Abstract
The B cell adaptor containing src homology 2 domain (BASH; also termed BLNK or SLP-65), is crucial for B cell antigen receptor (BCR)-mediated activation, proliferation, and differentiation of B cells. BCR-mediated tyrosine-phosphorylation of BASH creates binding sites for signaling effectors such as phospholipase Cγ (PLCγ)2 and Vav, while the function of its COOH-terminal src homology 2 domain is unknown. We have now identified hematopoietic progenitor kinase (HPK)1, a STE20-related serine/threonine kinase, as a protein that inducibly interacts with the BASH SH2 domain. BCR ligation induced rapid tyrosine-phosphorylation of HPK1 mainly by Syk and Lyn, resulting in its association with BASH and catalytic activation. BCR-mediated activation of HPK1 was impaired in Syk- or BASH-deficient B cells. The functional SH2 domain of BASH and Tyr-379 within HPK1 which we identified as a Syk-phosphorylation site were both necessary for interaction of both proteins and efficient HPK1 activation after BCR stimulation. Furthermore, HPK1 augmented, whereas its kinase-dead mutant inhibited IκB kinase β (IKKβ) activation by BCR engagement. These results reveal a novel BCR signaling pathway leading to the activation of HPK1 and subsequently IKKβ, in which BASH recruits tyrosine-phosphorylated HPK1 into the BCR signaling complex.
Keywords: antigen receptor signaling, BCR, BLNK, SH2 domain, IκB kinase
Introduction
Stimulation of B cells through the B cell antigen receptor (BCR) provokes, depending on their developmental stage, such diverse responses as proliferation, differentiation, cell-cycle arrest, or apoptosis. BCR signaling is initiated by activation of cytoplasmic protein tyrosine kinases (PTKs) including Src family kinases, Syk, and Btk. Activated PTKs then phosphorylate and enhance the enzymatic activities of various signaling intermediates including phospholipase Cγ (PLCγ)2, phosphoinositide 3-kinase, and Vav, which in turn transmit the signals into distinct pathways leading to the activation of nuclear transcription factors such as AP-1, nuclear factor (NF)-AT, and NF-κB 1 2 3 4. Distinct signaling pathways appear to be activated selectively, depending on the developmental stage, activation state, or tolerance status of B cells and the nature of the pertinent antigen 5 6. However, it remains to be understood how a receptor signals to several pathways and how the specific pathways are selected.
A recently identified novel B cell–specific protein termed B cell adaptor containing src homology 2 domain (BASH; also known as BLNK or SLP-65) has been shown to be an adaptor that plays a central role in BCR–signal transduction 7 8 9. BASH is structurally and functionally related to the adaptor protein SLP-76 of hematopoietic cells which serves a crucial function in signal transduction from TCR, Fcε-, and collagen-receptors in T cells, mast cells, and platelets 10. BASH consists of an NH2-terminal acidic domain containing tyrosine-based SH2 domain–binding motifs, a central domain containing proline-rich motifs and a COOH-terminal SH2 domain. After tyrosine phosphorylation primarily by Syk, BASH associates with PLCγ, Vav, and Btk through their SH2 domains, with Grb2 through its SH2 and SH3 domains and with Nck and Syk. Shortly after BCR stimulation, translocation of BASH to the membrane fraction of cells can be observed. The NH2-terminal and the central domains of BASH are presumably the binding sites for the aforementioned SH2 and SH3 domain–containing proteins, whereas the binding partners of the COOH-terminal SH2 domain remain unknown 7 8 9 11 12 13. Experiments using normal or BASH/BLNK-deficient B cell lines indicated that BASH mediates phosphorylation and activation of PLCγ2 by Syk and Btk, elevation of intracellular calcium concentration ([Ca2+]i), NF-AT activation, and activation of mitogen-activated protein kinases (MAPKs), extracellular signal–regulated kinase (ERK), JNK, and p38, after BCR engagement 7 12 13. Thus, BASH is proposed to function as a scaffold protein for various signaling effectors and to recruit them into a BCR signaling complex containing PTKs at the plasma membrane. Targeted disruption of the BASH gene in mice resulted in a partial block of early B cell development, absence of mature B and peritoneal B-1 cells, defective activation and proliferation of B cells upon BCR ligation in vitro, decreased serum Ig levels, and impaired T cell–independent immune responses. This phenotype can be ascribed to BCR and pre-BCR signaling deficiencies 14 15 16 17.
In an alternative approach to understand the mechanism how BASH mediates BCR signal transduction, we have searched for proteins that interact with the SH2 domain of BASH after BCR stimulation. Here we report the identification of hematopoietic progenitor kinase (HPK)1 as a BASH SH2 domain–associated protein. HPK1 is a hematopoietic cell-specific serine/threonine kinase and belongs to the GCK family of STE20-related kinases. The HPK1 kinase domain which occupies the NH2-terminal third of the protein is followed by four proline-rich motifs clustered in a central region. Ectopic expression of HPK1 in fibroblasts resulted in the activation of the JNK/SAPK MAPKs 18 19 and IκB kinases (IKKs; reference 20). The upstream receptors and the mechanism of activation for HPK1 in hematopoietic cells are only starting to emerge. SH3 domain–containing adaptor proteins such as Grb2, Crk, CrkL, SH3P7/HIP-55, and HS1 have been shown to interact with the proline-rich motifs of HPK1 21 22 23 24. Recently, Liou et al. have demonstrated that HPK1 is activated in response to TCR and BCR engagement 25. This activation is dependent, to various extents, on Src and Syk family PTKs and the adaptor proteins LAT, SLP-76, BLNK/BASH, Grb2, and Grap. This study also indicated that HPK1 is a negative regulator for the ERK pathway in TCR signaling. Furthermore, Liu et al. demonstrated activation of HPK1 in response to TCR stimulation and subsequent binding to Grb2-related adaptor Gads 26.
Here we present a mechanistic model that links BCR stimulation to the activation of HPK1 in B cells. Our data suggest that BCR activation induces phosphorylation of HPK1 on tyrosine-379, which then results in binding of HPK1 to the SH2 domain of BASH. We have also demonstrated that HPK1 does not affect ERK activation, but notably upregulates IκB kinase β (IKKβ) activation in the BCR signaling pathway.
Materials and Methods
Cells and Antibodies.
Stable transfectants of WEHI231 and its subclone (WEHI231.5, a gift of Dr. T. Tsubata, Medical Research Institute, Tokyo Medical and Dental University, Tokyo, Japan) were maintained as described previously 27. Spleen cells were prepared from C57BL/6 mice and the splenic B cells were enriched by killing T cells with culture supernatant of hybridoma T-24 (anti-Thy1 MoAb) and rabbit complement 16. WEHI231 and splenic B cells were stimulated with 10 μg/ml goat F(ab′)2 fragment anti–mouse IgM Ab (μ chain specific; Jackson ImmunoResearch Laboratories). Wild-type (WT) and Syk-, Lyn-, Btk-, or BASH/BLNK-deficient DT40 cells (gifts of Drs. T. Kurosaki and M. Ishiai, Institute for Liver Research, Kansai Medical University, Osaka, Japan) were maintained and stimulated with 25 μg/ml anti–chicken IgM MoAb (M4) as described previously 28 29 12. COS7 and 293T cells were cultured in DMEM supplemented with 10% FCS and 50 μm 2-ME. Rabbit antisera against HPK1 (amino acids 1–18, #2) or HPK1 (amino acids 811–825, #7) were used for immunoprecipitation and immunoblot analysis, respectively 21. Rabbit antiserum against BASH 16 was used for immunoprecipitation, while goat Ab (anti-BLNK, C-19, Santa Cruz Biotechnology, Inc.) was used for immunoblot analysis. mAbs against the T7 epitope, phosphotyrosine (PY20), the HA epitope (12CA5), and the Flag epitope (M5) were purchased from Novagen, Transduction Laboratories, Boehringer Mannheim, and Sigma-Aldrich, respectively. Rabbit anti-Lyn 44, anti-Syk (LR), anti-IKKα/β (H-470), anti c-Rel (C), and anti-SP1 (PEP2) Abs were from Santa Cruz Biotechnology, Inc. Goat anti-GST Ab was from Amersham Pharmacia Biotech. Control mouse IgG was from Zymed Laboratories.
Expression Constructs and Transfection.
Mouse BASH cDNA 16 was truncated at a StuI site (BASHΔSH2) or mutated by PCR-based site-directed mutagenesis (Quickchange; Stratagene) to substitute Lys for Arg-373 in the SH2 domain (BASH[R373K]). The mutated or WT BASH cDNAs were ligated in-frame with an NH2-terminal T7-epitope tag into pAT7neo expression vector 30. WEHI231 cells were electroporated (240 V, 900 μF) with either pAT7-BASH or pAT7-BASHΔSH2, selected with G418 (1.2 mg/ml), and stable clones expressing exogenous BASH proteins at high level were established. One of the selected clones expressing exogenous BASH (B7) was further transfected with HPK1 expression vectors, which are composed of pCAG-Puro vector 30 and either of the inserts of pcDNA3-HPK1:HA or pcDNA3-HPK1(K46E):HA 18, and selected with puromycin (1 μg/ml; Sigma-Aldrich) to establish stable transfectants. pMT2-HPK1 was described previously 18. The insert of the pcDNA3-HPK1:HA was recloned into pCAGGS expression vector 31 resulting in pCAGGS-HPK1:HA. Mutation of HPK1 Tyr-379 into Phe in pcDNA3-HPK1:HA gave rise to pcDNA3-HPK1(Y379F):HA. A BamHI fragment of pcDNA3-HPK1:HA containing a COOH-terminal part of HPK1 (residues 327–827) and an HA-epitope tag was inserted into a BamHI site of pGEX-3X (Amersham Pharmacia Biotech) to make pGST-HPK1-C. pME-Lyn and pME-Syk were described previously 32. Expression vectors for glutathione S-transferase (GST)-IκBα and Flag epitope-tagged IKKβ, respectively, were gifts of Dr. H. Nakano (Juntendo University, Tokyo, Japan; reference 33). pSRα-HA-MAPK encoding Xenopus ERK2 and pGST-Elk1 were gifts of Drs. E. Nishida (Kyoto University, Kyoto, Japan) and T. Kurosaki, respectively. DT40 or DT40 mutant cells (5 × 106 per 0.25 milliliter per cuvette) were transiently transfected with the indicated plasmids (25–40 μg in total) by electroporation (975 μF, 240 V) and harvested after 36 h. COS7 or 293T cells (5 × 105) were transfected using a TransIT-LT1 transfection reagent (Pan Vera) and harvested after 36 h.
In Vitro Binding Assay with Calmodulin-binding Protein Fusion Proteins.
Sequences corresponding to the SH2 domain (residues 341–457) of mouse BASH were amplified by PCR from BASH and BASH (R373K) cDNAs and inserted into the pCAL-n-EK prokaryotic expression vector, giving rise to a calmodulin-binding protein (CBP)–BASH SH2 domain fusion protein (Affinity LIC cloning and protein purification kit; Stratagene). CBP fusion proteins were purified with calmodulin affinity resins in binding buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM CaCl2, 1 mM magnesium acetate). Anti-IgM–stimulated or unstimulated WEHI231.5 cells (107) were lysed in the binding buffer supplemented with 1% NP-40 and protease and phosphatase inhibitors (1 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 5 mM sodium orthovanadate, 10 mM NaF). The cell lysates were precleared with the CBP–SH2(R373K)-bound calmodulin affinity resins (5 μg) then incubated with 2 μg of the purified CBP–SH2 (WT) or CBP–SH2 (R373K) fusion proteins immobilized on the affinity resins for 1 h at 4°C. The resins were washed five times with the binding buffer supplemented with 0.4% NP-40 and the inhibitors and the bound proteins were subjected to SDS-PAGE followed by immunoblot analysis.
GST and GST-HPK1-C proteins were produced in the recombinant bacterial strain TKB1 harboring an Elk tyrosine kinase gene or the parental TK-deficient BL21(DE3) cells (Stratagene) transformed with either pGEX-3X or pGST-HPK1-C, as follows. Recombinant protein expression was induced by 0.4 mM isopropylthiogalactopyranoside and cells were sonicated in bacterial lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DTT, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM PMSF, 1 mM sodium orthovanadate). The lysates were cleared by centrifugation at 12,000× g for 20 min and GST proteins were purified on glutathione sepharose 4B (Amersham Pharmacia Biotech). Purified GST proteins (0.4 μg) were precleared and incubated in binding buffer with resin-immobilized CBP–SH2 domain fusion proteins (0.5 μg) in the presence of protease and phosphatase inhibitors, and the bound proteins were analyzed as above.
Immunoprecipitation, Immunoblot Analysis, and In Vitro Kinase Assay.
For immunoprecipitations, cells were lysed on ice in lysis buffer (1% NP-40, 50 mM Tris, pH 7.8, 150 mM NaCl, 2 mM EDTA) containing protease and phosphatase inhibitors. Precleared lysates were incubated with appropriate Abs and protein G-sepharose beads (4 Fast Flow; Amersham Pharmacia Biotech). After extensive washing with lysis buffer (with 0.5% NP-40), the immunoprecipitates were subjected to immunoblot analysis using appropriate Abs, which were visualized using chemiluminescense (ECL kit; Amersham Pharmacia Biotech). For in vitro kinase assays, the washed immunocomplexes were divided into equal parts and half of the precipitates was subjected to the kinase reaction while the other half was subjected to immunoblotting to estimate the amounts of precipitated proteins. For HPK1 kinase assays, immunoprecipitates were washed three times with lysis buffer, twice with TNE buffer (50 mM Tris, pH 7.6, 150 mM NaCl, and 2 mM EDTA) and once with kinase buffer (50 mM Tris, pH 7.6, 8 mM MgCl2, 2 mM MnCl2, and 1 mM DTT). Autophosphorylation reactions were performed at 30°C for 20 min in the kinase buffer (30 μl) containing 10 μCi of [γ-32P] ATP. For transphosphorylation, 10 μg of myelin basic protein (MBP; Sigma-Aldrich) were added as a substrate in the presence of 5 μM of cold ATP. The reactions were stopped by addition of 5× SDS sample buffer, then samples were analyzed by SDS-PAGE, followed by autoradiography of dried gels. IKKβ kinase assays was performed as described previously 20 with 5 μg of GST-IκBα protein as a substrate in the presence of 13 μM cold ATP. ERK2 kinase assay was performed as described previously 34.
Nuclear extracts were prepared as follows: 4 × 106 cells per sample were resuspended in 150 μl buffer A (10 mM Hepes, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, and protease/phosphatase inhibitors) and placed on ice for 15 min. NP-40 (0.6% final) was added to the cell suspensions, which were then mixed vigorously for 10 s and centrifuged at 12,000 rpm for 1 min. The nuclear pellet was washed three times with buffer A and extracted with 50 μl buffer C (20 mM Hepes, pH 7.9, 25% glycerol, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and protease/phosphatase inhibitors) by vigorous agitation for 15 min at 4°C. The extracts were cleared by centrifugation for 5 min and the supernatants (20 μl per lane) were subjected to SDS-PAGE, followed by immunoblot analysis.
All data shown are representative of two to four independent experiments with essentially identical results.
Results and Discussion
BCR-induced Tyrosine-Phosphorylation of HPK1 and its Binding to the BASH SH2 Domain.
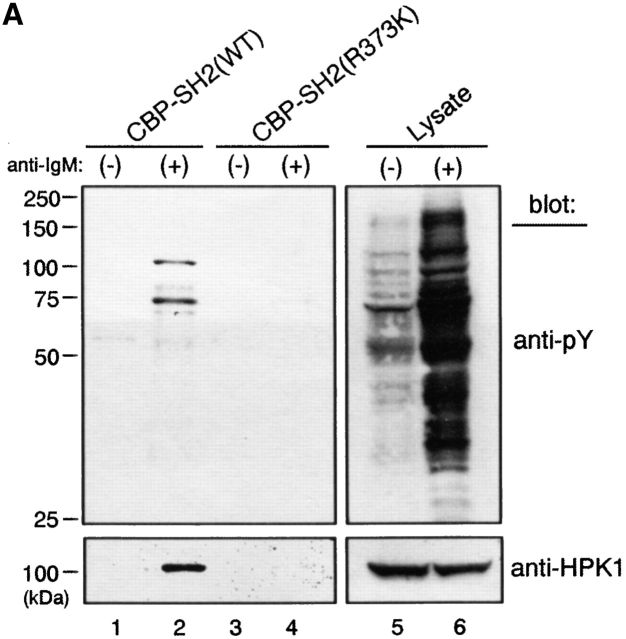
To identify proteins interacting with the SH2 domain of BASH, lysates of WEHI231 B lymphoma cells either unstimulated or after BCR ligation were first incubated with CBP fused to a nonfunctional mutant of the BASH SH2 domain (CBP-SH2[R373K]) to absorb nonspecific-binding proteins. Unbound material was then tested for its ability to bind to a CBP-fusion protein containing the WT BASH SH2 domain. Among the bound proteins, tyrosine-phosphorylated species were identified by antiphosphotyrosine immunoblotting (Fig. 1 A, top). Two phosphoproteins of an approximate molecular weight of 100 and 74 kD were found to specifically interact with the WT BASH SH2 domain only after BCR stimulation (lanes 1 and 2). Very little phosphoprotein interacted with the mutant SH2 domain, indicating high efficiency of the preabsorption procedure (lanes 3 and 4). Testing with various antibodies directed against known signaling entities identified the 100-kD phosphoprotein as the HPK1 (Fig. 1 A, bottom), while the identity of the 74-kD protein remains to be determined.
Figure 1.
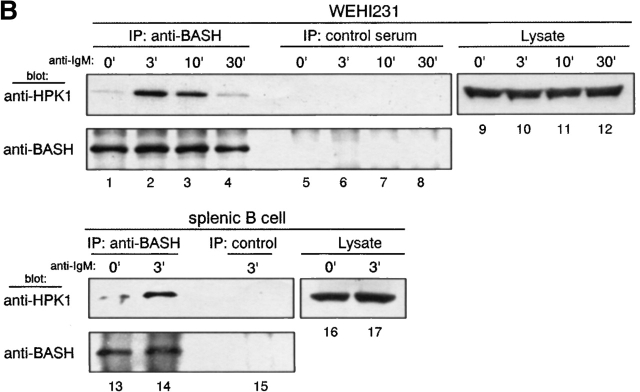
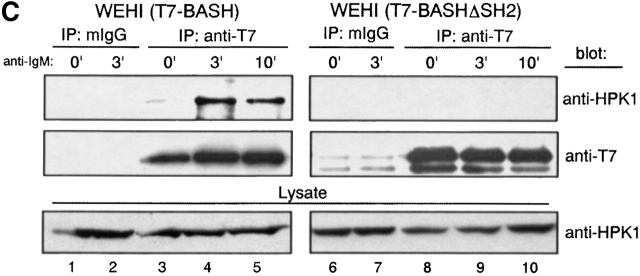
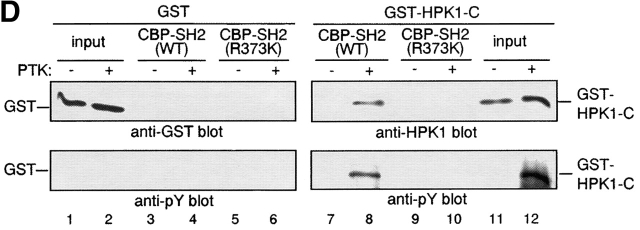
Direct binding of tyrosine phosphorylated HPK1 to the SH2 domain of BASH in response to BCR stimulation. (A) BCR engagement induced binding to the BASH SH2 domain in vitro of phosphoproteins including HPK1. WEHI231.5 cells were stimulated for 3 min with anti–IgM F(ab′)2 fragment (+) or left unstimulated (−). Lysates from these cells were precleared through binding to beads coupled with recombinant CBP fusion proteins of mutated (R373K) BASH SH2 domain, then incubated with the beads with the fusion proteins of the WT (lanes 1 and 2) or mutated (R373K) BASH SH2 domain (lanes 3 and 4). Bound material was eluted from the fusion protein, separated by SDS-PAGE, and subjected to immunoblotting with antiphosphotyrosine (anti-pY) Ab (top). Subsequently, the membrane was reprobed with anti-HPK1 #7 Ab (bottom). Whole cell lysates were included for visualization of all tyrosine-phosphorylated proteins (lanes 5 and 6). (B) Interaction of endogenous BASH with HPK1 in B cells. WEHI231.5 (1.5 × 107) or splenic B cells (6.8 ×107) were stimulated with anti–IgM F(ab′)2 fragment for the indicated periods of time (min). Lysates from these cells were immunoprecipitated (IP) with rabbit anti-BASH serum (lanes 1–4, 13, and 14) or control antiserum against MIST peptide (unpublished data; lanes 5–8 and 15), separated on SDS-PAGE, and then immunoblotted with anti-HPK1 #7 Ab (top). The same membranes were reprobed with anti-BASH (C-19) Ab to visualize the immunoprecipitated BASH proteins (bottom). The presence of HPK1 in whole cell lysates was ascertained by immunoblotting with the anti-HPK1 #7 Ab (lanes 9–12, 16, and 17). (C) In vivo interaction of HPK1 and BASH is dependent on the BASH SH2 domain. Stably transfected WEHI231 cells (107) expressing either T7 epitope–tagged WT BASH (T7-BASH) or a BASH SH2 domain truncation mutant (T7 BASHΔSH2) were stimulated with anti–IgM F(ab′)2 fragment for the indicated periods of time (min). Lysates from these cells were immunoprecipitated with either anti-T7 Ab or control mouse IgG (mIgG), and the precipitates were immunoblotted with anti-HPK1 #7 Ab (top), then reprobed with anti-T7 Ab (middle). Anti-HPK1 immunoblotting of the lysates demonstrated comparable levels of HPK1 proteins between the transfectants (bottom). (D) Binding of recombinant HPK1 to the BASH SH2 domain depends on HPK1 tyrosine phosphorylation. GST and GST-HPK1-C proteins produced in bacterial strains containing (+) or not containing (−) an active PTK were affinity purified and incubated with immobilized CBP-BASH SH2 domain fusion proteins (as described in A). Bound proteins were eluted and immunoblotted with anti-GST (top, lanes 3–6), anti-HPK1 #7 (top, lanes 7–10), and anti-pY (bottom) Abs. One-twenty-seventh of the GST fusion proteins subjected to the binding reaction was loaded on the same gel (“input,” lanes 1, 2, 11, and 12). The electrophoretic mobility of GST and GST-HPK1-C proteins is indicated.
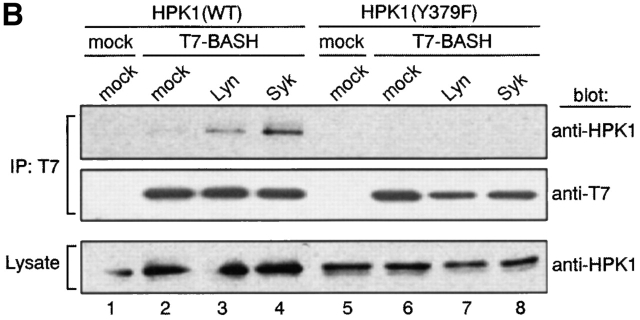
To address whether the interaction of BASH and HPK1 occurs at endogenous expression levels in vivo, WEHI231 cells and purified splenic B cells were treated with anti–IgM Ab, and lysed. Then the cell lysates were subjected to immunoprecipitation with anti-BASH antiserum or irrelevant antiserum as a negative control, and subsequently immunoblotted with anti-HPK1 antiserum (Fig. 1 B). In both cell types, HPK1 clearly coprecipitated with BASH and the amount of the precipitated HPK1 was markedly increased 3 min after BCR stimulation (lanes 1–4, 13, and 14). To confirm that the endogenous interaction between BASH and HPK1 is mediated through the SH2 domain of BASH, stably transfected WEHI231 cells expressing either T7 epitope-tagged WT BASH (T7-BASH) or a truncated form lacking the SH2 domain (T7-BASHΔSH2) were subjected to anti-T7 immunoprecipitation. As shown in Fig. 1 C, endogenous HPK1 coprecipitated with T7-BASH and again a marked enhancement of the interaction was observed after BCR ligation (lanes 3–5). HPK1 failed to interact with the BASHΔSH2 mutant (lanes 8–10). These results indicate that BASH forms a complex with HPK1 through its SH2 domain in B cells, which is strongly and transiently enhanced upon BCR engagement.
To determine whether the interaction of HPK1 and the BASH SH2 domain is direct, we prepared a GST fusion protein encompassing the noncatalytic domain of HPK1 (residues 327–827) from Escherchia coli in tyrosine-phosphorylated and nonphosphorylated form (see Fig. 1 D, lanes 11 and 12) and repeated the pull-down assay described above with these reagents. GST-HPK1 was bound to the BASH SH2 domain, which was dependent on tyrosine-phosphorylation (lanes 7 and 8), while no binding to the nonfunctional SH2 domain mutant (R373K) was observed (lanes 9 and 10). A control GST protein was bound to neither of the SH2 domains (lanes 3–6). These results clearly indicate a direct interaction between tyrosine-phosphorylated HPK1 and the SH2 domain of BASH.
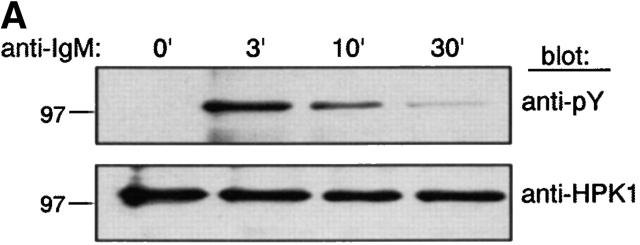
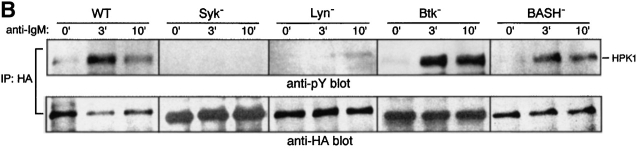
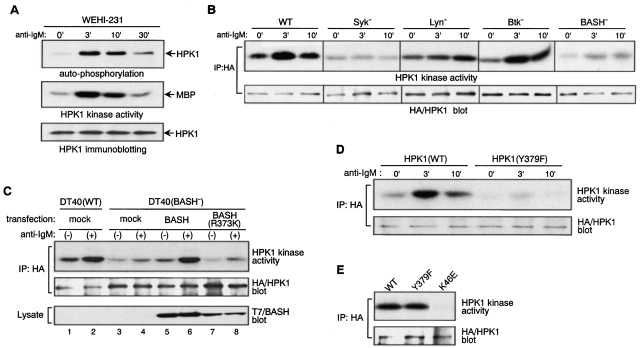
We next examined whether BCR stimulation induces HPK1 tyrosine-phosphorylation and if this phosphorylation is mediated by BCR-associated PTKs. In WEHI231 cells, BCR engagement markedly induced tyrosine phosphorylation of endogenous HPK1 peaking at 3 min after BCR ligation (Fig. 2 A). The same kinetics of HPK1 phosphorylation was observed in WEHI279 cell, an independent B cell lymphoma (data not shown). In addition, transiently expressed HPK1 was also tyrosine-phosphorylated in DT40 chicken B cells, the level of which was greatly augmented by BCR ligation (Fig. 2 B, first panel). BCR-induced HPK1 phosphorylation was undetectable in Syk-deficient DT40 cells, greatly reduced but detectable in Lyn-deficient DT40 cells and unaffected in Btk- or BASH-deficient DT40 cells. This result indicates that Syk is essential for BCR-induced tyrosine-phosphorylation of HPK1, and that Lyn strongly upregulates the HPK1 tyrosine phosphorylation level presumably through augmenting the catalytic activity of Syk 35 or by direct phosphorylation of HPK1 which would then be dependent on a preceding phosphorylation by Syk 36.
Figure 2.
BCR-mediated tyrosine-phosphorylation of HPK1 and its effect on interaction with BASH. (A) HPK1 is phosphorylated on tyrosine after BCR engagement. WEHI231 cells (107) were stimulated with anti–IgM F(ab′)2 fragment for the indicated periods of time (min) and HPK1 was immunoprecipitated with anti-HPK1 #2 Ab. The precipitates were immunoblotted with anti-pY Ab (top) and reprobed with anti-HPK1 #7 Ab (bottom). (B) BCR-mediated phosphorylation of HPK1 in PTK- or BASH-deficient DT40 cell lines. 1.5 × 107 WT or mutant DT40 cells were transiently transfected with 75 μg pCAGGS-HPK1:HA. After 36 h, the cells were stimulated with anti–chicken IgM Ab for the indicated periods of time (min). HA-tagged HPK1 proteins were immunoprecipitated with anti-HA Ab and immunoblotted with anti-pY Ab (top), then reprobed with anti-HA Ab (bottom). (C) Tyrosine-phosphorylation of HPK1 is required for the association of HPK1 with BASH. pMT2-HPK1 (2 μg) and either (1 μg) of pAT7-BASH, pAT7-BASH(R373K), or empty vector pAT7 (mock) were cotransfected transiently with either (2 μg) of pME-Lyn, pME-Syk, or empty vector pME (mock) into COS7 cells. The cells were lysed and BASH proteins were immunoprecipitated using anti-T7 Ab. One-half of the precipitate was immunoblotted using anti-HPK1 #7 (top), while the other half was immunoblotted with anti-pY Ab (left, upper middle), then reprobed with anti-T7 Ab (left, lower middle; right, middle). Equal HPK1 expression levels were visualized by immunoblotting of the cell lysates with anti-HPK1 #7 Ab (bottom).
In contrast to our results, Liou et al. detected no tyrosine-phosphorylation on HPK1 in response to TCR stimulation 25. Very recently, Liu et al. have demonstrated HPK1 tyrosine-phosphorylation in a murine T cell hybridoma after TCR activation 26. Thus, the extent of HPK1 tyrosine phosphorylation upon antigen receptor stimulation may substantially differ among lymphoid cell types.
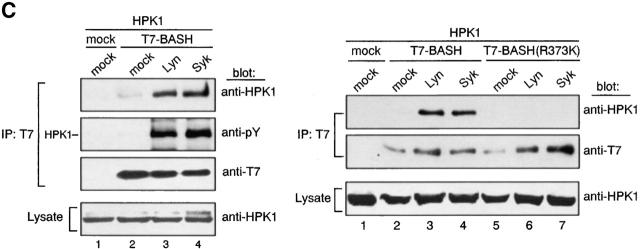
Transient expression of either Lyn or Syk in COS7 cells resulted in tyrosine-phosphorylation of HPK1 (Fig. 3 A, lanes 2 and 3), although Lyn was not able to phosphorylate HPK1 in the absence of Syk in DT40 cells after BCR ligation (Fig. 2 B). This result likely reflects a deregulated kinase activity of Lyn and possibly a relaxed dependence on compartmentalization in this overexpression system (Fig. 3 A, lane 3; unpublished data). To examine whether the PTK-mediated phosphorylation of HPK1 is involved in its association with BASH, expression vectors for HPK1, BASH, and either Lyn or Syk were cotransfected into COS7 cells, and binding of HPK1 to BASH was determined by immunoprecipitation analysis (Fig. 2 C). When coexpressed with Lyn or Syk, a considerable amount of tyrosine-phosphorylated HPK1 coprecipitated with WT BASH (Fig. 2 C, left, lanes 3 and 4). Minimal coprecipitation of HPK1 in the absence of exogenous PTKs was probably due to phosphorylation by undefined PTKs present in COS7 cells (Fig. 2 C, left, lane 2). No binding of HPK1 to the BASH SH2 domain mutant BASH (R373K) was detected irrespective of the presence of Lyn and Syk (Fig. 2 C, right, lanes 6 and 7; compare to lanes 3 and 4). These results indicated that PTK-mediated phosphorylation is a prerequisite for the interaction of HPK1 with the BASH SH2 domain.
Figure 3.
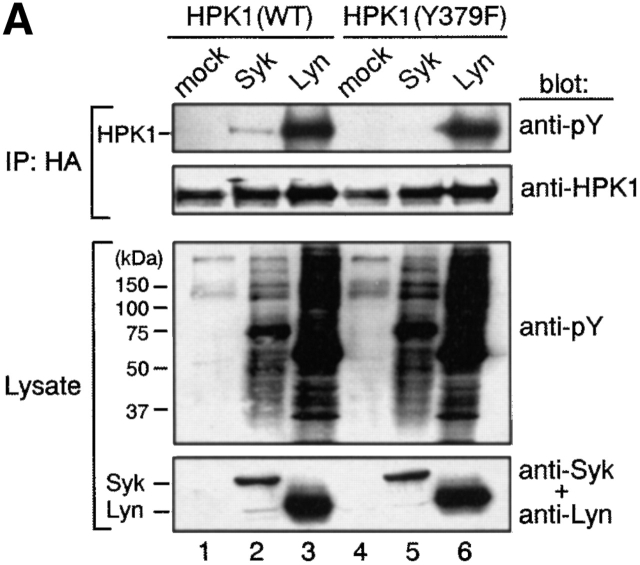
Phosphorylation at Tyr-379 within HPK1 is essential for its interaction with BASH. (A) Tyr-379 within HPK1 is a Syk-phosphorylation site. pcDNA-HPK1:HA or pcDNA-HPK1(Y379F):HA (1.2 μg each) was cotransfected transiently with pME-Lyn, pME-Syk, or empty vector (mock) (2.4 μg each) into COS7 cells. HPK1 proteins were immunoprecipitated with anti-HA Ab and immunoblotted with anti-pY (top), then reprobed with anti-HPK1 #7 Ab (upper middle). Whole cell lysates were immunoblotted with anti-pY Ab (lower middle) and anti-Syk plus anti-Lyn Abs (bottom) to visualize expression of the PTKs. (B) Tyr-379 is essential for binding HPK1 to BASH. HPK1(WT) or HPK1(Y379F) was coexpressed transiently with T7-BASH and either Lyn or Syk in COS7 cells. BASH was immunoprecipitated, and coprecipitated HPK1 proteins were analyzed as described in Fig. 2 C.
To further characterize the interaction between HPK1 and BASH, we next aimed to identify the relevant phosphorylation site within HPK1 that mediates binding to the BASH SH2 domain. A possible site was Tyr-379 located within a DDDYDDV motif (residues 376–382) just NH2-terminal of the second proline-rich region of HPK1. The sequence context of this motif resembles an ExDYEDV sequence within the HS1 protein that has previously been demonstrated to be an excellent substrate for Syk kinase and upon phosphorylation a binding site for the SH2 domains of Src family PTKs 37 38. Upon coexpression in COS7 cells, Syk failed to phosphorylate a HPK1 mutant (HPK1[Y379F]) in which Tyr-379 was substituted by Phe (Fig. 3 A, top, compare lanes 2 and 5; note that Syk expression [bottom] and total protein phosphorylation in the lysate [lower middle] are equivalent). This indicates that Tyr-379 is a major target site of Syk phosphorylation. Lyn-mediated phosphorylation of HPK1 was apparently not affected by the Y379F mutation, possibly because multiple tyrosine residues in HPK1 are phosphorylated by Lyn (Fig. 3 A, lanes 3 and 6). In contrast to WT HPK1 (Fig. 3 B, lanes 3 and 4), HPK1(Y379F) was not bound to BASH in transfected COS7 cells, even when coexpressed with Lyn or Syk (Fig. 3 B, lanes 7 and 8). These results indicate that Tyr-379 within HPK1 is essential for binding to BASH and thus strongly suggest that the DDDYDDV sequence containing the phosphorylated Tyr-379 is the binding site for the BASH SH2 domain. It has been shown that the similar motifs, QEVYDDV and DEVYDDV of the FYB/SLAP protein, are selectively phosphorylated on tyrosine by Fyn-T kinase. The tyrosine phosphorylated version mediates binding to the SLP-76 SH2 domain which is closely related to the SH2 domain of BASH 39 40. Taken together with our result, the phosphopeptide pYDDV is likely to be a specific SH2 domain ligand of SLP family proteins.
BCR-induced Activation of HPK1 through Tyrosine-Phosphorylation and Binding to BASH SH2 Domain.
The finding that BCR engagement induces tyrosine-phosphorylation of HPK1 and its binding to BASH prompted us to investigate whether the association with BASH contributes to the activation of HPK1. First, we examined whether endogenous HPK1 is activated by BCR engagement. In WEHI231 (Fig. 4 A) and WEHI279 cells (data not shown), HPK1 kinase activity was markedly elevated after BCR ligation, as assessed in vitro by both autophosphorylation and transphosphorylation of a substrate protein. The kinetics of HPK1 activation mirrored that of tyrosine-phosphorylation shown in Fig. 2 A, suggesting that PTK-mediated phosphorylation is involved in the upregulation of HPK1 activity during BCR signaling. The kinetics of BCR-mediated activation of transiently expressed HPK1 was roughly assessed in WT and various PTK-deficient DT40 mutants cells (Fig. 4 B). In DT40 cells, HPK1 kinase activity was clearly elevated after BCR ligation, as recently reported by Liou et al. 25. BCR-mediated HPK1 activation was absent from Syk-deficient cells, while in Btk-deficient cells it was comparable to WT DT40 cells, indicating that Syk, but not Btk, is essential for HPK1 activation. In Lyn-deficient cells, HPK1 activation was delayed but reached a significant level during the later phase of BCR stimulation. Together with the observation that a very low level of HPK1 tyrosine-phosphorylation was induced in Lyn-deficient cells (Fig. 2 B), this suggested that Lyn-mediated massive phosphorylation was not essential for BCR-mediated HPK1 activation but facilitated HPK1 activation.
Figure 4.
BCR-induced activation of HPK1 is largely dependent on its interaction with BASH. (A) HPK1 kinase is activated in response to BCR stimulation. WEHI231 cells (5 × 106) were stimulated with anti–IgM F(ab′)2 fragment for the indicated periods of time (min). Cells were lysed, HPK1 was immunoprecipitated using anti-HPK1 #2 Ab, and its in vitro kinase activity was determined. Reaction products were separated by SDS-PAGE and visualized by autoradiography. HPK1-autophosphorylation (top) and its kinase activity toward the exogenous substrate MBP (middle) are shown. Precipitated HPK1 proteins were visualized by immunoblotting with anti-HPK1 #7 Ab (bottom). (B) Activation of HPK1 in WT and mutant DT40 cells. WT or various mutant DT40 cell lines (1.5 × 107 cells each) were transfected with pCAGGS-HPK1:HA (75 μg) and stimulated with anti–chicken IgM Ab. HA-tagged HPK1 proteins were immunoprecipitated using anti-HA Ab and their in vitro kinase activities toward MBP were determined as described in A (top). Precipitated HPK1 was visualized by immunoblotting with anti-HA Ab (bottom). (C) The BASH SH2 domain mediates BCR-induced HPK1 activation. WT and BASH-deficient (BASH−) DT40 cells were transiently transfected with pCAGGS-HPK1:HA (30 μg) in combination with 60 μg of either T7-BASH, T7-BASH (R373K), or empty vector (mock). Transfected cells were either left unstimulated (−) or stimulated with anti–chicken IgM Ab for 3 min (+) and lysed. HPK1 was immunoprecipitated using anti-HA Ab and subjected to an in vitro kinase assay (top) and immunoblotting (middle) as described in B. BASH protein expression levels were visualized by immunoblotting of the lysates with anti-T7 Ab (bottom). (D) Tyr-379 is critical for activation of HPK1 by BCR stimulation. DT40 cells were transfected transiently with pcDNA3-HPK1:HA or pcDNA3-HPK1(Y379F):HA vector (75 μg each). Cells were stimulated and HPK1 kinase activity was assayed as in B. (E) Spontaneous kinase activity of ectopically expressed HPK1(Y379F) is comparable to WT HPK1. pcDNA3-HPK1:HA, pcDNA3-HPK1(Y379F):HA, or pcDNA3-HPK1(K46E):HA DNA (3 μg each) was transfected into 293T cells and kinase activity of HPK1 was assayed as described in B.
In BASH-deficient DT40 cells, the BCR-mediated HPK1 activation was substantially reduced (Fig. 4 B), although its tyrosine-phosphorylation level was comparable to that in WT DT40 cells (Fig. 2 B). Therefore, BASH contributed greatly to the activation, but not to the tyrosine-phosphorylation, of HPK1 by BCR. To determine the role of the SH2 domain of BASH in the activation of HPK1, BASH-deficient cells were transiently reconstituted with either WT BASH or its SH2 domain-mutant, BASH (R373K), and BCR-mediated activation of cotransfected HPK1 was measured by in vitro kinase assay. As shown in Fig. 4 C, WT BASH restored the activation of HPK1 by the BCR (lanes 5 and 6), while BASH (R373K) did not (lanes 7 and 8), indicating that a functional SH2 domain is a prerequisite for BASH-mediated HPK1 activation. Furthermore, the Syk phosphorylation–deficient mutant HPK1 (Y379F) was barely activated in DT40 cells after BCR ligation (Fig. 4 D). When expressed in 293T cells, spontaneous kinase activity of HPK1(Y379F) was comparable to that of WT HPK1, in contrast to that of a kinase-dead mutant of HPK1 (K46E), indicating that the Y379F mutation did not affect the intrinsic kinase activity of HPK1 (Fig. 4 E). Taken together, these data indicated that the BCR-induced activation of HPK1 is largely dependent on binding to the BASH SH2 domain, which is mediated by Tyr-379 after inducible phosphorylation by Syk.
Although our results clearly indicated that BASH mediates, through direct interaction, the activation of HPK1 after BCR ligation, the exact mechanism for this observation remains to be elucidated. Many protein kinases are known to be activated by translocation to the plasma membrane, and BASH has been shown to translocate to the membrane after BCR stimulation 7. Therefore, it is tempting to speculate that BASH mediates the translocation of HPK1 to the plasma membrane, where activation of HPK1 kinase takes place. However, we found a substantial amount of HPK1 protein constitutively associated with the membrane fraction of WEHI231 cells which was independent of BCR stimulation (data not shown). Therefore, BCR-induced activation of HPK1 is not likely to result from membrane translocation of HPK1. It is equally unlikely that BASH brings HPK1 close to the BCR-associated PTKs for activation, because BCR-induced tyrosine-phosphorylation of HPK1 was not abrogated in the absence of BASH (Fig. 2 B). Alternatively, binding of the BASH SH2 domain to Y379 of HPK1 may cause a conformational change, which leads to the activation of the kinase. Another possibility would be that BASH recruits an unknown enzyme to HPK1 which modifies HPK1 to become active. Further studies are necessary to test these possibilities.
While our results indicated that BASH greatly contributes to the activation of HPK1 during BCR signaling, HPK1 activation was not completely abolished in the absence of BASH (Fig. 4B and Fig. C). In line with this notion, we observed a weak activation of HPK1(Y379F), a mutant that fails to associate with BASH, after BCR ligation (Fig. 4 D). Therefore, BASH-independent pathways leading to HPK1 activation are likely to exist. Liou et al. reported a notable reduction, although not entire abrogation, of BCR-mediated HPK1 activation in DT40 cells lacking the two mutually related adaptor proteins Grb2 and Grap, indicating that Grb2 and Grap redundantly contribute to BCR-mediated activation of HPK1 25. In support of this observation, previous reports demonstrated binding of the Grb2 SH3 domains to proline-rich motifs within the HPK1 hinge region 21 22. Taken together, these data imply that BASH and Grb2/Grap adaptors are independently able to activate, albeit not fully, HPK1 and that both adaptor types may act synergistically to achieve efficient HPK1 activation. In addition, Grb2 was shown to associate with BASH through either its SH2 or SH3 domains 8 11. In summary, it is possible that HPK1, BASH, and Grb2/Grap upon BCR stimulation form a trimolecular complex which might result in the efficient activation of HPK1 in a physiological context in B cells.
HPK1 Is a Positive Regulator of IKKβ Activation by BCR.
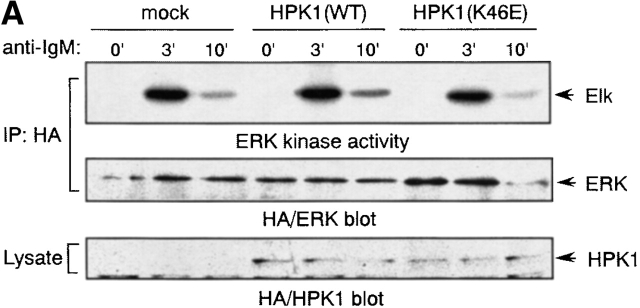
Previous reports have demonstrated that ectopic expression of HPK1 results in the activation of JNK/SAPK and IKKs in fibroblasts 18 19 20 and of NF-κB in T cells, myeloid progenitors, and epithelial cells 41. Liou et al. demonstrated that HPK1 is a negative regulator of TCR-induced ERK2 and AP-1 activation in T cells 25. To characterize the downstream effectors of HPK1 in BCR signaling, we examined the effect of HPK1 overexpression in DT40 cells on BCR-induced activation of such kinases. The activation states of the transiently expressed kinases were determined by in vitro kinase activities towards specific substrate proteins. The activity of exogenous ERK2 was markedly induced by anti–IgM Ab stimulation in DT40 cells, but was unaffected by the presence of exogenous WT or kinase-dead HPK1 (Fig. 5 A), even when the cells were stimulated suboptimally (1, 3, or 10 μg/ml of anti–IgM Ab, data not shown). Therefore, HPK1 is unlikely to play a major role in the regulation of ERK activity in response to BCR signaling, which is in contrast to its reported effect on TCR signaling. We failed to detect any induction of the kinase activity of exogenous JNK towards a GST-c-Jun fusion protein substrate after BCR ligation or stimulation with PMA plus ionomycin in our transient DT40 cell expression system (data not shown), although endogenous JNK has been shown to be activated by the same stimuli in DT40 cells 34 42. Thus, the involvement of HPK1 in BCR-mediated JNK activation remains to be clarified.
Figure 5.
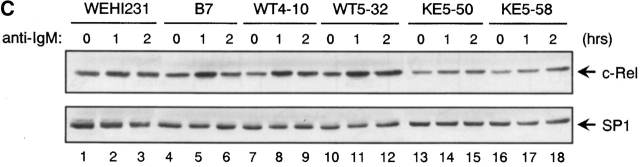
HPK1 positively regulates BCR-induced IKKβ activation. Expression vector (40 μg) for HA-epitope tagged ERK2 (A) or Flag-epitope tagged IKKβ (B) and either 40 μg of pcDNA-HPK1:HA (HPK[WT]), pcDNA-HPK1(K46E):HA [HPK(K46E)], pCAT7-HPK1-ΔN (HPK1-ΔN) 41, or empty vector (mock) were cotransfected into DT40 cells (107). After stimulation with anti–IgM Ab for the indicated periods of time (min), exogenous ERK2 (A) or IKKβ (B) were immunoprecipitated with antibodies directed against their corresponding tags and their catalytic activities were measured in vitro using the specific substrate proteins, GST-Elk1 (A) or GST-IκBα (B), respectively (top). The amount of the precipitated kinases was estimated by immunoblotting with anti-HA Ab (A, middle), anti-IKKβ Ab (B, middle, left), or anti-Flag Ab (B, middle, right). Expression levels of HPK1 proteins were visualized by immunoblotting with anti-HA Ab (bottom panels in A and B, left) or anti-HPK1 #7 Ab (B, bottom, right). In A, HA-epitope tagged HPK1 was also immunoprecipitated with ERK2. However, we confirmed that HPK1 is unable to phosphorylate GST-Elk1 as assessed in a separate in vitro kinase assay (data not shown). (C) Nuclear extracts were prepared from anti-IgM–stimulated (1 or 2 h) or unstimulated (0) cells of WEHI231, a WEHI231-stable transfectant highly expressing T7-tagged BASH (B7), B7-stable transfectants highly expressing either WT HPK1 (WT4-10 and WT5-32), or HPK1(K46E) (KE5-50 and KE5-58), and subjected to immunoblot analysis with anti-c-Rel antibody (top). A part of the blot was probed with anti-SP1 antibody to control for the amount and integrity of the nuclear extracts used (bottom).
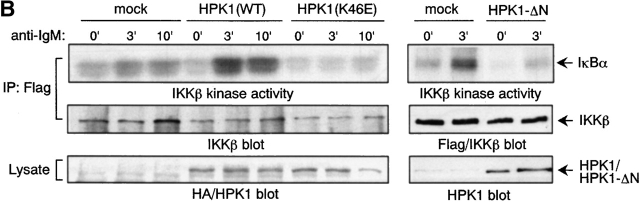
It has recently been reported that the IKK complex is activated by anti–IgM Ab stimulation of primary B cells 43. We have observed BCR-mediated activation of the endogenous IKK complex in a mouse B cell line, BAL-17. While we barely detected the activation of transfected exogenous IKKα in BAL-17 cells and DT40 cells (data not shown), we could readily detect BCR-induced activation of IKKβ upon transfection in these cell lines. BCR-dependent IKKβ activation was strongly enhanced by transfection of WT HPK1, while basal level IKKβ activity was unchanged. On the contrary, a kinase-dead mutant of HPK1, HPK1(K46E), potently inhibited BCR-induced IKKβ activation (Fig. 5 B, left; data not shown). In addition, the noncatalytic COOH-terminal part of HPK1 (HPK1-ΔN) potently inhibited BCR-induced IKKβ activation in DT40 cells (Fig. 5 B, right). The HPK1 COOH terminus has recently been shown to inhibit NF-κB activation via the IKK complex in a dominant negative fashion 41. These results strongly suggest that HPK1 is a physiological positive regulator of BCR-mediated activation of IKK complex. This function is dependent on HPK1 kinase activity and is most likely mediated by molecular interactions of its COOH-terminal part.
It is known that activation of the IKK complex results in the phosphorylation of specific serine residues, subsequent ubiquitination, and proteasome-dependent proteolysis of IκB. NF-κB/Rel proteins liberated from IκB then translocate to the nucleus and function as a transcription factor. In WEHI231 cells, it was previously shown that NF-κB/Rel (a p50-c-Rel dimer) continuously translocates to the nucleus, as the result of serine-phosphorylation– and proteasome-independent, constitutive proteolysis of IκBα 44. Thus, we observed constitutively nuclear c-Rel and only little increase of c-Rel in the nucleus after 1 h stimulation by BCR ligation in WEHI231 cells (Fig. 5 C, lanes 1–3). We found that overexpression of BASH in WEHI231 cells resulted in an enhancement of the BCR-induced nuclear accumulation of c-Rel (lanes 4–6). The BASH-mediated c-Rel nuclear accumulation was inhibited by overexpression of HPK1(K46E) (lanes 13–18) but not by WT HPK1 (lanes 7–12). This result suggests that BCR-induced nuclear translocation of c-Rel is upregulated by BASH through kinase activity of HPK1 and is consistent with our results indicating that HPK1 mediates BCR-induced IKKβ activation.
Collectively, we have identified a role of HPK1 in a selected signaling pathway leading to the IKK activation downstream of the BCR, which underscores the functional significance of HPK1 activation in BCR signal transduction. Obviously, further studies are needed to identify the direct target of HPK1 in the IKK activation pathway.
Concluding Remarks.
Here we provide evidence for an inducible interaction between the BASH SH2 domain and tyrosine-phosphorylated HPK1 in response to BCR ligation in vitro and in vivo. The interaction was dependent on tyrosine phosphorylation of HPK1 at a Syk consensus phosphorylation site (Tyr-379) and the presence of a functional BASH SH2 domain, both of which were also a prerequisite for the full BCR-mediated activation of HPK1 kinase activity. Our results are consistent with the results of a study which demonstrated an interaction of BLNK/BASH and HPK1 using a Syk-driven yeast two-hybrid system. The interaction was dependent on Syk kinase activity, a functional BLNK/BASH SH2 domain, and Tyr-379 of HPK1 (personal communication by K. Sauer et al.). In addition, in vivo SLP-76 interacted with HPK1 in T cells. TCR-mediated full activation of HPK1 in Jurkat T cells required the presence of an intact SLP-76 SH2 domain and Tyr-379 of HPK1 (personal communication). Together, these results indicate that HPK1 may be a common target of the conserved SH2 domains of SLP family hematopoietic adaptor proteins (including Clnk/MIST, references 45 and 46) in immunoreceptor signaling and reveal a novel function of these adaptors.
It has previously been shown that, upon BCR stimulation, tyrosine-phosphorylated BLNK/BASH interacts with the SH2 domains of PLCγ2 and Btk, and mediates PLCγ2 activation and the resulting calcium signaling 12 13. Recently, it has been shown that Btk and PLCγ2 are necessary for BCR-induced degradation of IκBα, as well as nuclear translocation and transcriptional activation of NF-κB in DT40 cells 43 47 48. Btk dependence of BCR-induced IKK activation, IκBα degradation, and nuclear translocation of NF-κB was also demonstrated using Btk-deficient mouse splenic B cells 43 47. Taken together with our results, BASH mediates activation of two independent pathways downstream of the BCR through distinct intramolecular sites; NH2-terminal phospho-tyrosine containing motifs function in Btk-mediated PLCγ2 activation, while the SH2 domain accomplishes HPK1 activation. Both pathways have now been shown to contribute to the activation of the IKK complex and NF-κB. However, each pathway can be activated independently by stimuli other than BCR ligation, and each pathway targets multiple and distinct downstream effectors. Therefore, BASH-mediated simultaneous activation of the two pathways may be critical for efficient linkage between BCR and IKK activation. In addition, this type of dual regulation system may contribute to the fine tuning of receptor signaling.
Acknowledgments
We would like to thank Drs. Tomohiro Kurosaki, Masamichi Ishiai, Hiroyasu Nakano, Eisuke Nishida, and Takeshi Tsubata for providing us the materials, and Dr. Karsten Sauer for the exchange of unpublished information.
This work was supported by grants to D. Kitamura and S. Tsuji from the Ministry of Education, Science, Sports, and Culture in Japan and Japan Society for the Promotion of Science, and to R. Goitsuka from Japan Science and Technology Corporation.
Footnotes
S. Tsuji's present address is Department of Immunology, Kinki University School of Medicine, Osaka-Sayama, Osaka 589-8511, Japan.
Abbreviations used in this paper: BASH, B cell adaptor containing SH2 domain; BCR, B cell antigen receptor; CBP, calmodulin-binding protein; ERK, extracellular signal–regulated kinase; HPK, hematopoietic progenitor kinase; IKKβ, IκB kinase β; MAPK, mitogen-activated protein kinase; MBP, myelin basic protein; NF, nuclear factor; PLCγ, phospholipase Cγ; PTK, protein tyrosine kinase; WT, wild-type.
References
- Reth M., Wienands J. Initiation and processing of signals from the B cell antigen receptor. Annu. Rev. Immunol. 1997;15:453–479. doi: 10.1146/annurev.immunol.15.1.453. [DOI] [PubMed] [Google Scholar]
- DeFranco A.L. The complexity of signaling pathways activated by the BCR. Curr. Opin. Immunol. 1997;9:296–308. doi: 10.1016/s0952-7915(97)80074-x. [DOI] [PubMed] [Google Scholar]
- Kurosaki T. Genetic analysis of B cell antigen receptor signaling. Annu. Rev. Immunol. 1999;17:555–592. doi: 10.1146/annurev.immunol.17.1.555. [DOI] [PubMed] [Google Scholar]
- Campbell K.S. Signal transduction from the B cell antigen-receptor. Curr. Opin. Immunol. 1999;11:256–264. doi: 10.1016/s0952-7915(99)80042-9. [DOI] [PubMed] [Google Scholar]
- Dolmetsch R.E., Lewis R.S., Goodnow C.C., Healy J.I. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- Healy J.I., Dolmetsch R.E., Timmerman L.A., Cyster J.G., Thomas M.L., Crabtree G.R., Lewis R.S., Goodnow C.C. Different nuclear signals are activated by the B cell receptor during positive versus negative signaling. Immunity. 1997;6:419–428. doi: 10.1016/s1074-7613(00)80285-x. [DOI] [PubMed] [Google Scholar]
- Fu C., Turck C.W., Kurosaki T., Chan A.C. BLNKa central linker protein in B cell activation. Immunity. 1998;9:93–103. doi: 10.1016/s1074-7613(00)80591-9. [DOI] [PubMed] [Google Scholar]
- Wienands J., Schweikert J., Wollscheid B., Jumaa H., Nielsen P.J., Reth M. SLP-65a new signaling component in B lymphocytes which requires expression of the antigen receptor for phosphorylation. J. Exp. Med. 1998;188:791–795. doi: 10.1084/jem.188.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goitsuka R., Fujimura Y., Mamada H., Umeda A., Morimura T., Uetsuka K., Doi K., Tsuji S., Kitamura D. BASH, a novel signaling molecule preferentially expressed in B cells of the bursa of Fabricius . J. Immunol. 1998;161:5804–5808. [PubMed] [Google Scholar]
- Myung P.S., Boerthe N.J., Koretzky G.A. Adapter proteins in lymphocyte antigen-receptor signaling. Curr. Opin. Immunol. 2000;12:256–266. doi: 10.1016/s0952-7915(00)00085-6. [DOI] [PubMed] [Google Scholar]
- Fu C., Chan A.C. Identification of two tyrosine phosphoproteins, pp70 and pp68, which interact with phospholipase Cγ, Grb2, and Vav after B cell antigen receptor activation. J. Biol. Chem. 1997;272:27362–27368. doi: 10.1074/jbc.272.43.27362. [DOI] [PubMed] [Google Scholar]
- Ishiai M., Kurosaki M., Pappu R., Okawa K., Ronko I., Fu C., Shibata M., Iwamatsu A., Chan A.C., Kurosaki T. BLNK required for coupling Syk to PLCγ2 and Rac1-JNK in B cells. Immunity. 1999;10:117–125. doi: 10.1016/s1074-7613(00)80012-6. [DOI] [PubMed] [Google Scholar]
- Hashimoto S., Iwamatsu A., Ishiai M., Okawa K., Yamadori T., Matsushita M., Baba Y., Kishimoto T., Kurosaki T., Tsukada S. Identification of the SH2 domain binding protein of Bruton's tyrosine kinase as BLNK. Functional significance of Btk-SH2 domain in B-cell antigen receptor-coupled calcium signaling. Blood. 1999;94:2357–2364. [PubMed] [Google Scholar]
- Jumaa H., Wollscheid B., Mitterer M., Wienands J., Reth M., Nielsen P.J. Abnormal development and function of B lymphocytes in mice deficient for the signaling adaptor protein SLP-65. Immunity. 1999;11:547–554. doi: 10.1016/s1074-7613(00)80130-2. [DOI] [PubMed] [Google Scholar]
- Pappu R., Cheng A.M., Li B., Gong Q., Chiu C., Griffin N., White M., Sleckman B.P., Chan A.C. Requirement for B cell linker protein (BLNK) in B cell development. Science. 1999;286:1949–1954. doi: 10.1126/science.286.5446.1949. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Nittono R., Okamoto N., Tsuji S., Hara Y., Goitsuka R., Kitamura D. The B cell-restricted adaptor BASH is required for normal development and antigen receptor-mediated activation of B cells. Proc. Natl. Acad. Sci. USA. 2000;97:2755–2760. doi: 10.1073/pnas.040575697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Tan J.E., Wong E.P., Manickam A., Ponniah S., Lam K.P. B cell development and activation defects resulting in xid-like immunodeficiency in BLNK/SLP-65-deficient mice. Int. Immunol. 2000;12:397–404. doi: 10.1093/intimm/12.3.397. [DOI] [PubMed] [Google Scholar]
- Kiefer F., Tibbles L.A., Anafi M., Janssen A., Zanke B.W., Lassam N., Pawson T., Woodgett J.R., Iscove N.N. HPK1, a hematopoietic protein kinase activating the SAPK/JNK pathway. EMBO J. 1996;15:7013–7025. [PMC free article] [PubMed] [Google Scholar]
- Hu M.C., Qiu W.R., Wang X., Meyer C.F., Tan T.H. Human HPK1, a novel human hematopoietic progenitor kinase that activates the JNK/SAPK kinase cascade. Genes Dev. 1996;10:2251–2264. doi: 10.1101/gad.10.18.2251. [DOI] [PubMed] [Google Scholar]
- Hu M.C., Wang Y., Qiu W.R., Mikhail A., Meyer C.F., Tan T.H. Hematopoietic progenitor kinase-1 (HPK1) stress response signaling pathway activates IκB kinases (IKK-α/β) and IKK-β is a developmentally regulated protein kinase. Oncogene. 1999;18:5514–5524. doi: 10.1038/sj.onc.1202740. [DOI] [PubMed] [Google Scholar]
- Anafi M., Kiefer F., Gish G.D., Mbamalu G., Iscove N.N., Pawson T. SH2/SH3 adaptor proteins can link tyrosine kinases to a Ste20-related protein kinase, HPK1. J. Biol. Chem. 1997;272:27804–27811. doi: 10.1074/jbc.272.44.27804. [DOI] [PubMed] [Google Scholar]
- Ling P., Yao Z., Meyer C.F., Wang X.S., Oehrl W., Feller S.M., Tan T.H. Interaction of hematopoietic progenitor kinase 1 with adapter proteins Crk and CrkL leads to synergistic activation of c-Jun N-terminal kinase. Mol. Cell. Biol. 1999;19:1359–1368. doi: 10.1128/mcb.19.2.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensenat D., Yao Z., Wang X.S., Kori R., Zhou G., Lee S.C., Tan T.H. A novel src homology 3 domain-containing adaptor protein, HIP-55, that interacts with hematopoietic progenitor kinase 1. J. Biol. Chem. 1999;274:33945–33950. doi: 10.1074/jbc.274.48.33945. [DOI] [PubMed] [Google Scholar]
- Nagata Y., Kiefer F., Watanabe T., Todokoro K. Activation of hematopoietic progenitor kinase-1 by erythropoietin. Blood. 1999;93:3347–3354. [PubMed] [Google Scholar]
- Liou J., Kiefer F., Dang A., Hashimoto A., Cobb M.H., Kurosaki T., Weiss A. HPK1 is activated by lymphocyte antigen receptors and negatively regulates AP-1. Immunity. 2000;12:399–408. doi: 10.1016/s1074-7613(00)80192-2. [DOI] [PubMed] [Google Scholar]
- Liu S.K., Smith C.A., Arnold R., Kiefer F., McGlade C.J. The adaptor protein Gads (Grb2-related adaptor downstream of Shc) is implicated in coupling hemopoietic progenitor kinase-1 to the activated TCR. J. Immunol. 2000;165:1417–1426. doi: 10.4049/jimmunol.165.3.1417. [DOI] [PubMed] [Google Scholar]
- Fukuda T., Kitamura D., Taniuchi I., Maekawa Y., Benhamou L.E., Sarthou P., Watanabe T. Restoration of surface IgM-mediated apoptosis in an anti-IgM-resistant variant of WEHI-231 lymphoma cells by HS1, a protein-tyrosine kinase substrate. Proc. Natl. Acad. Sci. USA. 1995;92:7302–7306. doi: 10.1073/pnas.92.16.7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M., Sabe H., Hata A., Inazu T., Homma Y., Nukada T., Yamamura H., Kurosaki T. Tyrosine kinases Lyn and Syk regulate B cell receptor-coupled Ca2+ mobilization through distinct pathways. EMBO J. 1994;13:1341–1349. doi: 10.1002/j.1460-2075.1994.tb06387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M., Kurosaki T. A role for Bruton's tyrosine kinase in B cell antigen receptor-mediated activation of phospholipase C-γ2. J. Exp. Med. 1996;184:31–40. doi: 10.1084/jem.184.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimura T., Goitsuka R., Zhang Y., Saito I., Reth M., Kitamura D. Cell-cycle arrest and apoptosis induced by Notch1 in B cells. J. Biol. Chem. 2000;275:36523–36531. doi: 10.1074/jbc.M006415200. [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Yamanashi Y., Fukuda T., Nishizumi H., Inazu T., Higashi K., Kitamura D., Ishida T., Yamamura H., Watanabe T., Yamamoto T. Role of tyrosine phosphorylation of HS1 in B cell antigen receptor-mediated apoptosis. J. Exp. Med. 1997;185:1387–1392. doi: 10.1084/jem.185.7.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H., Shindo M., Sakon S., Nishinaka S., Mihara M., Yagita H., Okumura K. Differential regulation of IκB kinase α and β by two upstream kinases, NF-κB-inducing kinase and mitogen-activated protein kinase/ERK kinase kinase-1. Proc. Natl. Acad. Sci. USA. 1998;95:3537–3542. doi: 10.1073/pnas.95.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto A., Okada H., Jiang A., Kurosaki M., Greenberg S., Clark E.A., Kurosaki T. Involvement of guanosine triphosphatases and phospholipase C-γ2 in extracellular signal-regulated kinase, c-Jun NH2-terminal kinase, and p38 mitogen-activated protein kinase activation by the B cell antigen receptor. J. Exp. Med. 1998;188:1287–1295. doi: 10.1084/jem.188.7.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T., Takata M., Yamanashi Y., Inazu T., Taniguchi T., Yamamoto T., Yamamura H. Syk activation by the Src-family tyrosine kinase in the B cell receptor signaling. J. Exp. Med. 1994;179:1725–1729. doi: 10.1084/jem.179.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzene M., Brunati A.M., Marin O., Donella-Deana A., Pinna L.A. SH2 domains mediate the sequential phosphorylation of HS1 protein by p72syk and Src-related protein tyrosine kinases. Biochemistry. 1996;35:5327–5332. doi: 10.1021/bi9528614. [DOI] [PubMed] [Google Scholar]
- Brunati A.M., Donella-Deana A., James P., Quadroni M., Contri A., Marin O., Pinna L.A. Molecular features underlying the sequential phosphorylation of HS1 protein and its association with c-Fgr protein-tyrosine kinase. J. Biol. Chem. 1999;274:7557–7564. doi: 10.1074/jbc.274.11.7557. [DOI] [PubMed] [Google Scholar]
- Brunati A.M., Donella-Deana A., Ruzzene M., Marin O., Pinna L.A. Site specificity of p72syk protein tyrosine kinaseefficient phosphorylation of motifs recognized by Src homology 2 domains of the Src family. FEBS Lett. 1995;367:149–152. doi: 10.1016/0014-5793(95)00555-n. [DOI] [PubMed] [Google Scholar]
- Raab M., Kang H., da Silva A., Zhu X., Rudd C.E. FYN-T-FYB-SLP-76 interactions define a T-cell receptor ζ/CD3-mediated tyrosine phosphorylation pathway that up-regulates interleukin 2 transcription in T-cells. J. Biol. Chem. 1999;274:21170–21179. doi: 10.1074/jbc.274.30.21170. [DOI] [PubMed] [Google Scholar]
- Geng L., Laab M., Rudd C.E. Cutting edgeSLP-76 cooperativity with FYB/FYN-T in the up-regulation of TCR-driven IL-2 transcription requires SLP-76 binding to FYB at Tyr595 and Tyr651 . J. Immunol. 1999;163:5753–5757. [PubMed] [Google Scholar]
- Arnold R., Liou J., Drexler H.C.A., Weiss A., Kiefer F. Caspase mediated cleavage of hematopoietic progenitor kinase 1 (HPK1) converts an activator of NFκB into an inhibitor of NFκB. J. Biol. Chem. 2001;276:14675–14684. doi: 10.1074/jbc.M008343200. [DOI] [PubMed] [Google Scholar]
- Jiang A., Craxton A., Kurosaki T., Clark E.A. Different protein tyrosine kinases are required for B cell antigen receptor-mediated activation of extracellular signal-regulated kinase, c-Jun NH2-terminal kinase 1, and p38 mitogen-activated protein kinase. J. Exp. Med. 1998;188:1297–1306. doi: 10.1084/jem.188.7.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petro J.B., Rahman S.M.J., Ballard D.W., Khan W.N. Bruton's tyrosine kinase is required for activation of IκB kinase and nuclear factor κB in response to B cell receptor engagement. J. Exp. Med. 2000;191:1745–1753. doi: 10.1084/jem.191.10.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Seufzer B.J., Shumway S.D. Novel IκBα proteolytic pathway in WEHI231 immature B cells. Mol. Cell. Biol. 1998;18:19–29. doi: 10.1128/mcb.18.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M.Y., Davidson D., Yu J., Latour S., Veillette A. Clnk, a novel SLP-76-related adaptor molecule expressed in cytokine-stimulated hemopoietic cells. J. Exp. Med. 1999;190:1527–1534. doi: 10.1084/jem.190.10.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goitsuka R., Kanazashi H., Sasanuma H., Fujimura Y., Hidaka Y., Tatsuno A., Ra C., Hayashi K., Kitamura D. A BASH/SLP-76-related adaptor protein MIST/Clnk involved in IgE receptor-mediated mast cell degranulation. Int. Immunol. 2000;12:573–580. doi: 10.1093/intimm/12.4.573. [DOI] [PubMed] [Google Scholar]
- Bajpai U.D., Zhang K., Teutsch M., Sen R., Wortis H.H. Bruton's tyrosine kinase links the B cell receptor to nuclear factor κB activation. J. Exp. Med. 2000;191:1735–1744. doi: 10.1084/jem.191.10.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petro J.B., Khan W.N. Phospholipase C-γ2 couples Bruton's tyrosine kinase to the NF-κB signaling pathway in B lymphocytes. J. Biol. Chem. 2001;276:1715–1719. doi: 10.1074/jbc.M009137200. [DOI] [PubMed] [Google Scholar]