Abstract
Assembly of T cell receptor (TCR)α/β genes by variable/diversity/joining (V[D]J) rearrangement is an ordered process beginning with recombination activating gene (RAG) expression and TCRβ recombination in CD4−CD8−CD25+ thymocytes. In these cells, TCRβ expression leads to clonal expansion, RAG downregulation, and TCRβ allelic exclusion. At the subsequent CD4+CD8+ stage, RAG expression is reinduced and V(D)J recombination is initiated at the TCRα locus. This second wave of RAG expression is terminated upon expression of a positively selected α/β TCR. To examine the physiologic role of the second wave of RAG expression, we analyzed mice that cannot reinduce RAG expression in CD4+CD8+ T cells because the transgenic locus that directs RAG1 and RAG2 expression in these mice is missing a distal regulatory element essential for reinduction. In the absence of RAG reinduction we find normal numbers of CD4+CD8+ cells but a 50–70% reduction in the number of mature CD4+CD8− and CD4−CD8+ thymocytes. TCRα rearrangement is restricted to the 5′ end of the Jα cluster and there is little apparent secondary TCRα recombination. Comparison of the TCRα genes expressed in wild-type or mutant mice shows that 65% of all α/β T cells carry receptors that are normally assembled by secondary TCRα rearrangement. We conclude that RAG reinduction in CD4+CD8+ thymocytes is not required for initial TCRα recombination but is essential for secondary TCRα recombination and that the majority of TCRα chains expressed in mature T cells are products of secondary recombination.
Keywords: T cell receptor α chain, gene rearrangement, regulation of gene expression, T cell receptor editing, recombination activating gene
Introduction
During lymphocyte development immunoglobulin and TCR genes are assembled from germline V, D, and J gene segments by a site-specific recombination reaction 1. The V(D)J recombination reaction is mediated by the products of the lymphocyte specific recombination activating genes RAG1 and RAG2 which recognize and cleave recombination signal sequences located adjacent to the coding V, D, and J segments 2 3 4 5. T and B lymphocyte development requires V(D)J recombination; in the absence of RAG1 and RAG2 6 7 or factors that repair the double strand DNA breaks created during V(D)J recombination there is a complete block in the early stages of B and T cell development 8 9 10 11 12 13 14 15.
In thymocytes, V(D)J recombination is initiated at the TCRβ locus in CD4−CD8− double negative (DN) T cells 16. Once a TCRβ chain is expressed it combines with pre-Tα and CD3 components to produce the pre-TCR complex (for reviews, see references 17 and 18). Pre-TCR expression downregulates RAG expression and induces T cells to mature to the CD4+CD8+ double positive (DP) stage. Upon entering the DP stage there is a second wave of RAG expression and V(D)J recombination 19 20 21. Regulation of RAG expression in developing thymocytes has been studied by two groups 22 23. Transgenic experiments with large bacterial artificial chromosomes (BACs) that carry fluorescent protein indicator genes in place of the RAG genes, showed that a cis element 35–70 kb 5′ of RAG2 is required for the second wave of RAG expression 22. However, RAG2 −/− blastocyst reconstitution experiments indicated that T cell development could be rescued with as little as 9 kb of sequence upstream of RAG2 23. Thus, the cis requirements for RAG reinduction in DP thymocytes and the functional consequences of reinduction remain poorly defined.
In DP thymocytes V(D)J recombination is targeted to the TCRα locus. The TCRα/δ locus is a 1 megabase locus that contains the δ locus nested between the Vα and the Jα segments; there are 61 Jα segments spread over 70 kb of DNA 24 25 26. TCRα recombination is believed to begin at the 5′ end of the Jα cluster and progress to the 3′ Jαs during thymocyte maturation 27 28 29. This idea is indirectly supported by the finding of sterile transcripts emanating from the 5′ T early α promoter (TEA) in late DN thymocytes 30 31. Successful rearrangement and expression of TCRα genes is marked by an increase in cell surface CD3/TCR levels, but expression of a TCRα/β dimer is not sufficient to turn off RAG expression and V(D)J recombination. TCRα recombination and RAG expression persist until positive selection 32 33. Continued TCRα recombination in cells that express nonselected α/β TCRs might result in absence of allelic exclusion, and could theoretically interfere with clonal selection 34. Persistent recombination may nonetheless be advantageous if nonselected or self-reactive receptors are replaced by useful receptors thereby salvaging thymocytes that would otherwise be deleted. Indeed, in transgenic and gene targeted mice, secondary TCRα recombination efficiently replaces TCRs that cannot be positively selected 20 33 35 36 37. Despite the potential importance of secondary TCRα recombination for tolerance and repertoire diversification the extent to which secondary recombination contributes to the TCR repertoire in normal mice has not been determined.
Here we report on T cell development and TCRα recombination in mice that are unable to upregulate RAG expression in DP thymocytes. The results indicate that secondary V(D)J recombination makes a major contribution to the normal TCR repertoire.
Materials and Methods
Mice.
Clone m3e8A is an 80-kb yeast artificial chromosome (YAC) containing the RAG1 and RAG2 genes (RYAC), identified by screening the YAC library of St. Mary's Hospital Medical School 38 with primers specific for RAG1 (Genethon). YAC DNA was purified by pulsed field electrophoresis as described 39, and microinjected into the pronuclei of fertilized ova of RAG1 −/− mice (129/SvxCD1 F1) 6. Transgenic founders (RYII and RYIII) were bred for seven generations to C57Bl/6 RAG1 −/− mice (The Jackson Laboratory). The single-copy line RYII was also bred to RAG2 −/− mice (Taconic Farms) 7. The RAG2 −/− TCRβ mice were from Taconic Farms.
Flow Cytometry.
Antibodies used were: PE anti-CD25, biotin anti-CD44, fluorescein anti-CD3, PE–anti-HSA, PE anti-CD8, allophycocyanin (APC) anti-CD4, APC anti-CD8, PE anti-TCRβ, and APC anti-B220 (BD PharMingen). Biotinylated antibodies were visualized with streptavidin-RED613 (GIBCO BRL). Acquisition and analysis were performed with a FACSCalibur™ and CELLQuest™ (Becton Dickinson). Subpopulations of thymocytes and spleen cells were sorted using a FACS Vantage™ (Becton Dickinson) and final purity was >98%.
PCR.
For reverse transcription (RT)-PCR total RNA from 50,000 cells from thymus or spleen/lymph nodes was prepared with TRIzol, primed with oligo-dT, and reverse transcribed with Superscript II (GIBCO/BRL). RAG1 primers were 5′-CAACCAAGCTGCAGACATTCTAGCACTC-3′ and 5′-CACGTCGATCCGGAAAATCCTGGCAATG-3′. β-actin primers were 5′-TACCACTGGCATCGTGATGGACT-3′ and 5′-TTTCTGCATCCTGTCGGCAAT-3′. PCR was performed at 94°C 30 s, 62°C 60 s, 72°C 45 s, for 32 cycles. VJα genes were amplified from cDNA primed with the α chain constant region–specific primer 5′-ATCCATAGCCTTCATGTCCA-3′. PCR was nested using a Cα primer 5′-TCAAAGTCGGTGAACAGGCA-3′ and a degenerate Vα primer AGAAGGTGAAGCAGAGNNM as described 40 for two cycles at 94°C 30 s, 52°C 60 s, 72°C 60 s, followed by 40 cycles at 94°C 30 s, 55°C 60 s, 72°C 60 s. PCR products were cloned before sequencing. V-Jβ sequences were cloned and sequenced using the degenerate Vβ primers 5′-GGMCAYAVTGCTVTKTWCTGGTA-3′, 5′- AAYCATGAYAMMATGTACTGGTA-3′, 5′-CARGCHCCTTCGVTGDNYTGGTA-3′, and Cβ nested primers, 5′- TCAGGCAGTAGCTATAATTGCTCTC-3′, 5′-TTGCCATTCACCCACCAGCTC-3′, and 5′-GCTCAGCTCCACGTGGTCAG-3′. (M = A/C, R = A/G, W = A/T, Y = C/T, K = G/T, V = A/C/G, H = A/C/T, D = A/G/T, N = A/C/G/T). Jα and Vβ-Jβ segments were identified by comparison to the GenBank/EMBL/DDBJ database sequences with accession nos. M64239 and AE000663–5, respectively.
TCRα gene recombination was measured by PCR reactions using the degenerate Vα primer described above in conjunction with either a proximal Jα primer 5′-ACATGAGCTCACTGTCAGCT-3′ (3′ to Jα24.2) or a distal Jα primer 5′-TTACTTGGCTTCACTGTGAG-3′ (3′ to Jα57.9; see Fig. 4). PCR was at: 94°C 30 s, 55°C 60 s, 72°C 3 min for 25 cycles. PCR products were analyzed by electrophoresis in agarose and visualized by blotting and hybridizing with radiolabeled Jα probes 5′-AGGAGGGTCTGCGAAGCTCATCTTT-3′ (proximal) and 5′-ACCAATACAGGCAAATTAACCTTTG-3′ (distal). For single cell PCR, the stained cells and the cytometer sheath fluid were treated with 0.25 pg/ml RNase A to avoid contamination with RNA from lysed cells. Single CD25+CD44− DN T cells from RYIIRAG1 −/− and wild-type mice were sorted into 96-well plates containing 4 μl catch buffer (75 mM NaCl, 1 mM DTT, 4 units Promega RNAsin, 7 units Eppendorf Prime RNase inhibitor) per well and placed on dry ice. RT reactions were performed by addition of 7 μl of random hexamer solution (300 ng random hexamers, 2 pmole RAG1-specific primer: 5′-CTTGAGTCCCCGATGGGCAGTAAA-3′, 1.4% NP-40, 10 units Eppendorph Prime RNase inhibitor, water) followed by a 1-min incubation at 37°C followed by addition of 14 μl of RT reaction solution (5 μl of 5× first strand Superscript buffer, 1 mM dNTPs, 8 mM DTT, 14 units Promega RNAsin, 7 units Prime RNAsin, 0.5 μl Superscript II [GIBCO BRL]). RT was for 10 min at 25°C, followed by 30 min at 37°C, and the enzyme was destroyed by incubation at 90°C for 6 min. 2.5 μl (10%) of the cDNA for each cell was used for nested PCR reactions to amplify RAG1 cDNA or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a positive control. The primers for both RAG1 and GAPDH were designed to span introns to distinguish cDNA from genomic DNA. PCR primers: RAG1 external sense: 5′-GCTATCTCTGTGGCATCGAGTGTT-3′; RAG1 external antisense: 5′-AAA-GACTTTGGGTTTTAGC-3′; RAG1-nested-sense: 5′-GCCG-GGAGGCCTGTGGAG-3′; RAG1 nested antisense: 5′-CCG-TCGGGTGGATGGAGTCAA-3′; GAPDH external sense: 5′-GGTCATCATCTCCGCCCCTTCTG-3′; GAPDH exter-nal antisense: 5′-CACCCTGTTGCTGTAGCCGTATTC-3′; GAPDH nested sense: 5′-TTTGGCATTGTGGAAGGGCTCAT-3′; GAPDH nested antisense: 5′-TCGAAGGTGGAAG-AGTGGGAGTTG-3′. PCR cycle conditions were: 94°C 15 min, 40 cycles of 94°C 30 s, 55°C 30 s, 72°C 30 s, and finally a 7-min extension at 72°C. A total of 138 cells were assayed for RAG1 expression from the RYIIRAG1 −/− mice and 64 cells were assayed from wild-type mice. To ensure that difference between RYIIRAG1 −/− and control samples was not due to the fourfold decrease in RAG1 mRNA in RYIIRAG1 −/− cells we performed four separate PCR experiments on each RYIIRAG1 −/− cell and diluted control cell cDNA.
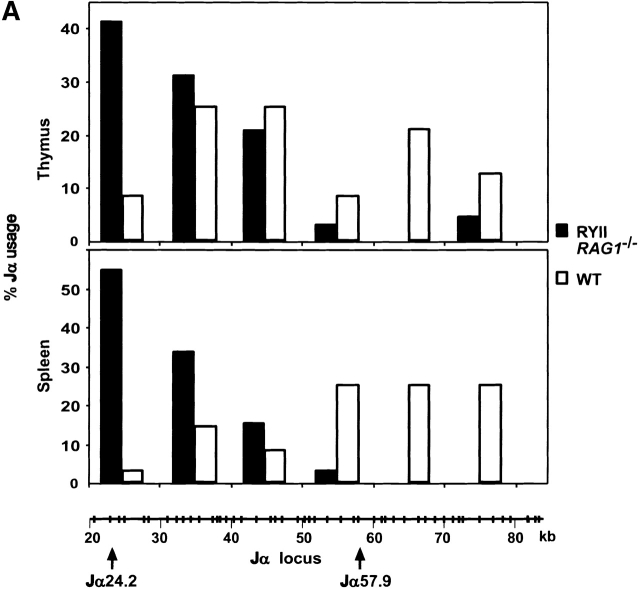
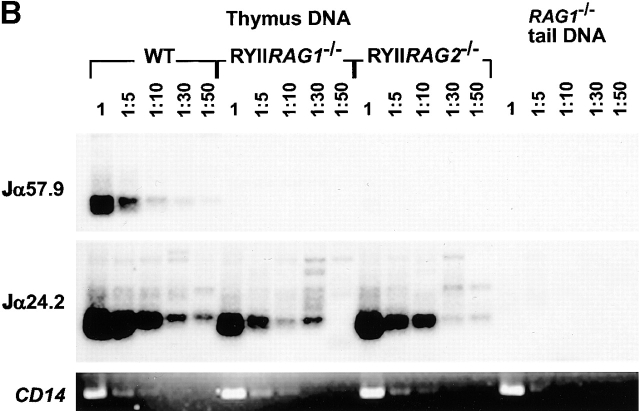
Figure 4.
5′ skewed Jα usage in RYIIRAG1 −/− and RYIIRAG2 −/− mice. (A) Percentage of TCRα mRNAs using Jαs found within the indicated 10-kb intervals in the Jα locus. The results represent 68 RYIIRAG1 −/− and 24 wild-type (WT) clones from the thymus, and 33 RYIIRAG1 −/− and 36 WT clones from the spleen. Schematic representation of the Jα locus is shown and position of Jαs from GenBank/EMBL/DDBJ accession no. M64239 with distances in kbs, the position of the PCR primers and probes used in B is shown. (B) V-Jα rearrangement measured by PCR on genomic DNA from total thymus. RAG1 −/− mouse tail DNA was used as a negative control. The first lane in each case represents ∼60 ng of template DNA diluted as shown. PCR on CD14 was used as loading control. Results are representative of two separate experiments with individual mouse samples.
Quantitative Southern Blotting.
Quantitative Southern blotting was performed exactly as described using the Jα19330.11, Jα42417.4, and Jα4.1 probes 28.
Immunofluorescence.
In situ staining for Nijmegen breakage syndrome (NBS)1 was as described previously 41.
Online Supplemental Data Section.
The online supplemental data contains two tables, online supplemental Tables S1 and S2. Online supplemental material is available at http://www.jem/org/cgi/content/full/194/4/471/DC1.
Results
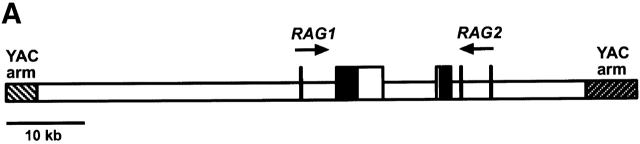
To clarify cis regulation of RAG expression in vivo we analyzed transgenic mice carrying an 80-kb YAC containing 33 kb of genomic sequence 5′ of RAG1 and 12 kb of sequence 5′ of RAG2 (RYAC; Fig. 1). The two lines reported here (RYII single-copy and RYIII three-copy) were maintained by breeding to RAG1 −/− or RAG2 −/− mice and are referred to as RYIIRAG1 −/−, RYIIRAG2 −/−, and RYIIIRAG1 −/−.
Figure 1.
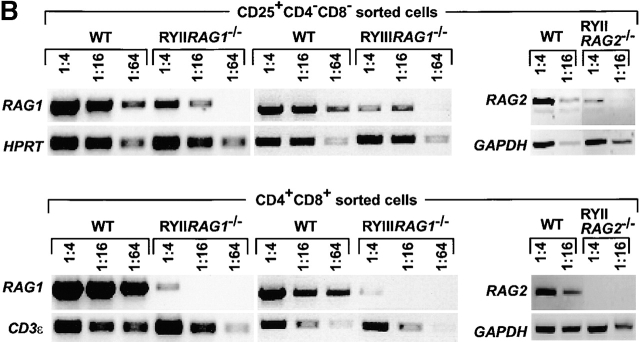
RAG1 is expressed in DN T cells in RYAC transgenic mice. (A) Diagrammatic representation of RYAC an 80-kb YAC containing the RAG genes, 12 kb of genomic DNA 5′ of RAG2, and 33 kb of genomic DNA 5′ of RAG1. (B) RAG1 expression in FACS® purified thymocyte subpopulations as measured by RT-PCR. HPRT, CD3ε, or GAPDH were used as loading controls for normalizing mRNA. The data shown are representative of five experiments. cDNA dilutions are indicated. WT, wild-type.
RAG Expression in DN Thymocytes.
To determine whether the RYAC directs expression of RAG1 and RAG2 in vivo we measured RAG1 mRNA levels in CD25+ DN thymocytes isolated from wild-type, RYIIRAG1 −/−, RYIIIRAG1 −/−, and RYIIRAG2 −/− mice by flow cytometry. Steady-state levels of RAG1 and RAG2 mRNA were estimated by semiquantitative RT-PCR (Fig. 1). CD25+ DN cells isolated from RYIIRAG1 −/−, RYIIIRAG1 −/−, and RYIIRAG2 −/− mice expressed RAG mRNA at three- to fourfold lower levels than wild-type CD25+ DN cells (Fig. 1 B). RAG indicator expression is variegated in mice that carry BAC transgenes similar to RYAC with a variable fraction of developing lymphocytes expressing the indicator transgene 22. To determine whether only a fraction of the DN cells in RYIIRAG1 −/− and RYIIIRAG1 −/− mice express RAG1 we conducted single-cell PCR experiments on purified CD44−CD25+ DN thymocytes. We found that the percentage of CD44−CD25+ thymocytes expressing RAG1 in RYIIRAG1 −/− was threefold lower than in wild-type mice (online supplemental Table S1). This heterogeneity in gene expression is consistent with variegation seen in BAC reporter mice that carry similar transgenes 22.
To determine whether the difference in RAG expression between RYAC and wild-type mice affects TCRβ recombination and expression we cloned and characterized TCRβ mRNAs from thymus. We find no significant differences in TCRβ V, D, or J usage and no significant differences in the nature of the joints between RYAC and wild-type mice (online supplemental Table S2). We conclude that the pattern of RAG1 and RAG2 expression in DN thymocytes in RYAC mice resembles that found in BAC transgenic mice and that this level of expression is sufficient for TCRβ V(D)J recombination.
RAG Expression in DP Thymocytes.
To determine whether RYAC directs regulated RAG expression in DP thymocytes we purified these cells from RYIIRAG1 −/−, RYIIIRAG1 −/−, RYIIRAG2 −/−, and wild-type control mice and measured steady-state levels of RAG1 and RAG2 mRNA by semiquantitative RT-PCR (Fig. 1 B). We found that there was little RAG1 or RAG2 expression in DP thymocytes in RYIIRAG1 −/−, RYIIIRAG1 −/−, and RYIIRAG2 −/− mice (60–120-fold less than in wild-type mice in five experiments). Thus, RYII and RYIII transgenic mice resemble previously characterized BAC transgenic mice in that a cis element that is not contained in RYAC is required for regulated RAG1 and RAG2 expression in DP thymocytes.
T Cell Development.
To determine the physiologic consequences of loss of RAG1 and RAG2 expression in DP cells we analyzed T cell development in RYIIRAG1 −/−, RYIIRAG2 −/−, and RYIIIRAG1 −/− mice. We found that the developmental profile of DN cells in RYIIRAG1 −/−, RYIIRAG2 −/−, and RYIIIRAG1 −/− mice was similar to that of wild-type thymocytes but that the number of thymocytes in the most mature DN subset (CD25−CD44−) was decreased (Fig. 2 A). There was also a corresponding, small but consistent, increase in the percentage of CD25+ CD44− T cells (the immediate precursors of CD25− CD44−T cells; Fig. 2 A) and a fourfold decrease in the percentage of these cells in the S or G2/M phase of the cell cycle in RYIIRAG1 −/− and RYIIIRAG1 −/− mice (as measured by analysis of DNA content after DAPI staining; data not shown). The accumulation of nonproliferating CD25+ CD44− thymocytes in the transgenic mice resembles the accumulation of these cells in RAG1 −/− mice and is consistent with variegated RAG expression in this stage of T cell development (see above).
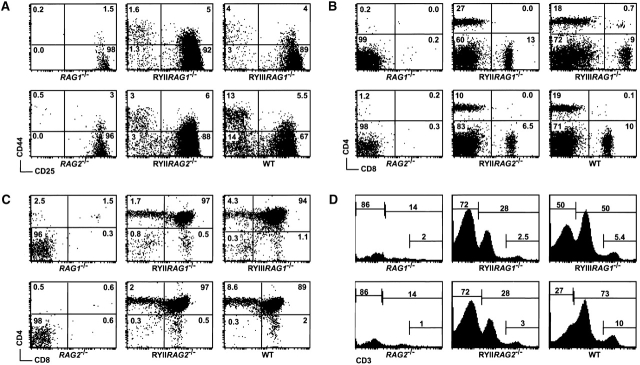
Figure 2.
T cell development in RAG1 −/−, RAG2 −/−, wild-type (WT), RYIIRAG1 −/−, RYIIRAG2 −/−, and RYIIIRAG1 −/− mice. (A) CD44 and CD25 staining profiles for DN thymocytes gated on CD4−CD8− HSAhigh cells. (B) CD4 and CD8 staining of splenocytes. (C) CD4 and CD8 staining of thymocytes. (D) CD3 staining of thymocytes. All plots are representative of 2–4 experiments. Numbers show the percentage of cells in each quadrant. The number of SP thymocytes and CD3high cells was 0.5–0.25 of wild-type controls in four separate experiments.
Despite the 98–99% reduction in RAG expression in DP thymocytes, T cell development in RAG1 −/− and RAG2 −/− mice appeared to be reconstituted by the RYAC transgene as measured by the number of cells in the thymus and spleen. In addition, CD4 and CD8 staining profiles in the thymus and the periphery were similar for RYIIRAG1 −/−, RYIIRAG2 −/−, or RYIIIRAG1 −/− transgenic mice and wild-type controls (Fig. 2B and Fig. C). However, the number of CD4+CD8− and CD4−CD8+ single positive (SP) cells was decreased two- to fourfold in the thymus of RYIIRAG1 −/−, RYIIRAG2 −/−, and RYIIIRAG1 −/− mice (Fig. 2 C). We conclude that T cell development can proceed to the SP stage with only minimal RAG mRNA expression in the DP compartment but fewer SP cells are produced. To examine TCR expression in developing T cells in RYIIRAG1 −/−, RYIIRAG2 −/−, and RYIIIRAG1 −/− mice we stained thymocytes for expression of the TCR–CD3 complex. Thymocytes can be divided into three groups based on low, medium, and high levels of CD3 expression. CD3low cells are the earliest DP cells and express TCRβ but not TCRα, CD3med cells express TCRα but have not yet completed positive selection, and CD3high cells have completed selection and as a result upregulate surface TCR expression. RYIIRAG1 −/−, RYIIRAG2 −/−, and RYIIIRAG1 −/− mice showed an altered distribution of CD3 expression: the majority of thymocytes in these mice were CD3low whereas this is only a minor population in the wild-type thymus (Fig. 2 D). In addition, we found that the transgenic mice displayed a two- to fourfold decrease in the number of CD3high cells, which is also consistent with the decrease in the number of SP thymocytes (Fig. 2 C, five experiments). Similar results were obtained by anti-TCRβ staining (not shown). We conclude that decreased RAG expression in DP thymocytes in RYIIRAG1 −/−, RYIIRAG2 −/−, and RYIIIRAG1 −/− mice leads to a decrease in the number of thymocytes expressing medium and high levels of TCR.
TCRα Recombination.
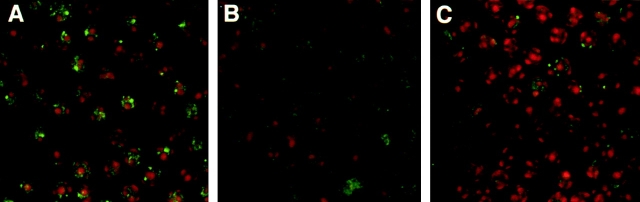
The relative decrease in RAG expression in DP thymocytes and the increase in the percentage of CD3low cells in RYAC mice suggest that there might be lower levels of TCRα recombination. Double stranded DNA break intermediates created during V(D)J recombination at the TCRα locus can be visualized in developing thymocytes by staining nuclei with antibodies to the NBS1 protein 41. To examine the extent of V(D)J recombination in RYIIRAG1 −/− thymocytes directly, we stained purified DP cells from these mice with antibodies to NBS1 and compared them to RAG2 −/− TCRβ transgenic and wild-type mice (Fig. 3). In agreement with previous results, 25% of wild-type DP thymocytes had NBS1 foci. These foci were not detectable in RAG2 −/− TCRβ mice, which have normal number of DP cells but do not undergo V(D)J recombination 21. In contrast, 5% of the DP thymocytes from RYIIRAG1 −/− mice showed NBS1 foci. Thus, the percentage of DP thymocytes with double stranded breaks in RYIIRAG1 −/− mice is decreased in a manner consistent with impaired TCRα recombination.
Figure 3.
NBS1 foci on DP T cells from RYIIRAG1 −/−, RAG2 −/−TCRβ transgenic, and wild-type mice. DP thymocytes were stained with anti-NBS1 (green) and counterstained with Topro-3 (red). (A) WT, (B) RAG2 −/−TCRβ, and (C) RYIIRAG1 −/−. 460–480 DP cells were examined for each mouse strain for the presence of foci.
To determine whether the TCRα genes expressed in RYAC transgenic mice differ from the wild-type controls we amplified and sequenced TCRα mRNAs from thymus and spleen (Fig. 4 A). Although the Vα genes expressed in RYIIRAG1 −/− mice and the VJα junctions were indistinguishable from controls, the Jαs used in these mice were highly biased to the proximal end of the Jα cluster (Fig. 4 A, and not shown). Whereas only 8% of the wild-type TCRα genes in the thymus used a Jα from the proximal 10 kb of this cluster, 41% of the TCRα expressed in the RYIIRAG1 −/− thymus used these 5′ most proximal Jαs (42; Fig. 4 A). Further, in wild-type mice 42% of the Jαs were from the distal half of the locus whereas only 7% of the TCRα genes expressed in RYIIRAG1 −/− mice carry Jαs from the distal part of the locus (42; Fig. 4 A). Proximal skewing of the Jαs was even more evident in T cells that had undergone selection and been exported to the spleen. In the wild-type, 75% of spleen T cells expressed TCRαs using distal Jαs whereas only 3% of the TCRαs cloned from RYII spleen T cells used distal Jαs (Fig. 4 A). We conclude that the TCRα genes expressed in RYIIRAG1 −/− mice are highly biased toward proximal Jα usage.
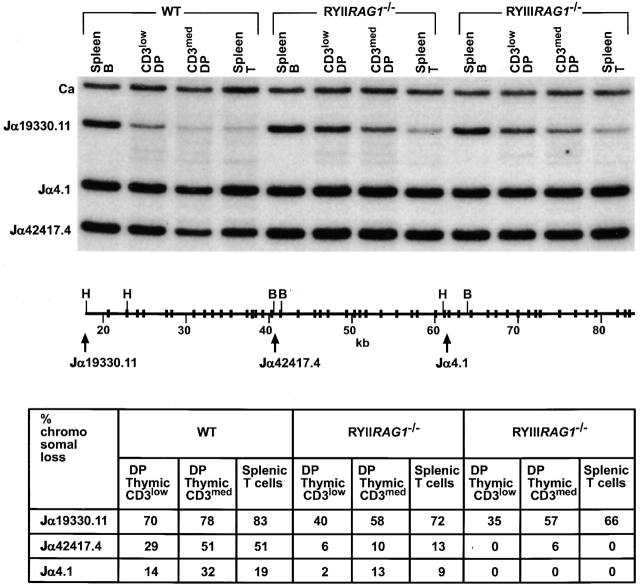
To determine whether skewed Jα usage is due to biased TCRα recombination we measured recombination by PCR using primers specific for the 5′ and 3′ ends of the Jα locus. Both Vα to 5′ Jα and Vα to 3′ Jα rearrangements were readily detected in wild-type mice but only Vα to 5′ Jα rearrangements were found in RYIIRAG1 −/− and RYIIRAG2 −/− mice (Fig. 4 B). Thus, there appears to be a relative absence of recombination to the 3′ Jα locus in RYIIRAG1 −/− and RYIIRAG2 −/− mice. We used Southern blotting on DNA purified from CD3low, med, high thymocytes with probes that hybridize to the 5′, middle, and 3′ ends of the Jα cluster to measure TCRα recombination directly 28. The amount of recombination was standardized with a Cα probe and quantified by phosphorimager analysis (Fig. 5). Overall, RYIIRAG1 −/− and RYIIIRAG1 −/− mice showed less Jα recombination than wild-type controls and almost all of the recombination was restricted to the 5′ portion of the Jα cluster (Fig. 5).
Figure 5.
Quantitation of V-Jα rearrangement by Southern blotting. CD4+ CD8+CD3low and CD3med populations were purified from thymus by flow cytometry; splenic T and B cells were purified from spleen. DNA was digested with BamHI (B) and HindIII (H). The probes are 5′ (Jα19330.11), middle (Jα42417.4), 3′ (Jα4.1), and Cα (reference 28). Schematic representation of the Jα locus showing only the relevant restriction sites; the position of the Jα probes is indicated with arrows. Jα hybridization to spleen B cell DNA was used to calculate the relative loss of Jα DNA in purified T cells. The results in the table are from one of two experiments, the variation between experiments was <5%.
Thymocytes must rearrange and express at least one TCRα gene to become CD3med; therefore the theoretical minimum V(D)J recombination that would allow a T cell to become CD3med is 50%. In wild-type mice Jα recombination was detected on 78–80% of the chromosomes in T cells reaching this stage in development, indicating that most CD3med cells have undergone more than a single V(D)J rearrangement (34 43 44 45; Fig. 5). In contrast, CD3med thymocytes in RYIIRAG1 −/− and RYIIIRAG1 −/− mice showed only 58 and 57% 5′ Jα recombination. Thus, most CD3med cells in RYIIRAG1 −/− and RYIIIRAG1 −/− mice have only attempted V(D)J recombination on one chromosome.
In wild-type mice TCRα recombination may begin with recombination to 5′ Jαs, and 3′ Jα recombination seems to increase as thymocytes progress to more mature stages in development 28. For example, there was 70% 5′, 29% middle, and 14% 3′ Jα recombination in CD3low DP cells and this increased to 83, 51, and 19%, respectively, in SP cells in wild-type mice (28; Fig. 5, bottom). RYIIRAG1 −/− and RYIIIRAG1 −/− mice showed a more drastic bias to 5′ Jα recombination in CD3low DP thymocytes and there was no significant additional recombination to the middle and 3′ part of the locus as thymocytes progressed in development (Fig. 5). We conclude that RYIIRAG1 −/− and RYIIIRAG1 −/− thymocytes differ from wild-type in that they do not recombine 3′ Jαs in the DP compartment.
Discussion
RAG Regulation.
RAG1 and RAG2 are closely linked genes that are believed to originate from a transposon which entered the vertebrate lineage at the time of the evolution of jawed fish 2 4 5 46 47 48 49 50 51 52 53. Expression of the RAG nuclease is highly restricted, but the regulation of RAG expression remains poorly defined.
In vitro analysis of cis regulation of the RAG1 promoter revealed only nonspecific basal promoter activity 54 55 56 57 58. In contrast, the RAG2 promoter displayed preferential activity in lymphoid cell lines that was PAX5 and GATA3 dependent but the activity was not developmentally restricted 59 60. Two systems have been used to study RAG transcription in vivo: RAG2 −/− blastocyst reconstitution and transgenic reporters 22 23. In the RAG2 −/− blastocyst reconstitution experiments an 18-kb genomic fragment extending from 9 kb upstream of the RAG2 promoter to 2.4 kb downstream of the 3′ UTR was able to reconstitute T cell development 23. These experiments suggested that all of the information required for RAG regulation might be found proximal to RAG2 23. In contrast, the transgenic reporter experiments showed that an element in the genomic region 35–70 kb upstream of the RAG2 promoter is required for RAG1 and RAG2 expression in DP T cells 22. Our results with RYAC confirm the presence of a distal regulatory element that regulates both RAG1 and RAG2 reinduction and also reconcile the apparent discrepancies between the two sets of in vivo experiments. The RYAC, which has only 12 kb of genomic sequence 5′ of the RAG2 promoter, resembles the 18-kb fragment used in the RAG2 −/− blastocyst system in that both DNA fragments reconstitute the T cell compartment. However, T cell reconstitution by RYAC occurs in absence of the second wave of RAG expression in DP T cells. Thus, the distal element identified in the transgenic reporter system could not have been detected by the T cell rescue experiments in the RAG2 −/− blastocyst system.
TCRα Recombination.
In wild-type mice, TCRα recombination is thought to proceed coordinately on both chromosomes without allelic exclusion 34 43 44 45 61. Only a small fraction of the TCRα genes in CD3med DP T cells are in the germline configuration, and there is an increase in the amount of 3′ Jα recombination as thymocytes mature, which is consistent with continuing chromosome loss due to continued TCRα recombination in the DP stage 27 29. In contrast, in RYAC mice, almost half of the TCRα genes in CD3med DP T cells are in the germline configuration; only 57% of the TCRα alleles are recombined. All CD3med DP T cells must express a TCRα gene, therefore 57% recombination is just over the theoretical limit of 50% required for TCRα expression in these cells. This indicates that most CD3med transgenic T cells undergo TCRα rearrangement on only one allele and suggests that TCRα recombination begins on one chromosome.
The 5′ portion of the TCRα locus is believed to be the first part of the Jα cluster to become available for recombination 27 28 29. Sterile transcription from the TEA is associated with accessibility to this part of the locus in the late DN stage of T cell development, and in the absence of the TEA the 5′ most Jαs are not recombined 30 31. The 3′ portion of the Jα cluster is thought to become accessible for recombination later in T cell development in a TEA-independent but TCRα enhancer–dependent fashion 26 62 63 64. Thus, the 5′ Jα recombination we find in RYAC transgenic mice might be accounted for by residual RAG protein in thymocytes transiting from the DN to the DP stage and the absence of 3′ Jα recombination due to the relative lack of RAG expression in DP thymocytes. Alternatively, the 1–2% of normal levels of RAG expression we find in DP T cells could be enough to recombine only 5′ Jα genes. In either case, the finding that only the 5′ Jα genes are rearranged in the absence of the normal second wave of RAG expression clearly demonstrates that TCRα recombination is ordered, and that the 5′ side of the Jα cluster is the first to become accessible to the recombinase 27 28 29 30.
The second wave of RAG expression in DP thymocytes is normally terminated during positive selection 20 32 33 65. Transgenic and gene targeted mice that carry nonselecting receptors undergo persistent TCRα locus secondary recombination 33 36 37. Additional support for the idea that there is continuing recombination in DP thymocytes comes from the finding that 3′ Jα rearrangements accumulate as normal thymocytes progress to the mature SP stage 28 37. RYAC transgenic thymocytes fail to activate the second wave of RAG expression and fail to accumulate 3′ Jα rearrangements that are indicative of secondary recombination. Despite this 5′ bias and apparent absence of continued V(D)J recombination, mature RYAC thymocytes show higher levels of Jα chromosome loss than immature CD3low DP thymocytes. We cannot rule out the possibility that the 1–2% of normal level RAG expression in RYAC DP thymocytes targets continuing 5′ Jα but not 3′ Jα recombination, but this seems unlikely. It seems more likely that the increase in chromosome loss in mature T cells in RYAC mice simply reflects selection for the few T cells that have randomly recombined both TCRα alleles because these cells have a higher probability of producing an in frame TCRα gene.
By comparing TCRα recombination in wild-type mice with RYAC transgenic mice that do not reinduce RAG expression, we can estimate the contribution of secondary TCRα recombination to the normal α/β T cell repertoire. Less than 10% of all TCRα genes expressed in mature RYAC T cells contain Jαs from the 3′ half of the Jα cluster. In contrast, 75% of all TCRα genes expressed by splenic T cells in wild-type mice carry Jαs from the 3′ half of the Jα cluster. Thus, at least 65% of the TCRα genes in wild-type mice appear to be products of secondary recombination, a much higher level than the 25% estimated for Igκ receptor editing in B cells 66. We conclude that the majority of the TCRα repertoire in the spleen of wild-type mice is the product of secondary V(D)J recombination and that secondary recombination makes a major contribution to the normal α/β TCR repertoire.
It has been proposed that γ/δ T cells are the evolutionary precursors of αβ T cells 67. The V(D)J recombination in RYAC α/β T cells is developmentally similar to that of the γ/δ cells in that it is mostly absent in the DP compartment. Unlike γ/δ T cells, in which the rearrangement is not ordered and shows no allelic exclusion 68, the simple pattern of rearrangement in RYAC α/β T cells results in virtual allelic exclusion of the TCRα locus. It leads, however, to a two- to threefold decrease in the number of SP thymocytes and a substantial decrease in the complexity of the α/β T cell receptor repertoire, as only a fraction of the available Jαs are used. We speculate that the cis regulatory element that induces the second wave of RAG expression is a late addition to the RAG locus that was selected in evolution at the expense of allelic exclusion because this element increased the diversity of the repertoire.
Acknowledgments
This paper is dedicated to the memory of our friend Eugenia Spanopoulou, who provided the panel of YAC clones with the RAG1 and RAG2 genes. We thank Frank Isdell and Michelle Genova for the FACS® sorting, and the members of the Nussenzweig laboratory, as well as Dr. Harald von Boehmer, Dr. A. Tarakhovsky, and Dr. M. Shlomchik for useful discussions.
This work was supported in part by grants from National Institutes of Health to M.C. Nussenzweig. M.C. Nussenzweig is a Howard Hughes Medical Institute investigator.
Footnotes
The online version of this article contains supplemental material.
Abbreviations used in this paper: APC, allophycocyanin; BAC, bacterial artificial chromosome; DN, double negative; DP, double positive; NBS, Nijmegen breakage syndrome; SP, single positive; RAG, recombination activating gene; RT, reverse transcription; TEA, T early α promoter; YAC, yeast artificial chromosome.
References
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Fugmann S.D., Lee A.I., Shockett P.E., Villey I.J., Schatz D.G. The RAG proteins and V(D)J recombinationcomplexes, ends, and transposition. Annu. Rev. Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- McBlane J.F., van Gent D.C., Ramsden D.A., Romeo C., Cuomo C.A., Gellert M., Oettinger M.A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- Oettinger M.A., Schatz D.G., Gorka C., Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- Schatz D.G., Oettinger M.A., Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- Mombaerts P., Iacomini J., Johnson R.S., Herrup K., Tonegawa S., Papaioannou V.E. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M., Alt F.W. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Barnes D.E., Stamp G., Rosewell I., Denzel A., Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- Difilippantonio M.J., Zhu J., Chen H.T., Meffre E., Nussenzweig M.C., Max E.E., Ried T., Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank K.M., Sekiguchi J.M., Seidl K.J., Swat W., Rathbun G.A., Cheng H.L., Davidson L., Kangaloo L., Alt F.W. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- Gao Y., Sun Y., Frank K.M., Dikkes P., Fujiwara Y., Seidl K.J., Sekiguchi J.M., Rathbun G.A., Swat W., Wang J. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- Gu Y., Seidl K.J., Rathbun G.A., Zhu C., Manis J.P., van der Stoep N., Davidson L., Cheng H.L., Sekiguchi J.M., Frank K. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A., Chen C., da Costa Soares V., Sanchez M., Sokol K., Nussenzweig M.C., Li G.C. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- Ouyang H., Nussenzweig A., Kurimasa A., Soares V.d.C., Li X., Cordon-Cardo C., Li W.-h., Cheong N., Nussenzweig M., Iliakis G. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination in vivo. J. Exp. Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Bogue M.A., Lim D.S., Hasty P., Roth D.B. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- Godfrey D., Kennedy J., Mombaerts P., Tonegawa S., Zlotnik A. Onset of TCR-beta gene rearrangement and role of TCR-beta expression during CD3−CD4−CD8− thymocyte differentiation. J. Immunol. 1994;152:4783–4792. [PubMed] [Google Scholar]
- von Boehmer H., Fehling H.J. Structure and function of the pre-T cell receptor. Annu. Rev. Immunol. 1997;15:433–452. doi: 10.1146/annurev.immunol.15.1.433. [DOI] [PubMed] [Google Scholar]
- Kruisbeek A.M., Haks M.C., Carleton M., Wiest D.L., Michie A.M., Zuniga-Pflucker J.C. Branching out to gain controlhow the pre-TCR is linked to multiple functions. Immunol. Today. 2000;21:637–644. doi: 10.1016/s0167-5699(00)01744-8. [DOI] [PubMed] [Google Scholar]
- Wilson A., Held W., MacDonald H. Two waves of recombinase gene expression in developing thymocytes. J. Exp. Med. 1994;179:1355–1360. doi: 10.1084/jem.179.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie H., Livak F., Schatz D., Strasser A., Crispe I., Shortman K. Multiple rearrangements in T cell receptor alpha chain genes maximize the production of useful thymocytes. J. Exp. Med. 1993;178:615–622. doi: 10.1084/jem.178.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y., Koyasu S., Nakayama K., Murphy K.M., Loh D.Y., Reinherz E.L., Alt F.W. Restoration of T cell development in RAG-2-deficient mice by functional TCR transgenes. Science. 1993;259:822–825. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- Yu W., Misulovin Z., Suh H., Hardy R.R., Jankovic M., Yannoutsos N., Nussenzweig M.C. Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5′ of RAG2. Science. 1999;285:1080–1084. doi: 10.1126/science.285.5430.1080. [DOI] [PubMed] [Google Scholar]
- Monroe R.J., Chen F., Ferrini R., Davidson L., Alt F.W. RAG2 is regulated differentially in B and T cells by elements 5′ of the promoter. Proc. Natl. Acad. Sci. USA. 1999;96:12713–12718. doi: 10.1073/pnas.96.22.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvin-Marche E., Hue I., Marche P.N., Liebe-Gris C., Marolleau J.P., Malissen B., Cazenave P.A., Malissen M. Genomic organization of the mouse T cell receptor V alpha family. EMBO J. 1990;9:2141–2150. doi: 10.1002/j.1460-2075.1990.tb07383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop B.F., Wilson R.K., Wang K., Vernooij B., Zallwer D., Kuo C.L., Seto D., Toda M., Hood L. Organization, structure, and function of 95 kb of DNA spanning the murine T-cell receptor C alpha/C delta region. Genomics. 1992;13:1209–1230. doi: 10.1016/0888-7543(92)90039-u. [DOI] [PubMed] [Google Scholar]
- Krangel M.S., Hernandez-Munain C., Lauzurica P., McMurry M., Roberts L.J., Zhong X.-P. Developmental regulation of the V(D)J recombination at the TCRα/δ locus. Immunol. Rev. 1998;165:131–147. doi: 10.1111/j.1600-065x.1998.tb01236.x. [DOI] [PubMed] [Google Scholar]
- Roth M.E., Holman P.O., Kranz D.M. Nonrandom use of J alpha gene segments. Influence of V alpha and J alpha gene location. J. Immunol. 1991;147:1075–1081. [PubMed] [Google Scholar]
- Petrie H., Livak F., Burtrum D., Mazel S. T cell receptor gene recombination patterns and mechanismscell death, rescue, and T cell production. J. Exp. Med. 1995;182:121–127. doi: 10.1084/jem.182.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.D., Pelkonen J., Hurwitz J.L. First T cell receptor alpha gene rearrangements during T cell ontogeny skew to the 5′ region of the J alpha locus. J. Immunol. 1990;145:2347–2352. [PubMed] [Google Scholar]
- Wilson A., de Villartay J.P., MacDonald H.R. T cell receptor delta gene rearrangement and T early alpha (TEA) expression in immature alpha beta lineage thymocytesimplications for alpha beta/gamma delta lineage commitment. Immunity. 1996;4:37–45. doi: 10.1016/s1074-7613(00)80296-4. [DOI] [PubMed] [Google Scholar]
- Villey I., Caillol D., Selz F., Ferrier P., de Villartay J.P. Defect in rearrangement of the most 5′ TCR-J alpha following targeted deletion of T early alpha (TEA)implications for TCR alpha locus accessibility. Immunity. 1996;5:331–342. doi: 10.1016/s1074-7613(00)80259-9. [DOI] [PubMed] [Google Scholar]
- Turka L.A., Schatz D.G., Oettinger M.A., Chun J.J., Gorka C., Lee K., McCormack W.T., Thompson C.B. Thymocyte expression of RAG-1 and RAG-2termination by T cell receptor cross-linking. Science. 1991;16:778–781. doi: 10.1126/science.1831564. [DOI] [PubMed] [Google Scholar]
- Borgulya P., Kishi H., Uematsu Y., von Boehmer H. Exclusion and inclusion of α and β T cell receptor alleles. Cell. 1992;69:529–537. doi: 10.1016/0092-8674(92)90453-j. [DOI] [PubMed] [Google Scholar]
- Malissen M., Trucy J.J., Jouvin-Marche E., Cazenave P., Scollay R., Malissen B. Regulation of TCR α and β gene allelic exclusion during T-cell development. Immunol. Today. 1992;13:315–322. doi: 10.1016/0167-5699(92)90044-8. [DOI] [PubMed] [Google Scholar]
- Malissen M., Trucy J., Letourneur F., Rebai N., Dunn D.E., Fitch F.W., Hood L., Malissen B. A T cell clone expresses two T cell receptor alpha genes but uses one alpha beta heterodimer for allorecognition and self MHC-restricted antigen recognition. Cell. 1988;55:49–59. doi: 10.1016/0092-8674(88)90008-6. [DOI] [PubMed] [Google Scholar]
- McGargill M.A., Derbinski J.M., Hogquist K.A. Receptor editing in developing T cells. Nat. Immunol. 2000;1:336–341. doi: 10.1038/79790. [DOI] [PubMed] [Google Scholar]
- Wang F., Huang C.-Y., Kanagawa O. Rapid deletion of rearranged T cell antigen receptor (TCR) Valpha-Jalpha segment by secondary rearrangement in the thymusrole of continuous rearrangement of TCR alpha chain gene and positive selection in the T cell repertoire formation. Proc. Natl. Acad. Sci. USA. 1998;95:11834–11839. doi: 10.1073/pnas.95.20.11834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier F.L., Keer J.T., Sutcliffe M.J., Henriques D.A., Mileham P., Brown S.D. Construction of a mouse yeast artificial chromosome library in a recombination-deficient strain of yeast. Nat. Genet. 1992;1:132–136. doi: 10.1038/ng0592-132. [DOI] [PubMed] [Google Scholar]
- Yannoutsos N., Ijzermans J.N., Harkes C., Bonthuis F., Zhou C.Y., White D., Marquet R.L., Grosveld F. A membrane cofactor protein transgenic mouse model for the study of discordant xenograft rejection. Genes Cells. 1996;1:409–419. doi: 10.1046/j.1365-2443.1996.d01-244.x. [DOI] [PubMed] [Google Scholar]
- Broeren C.P.M., Goerge G.M., Verjans G.M., Van Eden W., Kusters J.G., Lenstra J.A., Longtenberg T. Conserved nucleotide sequences at the 5′ end of T cell receptor variable genes facilitate polymerase chain reaction amplification. Eur. J. Immunol. 1991;21:569–575. doi: 10.1002/eji.1830210306. [DOI] [PubMed] [Google Scholar]
- Chen H.T., Bhandoola A., Difilippantonio M.J., Zhu J., Brown M.J., Tai X., Rogakou E.P., Brotz T.M., Bonner W.M., Ried T., Nussenzweig A. Response to RAG-mediated V(D)J cleavage by NBS1 and γ-H2AX. Science. 2000;290:1962–1964. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegert P., Gilfillan S. A conserved sequence block in the murine and human TCR Ja regionassessment of regulatory function in vivo. J. Immunol. 1999;162:3471–3480. [PubMed] [Google Scholar]
- Davis M.M., Bjorkman P.J. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Pircher H., Michalopoulos E.E., Iwamoto A., Ohashi P.S., Baenziger J., Hengartner H., Zinkernagel R.M., Mak T.W. Molecular analysis of the antigen receptor of virus-specific cytotoxic T cells and identification of a new V alpha family. Eur. J. Immunol. 1987;17:1843–1846. doi: 10.1002/eji.1830171226. [DOI] [PubMed] [Google Scholar]
- Uematsu Y. Preferential association of alpha and beta chains of the T cell antigen receptor. Eur. J. Immunol. 1992;22:603–606. doi: 10.1002/eji.1830220247. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., Mizuuchi K., Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- Landree M.A., Wibbenmeyer J.A., Roth D.B. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.R., Dai Y., Mundy C.L., Yang W., Oettinger M.A. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A.K., Guhathakurta A., Kleckner N., Haniford D.B. Tn10 transposition via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- Hiom K., Melek M., Gellert M. DNA transposition by the RAG1 and RAG2 proteinsa possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- Besmer E., Mansilla-Soto J., Cassard S., Sawchuk D.J., Brown G., Sadofsky M., Lewis S.M., Nussenzweig M.C., Cortes P. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol. Cell. 1998;2:817–828. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- Agrawal A., Eastman Q.M., Schatz D.G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Shockett P.E., Schatz D.G. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol. Cell. Biol. 1999;19:4159–4166. doi: 10.1128/mcb.19.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrin A.A., Fong I., Malkin L., Marsden P.A., Berinstein N.L. Cloning and characterization of the human recombination activating gene 1 (RAG1) and RAG2 promoter regions. J. Immunol. 1997;159:4382–4394. [PubMed] [Google Scholar]
- Kurioka H., Kishi H., Isshiki H., Tagoh H., Mori K., Kitagawa T., Nagata T., Dohi K., Muraguchi A. Isolation and characterization of a TATA-less promoter for the human RAG-1 gene. Mol. Immunol. 1996;33:1059–1066. doi: 10.1016/s0161-5890(96)00062-4. [DOI] [PubMed] [Google Scholar]
- Kitagawa T., Mori K., Kishi H., Tagoh H., Nagata T., Kurioka H., Muraguchi A. Chromatin structure and transcriptional regulation of human RAG-1 gene. Blood. 1996;88:3785–3791. [PubMed] [Google Scholar]
- Fuller K., Storb U. Identification and characterization of the murine Rag1 promoter. Mol. Immunol. 1997;34:939–954. doi: 10.1016/s0161-5890(97)00000-x. [DOI] [PubMed] [Google Scholar]
- Brown S.T., Miranda G.A., Galic Z., Hartman I.Z., Lyon C.J., Aguilera R.J. Regulation of the RAG-1 promoter by the NF-Y transcription factor. J. Immunol. 1997;158:5071–5074. [PubMed] [Google Scholar]
- Lauring J., Schlissel M.S. Distinct factors regulate the murine RAG-2 promoter in B- and T-cell lines. Mol. Cell. Biol. 1999;19:2601–2612. doi: 10.1128/mcb.19.4.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi H., Wei X.C., Jin Z.X., Fujishiro Y., Nagata T., Matsuda T., Muraguchi A. Lineage-specific regulation of the murine RAG-2 promoterGATA-3 in T cells and Pax-5 in B cells. Blood. 2000;95:3845–3852. [PubMed] [Google Scholar]
- Casanova J., Romero P., Widmann C., Kourilsky P., Maryanski J. T cell receptor genes in a series of class I major histocompatibility complex-restricted cytotoxic T lymphocyte clones specific for a Plasmodium berghei nonapeptideimplications for T cell allelic exclusion and antigen-specific repertoire. J. Exp. Med. 1991;174:1371–1383. doi: 10.1084/jem.174.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Munain C., Sleckman B.P., Krangel M.S. A developmental switch from TCR delta enhancer to TCR alpha enhancer function during thymocyte maturation. Immunity. 1999;10:723–733. doi: 10.1016/s1074-7613(00)80071-0. [DOI] [PubMed] [Google Scholar]
- McMurry M.T., Krangel M.S. A role for histone acetylation in the developmental regulation of V(D)J recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- Sleckman B.P., Bardon C.G., Ferrini R., Davidson L., Alt F.W. Function of the TCR alpha enhancer in alphabeta and gammadelta T cells. Immunity. 1997;7:505–515. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- Brandle D., Muller S., Muller C., Hengartner H., Pircher H. Regulation of RAG-1 and CD69 expression in the thymus during positive and negative selection. Eur. J. Immunol. 1994;24:145–151. doi: 10.1002/eji.1830240122. [DOI] [PubMed] [Google Scholar]
- Casellas R., Shih T.Y., Jasna R., Kleinewietfeld M., Nemazee D., Rajewsky K., Nussenzweig M.C. Contribution of receptor editing to the antibody repertoire. Science. 2001;291:1541–1544. doi: 10.1126/science.1056600. [DOI] [PubMed] [Google Scholar]
- Flajnik M.F. Churchill and the immune system of ectothermic vertebrates. Immunol. Rev. 1998;166:5–14. doi: 10.1111/j.1600-065x.1998.tb01248.x. [DOI] [PubMed] [Google Scholar]
- Sleckman B.P., Khor B., Monroe R., Alt F.W. Assembly of productive T cell receptor delta variable region genes exhibits allelic inclusion. J. Exp. Med. 1998;188:1465–1471. doi: 10.1084/jem.188.8.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]