Abstract
Notch signaling is known to differentially affect the development of lymphoid B and T cell lineages, but it remains unclear whether such effects are specifically dependent on distinct Notch ligands. Using a cell coculture assay we observed that the Notch ligand Delta-1 completely inhibits the differentiation of human hematopoietic progenitors into the B cell lineage while promoting the emergence of cells with a phenotype of T cell/natural killer (NK) precursors. In contrast, Jagged-1 did not disturb either B or T cell/NK development. Furthermore, cells cultured in the presence of either Delta-1 or Jagged-1 can acquire a phenotype of NK cells, and Delta-1, but not Jagged-1, permits the emergence of a de novo cell population coexpressing CD4 and CD8. Our results thus indicate that distinct Notch ligands can mediate differential effects of Notch signaling and provide a useful system to further address cell-fate decision processes in lymphopoiesis.
Keywords: cell-fate decision genes, lymphopoiesis, S17 cells, Notch signaling, B and T cells
Introduction
Notch receptors and their ligands are phylogenetically conserved transmembrane proteins which regulate cell-fate decision processes during development and postnatal life 1. In recent years, evidence has been accumulating which demonstrates that Notch signaling plays a critical role in the development of lymphoid B and T cell lineages 2 3. Signaling through activated Notch has been implicated in the decision processes underlying the functional bifurcation of CD4/CD8 T lymphocytes 4, the choice between αβ and γδ T cell receptors 5 and in maturation processes of CD4+ and CD8+ cells; 6 though, more recently, it has been shown that the absence of Notch-1 in thymocytes at an early stage of development does not appear to disturb subsequent T cell maturation 7. On the other hand, the constitutive activation of Notch-1 in mouse bone marrow cells was shown to block B lymphopoiesis while promoting T cell differentiation within the bone marrow 8. Conversely, transgenic mice with inactive Notch-1 in bone marrow and thymus display a severe impairment of T cell development while showing B cell development within the thymus 9. These observations strongly suggest that Notch signaling may influence the commitment of a common lymphoid progenitor 10 towards differentiation into the B or T/natural killer (NK) cell lineages. However, most of the above mentioned data reflect the effects of constitutively active forms of Notch, not allowing the analysis of putative differential actions induced by distinct Notch ligands. In mammalians these include two protein families, the first corresponding to homologues of Drosophila serrate, Jagged-1 and -2 11 12 13, and the second to homologues of Drosophila delta, Delta-like-1, 3, and 4 14 15 16. Jagged-1 and -2 are expressed in bone marrow and fetal liver stromal cells, in thymic epithelium and subsets of hematopoietic cells of different lineages, suggesting that Notch activation may be induced by interaction with these ligands in different hematopoietic environments 2. Several observations indicate that Jagged-1 and -2 act by expanding and maintaining primitive hematopoietic precursors while inhibiting or delaying their terminal differentiation 17 18 19 20 21 though recently, different views have emerged. Schroeder et al. 22 reported that Jagged-1 accelerates granulocytic differentiation of 32D cells, and the effects of Jagged-1 on proliferation and differentiation of human CD34+ cells appear to depend on the context of growth-promoting cytokines 23. However, these experiments were conducted in conditions that favor the differentiation into myeloid but not lymphoid lineages, thus failing to reveal what kind of specific effects may be caused by Jagged-1 and -2 in the differentiation processes of lymphoid cells. It is nonetheless accepted that they might also act in lymphopoeisis, as Jagged-1 and -2 are expressed in lymphoid tissues 2 and mice without functional Jagged-2 have abnormal thymic morphology and impaired differentiation of γδ T cells 24. Accordingly, it has been recently shown that Jagged-1 is involved in the regulation of CD4:CD8 cell ratio during thymic development 25.
Much less is known about the specific role of Delta ligands. The Delta-1 gene is expressed in the corticomedullary region of the newborn mouse thymus and in fetal and adult human hematopoietic tissues (unpublished observations and reference 26). Moreover, a soluble form of human Delta-1 was shown to delay the acquisition of differentiation markers by murine hematopoietic progenitors and to promote the expansion of primitive precursors 26. It would thus appear that different Notch ligands have redundant actions at the level of early hematopoietic precursors, promoting their expansion while preventing them to pursue terminal differentiation. However, other observations indicate that this might not be the case. Notch ligands have restricted patterns of expression during development and in adult tissues and knockout mice for Notch-1 and -2, Jagged-1 and -2, display distinct phenotypes 2. Therefore, it is conceivable that the plurality of Notch receptors and Notch ligands may translate into functional diversity of Notch actions in vivo.
To address this issue, we asked, specifically, whether the known inhibition of B cell development induced by active Notch 8 could depend on the action of specific Notch ligands. Making use of a cell coculture system previously known to favor the selective differentiation of hematopoietic progenitors into early B cells 27, we show that Delta-1 completely blocks the differentiation of progenitor cells into the B cell lineage, while promoting the emergence of a population of cells with characteristics of a T/NK cell precursor. In contrast, Jagged-1 apparently did not disturb the B or T/NK cell developmental potential of the original cells. Taken together, our data show that Jagged-1 and Delta-1 have differential effects in cell-fate decision processes in human lymphopoiesis in vitro, strongly suggesting that the interplay of Notch receptors with distinct ligands may underlay diverse biological outcomes of Notch signaling pathways.
Materials and Methods
Cord Blood CD34+ Cells.
Umbilical cord blood (CB) was collected in sterile heparinized tubes according to the guidelines approved by the Ethical Committee of the Lisbon Medical School. Mononuclear cells were isolated by density gradient centrifugation (Ficoll-Paque, Amersham Pharmacia Biotech) and CD34+ cells subsequently obtained using the MiniMACS™ separation column (Miltenyi Biotec). Technical procedures were done accordingly to the instructions included in the CD34 Progenitor Cell Isolation kit (QBEND/10). CD34+ expression on collected cells varied from 80 to 95% as assessed by flow cytometry. Collected cells were immediately processed for coculture experiments or, alternatively, aliquoted and stored at −80°C until use.
S17 Cell Line.
The murine stromal cell line S17 (provided by D. Rawlings, UCLA, Los Angeles, CA) was used as a stromal cell layer in coculture experiments with CB CD34+ cells exactly as described by Rawlings and colleagues 27, without added cytokines.
Delta-1 and Jagged-1 cDNAs, Retroviral Vectors, and Producer Cell Lines.
Full-length cDNAs encoding either the human Delta-1 or Jagged-1 (provided by G. Artavanis-Tsakonas, Harvard Medical School, Charlestown, MA) were cloned into the restriction sites of BamHI and EcoRI of the LZRS-linker-IRES-enhanced green fluorescent protein (eGFP) retrovirus (provided by H. Spits, The Netherlands Cancer Institute, Amsterdam, Holland). This vector permits the coexpression of cloned cDNAs and the marker gene eGFP, from a single bicistronic message 28 29. The open reading frame of Delta-1 was PCR amplified, from a plasmid containing the full length cDNA (GenBank accession number AF003522), using Delta-1 specific primers also containing appropriate restriction sites. The upstream primer contains a BamHI site: aaGGATCCaccatgggcagtcggtgcgcgct. The downstream primer contains an EcoRI site: ttGAATTC ttacacctcagttgctatgacgca (coding sequences are underlined, restriction sites are given in bold). Restriction and sequence analysis were performed to confirm proper amplification and ligation of the human Delta-1 open reading frame into the LZRS-polylinker-IRES-eGFP vector. High titres of empty (control GFP only) or recombinant virus were obtained after transfection of Phoenix-Ecotropic cell line (provided by G. Nolan, Stanford University, Stanford, CA). In brief, 2 d after transfection of the packaging cell line with a retroviral construct, selection started by addition of 2 μg/ml of puromycin (CLONTECH). 10–14 d after transfection, 15 × 106 puromycin selected cells were plated on T175 flasks (Becton Dickinson) in 30 ml of medium without puromycin 29. The following day the medium was replaced to remove dead cells and 24 h later retroviral supernatants were harvested, centrifuged, and frozen in aliquots at –80ºC until further use. To test the efficiency of the retroviral supernatants, NIH 3T3 cells were infected and GFP production monitored by flow cytometry after 48 h of infection (mean of 90% GFP+ cells).
Transduction of S17 Cells with Retroviruses Containing Delta-1 or Jagged-1 cDNAs.
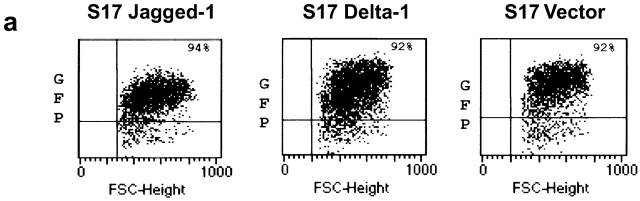
Aliquots of frozen retroviral supernatants containing either Delta-1, Jagged-1, or the empty virus (control) were thawed and incubated on ice after the addition of DOTAP (DOTAP liposomal transfection reagent; Boehringer Mannheim) at a final concentration of 100 μg/ml for 10 min. Subsequently, pellets of 105 of S17 cells were resuspended in the transfection mixture and incubated for 6–18 h at 37°C, 5% CO2 in 1 well of a 24-well, flat-bottomed plate (Nunclon; Nalge Nunc International). The transfection mixture was then removed and the cells transferred to a T25 flask (Falcon; Becton Dickinson) for further culture in RPMI (Sigma-Aldrich), 10% FBS (GIBCO BRL), and penicillin/streptomycin (p/s; GIBCO BRL). 48 h after transduction, S17 cells were analyzed for GFP expression by flow cytometry. Transduced cells were subsequently sorted until >90% of the cells expressed GFP (see Fig. 1 a) and used for coculture experiments with CB CD34+ cells.
Figure 1.
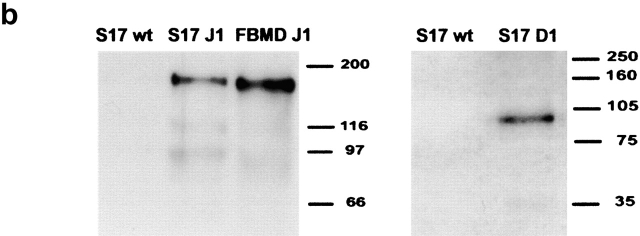
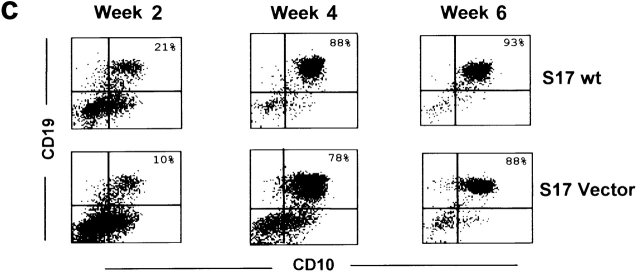
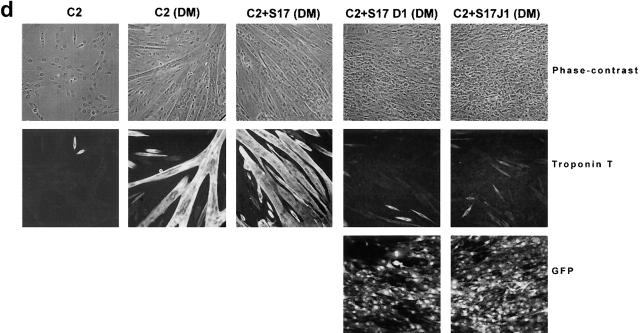
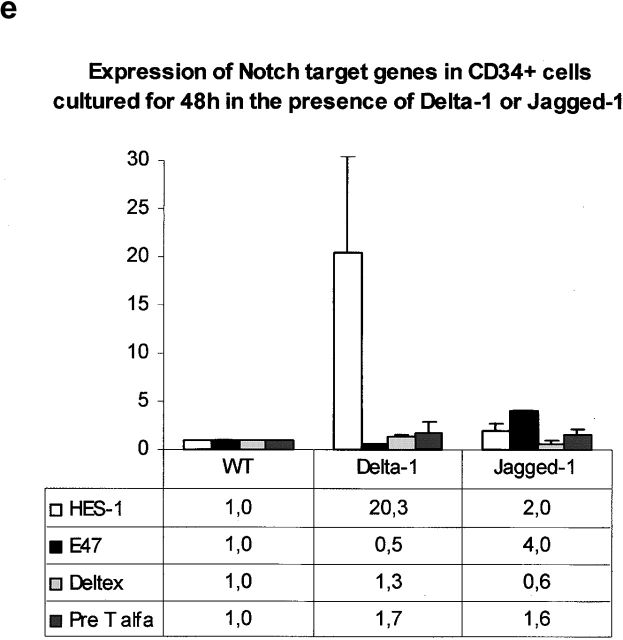
Analysis of GFP and protein expression on transduced S17 cells, of B cell differentiation in unmodified and vector-transduced S17 cells, and functional assays for Jagged-1 and Delta-1 proteins. (a) Flow cytometric analysis of GFP expression 3 wk after sorting for GFP+ transduced S17 cells. Quadrants were set as to exclude GFP-negative cells as determined by analysis of nontransduced cells. (b) Western blot analysis detection of human Jagged-1 and Delta-1 proteins in S17 cells transduced with vectors containing either Jagged-1 (S17 J1) or Delta-1 cDNAs (S17 D1) in the same samples as in a. FBMD J1-FBMD1 cells transduced with vector containing human Jagged-1 cDNA. Molecular weight markers are indicated on the right (kD). (c) Flow cytometric analysis of CD10 and CD19 expression in CD34+ cells cocultured with parental S17 cells (S17 wt, top) and S17 cells transduced with vector alone (S17 Vector, bottom) in three different time points. (d) Delta-1 and Jagged-1 expressed by S17 cells inhibits the differentiation of C2 cells. Pictures were taken with the confocal microscope Zeiss LSM-510. C2, proliferating C2 cells; DM, differentiation medium (reference 11); S17 D1, S17 cells transduced with Delta-1; S17 J1, S17 cells transduced with Jagged-1. Original magnification: ×20. (e) Expression levels of HES-1, Deltex, pre-T-α, and E47 in CD34+ cells cocultured with parental or transduced S17 cells for 48 h, as assessed by quantitative RT-PCR (n = 2 for E47 and n = 3 for other transcripts). Expression of each transcript was measured as a ratio with the GAPDH transcript and expressed in arbitrary units (each transcript in CD34+ cells cocultured with unmodified stroma = 1).
Coculture Assays.
24 h before their use in coculture experiments, 2 × 104 S17 cells in 1 ml of medium were plated in 24-well, flat-bottomed plates. Cocultures were initiated by seeding 5 × 104 CD34+ cells to the wells precoated with wild-type or transduced S17 stroma, and the cultures were maintained at 37°C, 5% CO2. Half of the culture medium (RPMI, 10% FBS, p/s) was replaced by fresh medium once a week. After the incubation periods (2, 4, and 6 wk), cells were harvested, counted, and used for subsequent analyses and experimental assays.
Differentiation of NK Cells.
After a period of 3 wk of coculture on a monolayer of S17 stroma, supernatant cells were harvested, counted, and tested for their capacity to develop into NK cells as described previously 30 31. In brief, 1–2 × 104 CB cells were further cultured in U-bottomed, 96-well plates in the presence of 10 ng/ml Flt-3 ligand (R&D Systems), 10 ng/ml IL-7 (R&D Systems), and 10 ng/ml IL-15 (R&D Systems). Similar cultures were started with fresh CB CD34+ cells to control for normal NK cell development in our system. The cultures were maintained on a humidified atmosphere at 37°C in 5% CO2 for 3 wk. The cells were then stained with fluorochrome-labeled antibodies (see below) and their antigenic phenotype analyzed by flow cytometry.
Flow Cytometry and mAbs.
Flow cytometry analyses were performed using a FACSCalibur™ (Becton Dickinson). Fresh CB progenitors or supernatant cells harvested at different time points, were stained with mAbs for 30 min, washed with a PBA solution consisting of PBS supplemented with 2% BSA (MERCK) and 0.01% NaN3 (Sigma-Aldrich), and analyzed by flow cytometry. For cytoplasmic staining of the CB cells, after membrane staining, the cells were fixed in 1% paraformaldehyde (Sigma-Aldrich) for 30 min. Next, the cells were washed with PBA and permeabilized by incubation with a solution of 0.25% saponin (Merck) for 2 min. The cells were washed again with PBA and incubated for 30 min with the antibodies of interest for detection of cytoplasmic expression. All steps of the staining procedure were carried out on ice and for both membrane and cytoplasmic staining, appropriate fluorochrome-conjugated, isotype-matched control Igs were used in all experiments. The following mouse anti–human mAbs (FITC-, PE-, PerCP-, or PE-Cy5 coupled) were used: CD34-FITC, CD38-PE, CD34-PE, CD14-FITC, and CD45-PE-Cy5 (purchased from CLB, Amsterdam, The Netherlands), CD7-FITC, CD4-FITC, CD19-PE, CD33-PE, CD56-PE, CD20-PE, CD3-PerCP, CD34-PerCP, CD8-PE, (all from Becton Dickinson), IgM-PE, and CD10-PE-Cy5 (BD PharMingen).
Functional Assays for Jagged-1 and Delta-1 Proteins.
The efficacy of transduced Delta-1 and Jagged-1 proteins in the activation of the Notch pathway was assessed in coculture experiments of the myoblast cell line C2 (a gift from A. Israel, Unité de Biologie Moléculaire de L'Expression Génique, Institut Pasteur, Paris, France). The functionality of the constructs was initially verified using NIH 3T3 cells transduced with either Jagged-1 or Delta-1 vectors cocultured with C2 cells as described by Jarriault and collaborators 32. As expected for a functional activation of Notch, myoblast differentiation was inhibited in the presence of NIH 3T3 cells transduced with Jagged-1 or Delta-1, as assessed by phase-contrast analysis and the expression of T-troponin (obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242) (immunocytochemical detection, data not shown). To verify whether these proteins, when expressed by S17 cells, maintained their functionality, similar experiments were done using transduced S17 cells instead of NIH 3T3 cells. As shown in Fig. 1 d a strong inhibition of myotube formation was observed when C2 myoblasts were cocultured with transduced S17 stromas for 4 d indicating that S17 cells are expressing functional Jagged-1 and Delta-1 proteins.
Western Blot Analysis.
Wild-type and retrovirally transduced S17 cells (with empty virus, Jagged-1 or Delta-1) were trypsinized and washed in PBS before protein extraction. Western blot analysis was done as described previously 33 using as primary antibodies goat-anti–Delta-1 (Santa Cruz Biotechnology, Inc.) and rat-anti–Jagged-1 (obtained from the Developmental Studies Hybridoma Bank; Fig. 1 b). The FBMD-1 cell line (provided by R. Ploemacher, Department of Hematology, Erasmus University, Rotterdam, Netherlands) transduced with the Jagged-1-retrovirus and known to express the Jagged-1 protein was used as a positive control for the anti–Jagged-1 antibody.
Real-Time PCR (TaqMan).
The expression of Notch-target genes HES-1, Deltex, pre–T-α and E47 was analyzed in CD34+ cells cultured in the presence of Delta-1 or Jagged-1 for 48 h using quantitative RT-PCR. Total RNA was extracted from cell pellets, lysed in GTC solution, and reverse transcribed using random hexamer-primers (Amersham Pharmacia Biotech) as described previously 34.
Primers and probes were designed using the Primer Express software (Applied Biosystems). Expression of each target gene was normalized using the endogenous gene hGAPDH (TaqMan PDAR reagent; Applied Biosystems). Primers and probes of genes under analysis were: HES-1 forward primer 5′ TGG AAA TGA CAG TGA AGC ACC T 3′; HES-1 reverse primer 5′ GTT CAT GCA CTC GCT GAA GC 3′; HES-1 probe CGC AGA TGA CGG CTG CGC TG 3′ (GenBank accession number AF264785); E47 forward primer 5′ AGA ACA CGT CAG CGG CTG A 3′; E47 reverse primer 5′ TAT TGG CCA TGC GCC TCT 3′; E47 probe 5′ TCC TCC AGG GAC AGC ACC TCG C 3′ (GenBank accession number M65214); Deltex forward primer 5′ CAT CAT CGA CCT GCA GTC CA 3′; Deltex reverse primer 5′ CGA CGA CGG GTC GTA GAA GT 3′; Deltex probe 5′ CAG GAC ACA GGC ACC ATG CGG 3′ (GenBank accession number XM012258); pre-TCR-α forward primer 5′ CAT CCT GGG AGC CTT TGG T 3′; pre-TCR-α reverse primer 5′ CCG GTG TCC CCC TGA GA 3′; and pre-TCR-α probe 5′ CCA TGC ATC TGT CAG GAG AGG CTT CTA CAG 3′ (GenBank accession number U36759). Probes were designed in exon/exon boundaries in order to avoid DNA amplification and were labeled at the 5′ end with the reporter dye molecule FAM and at the 3′ end with the quencher dye molecule TAMRA. All transcripts were amplified in 25 μl reactions using 2 μl of cDNA and the TaqMan Universal PCR Master Mix (Applied Biosystems) according to the manufacturer's instructions. Each reaction contained 300 nM of each primer and 200 nM of the labeled probe. Amplifications were done in an ABI Prism 5,700 thermocycler (Applied Biosystems) for 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 s at 95°C, and 1 min at 60°C. The CEM cell line was used as standard. For the GAPDH and each gene a logarithmic dilution of the CEM cDNA was done to allow quantification. Expression values of the wild-type samples at 48 h were used as calibrator. To analyze the intra-assay variation, 10 replicates of a 10−1 logarithmic dilution of the CEM cDNA were studied for the endogenous control and each one of the target genes. For each target gene the cycle threshold values of the 10 replicates had SD values <0.5. For each experiment all triplicate values of samples and standards had also SD of <0.5 and the mean values of the triplicates were taken as the final result.
Statistical Analysis.
Statistical analysis was performed using the nonpaired Student's t test.
Results
Delta-1 but not Jagged-1 Completely Inhibits the Differentiation of CD34+ Hematopoietic Progenitors into the B Cell Lineage.
To investigate whether distinct Notch ligands have different effects in human B cell differentiation, we have made use of a cell coculture system with the murine stromal cell line S17, which promotes the selective differentiation of CB CD34+ cells into the B cell lineage 27. Expression of Delta-1 or Jagged-1 in S17 cells was induced by transduction of retroviral vectors containing either human Jagged-1 or Delta-1 cDNAs. Protein expression was confirmed in sorted transduced-S17 cells by immunoblotting (Fig. 1 a and b) and immunocytochemical detection (data not shown). To determine whether retroviral transduction per se could affect the capacity of S17 cells to support B cell differentiation, cells transduced with a vector containing only IRES-eGFP sequences (vector alone) were cocultured with CB CD34+ cells during 6 wk. No significant differences were observed in the percentage of the CD19+ CD10+ cells emerging both in parental and transduced stroma at three different time points, indicating that retroviral infection does not disturb the well documented 27 capacity of these stromal cells to promote B cell differentiation (Fig. 1 c). To confirm that the exogenous Jagged-1 and Delta-1 proteins expressed by S17 cells were capable of activating Notch in Notch-expressing cells, parental and transduced S17 cells were used as a support layer for the differentiation of C2 myoblasts. We observed that both Jagged-1 and Delta-1 transduced stromas strongly inhibited the fusion of mononucleated myoblasts into multinucleated myotubes (Fig. 1 d). This phenotype is identical to that observed in the assays using transduced NIH 3T3 that were initially performed to verify the functionality of retroviral constructs (data not shown, see Materials and Methods), and indicative of activation of the Notch pathway mediated by both ligands 11 32 35 36. Analysis of genes targeted by Notch signaling (HES-1, Deltex, E47, and pre-Tα) was further done using quantitative RT-PCR. The study was done in CD34+ cells after 48 h of culture in the presence of Notch ligands and control stroma. As shown in Fig. 1 e, a marked increase in HES-1 expression was observed in CD34+ cells cultured in the presence of Delta-1, when compared with control stromal conditions. In contrast, lower levels of expression were observed for Deltex and pre-Tα. E47 expression was half of that of control cultures in the two samples analyzed for this gene. In Jagged-1 cultures, a modest increase in HES-1 expression was observed in contrast with E47 which was 5.2- and 2.9-fold in the two samples analyzed for this gene. Deltex expression was decreased whereas pre-Tα expression was 1.6 ± 0.6-fold (Fig. 1 e).
CD34+ cells were then cocultured with transduced stromas for 6 wk. Parental or vector-alone-transduced S17 cells were used as controls.
Supernatant cells collected at 2, 4, and 6 wk were counted and analyzed for the expression of cell differentiation antigens by flow cytometry. The increase in the total number of cells cultured with control or Jagged-1 S17 cells was similar at all time points, whereas the expansion of cells cultured on Delta-1 stroma was significantly lower (P < 0.025) (mean values at 6 wk: eightfold in Delta-1 versus 17-fold in Jagged-1 and 20-fold in parental stromas).
The phenotypic analysis of supernatant cells (Table ) showed an abrupt decrease in the percentage of CD34+ cells during the first 2 wk, and maintenance of CD38 at levels similar to those observed in fresh CD34+ cells, in all experimental conditions. As to the myeloid markers CD33 and CD14, it was observed that the percentage of CD33+ cells decreased progressively along the culture period, and the expression of CD14, though having reached 30% at week 4 for cells cultured in parental stroma, dropped to <7% at week 6 in all stromas tested (Table and Fig. 2). The absence of myeloid differentiation in this culture system is in agreement with previous findings using CB CD34+ cells cocultured with unmodified S17 cells 27. It is, however, worth noting that the decrease in CD14 and CD33 expression on cells cultured with Delta-1 stroma occurred earlier than with the other stromas. These differences reached statistical significance at weeks 2 and 4 for CD33 when compared with parental (week 2, P < 0.005; week 4, P < 0.05) and Jagged-1 (week 2, P < 0.05; week 4, P < 0.005) stromas, and at week 4 for CD14 when compared with Jagged-1 transduced stroma (P < 0.0005) (Table ).
Table 1.
CB CD34+ Cells Cocultured with S17 Cells Transduced with Different Notch Ligands
| Day 0 | Week 2 (n = 3) | Week 4 (n = 2–4) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| n = 4 | WT | Vector | Jagged-1 | Delta-1 | WT | Vector | Jagged-1 | Delta-1 | |
| CD34 | 87.1 ± 6.4 | 11.4 ± 5.4 | 16.6 ± 5.0 | 13.5 ± 6.5 | 18.6 ± 10.5 | 3.8 ± 3.3 | 2.6 ± 1.7 | 5.5 ± 3.1 | 5.0 ± 3.2 |
| CD38 | 97.1 ± 0.8 | 96.5 ± 2.7 | 94.6 ± 3.8 | 94.6 ± 3.8 | 97.3 ± 3.9 | 98.3 ± 2.0 | 93.9 ± 3.1 | 95.3 ± 3.6 | 95.9 ± 1.6 |
| CD10/CD19 | 1.9 ± 1.4 | 21.0 ± 5.7 | 10.1 ± 3.9 | 21.7 ± 15.6 | 0 ± 0 | 66.8 ± 18.1 | 58.8 ± 15.7 | 55.5 ± 12.3 | 0 ± 0 |
| CD33 | 77.9 ± 10.4 | 25.2 ± 3.6 | 24.4 ± 3.1 | 25.1 ± 9.1 | 10.4 ± 3.8 | 18.4 ± 10.6 | 17.4 ± 5.9 | 22.5 ± 4.5 | 7.4 ± 2.0 |
| CD14 | 1.4 ± 1.0 | 13.7 ± 5.9 | 16.4 ± 1.4 | 9.7 ± 6.9 | 5.7 ± 1.9 | 17.2 ± 13.5 | 10.7 ± 4.3 | 16.3 ± 3.1 | 4.2 ± 1.3 |
| CD7 | 10.1 ± 2.7 | ND | ND | ND | ND | 9.1 ± 4.4 | 7.6 | 18.9 ± 3.9 | 76.5 ± 1.0 |
| CD56 | 2.4 ± 0.7 | ND | ND | ND | ND | 1.5 ± 0.5 | 3.8 | 2.2 ± 0.7 | 6.0 ± 1.2 |
| CD4/CD8 | 0 ± 0 | ND | ND | ND | ND | 0 ± 0 | 0 | 0 ± 0 | 4.3 ± 2.5 |
| CD3 | 4.9 ± 2.0 | ND | ND | ND | ND | 1.5 ± 0.6 | 1.0 | 3.3 ± 1.4 | 3.6 ± 1.3 |
| Cyt CD3 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Week 6 (n = 2–4) | |||||||||
| WT | Vector | Jagged-1 | Delta-1 | ||||||
| CD34 | 0.5 ± 0.5 | 0 | 4.3 ± 0.5 | 5.0 | |||||
| CD38 | 98.9 ± 0.6 | 96.2 | 95.1 ± 0.1 | 96.0 | |||||
| CD10/CD19 | 94.5 ± 2.8 | 89.6 | 81.9 ± 8.2 | 0 ± 0 | |||||
| CD33 | 2.0 ± 1.5 | 1.5 | 4.7 ± 3.2 | 5.2 ± 1.8 | |||||
| CD14 | 1.0 ± 0.4 | 1.0 | 3.4 ± 3.4 | 2.7 ± 1.7 | |||||
| CD7 | 2.5 ± 1.5 | 5.0 | 7.2 ± 3.4 | 81.8 ± 3.8 | |||||
| CD56 | 3.2 ± 3.2 | 8.4 | 2.5 | 5.5 ± 1.2 | |||||
| CD4/CD8 | 0 ± 0 | 0 | 0 | 2.4 ± 2.4 | |||||
| CD3 | 2.0 ± 0.8 | 3.1 | 1.0 | 8.5 ± 6.5 | |||||
| Cyt CD3 | 0 ± 0 | 0 | 0 ± 0 | 66.2 ± 5.9 | |||||
Expression of cell differentiation antigens (% ± SE) on hematopoietic cells cocultured for 2, 4, and 6 wk with nontransduced and transduced S17 cells. Samples without SE, n = 1. Delta-1, S17 cells transduced with Delta-1 cDNA; Jagged-1, S17 cells transduced with Jagged-1cDNA; ND, not done. Vector, S17 cells transduced with vector alone; WT, parental S17 cell line.
Figure 2.
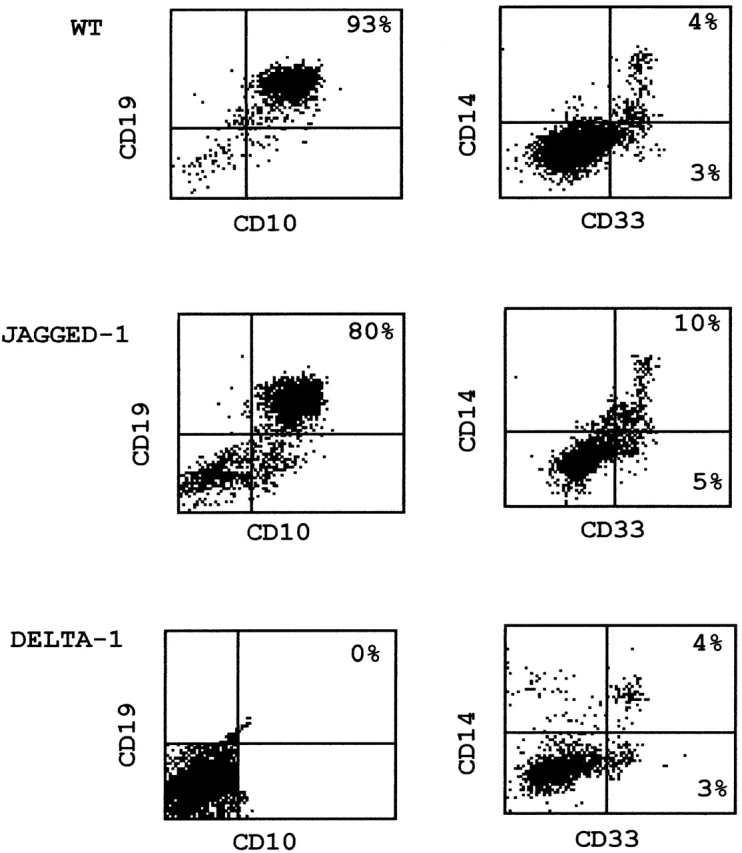
Phenotypic analysis of hematopoietic cell populations found after 6 wk of coculture of CD34+ cells with parental and transduced S17 stroma. Supernatant cells were counter-stained with anti-CD10/anti-CD19 for detection of pre-B cells (left) and with anti-CD14/anti-CD33 for detection of myeloid precursors (right). For the sake of simplicity, since results obtained with vector alone–transduced S17 cells were identical to those observed with parental stroma (see Table ), only the latter are depicted.
As to the expression of the B cell markers CD10 and CD19, a progressive increase in CD10+CD19+ cells was observed during the 6 wk culture of CB progenitors with the control and Jagged-1 stromas. At week 6, >80% of all cells coexpressed CD10 and CD19 (Table ), and expressed variable levels of CD20 although they were negative for IgM (data not shown). This phenotype is compatible with cells at an early stage of B cell differentiation and similar to what has been reported for cultures with unmodified S17 cells 27. However, when CB progenitors were cultured in the presence of Delta-1, no CD10+ CD19+ cells could be detected at any time point (Table and Fig. 2), suggesting a strong inhibitory effect of Delta-1 on early B cell differentiation.
In summary, these data show that, in marked contrast with Jagged-1, exposure of progenitor cells to Delta-1 induces a severe block in B cell development. Additionally, Jagged-1 and Delta-1 do not seem to impair the residual myeloid differentiation observed in this in vitro system.
Coculture of CD34+ Progenitors with Delta-1 Stroma Gives Rise to a Population of Cells with Characteristics of T Cell/NK Precursors.
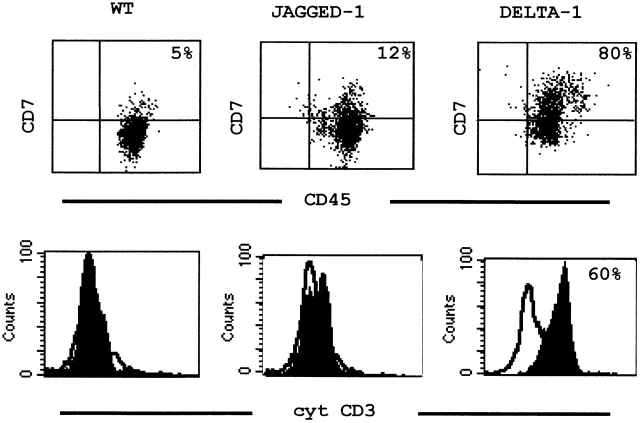
As shown above, the cell population emerging in cultures with Delta-1–expressing stroma did not contain any B cell precursors, nor did it possess features of precursor cells differentiating along the myeloid lineage. A small proportion of T cells (CD3+) and NK cells (CD56+) was found, also detectable in control and Jagged-1 stromas (Table ), which might have resulted from the expansion of contaminating mature cells present at the start of the culture (Table ). The analysis of CD7 and cytoplasmic CD3, two markers of T and NK cell precursors that appear before any other more mature antigens 37, showed that >76% of the cells developing upon coculture with Delta-1 stroma were CD7+, >60% of which coexpressed cytoplasmic CD3. This corresponds to an actual increase in absolute numbers of these cells, when compared with cultures in the other stromas (mean of 1.6 × 105 CD7+ cells in Delta-1 stroma versus 5 × 104 in Jagged-1 stroma). Furthermore, examination of CD4 and CD8 expression in cultures with Delta-1 transduced stroma revealed the presence of a de novo cell population, comprising between 2.4 and 6.8%, that coexpressed CD4 and CD8 (Table and Table ). This was in marked contrast with the findings in cultures with control (P < 0.001) and Jagged-1 (P < 0.0025) stromas where only a minor proportion of cells expressed CD7 at week 6, and no cytoplasmic CD3 or coexpression of CD4 and CD8 was observed (Table and Fig. 3). Therefore, these observations imply that Notch triggering by Delta-1, but not Jagged-1, induces the emergence of two cell populations; a predominant one bearing a phenotype of T and NK cell precursors and a minor one, with a phenotype similar to that observed in developing CD4+ CD8+ thymocytes.
Table 2.
CB CD34+ Cells Cultured with IL-15/IL-7/Flt-3L
| Sample | Antigens | Day 0 | 3w IL-15 | 3w Stroma | 3w Stroma + 3w IL-15 | ||||
|---|---|---|---|---|---|---|---|---|---|
| WT | Jagged-1 | Delta-1 | WT | Jagged-1 | Delta-1 | ||||
| 1 | CD7 | 10.5 | 88.2 | 22.6 | 19.1 | 67.5 | 27.8 | 39.5 | 82.4 |
| CD56 | 1.7 | 69.1 | 1.3 | 2.0 | 3.4 | 56.6 | 59.8 | 77.2 | |
| CD4/CD3 | 1.9 | 24.0 | 0.5 | 0.5 | 1.0 | 9.9 | 10.3 | 10.9 | |
| CD8/CD3 | 1.2 | 8.2 | 0 | 0.5 | 0.5 | 3.5 | 4.7 | 4.2 | |
| CD4/CD8 | 0 | 2.5 | 0 | 0 | 0.5 | 1.4 | 1.1 | 4.0 | |
| 2 | CD7 | 15.1 | 90.3 | 13.4 | 15.3 | 77.5 | 28.4 | 54.7 | 73.6 |
| CD56 | 4.5 | 55.5 | 1.0 | 1.9 | 7.0 | 32.4 | 53.1 | 62.9 | |
| CD4/CD3 | 5.5 | 14.1 | 1.4 | 3.4 | 14.3 | 3.7 | 13.1 | 14.5 | |
| CD8/CD3 | 2.5 | 7.2 | 0.6 | 1.2 | 7.2 | 2.2 | 9.7 | 3.5 | |
| CD4/CD8 | 0 | 1.7 | 0 | 0 | 3.0 | 0 | 0 | 2.8 | |
Data represent percentages of positive cells for expression of the indicated T and NK cell markers. Cellular expansion was identical for cultures with IL-15 but considerably lower for cultures with Delta-1 stroma as discussed in the Results. WT, parental S17 cell line; Jagged-1; S17 cells transduced with Jagged-1 cDNA; Delta-1, S17 cells transduced with Delta-1 cDNA.
Figure 3.
Expression of surface CD7 and cytoplasmic CD3 on progenitor cells cocultured with S17 cells. CB cells were cultured for 6 wk on parental (left), Jagged-1-transduced (middle), or Delta-1–transduced (right) stroma, and analyzed for expression of surface CD7 and cytoplasmic CD3. Histograms of cells stained with the indicated mAbs are depicted in black and cells stained with isotype-matched mouse IgG are depicted in white. Results are representative of two different experiments.
CB CD34+ Cells Cultured on Delta-1 Stroma Are Irreversibly Inhibited to Differentiate into B Cells but Can still Develop into NK Cells upon Culture with IL-15, IL-7, and Flt-3 Ligand.
Next, we tested whether the observed inhibition of B cell differentiation by the Delta-1 stroma was an irreversible phenomenon. With this purpose, CB CD34+ cells were cocultured with Delta-1–expressing stroma for 3 wk, and then transferred to nontransduced stroma and cultured for 3 more wk. No CD10+CD19+ cells were detected in these experimental conditions though, at the time of transference, a proportion of cells (11% in the 2 samples analyzed) still exhibited an immature phenotype as assessed by the expression of CD34 (data not shown). Therefore, after 3-wk culture on Delta-1 stroma, CB cells no longer retain the capacity to develop along the B cell lineage. To exclude the possibility that this was due to a general impairment of the progenitor cells to differentiate into any cell type, cells cocultured with Delta-1–expressing stroma for 3 wk were transferred to a cell suspension system and cultured for 3 more wk in the presence of IL-15, IL-7, and Flt-3L. This cytokine cocktail is known to favor the differentiation of hematopoietic precursors into the NK cell lineage 31. We found that those cells could develop into CD56+ cells, acquiring a phenotype similar to that observed for fresh CB CD34+ cells cultured for 3 wk in suspension with the same cytokines (Table and Fig. 4). A similar behavior was observed for cells cultured on parental and Jagged-1 stromas (Table ), indicating that neither Jagged-1 nor Delta-1 disturbed the capacity of this cytokine combination to either promote differentiation of precommitted NK-cell precursors or expand preexistent NK cells.
Figure 4.
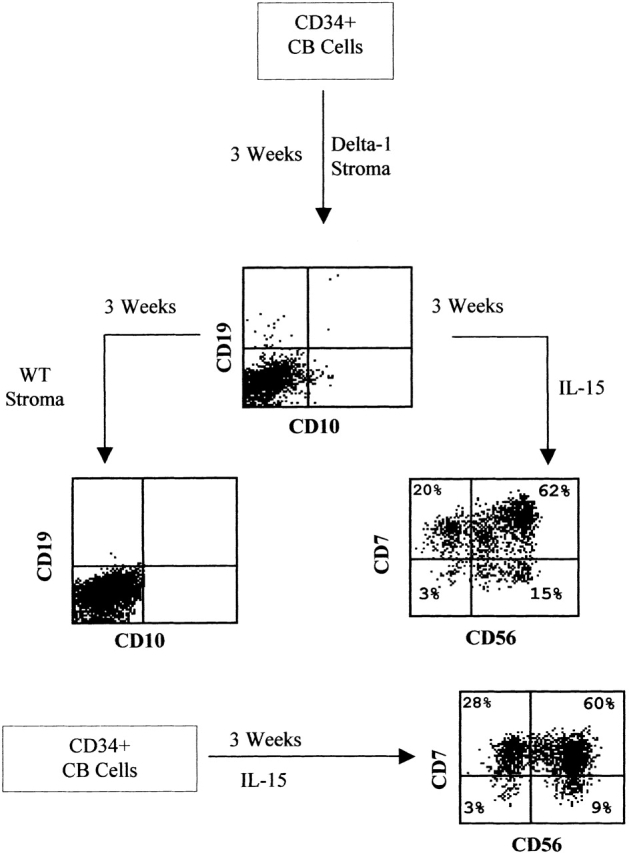
Differentiation capacity of CB progenitors into the B and NK cell lineages after 3 wk coculture with Delta-1–transduced S17 cells. Supernatant cells harvested after 3 wk culture on Delta-1 stroma were transferred to either parental stroma (wt) to test for their B cell differentiation potential, or to a cytokine cocktail consisting of IL-15, IL-7, and Flt-3L (IL-15) to test for their capacity to differentiate into NK cells. (Bottom) Expression of NK cell markers in CD34+ CB cells cultured for 3 wk in suspension with IL-15, IL-7, and Flt-3L.
Discussion
In this report we show that two distinct human Notch ligands, Delta-1 and Jagged-1, have differential effects in human lymphoid differentiation. In a culture system specially suited for the differentiation of hematopoietic progenitors into early B cells, Delta-1 signaling induces a complete and persistent block of B cell differentiation, while allowing the emergence of a cell population with a phenotype of T/NK cell progenitors. In contrast, Jagged-1 did not apparently interfere with the lymphoid developmental potential of hematopoietic progenitors.
These findings highlight a fundamental difference between the actions of two distinct Notch ligands, though constitutive Notch activation in mouse bone marrow cells has been previously shown to induce a persistent block of B cell maturation and the ectopic appearance of early T cells in the bone marrow environment 8. Therefore, the novel observation in the present work is that, in a ligand-dependent context, those effects appear to be ligand-type specific, rather than a common endpoint of Notch triggering by different ligands.
In our study, the block of B cell differentiation in cultures with Delta-1–expressing stromas was evident as early as the second week of culture, and seemingly an irreversible phenomenon, as immature supernatant cells when transferred from transduced to non-modified stroma failed to develop into CD19+CD10+ cells. The predominant cell population emerging in Delta-1–expressing stromas was characterized by the expression of CD7 and cytoplasmic CD3, features compatible with those of T/NK cell progenitors 37. This population remained phenotypically stable between the fourth and sixth wk of culture, and registered a moderate growth in the same period, suggesting that Delta-1, though possibly having an active role in the generation and maintenance of these cells, does not behave as a strong mitogenic factor. Importantly, the absolute numbers of CD7-expressing cells were significantly higher in Delta-1 cultures, when compared with those with other stromas. This indicates that Delta-1 induces a preferential expansion of CD7+ cells in this experimental system. Furthermore, when these cells were transferred to medium containing IL-15, IL-7, and Flt-3L, it became clear that rather than being frozen in that particular stage of differentiation they were capable of developing at least towards the NK cell lineage. This is in agreement with the observation that constitutive Notch activation in hematopoietic precursors establishes immortalized cell lines which are still capable of cytokine-dependent differentiation into lymphoid or myeloid cell lineages 38.
In contrast with Delta-1, Jagged-1–expressing stromas consistently gave rise to cells with an early B cell phenotype, similar to those observed in cultures with control stromas. Acquisition of CD56 was also observed when supernatant cells previously grown with Jagged-1 stroma were transferred to IL-15–containing medium. Thus, Jagged-1 does not appear to significantly disturb neither B cell nor NK potential of CD34+ progenitor cells. Jagged-1 is known to promote the survival of primitive fetal and bone marrow precursors 18 23 and to behave as a growth factor of human hematopoietic stem cells 39. The present data suggest that Jagged-1 also allows the survival and expansion of both early B and early T/NK cell progenitors in vitro. As Jagged-1 is expressed in bone marrow and thymic stromas (for a review, see references 2 and 40) a possibility is that Jagged-1 might contribute to the survival of developing cells belonging to different hematopoietic lineages in distinct environmental niches.
As to the biological mechanisms that may underlay the differential effects of Delta-1 and Jagged-1 on B cell development observed in this study, at least two (nonmutually exclusive) possibilities can be envisaged. One is that the observed absence of B cells in cultures with Delta-1–expressing stromas might partly be due to specific apoptosis of B cell progenitors. Supporting this idea is the recent finding that constitutive activation of Notch-1 induces cell-cycle arrest and apoptotic cell death in a chicken B cell line 41. Furthermore, a specific role for Delta-1 in apoptosis has been previously proposed in studies where Delta-1 was shown to induce apoptosis in monocytic cells 42. This effect could additionally explain our observation that CD33+ myeloid progenitors disappear faster in Delta-1 cultures when compared with other cultures. It is also known that Notch activation provides antiapoptotic signals in developing T cells 3. It thus appears that Notch signaling is capable of differentially regulating cell death in B and T lymphoid cell lineages. If so, the present data would further suggest that such differential effects might depend on Notch-triggering by distinct ligands, Delta-1 possibly mediating apoptosis in B cells only. Apart from a putative effect on cell apoptosis, another possibility is that Notch-triggering by Delta-1 (but not by Jagged-1) might deliver a B cell differentiation inhibitory signal to a common lymphoid precursor which would then choose the alternative T/NK-cell fate. Two candidate downstream targets of Notch activation have been convincingly implicated in the regulation of B and T cell development (for a review, see reference 43): HES-1, which is upregulated in cells undergoing T cell lineage specification and is required for the development of normal T cell numbers; and E47, a product of the E2A gene, which is essential for B cell specification. Since Notch signaling was shown to inhibit E47 activity 44, and mice lacking E2A display a complete block of B cell differentiation 45, it is possible that the inhibition of B cell development observed in this study might be caused by Delta-1–mediated repression of E47. It is in this respect noteworthy that, as opposed to Jagged-1, which is present in bone marrow and thymic stroma 2 40, Delta-1 appears to be expressed at low levels 19 46 or not at all 40 by bone marrow stromal cells, though it is detected in murine thymus (reference 26 and unpublished results). On the other hand, Delta-1 activation of Notch signaling is known to result in HES-1 transactivation 32, an effect that could explain the accumulation of cells with a phenotype of T/NK progenitors in Delta-1–expressing stromas as described in this paper. Our results on the expression of Notch target genes appear to support this idea. The expression of HES-1 in CD34+ cells cultured in the presence of Delta-1, when compared with cells cultured with unmodified stroma, is markedly upregulated in contrast with E47 (which was 0.5-fold in the two samples analyzed). It is conceivable that E47 might be functionally repressed by HES-1 in these cells as it is known that this protein associates with E47 inhibiting its DNA-binding activity 47. In contrast, the expression of Deltex, other known inhibitor of E47 was virtually unchanged in Delta-1 cultures. A different pattern was observed in CD34+ cells cultured in the presence of Jagged-1 where HES-1 expression was only modestly increased. Interestingly, our results on HES-1 expression in Jagged-1 cultures are reminiscent of those of Shawber et al. 35 and Nofziger et al. 36 in their studies on the role of Notch in muscle cell differentiation. These authors observed that Jagged-1, despite inhibiting myogenesis (as observed in this study), did not upregulate HES-1 expression in myoblast precursor cells 35. Furthermore, evidence was provided for the existence of a CBF-1 independent (and yet unknown) Notch signaling pathway induced by Jagged 1 36. Thus, it is possible that Jagged-1 might also act through a CBF-1 independent pathway in hematopoietic cells. An increased expression of E47 accompanied by a decrease in the expression of Deltex was observed in Jagged-1 cultures what is in agreement with the development of early B cells in these conditions. As to the expression of pre-Tα, although it has been reported that T cell lines 6 and bone marrow precursor cells 48 transduced with Notch IC upregulate its expression, we observed that transcripts of this gene were either unchanged or slightly increased both in Jagged-1 and Delta-1 cultures. As these results were obtained in a short time culture assay (when CD34+ cells have not yet acquired any T cell marker), a possibility is that upregulation of pre-Tα expression may occur at later time points of culture, when cells have reached a more advanced developmental stage. Studies aimed at clarifying this issue are now under way.
In conclusion, our results show that the differential effects of Notch signaling during B and T cell development may be mediated by distinct Notch ligands and introduce a novel in vitro system with which cell-fate decision phenomena in lymphopoiesis can be further addressed.
Acknowledgments
The authors are grateful to Dr. H. Spits for the LZRS-polylinker-IRES-eGFP retroviral vector and to Dr. António Coutinho (Instituto Gulbenkian de Ciência, Oeiras, Portugal) for the critical review of the manuscript and helpful comments.
This work was supported by a PRAXIS Program grant (SAU/14000/98) and A. Jaleco and H. Neves by PRAXIS postdoctoral and PhD fellowships, respectively.
Footnotes
Abbreviations used in this paper: CB, cord blood; eGFP, enhanced green fluorescent protein; NK, natural killer.
A.C. Jaleco and H. Neves contributed equally to this work.
References
- Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signalingcell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Milner L.A., Bigas A. Notch as a mediator of cell fate determination in hematopoiesisevidence and speculation. Blood. 1999;93:2431–2448. [PubMed] [Google Scholar]
- Osborne B., Miele L. Notch and the immune system. Immunity. 1999;11:653–663. doi: 10.1016/s1074-7613(00)80140-5. [DOI] [PubMed] [Google Scholar]
- Robey E., Chang D., Itano A., Cado D., Alexander H., Lans D., Weinmaster G., Salmon P. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–492. doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- Washburn T., Schweighoffer E., Gridley T., Chang D., Fowlkes B.J., Cado D., Robey E. Notch activity influences the αβ versus γδ T cell lineage decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- Deftos M.L., Huang E., Ojala E.W., Forbush K.A., Bevan M.J. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer A., Bakker T., Wilson A., Nicolas M., Ioannidis V., Littman D.R., Wilson C.B., Held W., MacDonald H.R., Radtke F. Inactivation of Notch1 in immature thymocytes does not perturb CD4 or CD8 T cell development. Nat. Immunol. 2001;2:235–241. doi: 10.1038/85294. [DOI] [PubMed] [Google Scholar]
- Pui J.C., Allman D., Xu L., DeRoco S., Karnell F.G., Bakkour S., Lee J.Y., Kadesch T., Hardy R.R., Aster J.C., Pear W.S. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- Radtke F., Wilson A., Stark G., Bauer M., Van Meerwijk J., MacDonald H.R., Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Kondo M., Weissman I., Akashi L.K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Lindsell C.E., Shawber C.J., Boulter J., Weinmaster G. Jaggeda mammalian ligand that activates Notch1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- Schawber C., Boulter J., Lindsell C.E., Weinmaster G. Jagged 2a serrate-like gene expressed during rat embryogenesis. Dev. Biol. 1996;80:370–376. doi: 10.1006/dbio.1996.0310. [DOI] [PubMed] [Google Scholar]
- Luo B., Aster J.C., Hasserjian R.P., Kuo F., Sklar J. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol. Cell. Biol. 1997;17:6057–6067. doi: 10.1128/mcb.17.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettenhausen B., Hrabe de Angelis M., Simon D., Guenet J., Gossier A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- Gray G.E., Mann R.S., Mitsiadis E., Henrique D., Carcangiou M.-L., Banks A., Leiman J., Ward D., Ish-Horowitz D., Artavanis-Tsakonas S. Human ligands of the Notch receptor. Am. J. Pathol. 1999;154:785–794. doi: 10.1016/S0002-9440(10)65325-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwoodie S.L., Henrique D., Harrison S.M., Bedington R.S.P. Mouse Dll3a novel divergent Delta gene which may complement the function of other Delta homologue during early pattern formation in the mouse embryo. Development. 1997;124:3065–3076. doi: 10.1242/dev.124.16.3065. [DOI] [PubMed] [Google Scholar]
- Li L., Milner L.A., Deng Y., Iwata M., Banta A., Graf L., Marcovina S., Friedman C., Trask B.J., Hood L., Torok-Storb B. The human homolog of rat Jagged, h-Jagged1, is expressed by marrow stroma and inhibits differentiation of 32D cells through interaction with Notch1. Immunity. 1998;8:43–55. doi: 10.1016/s1074-7613(00)80457-4. [DOI] [PubMed] [Google Scholar]
- Varnum-Finney B., Purton L.E., Yu M., Brashem-Stein C., Flowers D., Staats S., Moore K.A., Le Roux I., Mann R., Gray G. The Notch ligand, Jagged-1, influences the development of primitive hematopoietic precursor cells. Blood. 1998;11:4084–4091. [PubMed] [Google Scholar]
- Jones P., May G., Healy L., Brown J., Hoyne G., Delassus S., Enver T. Stromal expression of Jagged-1 promotes colony formation by fetal hematopoietic progenitor cells. Blood. 1998;92:1505–1511. [PubMed] [Google Scholar]
- Carlesso N., Aster J.C., Sklar J., Scadden D.T. Notch-1 induced delay of human hematopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. Blood. 1999;93:838–848. [PubMed] [Google Scholar]
- Tsai S., Fero J., Bartelmez S. Mouse Jagged 2 is differentially expressed in hematopoietic progenitors and endothelial cells and promotes the survival and proliferation of hematopoietic progenitors by direct cell-to-cell contact. Blood. 2000;96:950–957. [PubMed] [Google Scholar]
- Schroeder T., Just U. Notch signaling via RBP-J promotes myeloid differentiation. EMBO J. 2000;19:2558–2568. doi: 10.1093/emboj/19.11.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L., Lynch M., Silverman S., Fraser J., Boulter J., Weinmaster G., Gasson J.C. The Notch/Jagged pathway inhibits proliferation of human hematopoietic progenitors in vitro. Stem Cells. 1999;17:162–171. doi: 10.1002/stem.170162. [DOI] [PubMed] [Google Scholar]
- Jiang R., Lan Y., Chapman H.D., Shawber C., Norton C.R., Serreze D.V., Weinmaster G., Gridley T. Defects in limb, craniofacial, and thymic development in Jagged-2 mutant mice. Genes Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez E., Vicente A., Sacedón R., Muñoz J.J., Weinmaster G., Zapata A.G., Varas A. Distinct mechanisms contribute to generate and change the CD4:CD8 cell ratio during thymus developmenta role for the Notch ligand, Jagged1 J . Immunol. 2001;166:5898–5908. doi: 10.4049/jimmunol.166.10.5898. [DOI] [PubMed] [Google Scholar]
- Han W., Ye Q., Moore M.A.S. A soluble form of human Delta-like-1 inhibits differentiation of hematopoietic cells. Blood. 2000;95:1616–1625. [PubMed] [Google Scholar]
- Rawlings D.J., Quan S.G., Kato R. M., Witte O.N. Long-term culture system for selective growth of human B-cell progenitors. Proc. Natl. Acad. Sci. USA. 1995;92:1570–1574. doi: 10.1073/pnas.92.5.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleco A.C., Stegman A.P.A., Heemskerk M.H.M., Couwenberg F., Bakker A.Q., Weijer K., Spits H. Genetic modification of human B-cell developmentB-cell development is inhibited by the dominant negative helix loop helix factor Id3. Blood. 1999;94:2637–2646. [PubMed] [Google Scholar]
- Heemskerk M.H., Hooijberg E., Ruizendaal J.J., van der Weide M.M., Bakker A.Q., Schumacher T.N., Spits H. Enrichment of an antigen-specific T cell response by retrovirally transduced human dendritic cells. Cell. Immunol. 1999;195:10–17. doi: 10.1006/cimm.1999.1520. [DOI] [PubMed] [Google Scholar]
- Heemskerk M.H.M., Blom B., Nolan G., Stegmann A.P.A., Bakker A.Q., Weijer K., Res P.C.M., Spits H. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J. Exp. Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleco A.C., Blom B., Res P., Wijer K., Lanier L.L., Phillips J.H., Spits H. Fetal liver contains committed NK progenitors, but is not a site for development of CD34+ cells into T cells. J. Immunol. 1997;159:694–702. [PubMed] [Google Scholar]
- Jarriault S., Bail O., Hirsinger E., Pourquié O., Logeat F., Strong C.F., Brou C., Seidah N.G., Israel A. Delta-1 activation of Notch-1 signaling results in HES-1 transactivation. Mol. Cell. Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho M., Krauss R.D., Chiang L., Valcárcel J., Green M.R., Carmo-Fonseca M. Targetting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gameiro P., Vieira S., Carrara P., Silva A.L., Diamond J., Botelho de Sousa A., Mehta A., Prentice H.G., Guimarães J.E., Hoffbrand A.V. The PML-RARα transcript in long-term follow-up acute promyelocytic leukaemia (APL) patients. Hematologica. 2001;86:577–585. [PubMed] [Google Scholar]
- Shawber C., Nofziger D., Hsieh J.J.-D., Lindsell C., Bogler O., Hayward D., Weinmaster G. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- Nofziger D., Miyamoto A., Lyons K.M., Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- Spits H., Lanier L.L., Phillips J.H. Development of human T and natural killer cells. Blood. 1995;85:2654–2670. [PubMed] [Google Scholar]
- Varnum-Finney B., Xu L., Brashem-Stein C., Nourigat C., Flowers D., Bakkour S., Pear W.S., Bernstein I.D. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat. Med. 2000;11:1278–1281. doi: 10.1038/81390. [DOI] [PubMed] [Google Scholar]
- Karanu F.N., Murdoch B., Gallacher L., Wu D.M., Koremoto M., Sakano S., Bhatia M. The Notch ligand Jagged-1 represents a novel growth factor of human hematopoietic stem cells. J. Exp. Med. 2000;192:1365–1372. doi: 10.1084/jem.192.9.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand F.E., Eckfeldt C.E., Fink J.R., Lysholm A.S., Pribyl J.A.R., Shah N., LeBien T.W. Microenvironmental influences on human B-cell development. Immunol. Rev. 2000;175:175–186. [PubMed] [Google Scholar]
- Morimura T., Goitsuka R., Zhang Y., Saito I., Reth M., Kitamura D. Cell-cycle arrest and apoptosis induced by Notch1 in B cells. J. Biol. Chem. 2000;275:36523–36531. doi: 10.1074/jbc.M006415200. [DOI] [PubMed] [Google Scholar]
- Ohishi K., Varnum-Finney B., Flowers D., Anasetti C., Myerson D., Bernstein I.D. Monocytes express high amounts of Notch and undergo cytokine specific apoptosis following interaction with the Notch ligand, Delta-1. Blood. 2000;95:2847–2854. [PubMed] [Google Scholar]
- Rothenberg E.V. Stepwise specification of lymphocyte developmental lineages. Curr. Opin. Genet. Dev. 2000;10:370–379. doi: 10.1016/s0959-437x(00)00098-8. [DOI] [PubMed] [Google Scholar]
- Ordentlich P., Lin A., C-P. Shen C., Blaumueller K., Matsuno S., Artavanis-Tsakonas, Kadesch T. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol. 1998;18:2230–2239. doi: 10.1128/mcb.18.4.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G., Maandag E.C., Izon D.J., Amsen D., Kruisbeek A.M., Weintraub B.C., Krop I., Schlissel M.S., Feeney A.J., van Roon M. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- Karanu F.N., Murdoch B., Miyabayashi T., Ohno M., Koremoto M., Gallacher L., Wu D., Itoh A., Sakano S., Bhatia M. Human homologues of Delta-1 and Delta-4 function as mitogenic regulators of primitive human hematopoietic cells. Blood. 2001;97:1960–1967. doi: 10.1182/blood.v97.7.1960. [DOI] [PubMed] [Google Scholar]
- Sasai Y., Kageyama Y., Tagawa Y., Shigemoto R., Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- Allman D., Karnell F.G., Punt J.A., Bakkour S., Xu L., Myung P., Koretzky G.A., Pui J.C., Aster J.C., Pear W.S. Separation of Notch1 promoted lineage commitment and expansion/transformation in developing T cells. J. Exp. Med. 2001;194:99–106. doi: 10.1084/jem.194.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]