Abstract
The expression of the pre-T cell receptor α (pTa) gene occurs exclusively in immature T lymphocytes and is regulated by poorly defined mechanisms. We have analyzed the role of the upstream enhancer in pTa expression using conventional and bacterial artificial chromosome (BAC) reporter transgenes. The deletion of the enhancer completely abolished the expression of pTa BAC reporter in transgenic mice. Conversely, the combination of pTa enhancer and promoter targeted transgenes specifically to immature thymocytes, recapitulating the expression pattern of pTa. The core enhancer is conserved between mice and humans and contains a critical binding site for the transcription factor c-Myb. We also show that pTa promoter contains a conserved tandem E box site activated by E protein, HEB. These data establish the enhancer as a critical element regulating pTa gene expression and identify additional targets for c-Myb and E proteins in T cell development.
Keywords: transcription, thymus, c-Myb, basic helix-loop-helix proteins, HEB
Introduction
The development of T lymphocytes involves multiple steps of progressive cellular differentiation, selection and lineage commitment. Several of these steps occur at the earliest CD4−CD8− (double-negative [DN]) stage of T cell development in the thymus 1 2. First, thymic progenitors undergo full commitment to the T cell lineage and start rearrangement of the TCRβ, γ, and δ genes. Second, T cells with productive TCRβ rearrangement receive a selective survival signal from the pre-TCR at the so called β-selection checkpoint. Finally, the choice is made between the predominant T cell lineage expressing α/β TCR, and a specialized population of T cells expressing γ/δ TCR. Despite recent advances 3 4, transcriptional mechanisms regulating these processes are not fully understood.
Commitment to the T cell lineage depends on several transcription factors including GATA-3 5 and c-Myb 6. C-Myb is expressed in immature hematopoietic cells 7 and has been shown to activate several T cell–specific regulatory elements 8 9 10 11 12 13. Recently, it was demonstrated that T cell lineage commitment requires a signal from Notch receptors 14 15 and their downstream effector Hes1 16. Furthermore, T cell commitment requires the activity of basic helix-loop-helix (bHLH) transcription factors 17. In particular, bHLH proteins E2A and HEB regulate early T cell development at multiple steps 18 19. Members of the E protein family of bHLH factors, these proteins bind their target E box (CANNTG) elements as homo- or heterodimers, and are abundantly expressed in thymocytes 20. In addition to their role in T cell commitment, Notch signaling and E proteins activity appear to favor α/β over γ/δ T cell development 21 22. The identification of target genes and regulatory sites for these factors should establish the molecular basis of lineage commitment and stage-specific gene expression in T cell development.
The pre-TCRα (pTa) gene encodes a critical component of the pre-TCR complex in DN thymocytes undergoing β-selection 1. Signaling from the pre-TCR is required for the expansion of T cells with functional TCRβ products and for efficient TCRβ allelic exclusion, and is thought to promote commitment to the α/β T cell lineage 23. Consistent with its function, the expression of pTa is restricted to immature T cells in the thymus and in extrathymic sites of T cell development 24 25. In the thymus, pTa expression is highest in the DN population and decreases during differentiation into immature CD8+ single-positive (ISP) and then into CD4+CD8+ double-positive (DP) thymocytes. Subsequently, pTa expression is silenced at the transition to mature single-positive (SP) thymocytes and is absent from mature peripheral T cells or other cell types. Within the DN thymocyte subset, pTa is expressed at low levels in uncommitted thymic CD44+ CD25− (DN1) precursors. The major upregulation of pTa message coincides with commitment to T cell lineage at the CD44+CD25+ (DN2) stage 2 24. The expression reaches its peak in CD25+CD44− (DN3) cells undergoing β-selection and is downregulated in CD25−CD44− (DN4) postselection thymocytes. Therefore, the expression of pTa may serve as a marker and possibly one of the determinants of T cell commitment, representing an important model for the transcriptional regulation of early T call development.
Consistent with this notion, pTa was recently shown to be upregulated by Notch signaling 26 and by E proteins 22, particularly by HEB 27. However, the mechanism of these and other aspects of pTa transcriptional regulation remains to be investigated. To address this issue, we sought to identify elements responsible for tissue- and stage-specific expression of pTa, as well as important regulatory sites within these elements. We previously reported that the genomic region upstream of the mouse pTa gene contains specific DNase-hypersensitive sites and targets transgene expression to the thymus. Within this region, we identified a proximal promoter and an enhancer element located 4 kb upstream of the promoter 28. We now report that the enhancer is a critical element regulating pTa expression in immature T cells. We also identify conserved functional sites in the enhancer and promoter that are activated by c-Myb and HEB, respectively. These findings begin to establish the molecular basis for the cell- and stage-specific expression of pTa.
Materials and Methods
Transgenic Constructs.
A 160-kb bacterial artificial chromosome (BAC) clone containing the mouse pTa locus was modified using ET recombination as described by Stewart and colleagues 29 using the reagents provided by the authors. A fragment of the first pTa exon including part of the 5′ UTR, the initiation codon and most of the coding sequence (positions 570–651 of GenBank/EMBL/DDBJ entry U27268) was replaced by a fragment containing enhanced green fluorescent protein (EGFP; CLONTECH Laboratories, Inc.), BGH polyA signal, and an EM7-Sh ble prokaryotic Zeo r cassette (Invitrogen) flanked by FRT sites. The Zeo r cassette was subsequently removed using FLP-expressing bacterial strain 294-FLP. In the second round of recombination, the pTa upstream enhancer fragment (positions 1,366–1,697 of GenBank/EMBL/DDBJ entry AF132612) was replaced by the Zeo r cassette. Correct targeting and integrity of the BAC clones were confirmed by PCR and restriction analysis using conventional and pulsed-field gel electrophoresis. The EGFP-containing BAC clones were digested with NotI, which released an 85-kb fragment containing 30 kb 5′ and 45 kb 3′ of the pTa gene as determined by long-range mapping.
The enh-EGFP reporter construct contained the following fragments assembled in pBluescript: two 1.2-kb chicken β-globin insulator fragments 30; a 1.9-kb XbaI-EcoRI pTa enhancer fragment; a 0.27-kb pTa promoter fragment; EGFP gene; and BGH polyA signal. For the enh/Amut-EGFP construct, the mutation depicted in Fig. 5 A was introduced into the enhancer fragment before subcloning using QuikChange system (Stratagene), and verified by sequencing. The constructs were linearized with XhoI and NotI and purified from the vector backbone.
Figure 5.
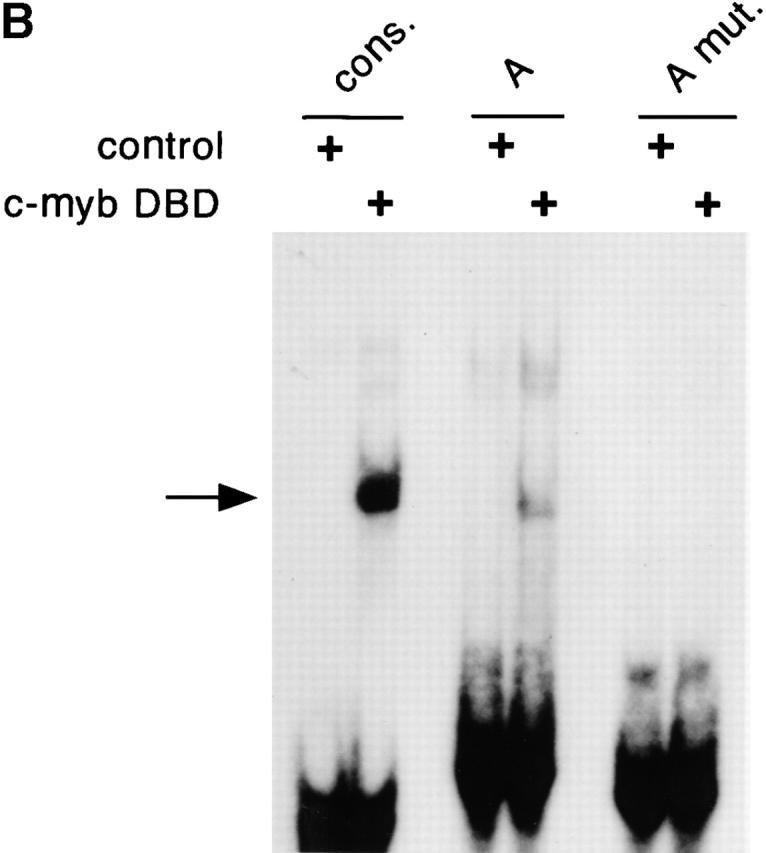
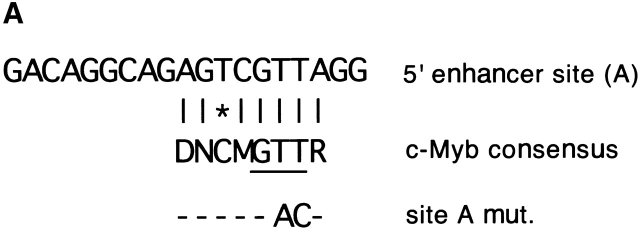
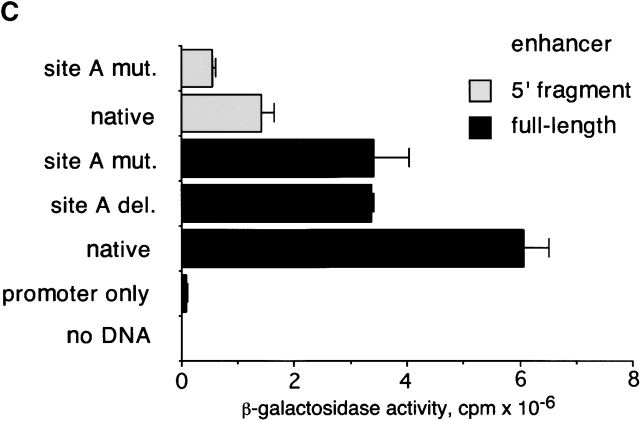
The binding of c-Myb to the 5′ enhancer site (site A). (A) Sequence of site A aligned to the c-Myb consensus binding sequence (reference 50) (D = not C, M = A or C, R = A or G, N = any base; the three bp core sequence is underlined). The mismatch is indicated by an asterisk. The mutation (mut.) introduced into the putative c-Myb binding site is shown. (B) Binding of the c-Myb DNA binding domain (DBD) to site A. In vitro translated B42 domain alone (control) or its fusion with c-Myb DBD were incubated with labeled oligonucleotides containing a synthetic consensus c-Myb binding site or native or mutated site A. Protein-DNA complexes were visualized by EMSA; the arrow indicates a c-Myb DBD/DNA complex. (C) Effect of the mutation in the c-Myb binding site on pTa enhancer activity. The mutation or deletion (del.) of site A was introduced into the full-strength minimal enhancer fragment (black boxes) or a partially disabled 5′ enhancer fragment (gray boxes). Constructs containing these fragments upstream of SV40 promoter/LacZ were transiently transfected into the pre-T cell line LR1, and β-galactosidase activity was determined.
For the hCD25-based reporter constructs, the following fragments were assembled using the SalI-SmaI fragment of pβGal vectors (CLONTECH Laboratories, Inc.) as a backbone: a 2.8-kb XbaI or a 0.25-kb BstEII-MluNI fragment containing pTa enhancer; a 0.5-kb HindIII-PstI pTa promoter fragment; and a 3-kb BamHI-SalI fragment of pKCR.H-2Kd.hIL-2R vector containing hCD25 cDNA within a promoterless β-globin exon-intron structure 31. The resulting constructs were linearized with SalI.
Analysis of Transgenic Mice.
The constructs were microinjected into fertilized oocytes of FVB mice, and transgenic offspring were identified by PCR using primers specific for EGFP or hCD25. Transgene copy number was determined by Southern hybridization of BamHI-digested genomic DNA with pTa promoter probe detecting germline and transgenic fragments of different size. Transgenic founders were analyzed directly, except for BAC transgenes, in which case hemizygous F1 animals were analyzed.
The lymphoid cells from transgenic mice were stained with direct antibody conjugates and analyzed using a FACSCalibur™ flow cytometer (BD PharMingen). For the analysis of EGFP expression, thymocytes were stained with mAb to CD3 (PE), CD8 (peridinine chlorophyll protein [PerCP]), and CD4 (allophycocyanin [APC]). For the analysis of EGFP expression in DN thymocyte subsets, cells were stained with mAb to CD25 (PE), CD44 (Cy-Chrome), and a cocktail of mAb to CD3, CD4, CD8, B220, and Mac-1 (APC). Splenocytes were stained with mAb to CD3 (PE) and B220 (APC). Bone marrow cells were stained with mAb to CD19 (PE) and B220 (APC). For the analysis of hCD25 expression, lymphocytes were stained with mAb to hCD25 (PE) and CD3 or B220 (FITC), or with mAb to hCD25 (APC), CD4 (PE), and CD8 (FITC).
Reporter Constructs.
A 0.15-kb PpuMI-PstI pTa promoter fragment or a 0.45-kb CD3δ promoter 32 were cloned into the promoterless β-galactosidase (LacZ) reporter vector pβGal-Basic (CLONTECH Laboratories, Inc.). In other constructs, tested fragments were inserted into the pβGal-promoter vector upstream of the SV40 early promoter and LacZ. These included a 0.25-kb BstEII-MluNI pTa enhancer fragment, a 0.6-kb CD3δ enhancer fragment 32, and oligonucleotides containing one or four copies of the 17-bp pTa enhancer site A (GACAGGCAGAGTCGTTA). Four copies of the 13-bp site A (GGCAGAGTCGTTA), either native or containing a mutation shown on Fig. 5 A, were inserted upstream of the SV40 TATA box and luciferase reporter in a modified pGL3-Basic luciferase reporter vector (Promega).
The following deletion fragments of the BstEII-MluNI enhancer 28 were used: 82–257 (a full-strength core fragment); 101–257 (deletion of the 5′ enhancer site); and 1–158 (a partially disabled fragment truncated at the 3′ end). A 0.3-kb human pTa enhancer fragment (position 39,422–39,728 of GenBank/EMBL/DDBJ entry HS475N16) was amplified by PCR from BAC clone RP3–475N16 (Research Genetics). Site-directed mutagenesis of promoter and enhancer fragments within reporter constructs was performed using QuikChange system, and the resulting constructs were verified by sequencing. Full-length mouse c-Myb or human HEB cDNA were cloned into the pCAGGS mammalian expression vector 33.
Transfection and Reporter Assays.
Cell lines LR1 (pTa-positive pre-T cell lymphoma; reference 28) and BW5147 (pTa-negative T cell lymphoma) were transfected in duplicate using Fugene 6 reagent (Roche Molecular Biochemicals). The β-galactosidase and/or luciferase activities were determined 24 h later using the corresponding chemiluminescent assays (Tropix). The ranges of duplicate samples were <15% of mean values. Data represent either mean cpm ± range or the ratio of mean cpm values. For the cotransfection experiments, LR1 cells were transfected with a β-galactosidase reporter construct, the expression vector (HEB or empty vector), and pGL3-control luciferase expression vector (Promega). The activity of β-galactosidase was normalized by the luciferase activity in the same lysates.
For stable transfection, BW5147 cells were electroporated in the presence of linearized pCAGGS/c-Myb and neo r cassette vector KT3NP4 at a 10:1 ratio. After 2 d of culture, cells were selected in the presence of 1 mg/ml G418 in 96-well plates, and individual clones were expanded. For the detection of c-Myb protein, total cell lysates prepared from 5 × 105 cells were analyzed by Western blotting using the c-Myb polyclonal antibody M-19 (Santa Cruz Biotechnology, Inc.).
Electrophoretic Mobility Shift Assay.
The assay was performed as described 28. A fragment of c-Myb cDNA encoding the complete DNA binding domain (amino acids 22–203 of c-Myb) was amplified by PCR and cloned in-frame with a V5 epitope tag and B42 activation domain into the pYESTrp2 vector (Invitrogen) downstream of the T7 promoter. This construct or the pYESTrp2 vector alone was translated in vitro using the T7 TnT reticulocyte lysate system (Promega). The integrity and size of the translation products were confirmed by Western blotting using an anti-V5 antibody (Invitrogen). Double-stranded oligonucleotide probes included a synthetic consensus c-Myb binding site (Santa Cruz Biotechnology, Inc.), pTa enhancer site A (three different probes containing site A sequence GACAGGCAGAGTCGTTA were used with similar results) or mutated site A (GACAGGCAGAGTCGACAGGGACACCTGCCTC).
Results
The Upstream Enhancer Is Necessary for pTa Expression.
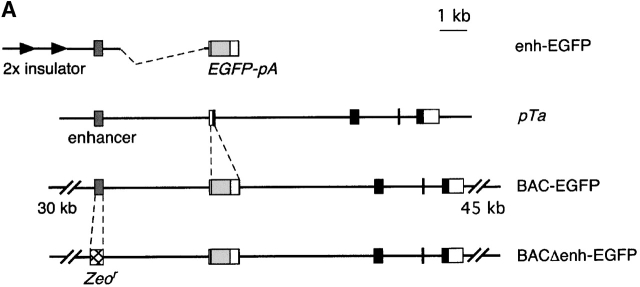
To explore the role of the upstream enhancer in pTa expression, we used a recently developed approach based on bacterial artificial chromosome (BAC) modification by homologous recombination. To create a reporter reflecting pTa locus transcription, we introduced the gene for EGFP into the first exon of the mouse pTa locus in a BAC clone (Fig. 1 A). Subsequenty, we replaced the core 0.3 kb upstream enhancer with a 0.45-kb prokaryotic Zeocin resistance (Zeo r) cassette. As the Zeo r fragment lacks any eukaryotic regulatory sequences, it is unlikely to interfere with the transcription of pTa locus in cells. The EGFP-modified BAC fragments with or without the enhancer deletion (constructs BAC-EGFP and BACΔenh-EGFP, respectively) were injected into mice, and transgenic progeny were analyzed for EGFP expression by flow cytometry.
Figure 1.
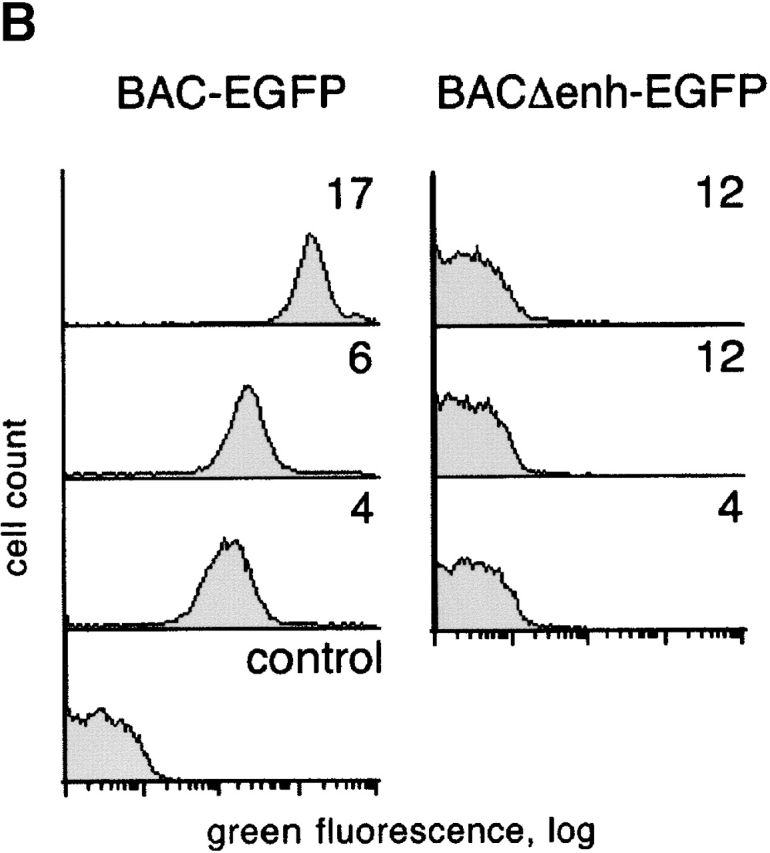
Transgenic analysis of the pTa enhancer function. (A) Map of the pTa locus and of the transgenic constructs used. The coding regions, UTR and the upstream enhancer of pTa are shown as black, white, and dark gray boxes, respectively. All constructs contained a reporter cassette consisting of EGFP cDNA (light gray box) and polyadenylation signal (EGFP-pA). The conventional transgenic construct (enh-EGFP) contained pTa enhancer and promoter fragments flanked at the 5′ end by two copies of the chicken β-globin insulator element (arrows). In the BAC modification scheme, EGFP-pA was introduced into the first pTa exon within a BAC clone. The resulting BAC fragment was used as a 85-kb transgene (BAC-EGFP) or further modified to yield construct BACΔenh-EGFP, in which a 0.3 kb upstream enhancer fragment was replaced by a 0.45-kb prokaryotic Zeor cassette (hatched box). (B) Expression of the EGFP reporter in thymocytes of BAC transgenic mice. Thymocytes were analyzed by four-color flow cytometry, and histograms of green fluorescence in DN3 subset are shown. The progeny of three independent transgenic lines for each construct were analyzed along with a nontransgenic mouse (control); transgene copy number of each line is indicated.
Fig. 1 B illustrates EGFP expression in DN3 thymocytes, which represent the primary pTa-expressing population 24. In the lines carrying a wild-type BAC transgene modified with EGFP, we observed homogeneous EGFP expression with the levels proportional to the transgene copy number. In contrast, no EGFP signal was detected in DN3 cells or any other lymphocyte population in transgenic lines lacking the enhancer. All in all, four transgenic lines were examined for each construct, yielding the same results. A similar replacement of another region upstream of pTa had no effect on EGFP expression in thymocytes, thus ruling out any nonspecific effect of Zeo r cassette insertion (unpublished data). We therefore conclude that the upstream enhancer is indispensable for the expression of the pTa locus in developing thymocytes.
The Enhancer and Promoter Recapitulate the Expression Pattern of pTa.
In a complementary approach, we created a conventional transgenic construct containing pTa enhancer and promoter fragments upstream of EGFP (construct enh-EGFP in Fig. 1 A). Our previous studies revealed that the upstream pTa region is subject to position effects in transgenic mice, resulting in a low frequency of founders expressing the transgene 28. To blunt the effects of transgene integration site, we included two chicken β-globin insulator elements at the 5′ end of the construct. The insulator elements are thought to facilitate the expression of a transgene by blocking the silencing effects of neighboring heterochromatin 30. Indeed, EGFP expression was detected in the thymuses of all six enh-EGFP founders examined, four founders demonstrating high expression levels (Table ). Although heterocellular expression reminiscent of position-effect variegation 34 was observed in enh-EGFP transgenic mice, all founders exhibited a similar pattern of EGFP expression in lymphocyte subsets.
Table 1.
The Expression of EGFP Reporter in enh-EGFP Transgenic Mice
| Founder | Transgene copy number | Percent positive | Fluorescence intensity |
|---|---|---|---|
| enh-EGFP | |||
| I | 1 | 18 | 37 |
| II | 2 | 10 | 111 |
| III | 2 | 43 | 1,245 |
| IV | 2 | 67 | 3,858 |
| V | 2 | 94 | 2,151 |
| VI | 8 | 82 | >9,000 |
| enh/Amut-EGFP | |||
| I | 2 | <1 | − |
| II | 4 | <1 | − |
| III | 6 | <1 | − |
| IV | 9 | 98 | 4,362 |
| V | 10 | <1 | − |
| VI | 11 | 19 | 248 |
Transgenic thymocytes were analyzed by four-color flow cytometry, and data for DN3 subset are presented. The percentage of EGFP-positive cells and the mean fluorescence intensity of the positive peak were determined using fixed fluorescence detector settings. The intensity of the control negative peak was 2. In the enh-EGFP founder VI, the intensity was above the detection range for the given settings.
Fig. 2 illustrates the expression in a representative founder (founder V in Table ) in comparison to the BAC-EGFP transgenic mouse. The BACΔenh-EGFP transgenic mice lacked any detectable EGFP expression in any lymphocyte subset, and a representative expression profile is shown as a negative control. As shown in the figure, the expression patterns of enh-EGFP and BAC-EGFP constructs were essentially identical and consistent with the expression of pTa itself. Within the DN thymocyte subset, the expression was low and heterogeneous in the earliest DN1 T cell precursors, then was upregulated in committed DN2 T cells, reached the highest level in the DN3 subset and decreased afterwards. Within the T cell lineage, the expression was the highest in DN thymocytes and was gradually decreasing during thymocyte maturation into ISP, large DP, small DP, and SP thymocytes. Importantly, mature peripheral T cells manifested a dramatic downregulation of EGFP compared with thymocytes, whereas cells of other lineages such as developing and mature B cells were completely EGFP negative. These data suggest that the pTa enhancer, in conjunction with its cognate promoter, can specify expression pattern similar to that of the 85 kb pTa genomic clone and of the pTa itself. Therefore, the enhancer appears sufficient for the correct tissue- and stage-specific expression of pTa.
Figure 2.
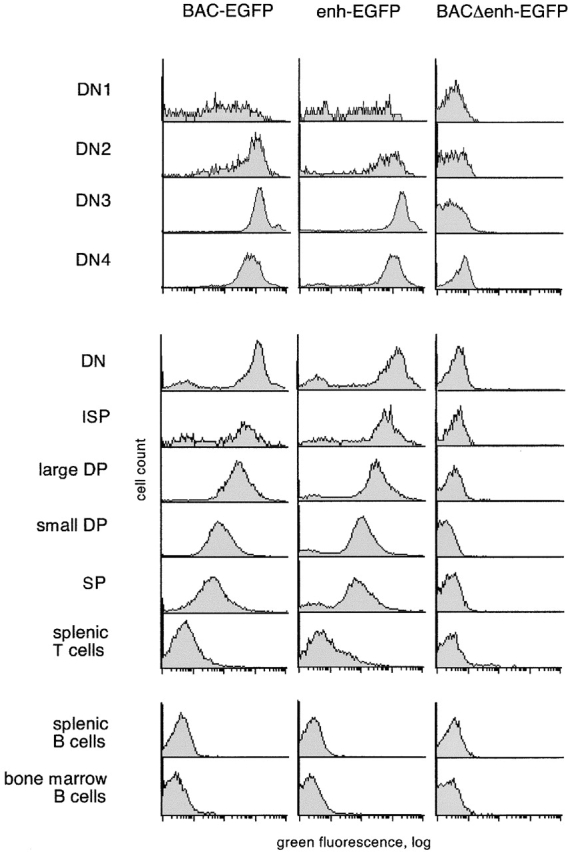
The expression profile of EGFP reporter driven by pTa genomic fragments. Lymphocytes from transgenic mice were analyzed by multicolor flow cytometry, and histograms of green fluorescence in the indicated lymphocyte subsets are shown. The green fluorescence observed in the BACΔenh-EGFP mice is identical to that of the wild-type mice, revealing no detectable EGFP expression. The top panel represents subpopulations of DN thymocytes, the middle panel represents subsets of thymocytes and peripheral T cells, and the bottom panel represents developing (bone marrow) and mature (splenic) B cells. The lymphocyte subsets were defined as follows: thymic DN (CD3−CD4−CD8−), ISP (CD3−CD4−CD8+), large DP (large CD3−CD4+CD8+), small DP (small CD3−/lowCD4+CD8+), SP (CD3highCD4+CD8− and CD3high CD4−CD8+), splenic T cells (CD3high), splenic B cells (B220+CD3−), and bone marrow B cells (B220+CD19+). For thymic DN subsets, thymocytes were stained with a lineage cocktail (CD3/CD4/CD8/B220/Mac1), and lineage-negative cells were gated as DN1 (CD44+CD25−), DN2 (CD44+CD25+), DN3 (CD44−CD25+), and DN4 (CD44− CD25−).
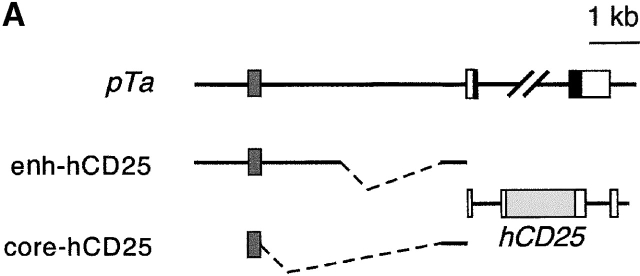
The only discrepancy between the observed versus the expected EGFP expression pattern was the lack of major EGFP downregulation in SP thymocytes and the presence of a minor fraction of peripheral T cells bearing trace levels of EGFP. Because such delayed EGFP downregulation was also observed in BAC-EGFP transgenic mice, it is likely to be caused by the unusual stability of EGFP rather than by an improper regulation of expression. Indeed, the DP/SP transition may not involve extensive proliferation occurring at the earlier differentiation steps, thus preventing EGFP dilution by cell division. To directly address this point, we created transgenic mice bearing an alternative reporter, the human CD25 protein. This reporter does not interfere with mouse lymphoid development 31 and can faithfully reproduce transgene downregulation in mature B lymphocytes 35. As shown in Fig. 3 A, we placed a larger pTa enhancer fragment or the core enhancer (constructs enh-hCD25 and core-hCD25, respectively) upstream of the pTa promoter driving hCD25 minigene expression.
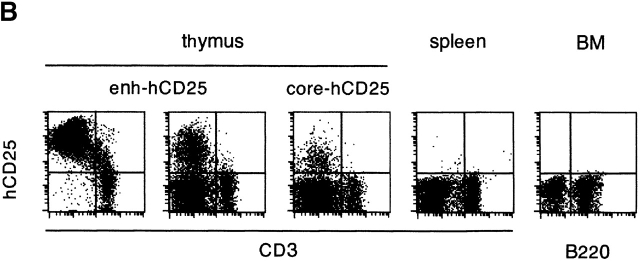
Figure 3.
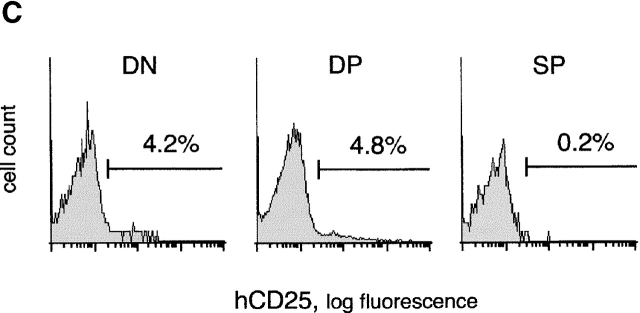
The expression profile of hCD25 reporter driven by pTa regulatory regions. (A) Map of the transgenic constructs used. The constructs contained a larger enhancer fragment (enh-hCD25) or the core enhancer (core-hCD25) upstream of the pTa promoter and human CD25 (hCD25) minigene. (B) Expression of the hCD25 reporter in transgenic mice. Lymphocytes were analyzed by two-color flow cytometry using mAb to hCD25 and to CD3 or B220. Shown are dot plots of thymocytes from two enh-hCD25 founders and a single core-hCD25 founder, and representative dot plots of splenocytes and bone marrow (BM) cells from an enh-hCD25 founder. (C) Expression of the hCD25 reporter in thymocyte subsets of the core-hCD25 transgenic founder. Lymphocytes were stained with mAb to CD4, CD8, and hCD25, and the histograms of hCD25 fluorescence in DN (CD4−CD8−), DP (CD4+CD8+), and SP (CD4−CD8+ and CD4+CD8−) thymocytes are shown. The percentage of hCD25-positive cells in each subset is indicated; the nontransgenic controls contained <0.4% positive cells in each subset.
As expected, in the absence of insulator elements these constructs were subject to severe position effects. Thus, 3 out of 8 enh-hCD25 founders and only 1 out of 10 core-hCD25 founders manifested detectable hCD25 expression in the thymus, the fraction of positive cells ranging widely between the founders. Nevertheless, the hCD25 expression patterns of both reporter constructs were similar and highly specific for immature thymocytes. As shown in Fig. 3 B, hCD25 expression was confined to CD3−/low thymocytes, while CD3high mature thymocytes were largely hCD25 negative. Accordingly, multicolor staining revealed hCD25 expression in DN and DP but not SP thymocytes, as illustrated in Fig. 3 C for the core-hCD25 founder. Moreover, no expression whatsoever could be detected in mature peripheral T cells or in other cell types in the spleen and bone marrow. We conclude that the pTa enhancer and promoter can properly downregulate the reporter expression in SP thymocytes and peripheral T cells, consistent with their primary role in determining the specificity of pTa expression.
The Enhancer Is Conserved between Mice and Humans.
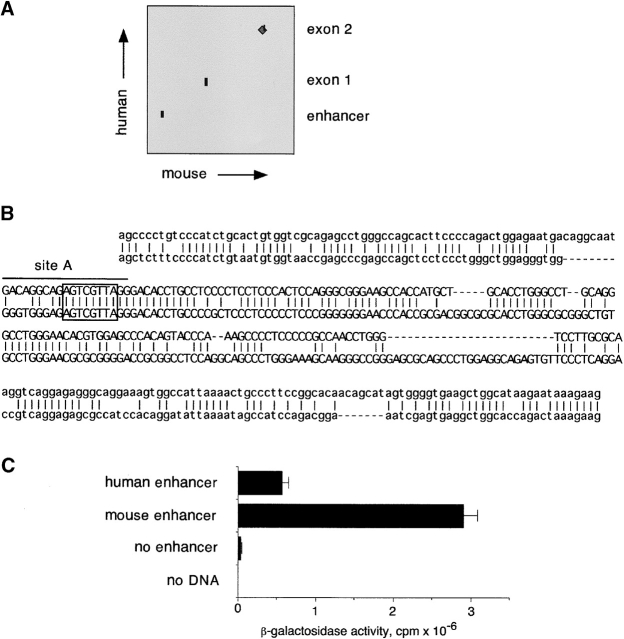
To confirm the importance of enhancer in the regulation of pTa expression, we examined its conservation across different species. To this end, we compared the sequence upstream of the mouse pTa gene 36 with the recently completed sequence of the human PTA locus. As shown in Fig. 4 A, the comparison revealed a conserved putative enhancer fragment located at the same position in the human gene. No other significant homology regions (except exons) were observed within the available sequences. Fig. 4 B illustrates the high degree of homology between the mouse and human enhancer fragments (∼60% identity in a 0.3 kb region). As demonstrated in Fig. 4 C, the human enhancer was active in a mouse pre-T cell line, albeit to a lesser extent than the homologous mouse enhancer fragment. A similarly weaker activity of the human enhancer was observed in the human T cell line MOLT4 (data not shown). The apparently lower intrinsic strength of the human enhancer may result from sequence divergence at its 3′ end, where activating sites are located in the mouse enhancer 28. Overall, the observed strong evolutionary conservation of the upstream pTa enhancer confirms its central role in the regulation of pTa gene expression.
Figure 4.
Identification of the human PTA enhancer. (A) Sequence comparison of the mouse and human pTa loci. The mouse (14 kb; GenBank/EMBL/DDBJ entries AF132612 and U27268) and human (20 kb; positions 25,001–45,000 of GenBank/EMBL/DDBJ entry HS475N16) sequences were aligned using BLAST 2 Sequences software (reference 49) with 20 bp window. Shown is the graphic output of BLAST 2 comparison highlighting conserved regions. (B) Alignment of the mouse (top) and human (bottom) enhancer sequences. The enhancer core as defined by deletion analysis (reference 28) is shown in uppercase letters. The position of a functional 5′ enhancer site (site A) is indicated, and a putative c-Myb binding site is boxed. (C) Activity of the human enhancer in pre-T cells. Constructs containing a 0.25 kb mouse pTa enhancer or a homologous human fragment upstream of SV40 promoter/LacZ were transiently transfected into the pre-T cell line LR1, and β-galactosidase activity was determined.
The Enhancer Contains a Critical Binding Site for c-Myb.
We have shown previously that a 19-bp deletion at the 5′ end of the enhancer core decreased its activity. The deleted site, termed site A, contained a sequence imperfectly matching the consensus binding site for the transcription factor c-Myb (Fig. 5 A). Genomic footprinting of the enhancer revealed a footprint corresponding to the putative c-Myb site (unpublished data). Moreover, the c-Myb site is perfectly conserved in the human enhancer (Fig. 4 B). To confirm the binding of c-Myb to site A, we conducted electrophoretic mobility shift assay (EMSA) using an in vitro translated complete c-Myb DNA-binding domain (DBD). Fig. 5 B illustrates binding of c-Myb DBD to the probes containing native site A, site A mutated in the predicted c-Myb binding core, or a consensus c-Myb binding site. Native, but not mutated site A formed a complex with the c-Myb DBD, while the synthetic consensus probe manifested stronger binding. Similarly, in a competition assay, the site A probe competed for the c-Myb binding at higher concentration than the consensus probe, whereas mutated site A or unrelated probes failed to compete at all (data not shown). These data suggest that the c-Myb DBD can specifically bind to site A, apparently with a lower affinity than to a consensus c-Myb binding site.
To test the involvement of c-Myb in the activity of site A, we introduced the same mutation into two enhancer fragments: a full-strength minimal enhancer and a partially disabled fragment truncated at the 3′ end. Fig. 5 C demonstrates that the mutation decreased the activity of both fragments in a pre-T cell line, causing the same effect as the deletion of site A. To confirm the in vivo importance of the c-Myb binding site, we introduced the same mutation into the enh-EGFP transgenic reporter construct, yielding construct enh/Amut-EGFP. As shown in Table , only two out of six transgenic founders expressed the mutated construct, compared with six out of six for the native construct. The expression pattern in the two enh/Amut-EGFP founders expressing EGFP was indistinguishable from that in enh-EGFP founders. Notably, the expression was observed only in the founders with high transgene copy number, and did not reach the level observed in the enh-EGFP founder with a comparable copy number. Thus, the mutation drastically decreased the frequency, and possibly the level, of the enhancer-driven reporter expression in transgenic mice. Altogether, these data suggest that the intact c-Myb binding site is critical for the optimal activity of the pTa enhancer.
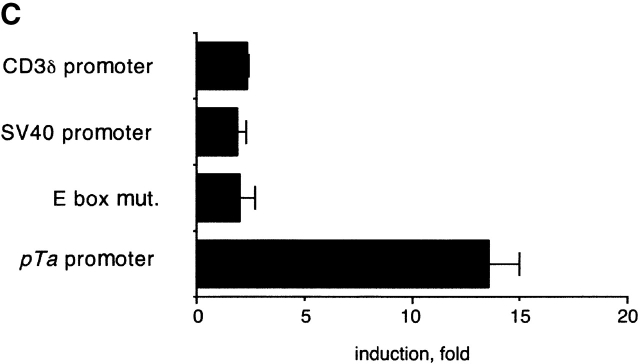
We observed previously that the pTa enhancer is preferentially active in the pTa-positive pre-T cell line LR1 compared with the pTa-negative T cell line BW5147. To test whether site A contributed to enhancer specificity, we transfected reporter constructs containing one or four copies of 17 bp site A upstream of an SV40 promoter into LR1 and BW5147 cells. As demonstrated in Fig. 6 A, site A reporter constructs caused stronger promoter induction in LR1 cells, in contrast to the control CD3δ enhancer. Accordingly, analysis of c-Myb protein expression in these cell lines revealed high c-Myb levels in LR1 cells compared with scarcely detectable levels in BW5147 cells (Fig. 6 B).
Figure 6.
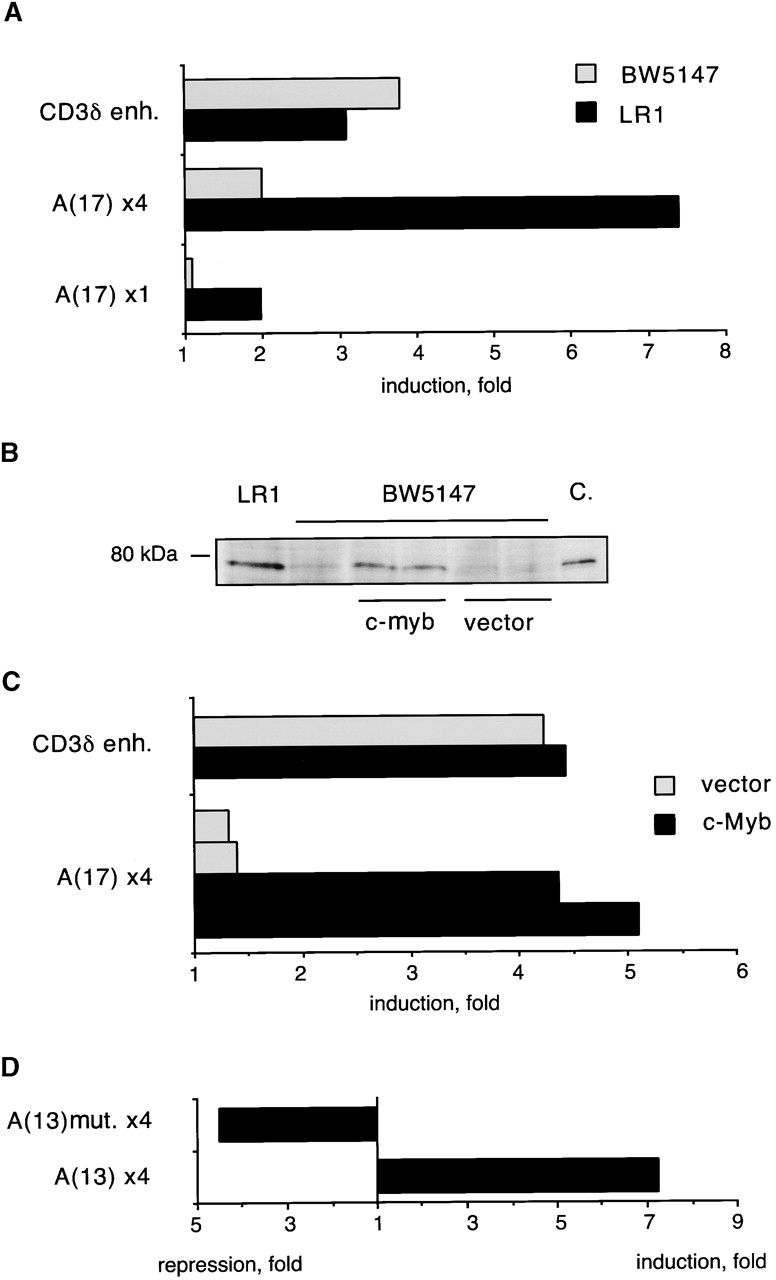
Activity of the site A in cells overexpressing c-Myb. (A) Activity of site A in T cell lines. LR1 and BW5147 cells were transiently transfected with reporters containing one or four copies of 17 bp site A, or a control CD3δ enhancer (enh.), upstream of SV40 promoter/LacZ. Data represent ratio of β-galactosidase activities of each reporter to the activity of the promoter alone. (B) Expression of c-Myb protein in T cell lines as determined by Western blotting. Protein lysates from LR1 and BW5147 lines, as well as from BW5147 clones stably transfected with c-Myb cDNA or expression vector alone, were analyzed. The in vitro translated full-length c-Myb protein was used as a control (C.). (C) Transactivation of site A in BW5147 cells overexpressing c-Myb. Two clones expressing c-Myb cDNA or the empty vector were transiently transfected with the reporter containing four copies of 17 bp site A, and the induction of SV40 promoter was determined as above. A representative transfection with the control CD3δ enhancer is also shown. (D) Activity of the native or mutated site A. The BW5147 clone overexpressing c-Myb was transiently transfected with constructs containing a 13 bp site A (either native or harboring a mutation in the c-Myb binding site) upstream of the SV40 TATA box/luciferase reporter. The induction or repression of the TATA box were determined as above.
To confirm the correlation between c-Myb expression and site A activity, we stably overexpressed c-Myb cDNA in BW5147 cells. As shown in Fig. 6 C, BW5147 clones overexpressing c-Myb supported higher activity of the multicopy site A reporter. Altogether, four c-Myb–expressing clones were tested with similar results. To ascertain that the activation of site A was occurring through the c-Myb binding site, we tested reporters containing four copies of 13 bp site A upstream of a TATA box and luciferase gene. Similar to the reporter described above, the 13 bp site A reporter was preferentially activated in LR1 cells and in BW5147 cells expressing c-Myb (Fig. 6 D, and data not shown). In contrast, site A containing a mutation in the c-Myb site was not activated, instead repressing the activity of the minimal promoter. These results confirm the transactivation of site A by c-Myb, and suggest that c-Myb might regulate the stage-specific activity of the pTa enhancer.
The pTa Promoter Contains an E Box Site Activated by HEB.
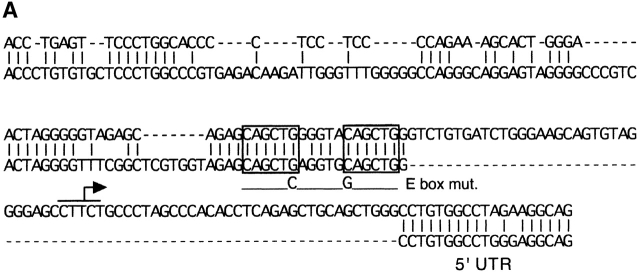
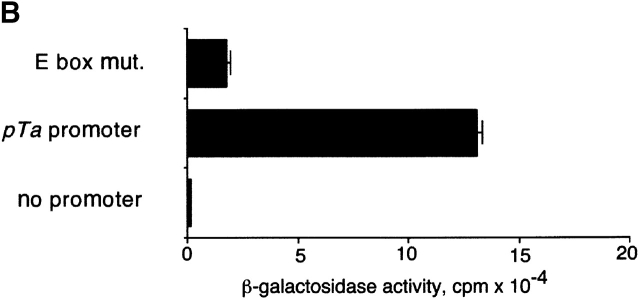
Recently, Herblot et al. demonstrated that pTa is activated by E box-binding transcription factor HEB, and suggested that the activation occurs through the pTa enhancer 27. However, we found that the sequential deletion or simultaneous mutation of E boxes within the enhancer had little effect on its activity in a pre-T cell line (unpublished data). To identify a possible alternative binding site for HEB in the pTa gene, we aligned the pTa core promoter region with the homologous human sequence (Fig. 7 A). In contrast to an overall poor sequence conservation, we noted a conserved site containing two E box elements. As illustrated in Fig. 7 A, we simultaneously introduced a point mutation into each E box. The mutation significantly decreased the mouse promoter activity in pre-T cells (Fig. 7 B), confirming the importance of intact E box elements for the promoter function.
Figure 7.
The identification of an E box-containing site within the pTa promoter. (A) Alignment of the mouse (top) and human (bottom) core promoter regions. The major transcription start site of the mouse pTa gene is indicated by an arrow. Two conserved E box elements are boxed; the introduced mutation (E box mut.) is shown at the bottom. (B) Effect of the E box mutation on the promoter activity. LacZ reporter constructs containing the native or mutated mouse pTa promoter, or a control promoterless construct, were transiently transfected into LR1 pre-T cells, and β-galactosidase activity was determined. (C) Activation of the pTa promoter after HEB overexpression. The indicated promoter/LacZ constructs were cotransfected with HEB cDNA-containing or empty expression vectors and a control luciferase reporter into LR1 cells. The induction of β-galactosidase activity in the presence of HEB as compared with the empty expression vector was determined for each reporter construct after normalization for luciferase activity. The mean induction ± SD of three independent experiments is shown.
The binding of T cell nuclear proteins to the pTa promoter E box site resulted in the formation of complexes containing E2A and HEB as revealed by antibody supershift experiments (data not shown). To confirm the binding of HEB to the pTa promoter, we tested the effect of HEB overexpression on the promoter activity in a pre-T cell line. As illustrated in Fig. 7 C, transient HEB overexpression significantly induced the native pTa promoter, apparently because the amount of endogenous HEB in LR1 cells is not saturating. In contrast, no induction by HEB was observed with the pTa promoter containing the E box site mutation, or with two control promoters. These data suggest that HEB is likely to activate pTa expression through a critical E box–containing site in the pTa promoter.
Discussion
This study was aimed at dissecting the regulatory elements of the pTa gene, which manifests a unique expression pattern specific for immature T cells. We tested the function of the upstream pTa enhancer by deleting it from a large BAC transgene containing a reporter-modified pTa locus. This approach to the study of lymphocyte-specific genes was pioneered by Yu et al., who used it to identify the regulatory elements of the RAG locus 37. We report that the deletion of the enhancer completely abolished pTa reporter expression, while similar deletion of another region had no effect (unpublished data). In a complementary approach, we tested the specificity of pTa enhancer linked to its cognate promoter in transgenic mice. Our data suggest that these constructs targeted reporter transgenes specifically to immature thymocytes in a pattern closely resembling that of pTa itself. Together with its strong evolutionary conservation described here, these data establish the enhancer as a critical regulatory element which is both necessary and sufficient for the correct pTa expression.
Several T cell–specific regulatory elements have been shown to function preferentially in developing rather than mature T cells. The E8III enhancer of the CD8 gene 38 and a distal genomic element of the RAG locus 37 are active specifically in DP thymocytes, but apparently not in the earlier DN T cells. The promoter and intronic enhancer of the human ADA gene supports strong reporter transgene expression in the thymus 39, particularly in immature cortical thymocytes 12. Nevertheless, significant reporter activity is observed in the spleen and bone marrow 39, consistent with the ubiquitous expression of the ADA gene. The proximal promoter of the p56 lck gene was shown to direct thymus-specific transgene expression 40. However, recent transgenic experiments using this promoter suggest that it maintains some activity in mature peripheral T cells 26 41 42. In particular, the EGFP reporter driven by lck proximal promoter manifested variably low levels in DN thymocytes, but equally high levels in DP thymocytes and in peripheral T cells 41 42. In contrast, the pTa enhancer and promoter supported a profound downregulation of EGFP expression in peripheral T cells. Moreover, the reporter protein (human CD25) with an apparently higher turnover rate revealed that the pTa regulatory elements are downregulated in SP thymocytes and are completely silent in mature T cells. Such specificity distinguishes the pTa enhancer and promoter from other known regulatory elements and warrants further study of their regulation.
In contrast to the BAC transgenes, the pTa enhancer-promoter constructs appeared to be strongly influenced by the site of integration in transgenic mice. The observed position effects could be partially overcome by the inclusion of heterologous insulator elements. It is likely that similar elements located elsewhere in the pTa locus augment its expression in the context of intact chromatin. Notably, the large enhancer fragment supported hCD25 transgene expression at a higher frequency than the core enhancer, although both constructs were equally efficient in transient transfections (data not shown). This suggests that an element modulating chromatin organization might be located adjacent to the enhancer. Indeed, the upstream DNase-hypersensitive region corresponding to the pTa enhancer contained two hypersensitive sites separated by 0.3–0.4 kb 28. The second site is likely to represent a matrix attachment region (MAR), which are often found in the vicinity of distal enhancers and can facilitate enhancer function by increasing chromatin accessibility 43.
Our analysis of the pTa enhancer revealed a binding site for the transcription factor c-Myb. A member of the tryptophan cluster family of transcription factors, c-Myb is essential for hematopoiesis 44 and was recently found indispensable for lymphoid development, and particularly for the establishment of the T cell lineage 6. Furthermore, c-Myb appears to regulate proliferation of DN thymocytes after β-selection 45. Important c-Myb binding sites were found in several T cell–specific regulatory elements including the CD4 promoter 8 and silencer 46, TCRγ 11 and TCRδ 9 enhancers, the ADA enhancer 12, the lck proximal promoter 10, and the T cell–specific site within RAG-2 promoter 13. Notably, the latter three elements are preferentially active in immature T cells. C-Myb is abundantly expressed in immature cortical thymocytes (which include DN and DP subsets), but not in medullary thymocytes (corresponding to SP subset) or in resting peripheral T cells 7. This pattern is consistent with the stage-specific expression of pTa in T cells. Importantly, it has been observed that c-Myb is expressed in early hematopoietic progenitors as well as in immature T cells, but not in immature B cells 47. Therefore, c-Myb is likely to be a major transcriptional activator in immature T cells, and may regulate both the stage- and tissue-specific gene expression.
Consistent with this notion, the c-Myb binding site is perfectly conserved in the mouse and human enhancer. In contrast, this site is completely deleted in the nonfunctional pTa enhancer homologue located at the mouse X chromosome, which otherwise exhibits high sequence identity with the enhancer 36. Indeed, the mutation of the c-Myb site in the pTa enhancer decreased both the enhancer activity in vitro and the frequency of transgene expression in vivo. The partial effect of the mutation can be explained by the apparently modular rather than cooperative nature of the pTa enhancer, so that even extensive deletions within the enhancer core do not completely abolish its activity 28. A similar decrease in the frequency of expression, with the minority of founders expressing nearly normal transgene levels, was observed following the mutation of a critical c-Myb binding site in the ADA enhancer 12. Thus, the c-Myb sites in these elements, although not absolutely required for the expression, appear to increase the probability of enhanceosome assembly and of efficient transcription. In transgenic analysis, this may manifest itself as the ability to overcome position effect variegation and to increase the frequency of expression. Although the mutation in the c-Myb site apparently did not alter the specificity of the p Ta transgene expression, we here show that c-Myb may contribute to the preferential pTa enhancer activity in immature T cell lines. These data, together with the presence of c-Myb in the developing but not in mature T cells, suggest that c-Myb might regulate the stage-specific expression of pTa.
In contrast to the majority of c-Myb binding sites described to date, the site in both mouse and human pTa enhancers contains a mismatch to the c-Myb consensus sequence and accordingly manifested lower binding capacity. Such an imperfect binding site may have arisen by chance, if its low affinity is sufficient for activation in the context of the pTa enhancer. Alternatively, a low-affinity binding site might require a higher concentration of c-Myb in the nucleus and consequently might render the enhancer more sensitive to c-Myb downregulation during T cell maturation. Be that as it may, these results call attention to the role of nonconsensus binding sites in transcriptional regulation.
Recently, pTa was shown to be activated by E proteins 22 and specifically by HEB 27, although the site of E protein activity has not been determined. We here identify a conserved tandem E box element within the pTa promoter, which could be activated by HEB. The mutation of this site decreased the promoter activity in pre-T cells, whereas the mutation of E boxes within the enhancer had no effect (unpublished data). Moreover, the tandem E box element in the promoter resembles a critical tandem E box site in the CD4 enhancer, a prototype target of E2A/HEB heterodimers in T cells 48. Thus, the promoter is likely to be a primary target for pTa upregulation by E proteins in immature T cells. Indeed, HEB is abundantly expressed in thymocytes, particularly in DN and DP rather than SP subsets 27. Moreover, the dynamic changes of bHLH protein complexes binding to this site, such as the displacement of SCL/LMO1 by HEB-containing complexes during T cell commitment, may contribute to the specificity of pTa expression.
In conclusion, we show that the pTa enhancer is a critical element regulating pTa expression in T cells. Moreover, in combination with the pTa promoter, the enhancer displays a unique specificity for immature T cells, displaying the highest activity in the DN population. These results establish a useful model for dissecting the transcriptional pathways involved in early T cell development and lineage commitment, such as the induction of pTa by Notch signaling 26. For practical purposes, the pTa regulatory elements described here may be useful for the targeting of transgenes specifically to immature T lymphocytes.
Acknowledgments
We thank Anne Harrington for oocyte injections, A. Francis Stewart, Youming Zhang, Tasuku Honjo, Kiflai Bein, Malcolm Logan, and Robert Kingston for the gifts of reagents, Mark Bedford, Yasumasa Ishida, and Jay Chung for helpful discussions, and Jianrong Lu, Jennifer Michaelson, Jan Pinkas, and Robert Weiss for critical reading of the manuscript.
B. Reizis was supported in part by a postdoctoral fellowship from the Cancer Research Institute.
Footnotes
Abbreviations used in this paper: APC, allophycocyanin; BAC, bacterial artificial chromosome; bHLH, basic helix-loop-helix; DBD, DNA-binding domain; DN, double-negative; DP, double-positive; EGFP, enhanced green fluorescent protein; EMSA, electrophoretic mobility shift assay; ISP, immature single-positive; pTa, pre-T cell receptor α; SP, single-positive.
References
- Fehling H.-J., von Boehmer H. Early alpha beta T cell development in the thymus of normal and genetically altered mice. Curr. Opin. Immunol. 1997;9:263–275. doi: 10.1016/s0952-7915(97)80146-x. [DOI] [PubMed] [Google Scholar]
- Rothenberg E.V. Stepwise specification of lymphocyte developmental lineages. Curr. Opin. Genet. Dev. 2000;10:370–379. doi: 10.1016/s0959-437x(00)00098-8. [DOI] [PubMed] [Google Scholar]
- DiSanto J.P., Radtke F., Rodewald H.-R. To be or not to be a pro-T? Curr. Opin. Immunol. 2000;12:159–165. doi: 10.1016/s0952-7915(99)00066-7. [DOI] [PubMed] [Google Scholar]
- Kuo C.T., Leiden J.M. Transcriptional regulation of T lymphocyte development and function. Annu. Rev. Immunol. 1999;17:149–187. doi: 10.1146/annurev.immunol.17.1.149. [DOI] [PubMed] [Google Scholar]
- Ting C.N., Olson M.C., Barton K.P., Leiden J.M. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- Allen R.D., III, Bender T.P., Siu G. C-myb is essential for early T cell development. Genes Dev. 1999;13:1073–1078. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ess K.C., Witte D.P., Bascomb C.P., Aronow B.J. Diverse developing mouse lineages exhibit high-level c-myb expression in immature cells and loss of expression upon differentiation. Oncogene. 1999;18:1103–1111. doi: 10.1038/sj.onc.1202387. [DOI] [PubMed] [Google Scholar]
- Siu G., Wurster A.L., Lipsick J.S., Hedrick S.M. Expression of the CD4 gene requires a Myb transcription factor. Mol. Cell. Biol. 1992;12:1592–1604. doi: 10.1128/mcb.12.4.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Munain C., Krangel M.S. Regulation of the T-cell receptor delta enhancer by functional cooperation between c-Myb and core-binding factors. Mol. Cell. Biol. 1994;14:473–483. doi: 10.1128/mcb.14.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Leung S., Bosselut R., Ghysdael J., Miyamoto N.G. Myb and Ets related transcription factors are required for activity of the human lck type I promoter. Oncogene. 1994;9:3609–3615. [PubMed] [Google Scholar]
- Hsiang Y.H., Goldman J.P., Raulet D.H. The role of c-Myb or a related factor in regulating the T cell receptor gamma gene enhancer. J. Immunol. 1995;154:5195–5204. [PubMed] [Google Scholar]
- Ess K.C., Whitaker T.L., Cost G.J., Witte D.P., Hutton J.J., Aronow B.J. A central role for a single c-myb binding site in a thymic locus control region. Mol. Cell. Biol. 1995;15:5707–5715. doi: 10.1128/mcb.15.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.-F., Lauring J., Schlissel M.S. c-Myb binds to a sequence in the proximal region of the RAG-2 promoter and is essential for promoter activity in T-lineage cells. Mol. Cell. Biol. 2000;20:9203–9211. doi: 10.1128/mcb.20.24.9203-9211.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H.R., Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Pui J.C., Allman D., Xu L., DeRocco S., Karnell F.G., Bakkour S., Lee J.Y., Kadesch T., Hardy R.R., Aster J.C., Pear W.S. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- Tomita K., Hattori M., Nakamura E., Nakanishi S., Minato N., Kageyama R. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 1999;13:1203–1210. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk M.H., Blom B., Nolan G., Stegmann A.P., Bakker A.Q., Weijer K., Res P.C., Spits H. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J. Exp. Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G., Engel I., Robanus Maandag E.C., te Riele H.P.J., Voland J.R., Sharp L.L., Chun J., Huey B., Pinkel D., Murre C. E2A deficiency leads to abnormalities in αβ T cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barndt R., Dai M.-F., Zhuang Y. A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during αβ thymopoiesis. J. Immunol. 1999;163:3331–3343. [PubMed] [Google Scholar]
- Bain G., Murre C. The role of E-proteins in B- and T-lymphocyte development. Semin. Immunol. 1998;10:143–153. doi: 10.1006/smim.1998.0116. [DOI] [PubMed] [Google Scholar]
- Washburn T., Schweighoffer E., Gridley T., Chang D., Fowlkes B.J., Cado D., Robey E. Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- Blom B., Heemskerk M.H.M., Verschuren M.C.M., vanDongen J.J.M., Stegmann A.P.A., Bakker A.Q., Couwenberg F., Res P.C.M., Spits H. Disruption of αβ but not γδ T cell development by overexpression of the helix-loop-helix protein Id3 in committed T cell progenitors. EMBO J. 1999;18:2793–2802. doi: 10.1093/emboj/18.10.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H., Aifantis I., Feinberg J., Lechner O., Saint-Ruf C., Walter U., Buer J., Azogui O. Pleiotropic changes controlled by the pre-T-cell receptor. Curr. Opin. Immunol. 1999;11:135–142. doi: 10.1016/s0952-7915(99)80024-7. [DOI] [PubMed] [Google Scholar]
- Saint-Ruf C., Ungewiss K., Groettrup M., Bruno L., Fehling H.J., von Boehmer H. Analysis and expression of a cloned pre-T cell receptor gene. Science. 1994;266:1208–1212. doi: 10.1126/science.7973703. [DOI] [PubMed] [Google Scholar]
- Bruno L., Rocha B., Rolink A., von Boehmer H., Rodewald H.-R. Intra- and extra-thymic expression of the pre-T cell receptor alpha gene. Eur. J. Immunol. 1995;25:1877–1882. doi: 10.1002/eji.1830250713. [DOI] [PubMed] [Google Scholar]
- Deftos M.L., Huang E., Ojala E.W., Forbush K.A., Bevan M.J. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herblot S., Steff A.-M., Hugo P., Aplan P.D., Hoang T. SCL and LMO1 alter thymocyte differentiationinhibition of E2A-HEB function and pre-Tα chain expression. Nat. Immunol. 2000;1:138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- Reizis B., Leder P. Expression of the mouse pre-T cell receptor alpha gene is controlled by an upstream region containing a transcriptional enhancer. J. Exp. Med. 1999;189:1669–1678. doi: 10.1084/jem.189.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyrers J.P.P., Zhang Y., Testa G., Stewart A.F. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.H., Whiteley M., Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- Nishi M., Ishida Y., Honjo T. Expression of functional interleukin-2 receptors in human light chain/Tac transgenic mice. Nature. 1988;331:267–269. doi: 10.1038/331267a0. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K., van der Elsen P., Bier E., Maxam A., Terhorst C. A T cell specific enhancer is located in a DNAse I-hypersensitive area at the 3′ end of the CD3-δ gene. EMBO J. 1988;7:2401–2407. doi: 10.1002/j.1460-2075.1988.tb03085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Yamamura K.-I., Miyazaki J.-I. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Robertson G., Garrick D., Wu W., Kearns M., Martin D., Whitelaw E. Position-dependent variegation of globin transgene expression in mice. Proc. Natl. Acad. Sci. USA. 1995;92:5371–5375. doi: 10.1073/pnas.92.12.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martensson I.L., Melchers F., Winkler T.H. A transgenic marker for mouse B lymphoid precursors. J. Exp. Med. 1997;185:653–661. doi: 10.1084/jem.185.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis B., Lee J.T., Leder P. Homologous genomic fragments in the mouse pre-T cell receptor alpha (pTa) and Xist loci. Genomics. 2000;63:149–152. doi: 10.1006/geno.1999.6068. [DOI] [PubMed] [Google Scholar]
- Yu W., Misulovin Z., Suh H., Hardy R.R., Jankovic M., Yannoutsos N., Nussenzweig M.C. Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5′ of RAG2. Science. 1999;285:1080–1084. doi: 10.1126/science.285.5430.1080. [DOI] [PubMed] [Google Scholar]
- Ellmeier W., Sunshine M.J., Losos K., Littman D.R. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity. 1998;9:485–496. doi: 10.1016/s1074-7613(00)80632-9. [DOI] [PubMed] [Google Scholar]
- Aronow B.J., Silbiger R.N., Dusing M.R., Stock J.L., Yager K.L., Potter S.S., Hutton J.J., Wiginton D.A. Functional analysis of the human adenosine deaminase gene thymic regulatory region and its ability to generate position-independent transgene expression. Mol. Cell. Biol. 1992;12:4170–4185. doi: 10.1128/mcb.12.9.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.M., Forbush K.A., Perlmutter R.M. Functional dissection of the lck proximal promoter. Mol. Cell. Biol. 1992;12:2758–2768. doi: 10.1128/mcb.12.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckland J., Pennington D.J., Bruno L., Owen M.J. Co-ordination of the expression of the protein tyrosine kinase p56lck with the pre-T cell receptor during thymocyte development. Eur. J. Immunol. 2000;30:8–18. doi: 10.1002/1521-4141(200001)30:1<8::AID-IMMU8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Shimizu C., Kawamoto H., Yamashita M., Kimura M., Kondou E., Kaneko Y., Okada S., Tokuhisa T., Yokoyama M., Taniguchi M. Progression of T cell lineage restriction in the earliest subpopulation of murine adult thymus visualized by the expression of lck proximal promoter activity. Int. Immunol. 2001;13:105–117. doi: 10.1093/intimm/13.1.105. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Forrester W.C., Fernandez-Herrero L.A., Laible G., Dull M., Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- Mucenski M.L., McLain K., Kier A.B., Swerdlow S.H., Schreiner C.M., Miller T.A., Pietryga D.W., Scott W.J., Potter S.S. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- Pearson R., Weston K. c-Myb regulates the proliferation of immature thymocytes following β-selection. EMBO J. 2000;19:6112–6120. doi: 10.1093/emboj/19.22.6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R.D., III, Kim H.K., Sarafova S.D., Siu G. Negative regulation of CD4 gene expression by a HES-1-c-Myb complex. Mol. Cell. Biol. 2001;21:3071–3082. doi: 10.1128/MCB.21.9.3071-3082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi K., Traver D., Miyamoto T., Weissman I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Sawada S., Littman D.R. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol. Cell. Biol. 1993;13:5620–5628. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T.A., Madden T.L. Blast 2 sequences - a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Tanikawa J., Yasukawa T., Enari M., Ogata K., Nishimura Y., Ishii S., Sarai A. Recognition of specific DNA sequences by the c-myb protooncogene productrole of three repeat units in the DNA-binding domain. Proc. Natl. Acad. Sci. USA. 1993;90:9320–9324. doi: 10.1073/pnas.90.20.9320. [DOI] [PMC free article] [PubMed] [Google Scholar]