Abstract
The Leishmania mexicana PFR2 locus encodes a component of the paraflagellar rod (PFR), a flagellar structure found only in the insect stage of the life cycle. PFR2 mRNA levels are 10-fold lower in the mammalian stage than in the insect stage. Nuclear run-on experiments indicate that the change in PFR2 mRNA abundance is achieved posttranscriptionally. Deletion and block substitution analysis of the entire 1,400-nucleotide 3′ untranslated region (UTR) of PFR2C led to the identification of a regulatory element contained within 10 nucleotides of the 3′ UTR, termed the PFR regulatory element (PRE), that is necessary for the 10-fold regulation of PFR2 mRNA levels. Comparison of the half-lives of PFR2 transcripts, identical except for the presence or absence of the PRE, revealed that the PRE acts by destabilizing the PFR2 mRNA in amastigotes. The PRE was inserted into a construct which directs the constitutive expression of a chimeric PFR2 transcript. Insertion of the PRE resulted in regulated expression of this transcript, demonstrating that the regulatory element is sufficient for promastigote-specific expression. Since the PRE is present in the 3′ UTR of all L. mexicana PFR genes examined so far, we propose that it serves a means of coordinating expression of PFR genes.
In tropical areas where phlebotomine sand fly vectors are endemic, protozoan parasites of the genus Leishmania cause widespread human disease. As Leishmania parasites cycle between the insect vector and the mammalian host, they differentiate into morphologically and biochemically distinct stages that are adapted for survival in the distinct environment of each host. Insect-stage promastigotes can be readily distinguished from mammalian-stage amastigotes by the presence of a long flagellum emerging anteriorally. The flagellum is the motility organelle of promastigotes and infectious metacyclic promastigotes (1, 26). Only a rudimentary nonemergent flagellum is present in amastigotes (25). The flagellum therefore affords a unique opportunity to understand stage-specific regulatory mechanisms employed by Leishmania.
Trypanosomatid protozoans such as Leishmania have evolved cellular pathways that are fundamentally different from those of organisms that have been studied more extensively, such as bacteria, yeasts, and mammals (27). In trypanosomes, mature mRNAs are formed by processing of polycistronic pre-mRNAs. This occurs by trans splicing of a capped 39-nucleotide (nt) miniexon near the 5′ end of the coding sequence (28). Polycistronic transcription units often contain mRNAs whose steady-state levels are vastly different or mRNAs that accumulate at different stages of the life cycle. Coupled with a failure to identify RNA polymerase II promoters associated with Leishmania genes, this has led to a paradigm in which posttranscriptional mechanisms for regulation of gene expression predominate (29). The Leishmania mexicana genes PFR1 and PFR2, which encode the major structural components of the paraflagellar rod (PFR), conform to this posttranscriptional regulation paradigm.
The PFR, restricted to the flagella of kinetoplastids, euglenoids, and some dinoflagellates, is a massive cytoskeletal structure that runs the length of the flagellum next to the axoneme once it emerges from the flagellar pocket (32). The PFR is essential for flagellar motility in Leishmania promastigotes; however, it is absent from the attenuated flagellum of amastigotes (1, 26). Two major protein components of the PFR have been identified in many trypanosomatid species and are referred to here as PFR1 and PFR2. The genetic loci share a common organization, being composed of tandem arrays of four and three genes, respectively (23). Steady-state mRNA levels of PFR1 and PFR2 are about 10-fold greater in promastigotes, which possess a PFR, than in amastigotes, which lack a PFR. Genes flanking the PFR1 and PFR2 arrays do not display this regulation, which suggests either the presence of specialized regulated promoters for the PFR genes or a posttranscriptional means of regulation (23).
Unlike the regulation of gene expression in most prokaryotic and eukaryotic organisms, which occurs primarily at the level of transcription, gene regulation in trypanosomes is largely posttranscriptional, occurring at the level of trans splicing, polyadenylation, mRNA stability, translation, and protein stability (12, 29, 30). However, transcriptional regulation in trypanosomes has been observed in only a few cases of specialized polymerase I promoters, such as in the genes that encode variable surface glycoproteins and procyclic acidic repetitive proteins (13). In contrast to the dearth of evidence for transcriptional control in Leishmania, there are many examples of trypanosomatid genes whose mRNA levels are modulated by posttranscriptional mechanisms. Sequences within the 3′ untranslated region (UTR) (2, 4, 10, 14, 15, 33), the 5′ UTR (21), or the coding sequence of an mRNA (33) can contribute to the regulation of a particular mRNA's abundance.
Sequences outside the mature mRNA can also control differential expression of Leishmania genes. For example, Brooks et al. have shown that stage-regulated differential gene expression of the cysteine protease gene array is dependent upon the presence or absence of short sequence elements in the respective intercistronic region and that these sequence elements influence processing of precursor mRNA (3). These regulatory sequences presumably influence events in the maturation of mRNA such as trans splicing.
The expression of many proto-oncogenes, cytokines, and lymphokines in higher eukaryotes is controlled at the level of mRNA stability. AU-rich elements (AREs), present in the 3′ UTRs of these mRNAs, control the decay rates of these transcripts by modulating poly(A)-deadenylation rates and subsequent decay of the mRNA (5, 22). Recently, a yeast transcript, TIF51A, was shown to be subject to regulation by an ARE present in its 3′UTR. Both yeast and mammalian AREs promoted deadenylation-dependent mRNA decay in the yeast system, suggesting conservation of this decay process from yeasts to mammals (31). In trypanosomatids, a specific ARE has been shown to control the stage-regulated expression of the Trypanosoma cruzi mucin gene family (8). This suggests that AREs might be involved in regulation of other trypanosomatid gene families.
We report here that expression of the L. mexicana PFR2 genes is regulated posttranscriptionally by modulation of mRNA decay rates, and we have identified a 10-nt AU-rich regulatory element in the 3′ UTR of the PFR2C gene that is both necessary and sufficient for the stage-specific regulation of PFR2 mRNA.
MATERIALS AND METHODS
Parasites and cell culture.
Promastigotes of L. mexicana (WHO strain MNYC/BZ/62/m379) were cultured in M199 medium containing 5% (vol/vol) fetal bovine serum and 5% (vol/vol) bovine embryonic fluid at 26°C as described previously (18). All amastigotes of L. mexicana used in this study were from axenic cultures and were obtained by shifting the incubation conditions of promastigotes to 33°C and pH 5.5 in a modified UM 54 medium as described previously (23). Amastigote cultures were maintained by serial dilutions (1:25) every 3 to 4 days. Axenic amastigotes were harvested at least 5 days after initial differentiation for RNA and protein analysis. Logarithmic-phase cultures (5 × 106 to 9 × 106 cells/ml) were used in all experiments. The Δpfr2 line used in this work was the previously described L. mexicana PFR2 knockout line, 13.2 (26).
Transfection.
Methods for electroporation of DNA into Leishmania and selection of transfectants were described previously (18). Puromycin was the selective drug for all experiments described in this work and was maintained at concentrations of 10 μM in liquid culture and 20 μM on selective plates. Leishmania lines generated from three to five independent transformants were assayed for regulation.
RNA analysis.
Total Leishmania RNA (5 μg) was isolated and was fractionated by electrophoresis on 1.2% (wt/vol) agarose gels containing formaldehyde as described previously (19). The RNA was transferred to Hybond-N nylon membranes (Amersham) in 20× SSPE (0.2 M sodium phosphate [pH 7.4], 0.2 M EDTA, 3 M NaCl). Prehybridization and hybridization were carried out at 65°C in a hybridization oven using a buffer containing 50% formamide, 5× Denhardt's solution (0.1% Ficoll, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin), 5× SSPE, 0.1% sodium dodecyl sulfate, 0.2 mg of denatured salmon sperm DNA per ml, and 0.3 mg of yeast tRNA per ml. Radiolabeled antisense PFR2 RNA probes were generated by in vitro transcription of EcoRI-linearized pPFR2 (20) with bacteriophage T3 RNA polymerase. PFR2 mRNA levels were determined with a phosphorimager and normalized to rRNA levels by reprobing membranes with a radiolabeled ribosomal small-subunit-RNA probe prepared by in vitro transcription of EcoRI-linearized pLmrRNA, which is described below.
In vitro nuclear run-on assay.
Promastigotes and amastigotes were grown to mid-log phase (5 × 106 to 1 × 107 parasites/ml) before nuclei were isolated. Intact nuclei were isolated from approximately 2 × 109 cells or about 100 to 200 ml of culture. The parasites were harvested by centrifugation at 1,000 × g in a table-top centrifuge and washed once in cold PS buffer (44 mM NaCl, 57 mM Na2HPO4, and 3 mM KH2PO4 at pH 8.0) (7). The pellet was resuspended in 1 ml of cold PS to a concentration of 2 × 109 parasites/ml and placed on ice. NP-40 was added to a final concentration of 0.8%. Lysis of the parasite plasma membrane but not the nuclear membrane was verified by phase-contrast microscopy. The nuclei were pelleted and washed twice in nucleus wash buffer (100 mM HEPES [pH 7.5], 50 mM NaCl, 50 mM KCl, 1 mM EGTA, 1 mM dithiothreitol, 1 mM spermidine). Nuclei were resuspended in 100 μl of nucleus storage buffer (20% glycerol, 100 mM HEPES [pH 7.5], 50 mM NaCl, 50 mM KCl, 2 mM MgCl2) and stored at −70°C after quick-freezing in liquid N2.
Reactions were run using 2 × 109 nuclei per reaction. The reaction was initiated by the addition of 200 μl of elongation buffer (100 mM HEPES (pH 7.5), 50 mM NaCl, 50 mM KCl, 2 mM MgCl2, 5 μM dithiothreitol, 0.5 mM spermine, 1 mM spermidine, 1 mM putrescine, 20% glycerol, 1 mM ATP, 1 mM GTP, 1 mM UTP, 0.1 mM creatine phosphate, 10 μg of creatine kinase, 160 U of RNasin, 500 μCi of [α-32P]CTP at 800 Ci/mmol) and incubation at 30°C. After 10 min the reaction was stopped by the addition of 20 U of DNase I followed by 200 μl of 20 mM Tris (pH 7.5), 20 mM EDTA, 1 mg of proteinase K per ml, and 1% sodium dodecyl sulfate and incubated for 10 min at 37°C. Nascent RNA was isolated by phenol-chloroform (1:1) extraction and ethanol precipitation. Labeled nascent RNA was hybridized to filters containing plasmid DNA. The plasmid DNA was prepared as follows. Five hundred nanograms of pfla2 (and equal molar amounts of all other plasmids) was diluted in a total volume of 300 μl of H2O, denatured by the addition of 30 μl of 2 M NaOH for 5 min, and neutralized by the addition of 30 μl of 3 M sodium acetate. Denatured plasmid DNAs were slotted onto Nytran membranes (Schleicher & Schuell) and fixed to the membrane by UV cross-linking. Prehybridization of membranes and washes were as previously described (19). The entire quantity of radiolabeled RNA obtained per 2 × 109 nuclei was allowed to hybridize at 42°C for 72 h. A phosphorimager was used to quantify radiolabeled RNA. Hybridization values were corrected for a low level of hybridization to the vector backbone, pBluescript II SK(+) (Stratagene), by performing the hybridization with equal molar amounts of each plasmid and subtracting the value obtained by hybridization to pBluescript. After subtraction, the phosphorimager values were divided by the size of the insert for each clone to arrive at a value representing transcription per kilobase. Transcription across the PFR2 coding sequences was normalized to take into account the presence of three gene copies. To compare results from different experiments, all values were normalized to those for transcription of β-tubulin, whose steady-state mRNA levels are expressed constitutively (16). Plasmids p2.5H, p1.5SM, pfla2, p3.0X, and p1.9X, which contain subcloned portions of the PFR2 locus, have been described (23). Plasmid pLT-β1, which contains β-tubulin sequences, is described below.
Plasmid construction.
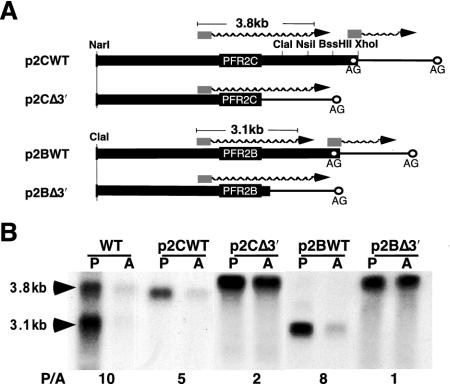
All transfected plasmids, except for pLocus, were constructed in the pX63PAC backbone, a Leishmania expression vector which confers resistance to puromycin (18). Plasmid pLocus contains a segment of about 23 kb of L. mexicana genomic DNA that spans the PFR2 locus from a ClaI site 7 kb downstream of the PFR2C stop codon to an Asp718 site about 7 kb upstream of the PFR2A start codon (see Fig. 2), housed in pBluescript II SK(+). Plasmid pNC2C contains a 10.5-kb segment of the L. mexicana PFR2 locus, extending from a NarI site 6 bp downstream from the PFR2B stop codon to a ClaI site about 7 kb downstream of the PFR2C stop codon, cloned between the HincII and ClaI sites of pBluescript II SK(+). The NarI 5′ overhang was filled in with the Klenow fragment of DNA polymerase I to enable blunt end ligation to the HincII site in pBluescript II SK(+). Plasmid pPFR2C was created by excising the 10.5-kb Leishmania DNA fragment of pNC2C with polylinker Asp718 and EcoRV sites, filling the 5′ overhang of the Asp718 site with Klenow, and blunt-end ligating it to BamHI-cut and Klenow-treated pX63PAC. The PFR2C and PAC genes were in the same relative orientation in pPFR2C (see Fig. 2). Plasmid p2CWT was made by excising a 5.5-kb XhoI fragment from pNC2C, filling in the termini with Klenow fragment, and blunt-end ligating it to BamHI-cut and Klenow-treated pX63PAC such that the PFR2C and PAC genes were in the same relative orientation (see Fig. 2). Plasmid p2CH is identical to p2CWT except that a HindIII site was inserted two nucleotides downstream of the pNC2C stop codon by a PCR-based mutagenesis strategy analogous to that described previously (26). Plasmid p2CΔ3′, a derivative of p2CH that lacks 3′ sequences downstream of the HindIII site, was made by deleting a 2-kb HindIII-BglII fragment of p2CH. Plasmid p2BΔ3′ is identical to plasmid pX63PAC-PFR2B, which was described previously (26). This plasmid contains only 200 nt of the PFR2B 3′ UTR. Plasmid p2BWT was made by inserting a 1.4-kb ClaI-SnaBI fragment from plasmid p3.8Cla that contained PFR2B 3′ flanking sequence into plasmid p2BΔ3′ (23).
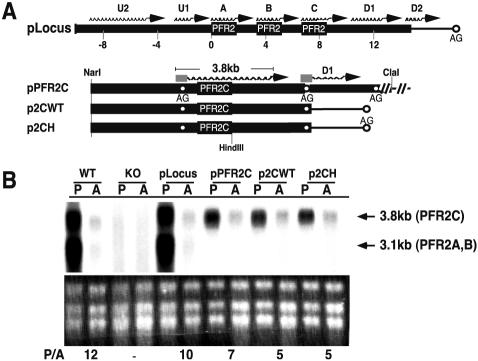
FIG. 2.
Episomal expression of PFR2 genes recapitulates wild-type regulation in the Δpfr2 background. (A) Schematic representation of various constructs used in stable-transfection experiments. Thick lines represent Leishmania sequences, and thin lines represent sequences from the vector itself. U1 and U2 are upstream transcripts; D1 and D2 are downstream transcripts relative to the PFR2 genes. Wavy lines at the top of the genes represent the position of mRNA synthesized from respective constructs. Open circles denote the position of the splice acceptor sites (AG). The splice acceptor sites for the PFR2C transcript and the D1 transcript are indicated under the position of the respective mRNA. A rectangular box at the 5′ end of the transcript denotes the miniexon. (B) Northern analysis of total RNA from promastigotes (P) and amastigotes (A) of wild-type L. mexicana (WT), the Δpfr2 strain (KO), and the Δpfr2 strain containing the indicated plasmids probed with PFR2 coding sequence as described in the text. Ethidium bromide-stained rRNA is shown below the autoradiogram. Numbers below rRNA are the ratio of PFR2 mRNA in promastigotes to that in amastigotes (P/A). This ratio was generated using PFR2 mRNA levels determined by using a phosphorimager and then normalizing to ethidium bromide-stained rRNA quantified by densitometry.
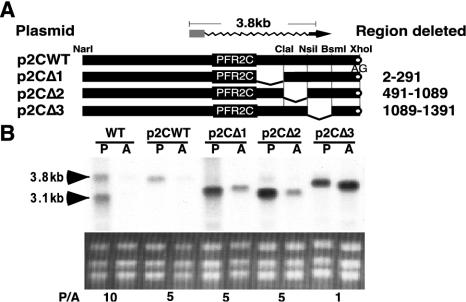
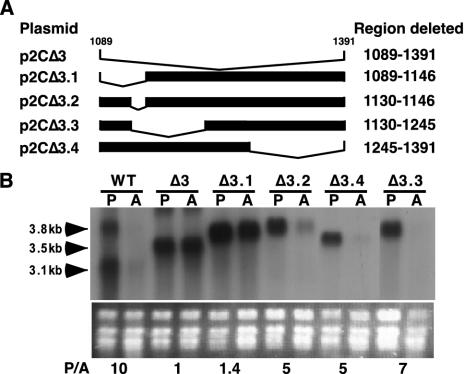
Plasmid p2CΔ1 was derived from p2CH by deletion of a 489-bp HindIII-ClaI fragment in the 3′ UTR of PFR2C. Plasmid p2CΔ2 was derived from plasmid p2CWT by deletion of a 595-bp ClaI-NsiI fragment in the 3′ UTR of PFR2C. Plasmid p2CΔ3 was derived from p2WT by deletion of a 305-bp NsiI-BsmI fragment in the 3′ UTR of PFR2C (see Fig. 4). Plasmid p2CΔ3.1 is a derivative of p2CWT with sequences deleted between an NsiI site at position 1089 and an NaeI site at position 1146. Plasmid p2CΔ3.2 is a derivative of p2CWT with sequences deleted between NaeI sites at positions 1130 and 1146. Plasmid p2CΔ3.3 is a derivative of p2CWT with sequences deleted between an NaeI site at position 1130 and an ApaLI site at position 1245 (see Fig. 5). Plasmid p2CΔ3.4 is a derivative of p2CWT with sequences deleted between an ApaLI site at position 1245 and a BsmI site at position 1391 (see Fig. 5). Each of these four deletions was verified by sequencing.
FIG. 4.
Deletion analysis of the PFR2C 3′ UTR. (A) Schematic representation of PFR2C cassettes. Symbols are as described in the legend to Fig. 2A. The position of the deletions is indicated by the absence of a solid bar. (B) Northern analysis of total RNA from promastigotes (P) and amastigotes (A) of the wild type (WT) and of the Δpfr2 strain containing the indicated plasmids probed with PFR2 coding sequence. Ethidium bromide-stained rRNA is shown below the autoradiogram. Numbers below the rRNA are ratios of PFR2 mRNA in promastigotes to that in amastigotes (P/A), determined as described in the legend to Fig. 2B.
FIG. 5.
Identification of regulatory sequences in region 3 of the 3′ UTR. (A) Schematic diagram of further deletions (Δ3.1, Δ3.2, Δ3.3, and Δ3.4) in region 3 of the PFR2C 3′ UTR. The position of the deletions is indicated by the absence of a solid bar. (B) Northern analysis of total RNA from promastigotes (P) and amastigotes (A) of the wild type (WT) and the Δpfr2 line containing the indicated plasmids (p2CΔ3, p2CΔ3.1, p2CΔ3.2., p2CΔ3.3, and p2CΔ3.4) probed with PFR2 coding sequence. Ethidium bromide-stained rRNA is shown below the autoradiogram. Numbers below the rRNA are ratios of PFR2 mRNA in promastigotes to that in amastigotes (P/A), determined as described in the legend to Fig. 2B.
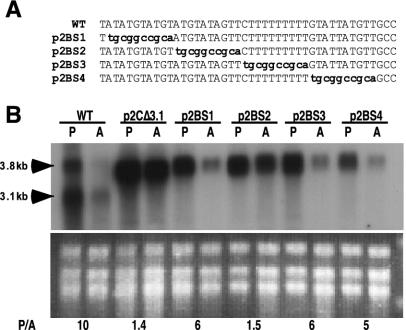
To facilitate block substitution mutagenesis in the PFR2C 3′ UTR, a 2-kb HindIII-Asp718 fragment from plasmid p7NEO (26) was ligated into HindIII and Asp718-digested pUC18, creating plasmid p3′UTR. The inserted fragment encompasses the PFR2C 3′ UTR from a HindIII site just downstream from the PFR2C stop codon to a XhoI site 2 kb further downstream. Block substitutions were generated in this plasmid by excising the 59-bp NsiI-NaeI fragment and replacing it with pairs of annealed oligonucleotides that spanned the region but contained the sequence AGCGGCCGCT, which contains a NotI cleavage site, in place of successive 10-nt segments. Additionally, each oligonucleotide replaced the C at position 1130 (relative to the PFR2C stop codon) with an A to eliminate the first of the two successive NaeI sites. Each oligonucleotide was phosphorylated at 37°C using polynucleotide kinase (1 U) and then annealed to the complementary oligonucleotide. The annealed oligonucleotide pairs which contained a 3′ NsiI overhang at one end and a 5′ NaeI overhang at the other end were then ligated to the NsiI- and NaeI-cut p3′UTR DNA. The oligonucleotide sequences are as follows: BS1A, 5′-TTGCGGCCGCAATGTATAGTTCTTTTTTTTTGTATTATGTTGCAGGCTGCTGCCCCCG; BS1B, 5′-CCGGCGGGGGCAGCAGCCTGCAACATAATACAAAAAAAAAGAACTATACATTGCGGCCGCAATGCA; BS2A, 5′-TATATGTATGTTGCGGCCGCACTTTTTTTTTGTATTATGTTGCAGGCTGCTGCCCCCG; BS2B, 5′-CCGGCGGGGGCAGCAGCCTGCAACATAATACAAAAAAAAAGTGCGGCCGCAACATACATATATGCA; BS3A: 5′-TATATGTATGTATGTATAGTTTGCGGCCGCAGTATTATGTTGCAGGCTGCTGCCCCCG; BS3B, 5′-CCGGCGGGGGCAGCAGCCTGCAACATAATACTGCGGCCGCAAACTATACATACATACATATATGCA; BS4A, 5′-TATATGTATGTATGTATAGTTCTTTTTTTTTTGCGGCCGCAGCAGGCTGCTGCCCCCG; and BS4B, 5′-CCGGCGGGGGCAGCAGCCTGCTGCGGCCGCAAAAAAAAAAGAACTATACATACATACATATATGCA.
Oligonucleotides BS1A and BS1B were annealed and ligated into plasmid p3′UTR to create plasmid p2BS1. To insert the block substitution into p2CWT, a unique 1.5-kb BstXI-BinI fragment spanning the substitution was excised and inserted in place of the corresponding fragment of p2CWT, creating plasmid p2BS1 (see Fig. 6). The block substitution plasmids p2BS2, p2BS3, and p2BS4 were made similarly. The plasmids were verified by sequencing.
FIG. 6.
Block substitution analysis of PFR2C 3′ UTR containing the putative regulatory region. (A) Position of the 10-nt substitution sequences, shown in lowercase, in each of the four block substitution constructs made as described in the text. (B) Northern analysis of total RNA from promastigotes (P) and amastigotes (A) of the wild type (WT) and the Δpfr2 line containing the indicated plasmids probed with PFR2 coding sequence. Ethidium bromide-stained rRNA is shown below the autoradiogram. Numbers below the rRNA are ratios of PFR2 mRNA in promastigotes to that in amastigotes (P/A), determined as described in the legend to Fig. 2B.
PCR was used to insert the sequence ATGTATAGTT into plasmid p2CΔ3′ at position 1014 downstream of the BamHI site. A 711-bp DraI site was excised from the plasmid and replaced with a DraI-digested PCR product generated by using two primers, RM1 and RM2, that span the DraI sites using p2CΔ3′ as a template. Primer RM1 contains three nucleotide changes that specify the sequence ATGTATAGTT at the aforementioned position. A fourth nucleotide change was required to eliminate an adjacent DraI site. The resultant plasmid, pΔM, was verified by sequencing (see Fig. 7): RM1, 5′-CCCTTTTAAATTAAAAATGAAGTTTTAAG*TCAATG*TAT*AGT*TTATATGAGT (nucleotides preceding asterisks have been changed for the creation of the regulatory element in the 3′ UTR and the elimination of the DraI site); RM2, 5′-GCACTTTTAAAGTTCTGCTATGTGGCGCGGTATTATCCCGTATTGACGCCGGGC.
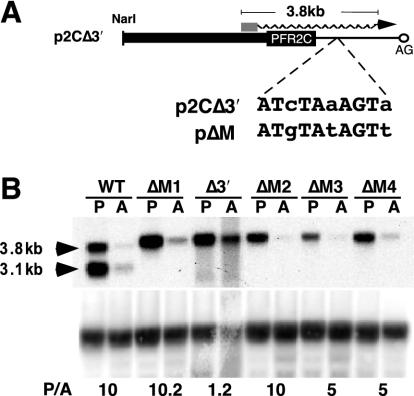
FIG. 7.
The 10-nt element is sufficient to confer promastigote-specific expression. (A) Schematic diagram showing the position of a 7-of-10 match to the 10-nt sequence in the plasmid p2CΔ3′. Symbols are as described in the legend to Fig. 2A. Nucleotides in lowercase were mutated, resulting in plasmid pΔM, which contains the 10-nt element. (B) Northern analysis of total RNA from promastigotes (P) and amastigotes (A) of the wild type (WT) and four Δpfr2 clonal lines harboring pΔM (ΔM1, ΔM2, ΔM3, and ΔM4) probed with PFR2 coding sequence. The blot was reprobed with an rRNA probe as described in the text. Numbers below the autoradiogram are ratios of PFR2 mRNA in promastigotes to that in amastigotes (P/A), determined as described in the legend to Fig. 2B except that the rRNA was quantified with a phosphorimager.
Plasmid pLmrRNA, which contains L. mexicana small-subunit rRNA sequence, was generated by PCR from 200 ng of L. mexicana genomic DNA by using primers 450H and 449H and cloned into the EcoRI and BamHI sites of pBluescript II SK(+): 450H, 5′-gggaattcCCCCCTGAGACTGTAACC; 449H, 5′-ggggatccGCGGTAATTCCAGCTCCAAAAG. Uppercase bases represent the small-subunit-rRNA sequence.
RESULTS
Expression of L. mexicana PFR2 is posttranscriptionally regulated.
PFR2 is encoded by a tandem array of three identical genes whose steady-state mRNA levels are about 10-fold higher in promastigotes than in amastigotes (23). Transcripts of two sizes are generated from these three genes, which differ in the size and sequence of their 3′ UTRs. A 3.1-kb transcript arises from the first two genes of the PFR2 array, PFR2A and -B, and a 3.8-kb transcript arises from the third gene in the array, PFR2C.
In order to determine whether the regulation of steady-state PFR2 mRNA levels was due to changes in the rate of mRNA synthesis, RNA polymerase density across the PFR2 locus was measured with an in vitro nuclear run-on assay (7). Plasmid clones spanning the PFR2 array extending through two adjacent upstream (U1 and U2) and downstream (D1 and D2) transcripts were hybridized to 32P-labeled RNA synthesized in isolated nuclei from 109 promastigotes or amastigotes (23). Plasmid pLT-β1, which contains the β-tubulin gene, was included for normalization among experiments. Table 1 summarizes the results of experiments with promastigote and amastigote nuclei. Transcription rates across the PFR2 locus were nearly constant, differing less than twofold in both promastigotes and amastigotes. This continuity of transcription is consistent with the model that the PFR2 locus is transcribed as a polycistronic precursor. The α-amanitin sensitivity profile of PFR2 transcription indicated that the PFR2 locus was transcribed by RNA polymerase II (data not shown). As there is a 10-fold difference in steady-state PFR2 mRNA levels, the differential expression of PFR2 mRNA in the two life cycle stages must be controlled by regulatory events that occur posttranscriptionally.
TABLE 1.
Relative rates of transcription across the PFR2 region
| DNA | Transcript | Transcription ratea in:
|
P/Ab | |
|---|---|---|---|---|
| Promastigotes | Amastigotes | |||
| p2.5H | U2 | 6.6 ± 1.9 | 4.4 ± 1.8 | 1.5 |
| p1.5SM | U1 | 5.7 ± 0.7 | 4.8 ± 1.6 | 1.2 |
| pfla2 | PFR2 | 6.6 ± 0.7 | 4.9 ± 2.8 | 1.4 |
| p3.0X | D1 | 7.3 ± 2.0 | 6.4 ± 2.4 | 1.1 |
| p1.9X | D2 | 4.5 ± 1.6 | 4.3 ± 0.5 | 1.1 |
In arbitrary units.
P/A, ratio of rate in promastigotes to rate in amastigotes.
The half-life of PFR2 mRNA is shorter in amastigotes.
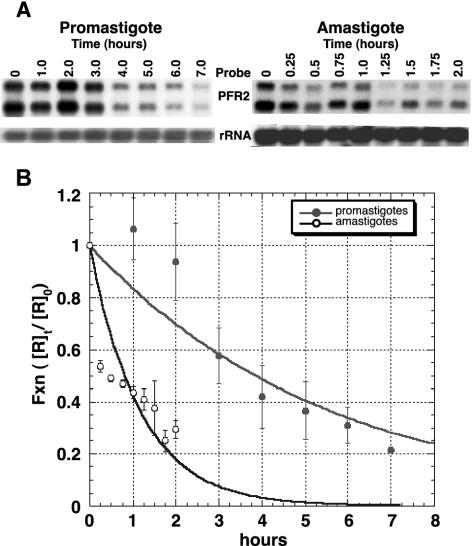
The decay rates of PFR2 mRNA in promastigotes and amastigotes were determined by using the RNA polymerase inhibitor actinomycin D. Total RNA was isolated after exposure of promastigotes or amastigotes to actinomycin D for various times. The amount of PFR2 mRNA was then determined by Northern blot analysis and quantified on a phosphorimager. A plot of the quantity of promastigote PFR2 mRNA versus the time course of actinomycin D incubation revealed the expected exponential decay pattern with an observed half-life of approximately 3.9 h (Fig. 1). In contrast, the PFR2 mRNA was degraded more rapidly in amastigotes with an observed half-life of about 0.8 h. These results indicate that changes in the rate of decay of mature PFR2 mRNA can account for the large differences in steady-state PFR2 mRNA levels during the life cycle. Therefore, regulatory elements governing the observed changes in the rate of decay must reside within the mature mRNA sequence.
FIG. 1.
Stability of PFR2 mRNA in promastigotes and amastigotes. Promastigotes and amastigotes were treated with actinomycin D (10 μg/ml) for the time indicated. Amastigotes received additional actinomycin D at 1 h. The zero time point was taken just prior to the addition of actinomycin D. Five micrograms of total RNA from each time point was fractionated, blotted, and probed with in vitro-synthesized RNA from pPFR2 as described in the text. (A) Representative Northern blots for the promastigote and amastigote time courses. The signals for the the PFR2 and rRNA probes are displayed. (B) PFR transcript abundance was quantitated by using a phosphorimager and was normalized to the small subunit of rRNA to control for loading. For each time point, the normalized values for PFR2 mRNA ([R]t) were divided by the normalized time zero value ([R]0) to obtain a value for the fraction of PFR2 mRNA remaining ([R]t/[R]0) which was plotted against time, in hours. The plotted data were fitted to the exponential decay expression: [R]t/[R]0 = e−kt, where k is a rate constant, t is time in hours, and e is the inverse natural log. For the promastigote data, k is 0.18 h−1; for amastigotes, k is 0.86 h−1. Values for the half-life were calculated by setting [R]t/[R]0 at 0.5 and solving for t. Each data point is the average for three independent experiments. Error bars show standard deviations.
In the course of these experiments, we found that amastigote PFR2 mRNA levels began to rise after 1 h of actinomycin D incubation. This phenomenon, which has been reported previously (4), suggests that actinomycin D might be unstable in amastigote media, since the effect could be eliminated by subsequent addition of more actinomycin D (Fig. 1).
Episomal expression of PFR2 genes recapitulates the wild-type regulation of mRNA levels.
In order to map the cis elements responsible for PFR2 regulation, we took advantage of L. mexicana knockout lines in which PFR2 genes have been deleted by homologous replacement. Using these clean genetic backgrounds, we tested whether it was possible to assay regulation of transcripts arising from episomal constructs in which PFR2 genes are linked to endogenous 5′ and 3′ flanking sequences. In initial experiments, plasmids containing the entire PFR2 locus (pLocus) or the PFR2C gene with either 7 kb (pPFR2C) or 2 kb (p2CWT and p2CH) of 3′ flanking sequence were introduced into Δpfr2 promastigotes (Fig. 2). The resultant lines were converted into axenic amastigotes, and RNAs from both promastigote and amastigote forms were analyzed by Northern blotting using the PFR2 coding region as a probe. Figure 2 shows the results of Northern blots obtained with a representative clonal line harboring each plasmid; however, at least three clonal lines were assayed in each case. L. mexicana Δpfr2 promastigotes containing pLocus directed expression of the 3.8- and 3.1-kb PFR2 transcripts as expected. The steady-state level of PFR2 mRNA was about 10-fold higher in promastigotes containing pLocus than in amastigotes. The two transcripts displayed equivalent regulation. Wild-type L. mexicana displayed a 12-fold regulation of PFR2 mRNA in control experiments (Fig. 2B). Leishmania harboring plasmids pPFR2C, p2CWT, or p2CH displayed levels of PFR2C mRNA that were at least fivefold higher in promastigotes than in amastigotes. Although Δpfr2 L. mexicana harboring the described plasmids did not fully duplicate the level of regulation observed with wild-type L. mexicana, the 5- to 10-fold regulation achieved in these cell lines recapitulates the authentic regulation sufficiently to enable mapping of the regulatory elements involved in PFR2 regulation. Furthermore, the size of each mRNA was identical to that of the wild-type mRNA, indicating that p2CWT and p2CH retained the native downstream splice site that specifies the site of 3′ polyadenylation.
3′ UTR sequences confer stage specificity to PFR2 mRNA accumulation.
The 3′ UTR participates in the posttranscriptional regulation of many trypanosomatid stage-specific transcripts. In order to determine whether sequences within the PFR2 3′ UTRs were required for the stage specificity of PFR2 expression, the expression pattern of PFR2 plasmids lacking the 3′ UTR and flanking sequences was compared with that of PFR2 expression plasmids that had intact 3′ flanking sequences. Plasmid p2CΔ3′ is a derivative of p2CWT that lacks all 3′ sequence associated with PFR2C. Plasmid p2BΔ3′ is a derivative of p2BWT that lacks sequences downstream of a ClaI site 200 bp downstream of the stop codon of PFR2B. The Δpfr2 lines harboring these constructs direct the constitutive high levels of expression of a chimeric PFR2 transcript possessing a 3′ UTR derived from vector sequences due to the presence of an adventitious splice acceptor site within the vector (Fig. 3B). In contrast, plasmids p2CWT and p2BWT, which contain the endogenous 3′ flanking sequences of PFR2C and PFR2B, directed expression of appropriately regulated PFR2 transcripts whose steady-state levels were five- to eightfold higher in promastigotes than in amastigotes. These experiments indicate that 3′ sequences are necessary for the regulation of PFR2 mRNA. Since the half-life of the mature PFR2 mRNA is subject to stage-specific control, the above data strongly implicate sequences within the 3′ UTR of the mature PFR2 mRNA functioning as a regulatory element.
FIG. 3.
The 3′ UTR is required for regulation of PFR2 mRNAs. (A) Schematic representation of PFR2 DNA cassettes. Symbols are as described in the legend to Fig. 2A. (B) Northern analysis of total RNA from promastigotes (P) and amastigotes (A) of wild-type L. mexicana (WT) and the Δpfr2 strain transfected with the indicated plasmids probed with PFR2 coding sequence. Numbers below the autoradiogram are ratios of PFR2 mRNA in promastigotes to that in amastigotes (P/A), determined as described in the legend to Fig. 2B.
Identification of a cis regulatory element in the 3′ UTR.
We chose to map the putative regulatory element in the PFR2C 3′ UTR. Initially, a series of three plasmids were derived from p2CWT. Plasmids p2CΔ1, p2CΔ2, and p2CΔ3 each lack a segment of the 1,400-nt PFR2C 3′ UTR but retain the intergenic region and endogenous downstream processing signal required for appropriate mRNA maturation (Fig. 4). Together, the three deletion constructs span the entire 3′ UTR. Northern analysis of L. mexicana Δpfr2 lines containing these 3′ UTR deletions revealed that lines harboring p2CΔ1 or p2CΔ2 retained the ability to produce an appropriately regulated PFR2 mRNA. In contrast, lines harboring p2CΔ3 failed to regulate PFR2 mRNA abundance (Fig. 4B). Instead, levels of PFR2 mRNA in amastigotes were elevated to the levels observed in promastigotes. This constitutive high-level expression of PFR2 mRNA indicates that the cis element required for regulation of PFR2C mRNA levels resides in region 3 of the PFR2C 3′ UTR. In each case, the size of the mRNA produced from the deletion constructs was in agreement with the predicted size, assuming that the wild-type polyadenylation site was used. Thus, the cis element does not appear to alter the site of polyadenylation.
To further map the regulatory sequences in region 3 of the 3′ UTR, four deletion constructs of p2CΔ3 were made based on unique restriction sites present in region 3. p2CΔ3.1 deletes a 60-bp NsiI-NaeI fragment, and p2CΔ3.2 deletes a 16-bp NaeI-NaeI fragment that is contained within the 60-bp deletion of p2CΔ3.1 (Fig. 5A). Plasmid p2CΔ3.3 removes a 115-bp NaeI-ApaLI fragment and p2CΔ3.4 deletes a 146-bp ApaLI-BsmI fragment of region 3 (Fig. 5A). Northern blot analysis of Δpfr2 lines containing these constructs (Fig. 5B) revealed that of the four constructs, only lines containing p2CΔ3.1 displayed a loss of regulation of PFR2 mRNA accumulation. This result indicates the presence of a regulatory element within the 60-nt region of the p2CΔ3.1 deletion. Furthermore, since p2CΔ3.2 overlaps 16 nt within the p2CΔ3.1 deletion but shows no loss of regulation, the regulatory element must be contained within a 44-nt segment of the p2CΔ3.1 deletion.
To further localize the regulatory sequence within the 3.1 region, a block substitution approach was used. Four constructs, each containing a contiguous 10-bp replacement made sequentially across the 44-bp region defined by p2CΔ3.1, were built into the p2CWT background and designated p2BS1, p2BS2, p2BS3, and p2BS4 (Fig. 6A). These constructs were transfected into Δpfr2 promastigotes, and PFR2 mRNA abundance was quantitated as in previous experiments. Lines containing p2BS2 displayed constitutive expression of PFR2, while lines containing the three other block substitutions retained the ability to regulate expression of PFR2 mRNA (Fig. 6B). These experiments indicate that the 10-nt RNA sequence AUGUAUAGUU contains a regulatory element required for stage-specific regulation of PFR2C mRNA.
A 10-nt element is sufficient to confer promastigote-specific expression.
To test whether the putative regulatory element was sufficient to generate regulation of mRNA accumulation, the 10-nt sequence defined by the BS2 block substitution was inserted into a construct which contained the PFR2 coding sequences fused to pBluescript II SK(+) vector sequences. This construct, p2CΔ3′ (Fig. 3), directs the expression of a constitutively expressed, chimeric PFR2 transcript possessing a 3′ UTR derived from vector sequence due to the presence of an adventitious splice acceptor site in the vector sequence. Within the vector-derived portion of the 3′ UTR is a sequence with a 7-of-10 nucleotide match to the BS2 sequence (Fig. 7). This sequence was converted into an exact match with the 10 nt defined by the BS2 block substitution in plasmid pΔM. Figure 7 shows that four clonal lines harboring pΔM (Fig. 7B, pΔM1-4) demonstrated regulated expression of PFR2 transcripts. This indicates that the putative regulatory element is sufficient for stage-specific regulation of PFR2 mRNA.
The 10-nt element alters the half-life of PFR2 mRNA in amastigotes.
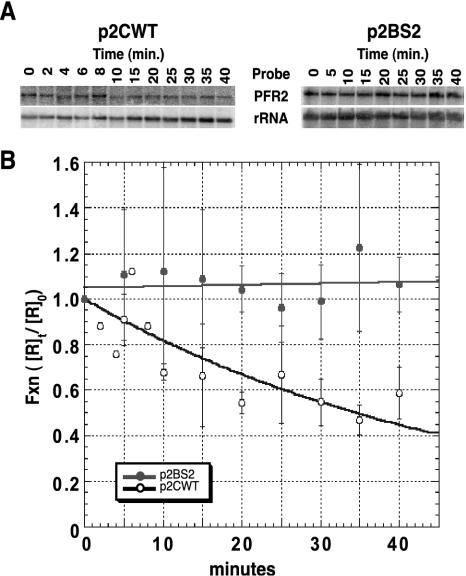
The experiments described above demonstrate that the 10-nt element is responsible for regulating PFR2 mRNA abundance. In order to demonstrate directly that the element functions by destabilizing the PFR2 mRNA in amastigotes, we measured the difference in stability of PFR2 mRNA in Δpfr2 L. mexicana amastigotes containing plasmids p2CWT compared to amastigotes containing p2BS2 (Fig. 8). Plasmid p2CWT directs expression of an appropriately regulated PFR2C mRNA, while p2BS2 directs expression of a constitutively regulated PFR2C mRNA due to a block substitution that disrupts the regulatory element. Quantitation of mRNA from the two amastigote lines incubated in the presence of actinomycin D for various times revealed a pronounced change in the stability of the PFR2 mRNA. The half-life of PFR2 mRNA in amastigotes containing p2BS2 could not be determined in this range of time values; however, the amount of RNA remains relatively constant throughout the experiment. The half-life of PFR2 mRNA in amastigotes containing p2CWT was determined to be 34.7 min—a value almost equal to the half-life of wild type PFR2 mRNA in amastigotes (Fig. 1). These data demonstrate that the regulatory element acts by destabilizing the PFR2 mRNA in amastigotes.
FIG. 8.
The regulatory element is responsible for destabilization of mRNA in amastigotes. Amastigote lines harboring either p2CWT or p2BS2 plasmids were treated with actinomycin D (10 μg/ml) for the time indicated. The value for the zero time point was taken just prior to the addition of actinomycin D. Two micrograms of total RNA from each time point was fractionated, blotted, and probed with in vitro-synthesized RNA from pPFR2 as described in the text. (A) Representative Northern blots for the time courses. The signals for the PFR2 and rRNA probes are displayed. (B) PFR2 transcript abundance was quantitated with a phosphorimager and was normalized to the value for small-subunit rRNA to control for loading. For each time point, the normalized values for PFR2 mRNA ([R]t) were divided by the normalized time zero value ([R]0) to obtain a value for the fraction of PFR2 mRNA remaining ([R]t/[R]0), which was plotted against time in minutes. The plotted data for p2CWT RNA was fitted to the exponential decay expression [R]t/[R]0 = e−kt, where k is a rate constant, t is time in minutes, and e is the inverse natural log. For the p2CWT data, k is 0.02 min−1. The value for the half-life of p2CWT was calculated by setting [R]t/[R]0 at 0.5 and solving for t. The results from three independent experiments were averaged. Error bars show standard deviations.
DISCUSSION
A negative regulatory circuit controls PFR2 expression.
The PFR2 mRNA is regulated 10-fold during the Leishmania life cycle in concert with the elaboration of the PFR within the flagellum (23). Based on the data in this report, this regulation can now be explained largely by changes in the half-life of the PFR2 mRNA during the Leishmania life cycle. We have identified a single sequence element, contained within 10 nt of the 3′ UTR of the PFR2C mRNA, that is responsible for PFR2C mRNA regulation. Deletion of this element from the PFR2C transcript resulted in an increase in amastigote mRNA abundance until it coincided with levels in promastigotes, thereby eliminating regulated expression. Insertion of this element into an unregulated transcript conferred regulation of the transcript by decreasing mRNA levels in amastigotes. Thus, within the contexts examined, it appears to be both necessary and sufficient for regulation of PFR2 mRNA levels. We refer to this element defined by the 10-nt block substitution, BS2, as the PFR regulatory element (PRE). To our knowledge, the PRE represents the only cis regulatory element controlling mRNA degradation that has been mapped to this resolution in Leishmania to date.
We have demonstrated in two ways that the PRE acts by destabilizing the PFR2 mRNA in amastigotes rather than by stabilizing the mRNA in promastigotes. First, elimination of the PRE results in elevation of steady-state PFR2 mRNA levels in amastigotes but has no effect on steady-state PFR2 mRNA levels in promastigotes. Second, direct measurement of the kinetics of mRNA degradation in amastigotes indicates an alteration in the rate of degradation that depends on the presence of the PRE. Thus, the element must be part of a negative regulatory circuit that exerts its effect in amastigotes. We did not detect any function of the PRE in promastigotes.
This negative regulatory circuit could be described most simply by postulating a trans-acting factor(s), present exclusively in amastigotes, whose interaction with the PRE results in the degradation of the PFR2 mRNA. Degradation might be initiated by a direct cleavage at the site of binding or by facilitating deadenylation of the poly(A) tail of the mRNA followed by the action of an exonuclease on the deadenylated transcript (17). However, negative regulatory circuitry could be envisioned involving a constitutively expressed trans-acting factor that interacts with the PRE. For example, a constitutively expressed trans-acting factor could be activated either by covalent modification exclusively in amastigotes or by recruitment of additional factors exclusive to amastigotes. Alternatively, regulation could be achieved by a constitutively expressed RNA binding protein if it were selectively sequestered in promastigotes but available for binding in amastigotes. This mechanism has been postulated to account for the ability of members of the ELAV protein family to selectively act on different transcripts containing AREs (11, 24). Therefore, although we believe that negative regulation must occur in amastigotes, it is not necessarily the case that the trans-acting factor that engages the element must be amastigote specific.
Global regulation of PFR genes by a common regulatory element.
In what other contexts does the PRE regulatory sequence occur? We searched the 3′ UTR sequences of other PFR mRNAs to determine whether all or part of the 10-nt sequence defined by the BS2 block substitution is present. We have found in each of five other PFR gene 3′ UTRs (PFR2A, PFR2B, PFR1C, PFR1D, and PFR4) an exact match to an identical 8-nt subset of the 10 nt altered in BS2 (Table 2). Unlike the position- and orientation-dependent activity of the 3′ UTR regulatory element of the regulated amastin mRNA in T. cruzi (6), the position dependence of the PRE is not conserved in the other PFR genes. The orientation dependence of the PRE has not been tested.
TABLE 2.
Location of the PRE in the 3′ UTRs of other PFR genesa
| L. mexicana gene | GenBank accession no. | Position in GenBank sequences | Distance (nt) from stop codon | Sequence |
|---|---|---|---|---|
| PFR2C | U45884 | 4492 | 1,098 | AUGUAuAGUu |
| PFR2A | NA | NA | 505 | AUGUAaAGUa |
| PFR2B | U45884 | 508 | 505 | AUGUAaAGUa |
| PFR1C | AY198411 | 2809 | 1,354 | AUGUAuAGUg |
| PFR1D | AY198411 | 6217 | 778 | AUGUAcAGUg |
| PFR4 | AY198410 | 3680 | 117 | AUGUAaAGUa |
| Consensus | AUGUAnAGUn |
NA, not available.
A search for this 8-nt conserved element in the L. major Friedlin genomic sequences deposited in GenBank revealed its presence at a frequency of 1:520,296 nt, in good agreement with the expected frequency of this 8-nt sequence in a genome with a G+C content of 63% (1:296,308). The occurrence of the PRE in each of the available L. mexicana PFR gene 3′ UTRs at a frequency that is about 370 times greater than its overall frequency in the L. major genome, is unlikely to be a chance occurrence. Rather, it suggests that the PRE element functions in the coordinate regulation of a group of functionally related genes. Northern analysis on L. mexicana total RNA from seven of these genes identified in the database search shows regulated mRNA accumulation analogous to PFR2C (T. R. Holzer, K. K. Mishra, and J. H. LeBowitz, unpublished data). These genes include the PFR1 and PFR4 genes, where the PRE is conserved between the two species. As expected, several of the other genes have been implicated in flagellar structure or biology.
A protein that specifically recognizes AU-rich instability elements involved in the stage-regulated expression of the mucin-type gene family of T. cruzi has been identified (9). AREs, a family of functionally and structurally distinct sequence motifs such as the AUUUA pentamer, the UUAUUUA(U/A)(U/A) nonamer, and stretches of U-rich domain that range in size from 50 to 150 bp, are destabilizing elements that are present in 3′ UTRs of higher eukaryotic early-response-gene mRNAs, including cytokines, lymphokines, and proto-oncogenes (for a review, see reference 5). In the case of the T. cruzi mucin SMUG gene, an RNA-binding protein, TcUBP, has been identified that is believed to be the trans-acting factor responsible for the 6- to 10-fold-lower levels of SMUG mRNA in trypomastigotes compared to epimastigotes. TcUBP-1 appears to be involved in the destabilization of SMUG mRNA. TcUBP-1 levels are 10-fold higher in amastigotes and trypomastigotes than in epimastigotes. In fact, overexpression of the protein in epimastigotes results in a lowering of SMUG mRNA in that stage (9). Thus, TcUBP appears to participate in a negative regulatory circuit similar to that observed for the L. mexicana PFR2 gene.
Can the PRE be a member of the AU-rich family of regulatory elements? The T. cruzi AREs identified in the mucin gene family bear a striking resemblance to their mammalian counterparts. In contrast, the PRE, although it is AU rich and is imbedded in an AU-rich sequence in PFR2C, has not previously been reported as a member of the ARE family. However, the sequence AUGUA, which is contained in the PRE, has been shown to compete effectively with the canonical AU-rich pentamer, AUUUA, for binding to the mammalian AU-rich binding protein HuR (24). The PRE may represent a novel member of the ARE family. In this case, given that a Leishmania homolog of the TcUBP-1 family has been reported, it or a relative might interact with the PRE. We are currently testing this possibility.
Acknowledgments
We thank James Forney, Steve Broyles, and Barbara Golden (Department of Biochemistry, Purdue University) for their many helpful discussions of this work.
This work was supported by National Institute of Health grant A147909 and National Science Foundation grant 9724752. J.H.L. was a Burroughs Welcome Fund new investigator in Molecular Parasitology.
This is paper 16976 from Purdue Agriculture Research Programs.
REFERENCES
- 1.Bastin, P., T. Sherwin, and K. Gull. 1998. Paraflagellar rod is vital for trypanosome motility. Nature 391:548. [DOI] [PubMed] [Google Scholar]
- 2.Boucher, N., Y. Wu, C. Dumas, M. Dube, D. Sereno, M. Breton, and B. Papadopoulou. 2002. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3′-untranslated region element. J. Biol. Chem. 277:19511-19520. [DOI] [PubMed] [Google Scholar]
- 3.Brooks, D. R., H. Denise, G. D. Westrop, G. H. Coombs, and J. C. Mottram. 2001. The stage-regulated expression of Leishmania mexicana CPB cysteine proteases is mediated by an intercistronic sequence element. J. Biol. Chem. 276:47061-47069. [DOI] [PubMed] [Google Scholar]
- 4.Charest, H., W. W. Zhang, and G. Matlashewski. 1996. The developmental expression of Leishmania donovani A2 amastigote-specific genes is post-transcriptionally mediated and involves elements located in the 3′-untranslated region. J. Biol. Chem. 271:17081-17090. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 6.Coughlin, B. C., S. M. Teixeira, L. V. Kirchhoff, and J. E. Donelson. 2000. Amastin mRNA abundance in Trypanosoma cruzi is controlled by a 3′-untranslated region position-dependent cis-element and an untranslated region-binding protein. J. Biol. Chem. 275:12051-12060. [DOI] [PubMed] [Google Scholar]
- 7.Curotto de Lafaille, M. A., A. Laban, and D. F. Wirth. 1992. Gene expression in Leishmania: analysis of essential 5′ DNA sequences. Proc. Natl. Acad. Sci. USA 89:2703-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Orso, I., and A. C. Frasch. 2001. Functionally different AU- and G-rich cis-elements confer developmentally regulated mRNA stability in Trypanosoma cruzi by interaction with specific RNA-binding proteins. J. Biol. Chem. 276:15783-15793. [DOI] [PubMed] [Google Scholar]
- 9.D'Orso, I., and A. C. Frasch. 2001. TcUBP-1, a developmentally regulated U-rich RNA-binding protein involved in selective mRNA destabilization in trypanosomes. J. Biol. Chem. 276:34801-34809. [DOI] [PubMed] [Google Scholar]
- 10.Furger, A., N. Schurch, U. Kurath, and I. Roditi. 1997. Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol. Cell. Biol. 17:4372-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Good, P. J. 1995. A conserved family of elav-like genes in vertebrates. Proc. Natl. Acad. Sci. USA 92:4557-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham, S. 1995. Mechanisms of stage-regulated gene expression in Kinetoplastida. Parasitol. Today 11:217-223. [DOI] [PubMed] [Google Scholar]
- 13.Hotz, H. R., S. Biebinger, J. Flaspohler, and C. Clayton. 1998. PARP gene expression: control at many levels. Mol. Biochem Parasitol. 91:131-143. [DOI] [PubMed] [Google Scholar]
- 14.Hotz, H. R., C. Hartmann, K. Huober, M. Hug, and C. Clayton. 1997. Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 25:3017-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hotz, H. R., P. Lorenz, R. Fischer, S. Krieger, and C. Clayton. 1995. Role of 3′-untranslated regions in the regulation of hexose transporter mRNAs in Trypanosoma brucei. Mol. Biochem. Parasitol. 75:1-14. [DOI] [PubMed] [Google Scholar]
- 16.Huang, P. L., B. E. Roberts, D. M. Pratt, J. R. David, and J. S. Miller. 1984. Structure and arrangement of the beta-tubulin genes of Leishmania tropica. Mol. Cell. Biol. 4:1372-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irmer, H., and C. Clayton. 2001. Degradation of the unstable EP1 mRNA in Trypanosoma brucei involves initial destruction of the 3′-untranslated region. Nucleic Acids Res. 29:4707-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeBowitz, J. H. 1994. Transfection experiments with Leishmania. Methods Cell Biol. 45:65-78. [DOI] [PubMed] [Google Scholar]
- 19.LeBowitz, J. H., H. Q. Smith, L. Rusche, and S. M. Beverley. 1993. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 7:996-1007. [DOI] [PubMed] [Google Scholar]
- 20.Maga, J. A., T. Sherwin, S. Francis, K. Gull, and J. H. LeBowitz. 1999. Genetic dissection of the Leishmania paraflagellar rod, a unique flagellar cytoskeleton structure. J. Cell Sci. 112:2753-2763. [DOI] [PubMed] [Google Scholar]
- 21.Mahmood, R., J. C. Hines, and D. S. Ray. 1999. Identification of cis and trans elements involved in the cell cycle regulation of multiple genes in Crithidia fasciculata. Mol. Cell. Biol. 19:6174-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell, P., and D. Tollervey. 2000. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 10:193-198. [DOI] [PubMed] [Google Scholar]
- 23.Moore, L. L., C. Santrich, and J. H. LeBowitz. 1996. Stage-specific expression of the Leishmania mexicana paraflagellar rod protein PFR-2. Mol. Biochem. Parasitol. 80:125-135. [DOI] [PubMed] [Google Scholar]
- 24.Myer, V. E., X. C. Fan, and J. A. Steitz. 1997. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 16:2130-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan, A. A., and S. C. Pan. 1986. Leishmania mexicana: comparative fine structure of amastigotes and promastigotes in vitro and in vivo. Exp. Parasitol. 62:254-265. [DOI] [PubMed] [Google Scholar]
- 26.Santrich, C., L. Moore, T. Sherwin, P. Bastin, C. Brokaw, K. Gull, and J. H. LeBowitz. 1997. A motility function for the paraflagellar rod of Leishmania parasites revealed by PFR-2 gene knockouts. Mol. Biochem. Parasitol. 90:95-109. [DOI] [PubMed] [Google Scholar]
- 27.Stiles, J. K., P. I. Hicock, P. H. Shah, and J. C. Meade. 1999. Genomic organization, transcription, splicing and gene regulation in Leishmania. Ann. Trop. Med. Parasitol. 93:781-807. [DOI] [PubMed] [Google Scholar]
- 28.Sutton, R. E., and J. C. Boothroyd. 1986. Evidence for trans splicing in trypanosomes. Cell 47:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanhamme, L., and E. Pays. 1995. Control of gene expression in trypanosomes. Microbiol. Rev. 59:223-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vassella, E., R. Braun, and I. Roditi. 1994. Control of polyadenylation and alternative splicing of transcripts from adjacent genes in a procyclin expression site: a dual role for polypyrimidine tracts in trypanosomes? Nucleic Acids Res. 22:1359-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasudevan, S., and S. W. Peltz. 2001. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell 7:1191-1200. [DOI] [PubMed] [Google Scholar]
- 32.Vickerman, K. 1962. The mechanism of cyclical development in trypanosomes of the Trypanosoma brucei subgroup: a hypothesis based on ultrastructural observations. Trans. R. Soc. Trop. Med. Hyg. 56:487-495. [DOI] [PubMed] [Google Scholar]
- 33.Weston, D., A. C. La Flamme, and W. C. Van Voorhis. 1999. Expression of Trypanosoma cruzi surface antigen FL-160 is controlled by elements in the 3′ untranslated, the 3′ intergenic, and the coding regions. Mol. Biochem. Parasitol. 102:53-66. [DOI] [PubMed] [Google Scholar]