-cell precursors in two patients with both Hodgkin's disease and non-Hodgkin's lymphoma. N. Engl. J. Med. 1999;340:1239–1247. doi: 10.1056/NEJM199904223401604. [DOI] [PubMed] [Google Scholar]
- Müschen M., Rajewsky K., Bräuninger A., Baur A.S., Oudejans J.J., Roers A., Hansmann M.L., Kuppers R. Rare occurrence of classical Hodgkin's disease as a T cell lymphoma. J. Exp. Med. 2000;191:387–394. doi: 10.1084/jem.191.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone A., Cilia A.M., Gloghini A., Canzonieri V., Pastore C., Todesco M., Cozzi M., Perin T., Volpe R., Pinto A., Gaidano G. Establishment of HHV-8-positive and HHV-8-negative lymphoma cell lines from primary lymphomatous effusions. Int. J. Cancer. 1997;73:562–569. doi: 10.1002/(sici)1097-0215(19971114)73:4<562::aid-ijc18>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Speck S.H., Chatila T., Flemington E. Reactivation of Epstein-Barr virusregulation and function of the BZLF1 gene. Trends Microbiol. 1997;5:399–405. doi: 10.1016/S0966-842X(97)01129-3. [DOI] [PubMed] [Google Scholar]
- Feederle R., Kost M., Baumann M., Janz A., Drouet E., Hammerschmidt W., Delecluse H.J. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 2000;19:3080–3089. doi: 10.1093/emboj/19.12.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppi M., Barozzi P., Maiorana A., Artusi T., Trovato R., Marasca R., Savarino M., Ceccherini-Nelli L., Torelli G. Human herpes virus-8 DNA sequences in human immunodeficiency virus-negative angioimmunoblastic lymphadenopathy and benign lymphadenopathy with giant germinal center hyperplasia and increased vascularity. Blood. 1996;87:3903–3909. [PubMed] [Google Scholar]
- Lee H., Veazey R., Williams K., Li M., Guo J., Neipel F., Fleckenstein B., Lackner A., Desrosiers R.C., Jung J.U. Deregulation of cell growth by the K1 gene of Kaposi's sarcoma-associated herpes virus. Nat. Med. 1998;4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- Humphrey R.W., Davis D.A., Newcomb F.M., Yarchoan R. Human herpes virus 8 (HHV-8) in the pathogenesis of Kaposi's sarcoma and other diseases. Leuk. Lymphoma. 1998;28:255–264. doi: 10.3109/10428199809092681. [DOI] [PubMed] [Google Scholar]
- Linke B., Bolz I., Fayyazi A., von Hofen M., Pott C., Bertram J., Hiddemann W., Kneba M. Automated high resolution PCR fragment analysis for identification of clonally rearranged immunoglobulin heavy chain genes. Leukemia. 1997;11:1055–1062. doi: 10.1038/sj.leu.2400736. [DOI] [PubMed] [Google Scholar]
- Klein U., Goossens T., Fischer M., Kanzler H., Bräuninger A., Rajewsky K., Kuppers R. Somatic hypermutation in normal and transformed human B cells. Immunol. Rev. 1998;162:261–280. doi: 10.1111/j.1600-065x.1998.tb01447.x. [DOI] [PubMed] [Google Scholar]
- Küppers R., Rajewsky K., Hansmann M.L. Diffuse large cell lymphomas are derived from mature B cells carrying V region genes with a high load of somatic mutation and evidence of selection for antibody expression. Eur. J. Immunol. 1997;27:1398–1405. doi: 10.1002/eji.1830270616. [DOI] [PubMed] [Google Scholar]
- Goossens T., Klein U., Küppers R. Frequent occurrence of deletions and duplications during somatic hypermutationimplications for oncogene translocations and heavy chain disease. Proc. Natl. Acad. Sci. USA. 1998;95:2463–2468. doi: 10.1073/pnas.95.5.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzouk B.I., Srinivas S., Sample C.E., Singh V., Sixbey J.W. Epstein-Barr Virus DNA recombination and loss in sporadic Burkitt's lymphoma. J. Infect. Dis. 1996;173:529–535. doi: 10.1093/infdis/173.3.529. [DOI] [PubMed] [Google Scholar]
- Cleary M.L., Nalesnik M.A., Shearer W.T., Sklar J. Clonal analysis of transplant-associated lymphoproliferations based on the structure of the genomic termini of the Epstein-Barr virus. Blood. 1988;72:349–352. [PubMed] [Google Scholar]
- Chadburn A., Cesarman E., Liu Y.F., Addonizio L., Hsu D., Michler R.E., Knowles D.M. Molecular genetic analysis demonstrates that multiple posttransplantation lymphoproliferative disorders occurring in one anatomic site in a single patient represent distinct primary lymphoid neoplasms. Cancer. 1995;75:2747–2756. doi: 10.1002/1097-0142(19950601)75:11<2747::aid-cncr2820751119>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Pizzolo G., Vinante F., Agostini C., Zambello R., Trentin L., Masciarelli M., Chilosi M., Benedetti F., Dazzi F., Todeschini G. Immunologic abnormalities in angioimmunoblastic lymphadenopathy. Cancer. 1987;60:2412–2418. doi: 10.1002/1097-0142(19871115)60:10<2412::aid-cncr2820601010>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Thomas J.A., Hotchin N.A., Allday M.J., Amlot P., Rose M., Yacoub M., Crawford D.H. Immunohistology of Epstein-Barr virus-associated antigens in B cell disorders from immunocompromised individuals. Transplantation. 1990;49:944–953. doi: 10.1097/00007890-199005000-00022. [DOI] [PubMed] [Google Scholar]
- Babcock G.J., Decker L.L., Volk M., Thorley-Lawson D.A. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- Kurth J., Spieker T., Wustrow J., Strickler J.G., Hansmann M.-L., Rajewsky K., Küppers R. EBV-infected B cells in infectious mononucleosisviral strategies for spreading in the B cell compartment and establishing latency. Immunity. 2000;13:485–495. doi: 10.1016/s1074-7613(00)00048-0. [DOI] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Lam K.P., Kühn R., Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- Küppers R., Klein U., Hansmann M.L., Rajewsky K. Cellular origin of human B-cell lymphomas. N. Engl. J. Med. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- Caldwell R.G., Wilson J.B., Anderson S.J., Longnecker R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9:405–411. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- Gires O., Zimber-Strobl U., Gonnella R., Ueffing M., Marschall G., Zeidler R., Pich D., Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 1997;16:6131–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss H.D., Anagnostopoulos I., Herbst H., Grebe M., Ziemann K., Hummel M., Stein H. Patterns of cytokine gene expression in peripheral T-cell lymphoma of angioimmunoblastic lymphadenopathy type. Blood. 1995;85:2862–2869. [PubMed] [Google Scholar]
- Yamaguchi S., Kitagawa M., Inoue M., Tomizawa N., Kamiyama R., Hirokawa K. Cell dynamics and expression of tumor necrosis factor (TNF)-α, interleukin-6, and TNF receptors in angioimmunoblastic lymphadenopathy-type T cell lymphoma. Exp. Mol. Pathol. 2000;68:85–94. doi: 10.1006/exmp.1999.2297. [DOI] [PubMed] [Google Scholar]
- Kehrl J.H., Alvarez-Mon M., Delsing G.A., Fauci A.S. Lymphotoxin is an important T cell-derived growth factor for human B cells. Science. 1987;238:1144–1146. doi: 10.1126/science.3500512. [DOI] [PubMed] [Google Scholar]
- Boussiotis V.A., Nadler L.M., Strominger J.L., Goldfeld A.E. Tumor necrosis factor alpha is an autocrine growth factor for normal human B cells. Proc. Natl. Acad. Sci. USA. 1994;91:7007–7011. doi: 10.1073/pnas.91.15.7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irsch J., Nitsch S., Hansmann M.L., Rajewsky K., Tesch H., Diehl V., Jox A., Küppers R., Radbruch A. Isolation of viable Hodgkin and Reed-Sternberg cells from Hodgkin disease tissues. Proc. Natl. Acad. Sci. USA. 1998;95:10117–10122. doi: 10.1073/pnas.95.17.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jox A., Zander T., Küppers R., Irsch J., Kanzler H., Kornacker M., Bohlen H., Diehl V., Wolf J. Somatic mutations within the untranslated regions of rearranged Ig genes in a case of classical Hodgkin's disease as a potential cause for the absence of Ig in the lymphoma cells. Blood. 1999;93:3964–3972. [PubMed] [Google Scholar]
- Marafioti T., Hummel M., Foss H.D., Laumen H., Korbjuhn P., Anagnostopoulos I., Lammert H., Demel G., Theil J., Wirth T., Stein H. Hodgkin and Reed-Sternberg cells represent an expansion of a single clone originating from a germinal center B-cell with functional immunoglobulin gene rearrangements but defective immunoglobulin transcription. Blood. 2000;95:1443–1450. [PubMed] [Google Scholar]
- Vockerodt M., Soares M., Kanzler H., Küppers R., Kube D., Hansmann M.L., Diehl V., Tesch H. Detection of clonal Hodgkin and Reed-Sternberg cells with identical somatically mutated and rearranged VH genes in different biopsies in relapsed Hodgkin's disease. Blood. 1998;92:2899–2907. [PubMed] [Google Scholar]
- de Wildt R.M., van Venrooij W.J., Winter G., Hoet R.M., Tomlinson I.M. Somatic insertions and deletions shape the human antibody repertoire. J. Mol. Biol. 1999;294:701–710. doi: 10.1006/jmbi.1999.3289. [DOI] [PubMed] [Google Scholar]