Abstract
With the recent development of techniques for analyzing transmembrane thylakoid proteins by two-dimensional gel electrophoresis, systematic approaches for proteomic analyses of membrane proteins became feasible. In this study, we established detailed two-dimensional protein maps of Chlamydomonas reinhardtii light-harvesting proteins (Lhca and Lhcb) by extensive tandem mass spectrometric analysis. We predicted eight distinct Lhcb proteins. Although the major Lhcb proteins were highly similar, we identified peptides which were unique for specific lhcbm gene products. Interestingly, lhcbm6 gene products were resolved as multiple spots with different masses and isoelectric points. Gene tagging experiments confirmed the presence of differentially N-terminally processed Lhcbm6 proteins. The mass spectrometric data also revealed differentially N-terminally processed forms of Lhcbm3 and phosphorylation of a threonine residue in the N terminus. The N-terminal processing of Lhcbm3 leads to the removal of the phosphorylation site, indicating a potential novel regulatory mechanism. At least nine different lhca-related gene products were predicted by comparison of the mass spectrometric data against Chlamydomonas expressed sequence tag and genomic databases, demonstrating the extensive variability of the C. reinhardtii Lhca antenna system. Out of these nine, three were identified for the first time at the protein level. This proteomic study demonstrates the complexity of the light-harvesting proteins at the protein level in C. reinhardtii and will be an important basis of future functional studies addressing this diversity.
In all eukaryotic oxygenic photosynthetic organisms, light-harvesting chlorophyll a- or b-binding proteins (LHC proteins) function in the collection and transfer of light energy to the reaction centers of photosystem II (PSII) (Lhcb proteins) and photosystem I (PSI) (Lhca proteins). Additionally these proteins are also involved in light dissipation and energy quenching. Therefore, light-harvesting proteins are important components of the photosynthetic machinery that optimize photosynthetic function and minimize photooxidative damage in response to light quantity and quality. It has been known for several years that light-harvesting proteins are products of many genes. This concept is illustrated by a recent analysis of the Arabidopsis genome which revealed that the lhc gene family is composed of more than 20 genes (24). Besides the large number of lhc gene products, posttranslational modifications, such as phophorylation, contribute to even more complexity at the protein level (31, 45). Phosphorylation of the major Lhcb proteins of PSII is important in the process of state transitions. This process leads to a redistribution of excitation energy between PSII and PSI by reorganization of the antennae and thereby regulates energy flow between the photosystems. The importance of phosphorylation for state transitions is shown by the phenotype of the Chlamydomonas reinhardtii Stt7 mutant. This mutant is markedly affected in LHCII protein (LHC protein of PSII) phosphorylation and consequently is unable to perform state transitions. In keeping with the observed phenotype, this mutant is deficient in a chloroplast thylakoid-associated serine-threonine kinase (10).
Although C. reinhardtii has been used as a model system for elucidating the assembly and regulatory processes of the photosynthetic machinery, no thorough analysis of LHC protein composition has been performed for this alga. What is known about the LHC protein composition of C. reinhardtii? Through searching of Chlamydomonas databases, 10 genes that potentially encode Lhcb polypeptides that are associated with the major trimeric PSII antenna (Lhcbm) have been described (11). In addition, the products of two other genes (lhcb4 and lhcb5) that correspond to minor LHC proteins (CP29 and CP26) have been identified (42). At least seven distinct Lhca subunits have been proposed (3, 42). For six Lhca proteins, N-terminal amino acid sequences have been obtained by Edman amino acid sequencing (3). Separation of isolated PSI complexes from C. reinhardtii by two-dimensional gel electrophoresis has revealed the presence of about 18 LHCI protein (LHC protein of PSI) spots, thereby suggesting an even more extensive variability of Lhca proteins (20). To characterize the compositions of the lhca and lhcb gene products at the protein level and to monitor the posttranslational modifications of LHC proteins of C. reinhardtii, we performed a detailed proteomic study.
The genome project for the photosynthetic eukaryotic model organism, the green alga C. reinhardtii (more than 180,000 expressed sequence tag [EST] sequences have been obtained, and ninefold whole-genome shotgun coverage was released by the Department of Energy Joint Genome Institute in February 2003) provides excellent opportunities for applying systematic or functional proteomic approaches. Proteomic analysis has already become a powerful tool for the analysis of profiles of protein expression in vascular plants as well as Chlamydomonas (21, 44). Plant cell organelles, such as chloroplasts, offer excellent model systems for proteomic analysis, since an organelle or its subcompartments can be isolated easily in large amounts and at high purities. The standard technique for quantitative proteomic analysis combines protein separation by high-resolution (isoelectric focusing combined with sodium dodecyl sulfate [SDS]-polyacrylamide gel electrophoresis)two-dimensional gel electrophoresis (2-DE) with mass spectrometry (MS) or tandem MS (MS-MS) identification of selected protein spots.
The separation of hydrophobic intrinsic membrane proteins by 2-DE has long been a difficult task (40). However, with the recent development of procedures for the analysis of transmembrane thylakoid proteins by 2-DE, proteomic analysis of membrane proteins has become feasible (20). This procedure allowed the separation and identification of hydrophobic transmembrane proteins, such as the light-harvesting proteins. The technique was used to create 2-DE protein maps of thylakoid membrane proteins from wild-type and mutant strains of C. reinhardtii (20) as well as to monitor changes in the protein compositions of the thylakoid membranes during the physiological adaptation to iron deficiency (33).
In this study, we performed detailed 2-DE protein mapping of light-harvesting proteins associated with PSII (Lhcb) and PSI (Lhca) of Chlamydomonas. These polypeptides are membrane proteins that span the lipid bilayer with three α-helical transmembrane domains (28). This study represents the first detailed analysis of LHC proteins by 2-DE and MS-MS. In addition to protein separation by 2-DE, intact LHC proteins were extracted with organic solvents and separated by reversed-phase liquid chromatography (LC). Identification of vascular plant chloroplast transmembrane LHC proteins has been achieved by coupling LC with electrospray ionization MS and MS-MS (9, 16, 47).
The proteomic analysis performed in this study predicts the expression of at least nine different Lhca proteins and eight distinct Lhcb proteins at the protein level and supports the previous indications of the extensive variability of LHC proteins of C. reinhardtii. The finding that the N-terminal protein processing of Lhcbm3 leads to the removal of a phosphorylation site indicates a novel regulatory mechanism for state transitions.
MATERIALS AND METHODS
Strains and culture conditions.
Solid Tris-acetate-phosphate medium and high-salt minimal medium were prepared as described previously (17). Iron-deficient media were prepared as described by Moseley et al. (33). Liquid cultures were grown at 20 to 25°C with shaking at 200 to 250 rpm and light intensities of 50 to 100 μmol m−2 s−1. Strains were cultured on plates at 24 to 25°C.
Construction of recombinant strains and genetic analysis.
The recombinant strain Lep2 was generated by inserting a hemagglutinin (HA) epitope-tagged form of lhcbm6 (lhcII-1.1) (23) randomly into C. reinhardtii strain CC2454. Plasmid pBQ2.4, carrying a 2.4-kb EcoRI/HindIII fragment with exons 2 to 4 and 3′-flanking sequences of the lhcbm6 gene (25), was linearized with SacI, which cuts the plasmid at a single site within exon 2. This site was used to introduce the double-stranded oligonucleotide ocabhem1 with SacI-compatible 3′ overhangs, obtained by annealing single-stranded oligonucleotides ocabhem1a (5′-ACCCCTACGACGTCCCCGACTACGCTAGCT-3′) and ocabhem1b (5′-AGCGTAGTCGGGGACGTCGTAGGGGTAGCT-3′). The resulting plasmid, pAL1, carrying the insert, was sequenced to verify its correct orientation and in-frame insertion of the HA epitope. With this procedure, the peptide sequence YPYDVPDYAS was introduced between Ser31 and Ser32 of the deduced precursor protein sequence encoded by the lhcbm6 gene. To reconstruct the complete HA epitope-tagged lhcbm6 gene, the 3.0-kb HindIII fragment with exon 1 and the lhcbm6 promoter region was inserted into pAL1 that had been linearized with HindIII. The resulting plasmid, pAL2, was analyzed for correct insertion and orientation of the 3.0-kb fragment by DNA sequencing. Standard protocols were used for all DNA manipulations (39).
Cotransformation of strain CC2454 with plasmids pMN24 (13) and pAL2 was carried out by using particle gun transformation (26). After transformation, cells were placed on ammonium-free Sager-Granick 2 medium to select transformants containing intact nitrate reductase encoded by pMN24. These colonies were then screened for the expression of the tagged protein by Western blot analysis with the HA-specific mouse monoclonal antibody 12CA5 (BabCO, Richmond, Va.).
Generation of gametes, matings, and zygote analysis were performed as described previously (17).
Chlorophyll determination.
Precipitated protein and cell debris were removed by centrifugation, and the chlorophyll concentration was estimated by the method of Porra et al. (36).
Isolation of PSI complexes.
PSI particles were isolated as described previously (18). Determination of the protein concentration in solution was done by the bicinchoninic acid method according to the manufacturer's instructions (Sigma, Taufkirchen, Germany).
In vitro phosphorylation of thylakoid membranes and green gel electrophoresis.
In vitro phosphorylation of thylakoid membranes was performed as described previously (14), and green gel electrophoresis was conducted as described by Peter and Thornber (34).
2-DE, immunodetection, and trypsin digestion.
Preparation of samples for 2-DE, immunodetection, and trypsin digestion of excised protein spots were performed as described previously (20).
Tryptic hydrolysis of PSI complexes.
An aliquot of the sucrose gradient enriched in PSI particles, which corresponded to 10 μg of chlorophyll with a 0.05% (wt/vol) concentration of β-dodecyl maltoside, was heated for 10 min at 95°C. The liquid was evaporated by using a vacuum centrifuge, and the pellet was resuspended in 50 μl of a 50 mM hydrogen bicarbonate solution and incubated at 30°C for 15 min and then at 37°C for 16 h with 6 μg of trypsin (sequencing grade modified; Promega, Madison, Wis.). After hydrolysis with trypsin, the reaction mixture was lyophilized, and the pellet was resuspended in 40 μl of a 5:90:5 (vol/vol) methanol-water-formic acid solution (Lichrosolv [grade]; E. Merck AG, Darmstadt, Germany) and vortexed for 20 min. To prepare a hydrophobic column, approximately 10 μl of dry POROS-R2 (Applied Biosystems, Foster City, Calif.) material was resuspended in a 60% (vol/vol) methanol solution and loaded into the top of a C18 Zip-Tip column (Millipore, Eschborn, Germany) with a 5-μl bed volume. The column was packed by brief centrifugation in a tabletop microcentrifuge with an insert meant for holding nanospray needles. Alternatively, a 0.6-ml microcentrifuge tube with a small hole in the bottom and placed inside a 1.5-ml microcentrifuge tube could be used to hold the POROS-R2-containing Zip-Tip column. The liquid phase was then replaced by an aqueous phase by washing of the column packing twice with 40 μl of methanol-water-formic acid (5:90:5, vol/vol/vol). The sample was centrifuged in a tabletop centrifuge for 5 min, and the supernatant was removed and applied to the column packing by brief centrifugation. The column was washed twice with 40 μl of methanol-water-formic acid (5:90:5, vol/vol/vol), and the sample was eluted into a microcentrifuge tube with two 40-μl aliquots of a solution containing methanol-water-formic acid (60:35:5 [vol/vol]) by centrifugation in a tabletop centrifuge at 10,000 rpm (Hermle Z 160 M). The sample was lyophilized and resuspended in 10 μl of LC buffer A (see below). Typically, a 1:10 or 1:100 dilution was analyzed by LC-MS.
LC-MS.
Analyte sampling, chromatography, and production and acquisition of MS and MS-MS data were performed online by using fully automated instrument functions. A Famos 48-well plate autosampler (LC-Packings, Amsterdam, The Netherlands) was used to sample the tryptic digests in batch mode with a user-defined program to withdraw 1 μl from each sample. Solvents were purchased from Merck and were of LC grade. The aqueous phase (A) (0.1% [vol/vol] formic acid in 5:95 acetonitrile-water) and the organic phase (B) (0.1% [vol/vol] formic acid in 80:20 acetonitrile-water) were delivered by an Ultimate (LC-Packings) micropump in most instances as follows: 5% B in the first 8 min, 5 to 50% B from 8 to 38 min, 50 to 95% B from 38 to 39 min, 95% B from 39 to 49 min, 95 to 5% B from 49 to 50 min, and 5% B from 50 to 75 min. Solvents delivered by the micropump were split before reaching the column by a splitter at a flow ratio of 1:1,250, with the postsplitter flow rate set at 250 nl/min. The peptide mixture was fractioned on an LC-Packings PepMap C18 column (75-μm [inner diameter] by 150 mm) with a 3-μm particle size and a 100-Å-pore diameter (NAN-75-15-03-PM). The column eluent was monitored at 214 and 280 nm with a rapid-scanning spectrophotometer equipped with a 3-nl flow cell (LC-Packings UZ-N10 160015). The interface between the liquid phase and the gas phase was provided by a fused silica needle that had a 75-μm inner diameter, a length of approximately 12 cm, and a 30-μm aperture (FS 360-75-30; New Objective, Woburn, Mass.) and that was mounted on a nano-electrospray ionization source (New Objective). Atmospheric pressure electrospray ionization was accomplished by applying a voltage difference of +1.3 kV across the fused silica needle. The aperture of the fused silica needle was positioned 1 to 2 mm from the opening of the ion transfer capillary and slightly off axis to minimize the entrance of nonvaporized solvent into the LCQ Deca XP ion trap mass spectrometer (Thermofinnigan, San Jose, Calif.). No sheath gas was used.
Most mass spectra were acquired by using the repetitive “triple-play” sequence as recommended by the manufacturer, consisting of a full scan event for ions with mass-to-charge ratios of 400 to 1,200, a zoom scan event acquired within a mass-to-charge ratio window of 10 units centered around the chosen ion, and an MS-MS scan event. Ions were selected for the zoom scan and for the MS-MS scan automatically by using instrument software in the data-dependent manner, whereby ions of sequentially decreasing abundance were chosen and two scan events were allowed for any given ion in a 3-min time window. For complex samples, the zoom scan event was omitted and replaced with two MS-MS scan events. The tolerance for the selection of the MS-MS precursor ranged from 1.5 to 3.0 m/z (low to high m/z).
Sequest.
The measured MS-MS spectra were matched with tryptic peptide amino acid sequences from a small C. reinhardtii database consisting primarily of PSI or PSII polypeptide sequences (about 90 entries), from the translated Kazusa EST database (1, 2), from the EST database of the Chlamydomonas genome project (A. Grossman, J. Davies, N. Federspiel, E. Harris, P. Lefebvre, C. Silflow, D. Stern, and R. Surzycki, unpublished data), or from a genomic database (version 1.0 assembly). In addition, the data sets were partially compared with the genomic sequences for the presence of new LHC polypeptides. All databases were in the FASTA format. Cys modification by carbamidomethylation (+57 Da) was taken into account. Raw MS-MS data files that had a minimum total ion current of 105 and contained 15 or more fragment ions were selected. Known contaminants were filtered out. Computational analysis was done on a Dell personal computer. The tolerance window set for the grouping of raw MS-MS data files into input files for the Finnigan Sequest/Turbo Sequest software (revision 2.0; ThermoQuest, San Jose, Calif.) was set to 1.4 atomic mass units.
The Sequest algorithm was used to quickly identify and retrieve database sequences having at least one tryptic end, having a theoretical mass within ±1.25 atomic mass units of that measured for the precursor ion, and having a theoretical y- and b-ion fragmentation profile with a high degree of similarity to the experimentally measured MS-MS spectrum. The similarity between a measured MS-MS spectrum and a theoretical MS-MS spectrum, reported as the cross-correlation factor (Xcorr), and the difference between the unit-normalized Xcorr values of the first- and second-ranked sequences (ΔCorr) provided the preliminary criteria for assigning amino acid sequences to experimental MS-MS spectra. Sequences are reported here if the Xcorr values calculated for the measured and theoretical MS-MS spectra were equal to or above 1.5, 2.25, or 3.5 for singly, doubly, or triply charged precursor ions, respectively, and if the ΔCorr value exceeded 0.1 within the background of the Kazusa EST database (1, 2) or the genomic database.
Since the N termini of the mature proteins are not yet clear, a database subset containing sequences of the lhca or lhcb gene products from Chlamydomonas was created. This database contains amino acid sequences where the first N-terminal amino acids of respective gene products were excised in a stepwise manner. This step was repeated 45 times to create different N-terminal sequences. This database, containing different N termini, allowed us to search for N-terminal peptides. It was also checked for carbamylation.
In order to identify C. reinhardtii contigs, the translated ESTs were subjected to a WU-BLAST 2 search of different EST assemblies at http://www.biology.duke.edu/chlamy_genome/blast/blast_form.html.
Phylogenetic analyses.
A CLUSTAL W alignment of the Lhca protein sequences was performed by using the European Molecular Biology Laboratory-European Bioinformatics Institute computer server (http://www.ebi.ac.uk /clustalw/) with the default settings. Phylogenetic trees were inferred by using PAUP algorithms (PAUP version 4.0b8; Sinauer Associates, Sunderland, Mass.). The confidence of branching was assessed by using 1,000 bootstrap resamplings for distance analysis with the neighbor-joining method.
RESULTS
In a recent study, Hippler et al. established a procedure for separating thylakoid membrane proteins from C. reinhardtii by 2-DE (20). The successful separation of transmembrane proteins by 2-DE was achieved by combining the extraction of hydrophobic proteins with organic solvents and the subsequent solubilization of the precipitated proteins in a mixture of detergents together with urea and thiourea. The 2-DE separation was combined with the identification of LHC protein spots by MS-MS and immunobiochemical techniques (20). The immunodetected protein spots were investigated in this study, and thereby we established detailed 2-DE protein maps of C. reinhardtii Lhcb and Lhca proteins by extensive MS analysis (Fig. 1; see Fig. 4). Spots of interest were excised from the gel, digested with trypsin, and analyzed by coupling LC with ion trap MS (LC-MS and LC-MS-MS). We used reversed-phase LC as an online preparative method for separating tryptic peptides. Qualitative analysis and identification of proteins were performed by using data gathered from MS and MS-MS of peptides. To identify peptides from MS-MS data, we used Sequest (12) as a computer search algorithm for comparing the collision-induced (CID) fragmentation pattern of a precursor peptide with theoretical computer-generated fragmentation spectra deduced from a protein or DNA database. In this study, a Chlamydomonas EST database, a database containing genomic data, or a database containing a defined set of proteins from C. reinhardtii was used.
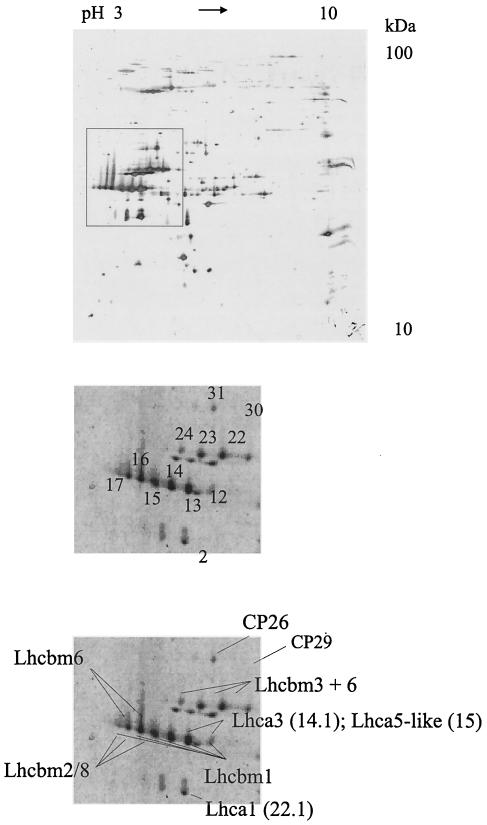
FIG. 1.
2-DE protein maps of major C. reinhardtii LHC proteins after separation of thylakoid membranes (200 μg of protein). The upper panel shows a 2-DE map of thylakoid membranes stained with silver. The inset in the upper panel indicates the region presented in the lower panels. Each of the numbered spots on the Coomassie blue-stained 2-DE map (lower panels) was excised and digested in a gel. The subsequent MS-MS analysis resulted in the identification of LHC proteins. The labeled protein spots indicate proteins that were unambiguously identified by the MS data. Lhcbm3 + 6, Lhcbm3 and Lhcbm6; Lhcbm2/8, Lhcbm2 or Lhcbm8.
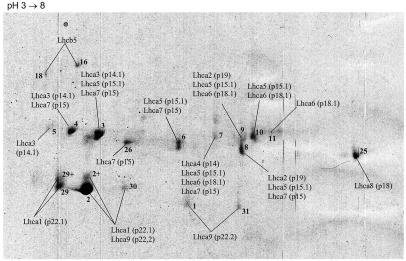
FIG. 4.
PSI 2-DE protein map. Coomassie blue-stained 2-DE-separated proteins (corresponding to 22 μg of chlorophyll) that copurified with PSI particles during sucrose gradient ultracentrifugation are shown. Each of the numbered spots was excised and digested in a gel. The resulting tryptic peptides were analyzed by using LC-MS and database searching. The Coomassie blue-stained protein spots are labeled according to the lhc gene products detected from the tryptic digests; the relative SDS-PAGE mobility identifiers are indicated in parentheses. Most of the peptides identified corresponded to lhca gene products. Spots 1, 5, 11, 25, 29, 29+, 30, and 31 were found to contain peptides corresponding to lhca gene products only. Previously unidentified lhca gene products are labeled as follows: Lhca9 (p22.2), Lhca2 (p19), and Lhca7 (p15).
Lhcb protein map.
When thylakoid membranes are separated by 2-DE and stained with Coomassie blue, we can differentiate two rows of Lhcb protein spots that are diverse in their apparent molecular masses (20). The upper row consists of spots 22 to 24, and the lower row consists of spots 12 to 17. We analyzed the tryptic digests of these spots by MS-MS and unambiguously identified eight distinct Lhcb proteins (Tables 1 and 2): Lhcbm1, Lhcbm3, and Lhcbm5 (identified in enriched PSI particles [Table 2]); Lhcbm6, Lhcbm2 or Lhcbm8, and Lhcbm4 or Lhcbm9 (identified from green gels; see below); and CP26 and CP29 (Table 1). Lhcbm1 is LhcII-4, Lhcbm2 is LhcII-3, Lhcbm3 is LhcII-1.3, Lhcbm5 is Lhcb3, Lhcbm6 is Lhcb1 (CabII-1), and Lhcbm8 is CabII-2 (11). For CP26 and CP29, we detected peptides from spots 31 and 30, respectively. Although the Lhcbm proteins are highly similar, we identified peptides that are redundant but also others that are unique for specific lhcbm gene products (Table 1) and thereby distinguish the corresponding protein clearly in the respective protein spot. For spots 22 to 24, the MS-MS data identify specific peptides corresponding to Lhcbm3. For spots 22 and 23, peptides specific for Lhcbm6 can be identified, whereas for spot 24, peptides specific for Lhcbm4 or Lhcbm6 can be found. The occurrence of the Lhcbm6 protein (Lhcb1 or CabII-1) in the upper and lower rows of spots confirms recent MS and immunochemical test results (20). Of note is the finding that a peptide-specific antibody directed against the N-terminal part of Lhcbm6 recognizes only the upper row of spots. For the lower row of spots, Lhcbm1-specific peptides can be distinguished in spots 12 to 17, Lhcbm2- or Lhcbm8-specific ones can be seen in spots 14 to 17, and an Lhcbm6-specific one can be seen in spots 16 and 17. Thus, spots 12 and 13 can be correlated with Lhcbm1.
TABLE 1.
Lhcb proteins identified by MS-MS data from protein spots excised from thylakoid 2-DE mapsa
| 2-DE Spot | Proteinb | Peptidec | Positionsd | ze | ΔM
|
Xcorr
|
||
|---|---|---|---|---|---|---|---|---|
| With Fe | Without Fe | With Fe | Without Fe | |||||
| 12 | Lhca7 (p15) | NPGSQADGSFLGFTEEFK | 141-158 | 2 | 1.5 | 3.52 | ||
| Lhca7 (p15) | GLENGYPGGR | 159-168 | 2 | 0.2 | 2.27 | |||
| Lhca3 (p14.1) | GSGDAAYPGGPFFNLFNLGK | 186-205 | 2 | 1.3 | 4.10 | |||
| Lhca3 (p14.1) | GPFQNLVEHLADPVNNNILTNFGK | 241-264 | 3 | 0.9 | 5.19 | |||
| Lhca3 (p14.1) | WLQYSEVIHAR | 86-96 | 2 | 1.0 | 3.09 | |||
| Lhcbm1 | VNGGPLGEGLDK | 158-169 | 2 | 0.0 | 0.8 | 3.03 | 2.8 | |
| Lhcbm1 | QPLQNLSDHLANPGTNNAFAYATK | 228-251 | 3 | 1.5 | 6.31 | |||
| Lhcbm1, 10 | ASTPDSFWYGPER | 2 | 0.9 | 1.1 | 2.72 | 2.37 | ||
| Lhcbm1-10 | YRELELIHAR | 3 (2) | 1.1 | 0.8 | 3.60 | 3.48 | ||
| 13 | Lhca7 (p15) | VGLGFPEWYDAGK | 95-107 | 2 | 0.1 | 3.41 | ||
| Lhca3 (p14.1) | GSGDAAYPGGPFFNLFNLGK | 186-205 | 2 | −0.6 | 3.85 | |||
| Lhca3 (p14.1) | GPFQNLVEHLADPVNNNILTNFGK | 241-264 | 3 | −0.9 | 6.29 | |||
| Lhca3 (p14.1) | WLQYSEVIHAR | 86-96 | 2 | 0.8 | 4.18 | |||
| Lhcbm1 | LYPGGSFDPLGLADDPDTFAELKVK | 170-194 | 2 | 0.4 | 2.36 | |||
| Lhcbm1 | VNGGPLGEGLDK | 158-169 | 2 | −0.2 | 0.0 | 3.16 | 2.84 | |
| Lhcbm1 | GPLQNLSDHLANPGTNNAFAYATK | 229-252 | 3 | 1.3 | 6.40 | |||
| Lhcbm1, 10 | ASTPDSFWYGPER | 2 | 1.0 | 1.1 | 2.57 | 2.48 | ||
| Lhcbm1-10 | YRELELIHAR | 3 | 0.8 | 4.09 | ||||
| Lhcbm1-10 | ELELIHAR | 2 | 0.9 | 2.59 | ||||
| 14 | Lhcbm1 | VNGGPLGEGLDK | 158-169 | 2 | 0.8 | −0.2 | 3.12 | 3.42 |
| Lhcbm1 | LYPGGSFDPLGLADDPDTFAELKVK | 170-194 | 3 | 1.4 | 3.84 | |||
| Lhcbm1, 10 | ASTPDSFWYGPER | 2 | 1.0 | 0.9 | 2.78 | 3.46 | ||
| Lhcbm2, 8 | ANGGPLGEGLDPLHPGGAFDPLGLADDPD TFAELK | 160-194 | 3 | 1.2 | 4.02 | |||
| Lhcbm1-10 | YRELELIHAR | 3 | 1.0 | 4.35 | ||||
| Lhcbm1-10 | ELELIHAR | 2 | 0.7 | 2.58 | ||||
| 15 | Lhcbm1 | VNGGPLGEGLDK | 158-169 | 2 | 0.4 | −0.3 | 2.65 | 3.28 |
| Lhcbm2, 8 | IEWYGPDRPK | 30-39 | 2 | 0.8 | 0.9 | 2.71 | 2.31 | |
| Lhcbm2, 8 | NGIPFGEAVWFK | 105-116 | 2 | 0.2 | 0.0 | 3.11 | 2.48 | |
| Lhcbm1-10 | ELELIHAR | 2 | 1.1 | 2.35 | ||||
| 16 | Lhcbm1 | VNGGPLGEGLDK | 158-169 | 2 | 0.7 | 2.98 | ||
| Lhcbm1-10 | YRELELIHAR | 2 | 0.7 | 3.51 | ||||
| Lhcbm1-10 | ELELIHAR | 2 | 0.8 | 2.28 | ||||
| Lhcbm2, 8 | IEWYGPDRPK | 30-39 | 2 | 0.9 | 0.3 | 2.50 | 2.48 | |
| Lhcbm2, 8 | NGIPFGEAVWFK | 105-116 | 2 | −0.1 | −0.4 | 2.63 | 2.83 | |
| Lhcbm2, 8 | GPIQNLDDHLANPTAVNAFAYATK | 221-244 | 3 | 1.4 | 5.76 | |||
| Lhcbm2, 8 | FLGPFSEGDTPAYLTGEFPGDYGWDTAGLS ADP ETFKR | 40-77 | 4 | 1.1 | 4.24 | |||
| Lhcbm2, 3, 4, 8, 10 | WAMLGALGCL(I)TPELLAK | 2 | 1.4 | 4.87 | ||||
| Lhcbm6 | WAMLGALGCQTPELLAK | 92-108 | 2 | 0.2 | 0.2 | 2.25 | 2.40 | |
| 17 | Lhcbm1 | VNGGPLGEGLDK | 158-169 | 2 | 0.6 | 0.9 | 2.66 | 3.04 |
| Lhcbm1, 10 | ASTPDSFWYGPER | 2 | 1.0 | 2.98 | ||||
| Lhcbm1-10 | YRELELIHAR | 3 (2) | −0.3 | 0.9 | 3.17 | 3.29 | ||
| Lhcbm1-10 | ELELIHAR | 2 | −0.1 | 0.7 | 2.25 | 2.35 | ||
| Lhcbm2, 8 | IEWYGPDRPK | 30-39 | 2 | 0.3 | 0.8 | 2.66 | 2.35 | |
| Lhcbm2, 3, 4, 5, 8, 10 | WAMLGALGCL(I)TPELLAK | 2 | 1.3 | 4.87 | ||||
| Lhcbm2, 8 | FLGPFSEGDTPAYLTGEFPGDYGWDTAGL SADPETFK | 40-76 | 3 | 0.6 | 6.66 | |||
| Lhcbm2, 8 | FLGPFSEGDTPAYLTGEFPGDYGWDTAGL SADP ETFKR | 40-77 | 3 | −1.2 | 4.48 | |||
| Lhcbm2, 8 | NGIPFGEAVWFK | 105-116 | 2 | −0.4 | 2.99 | |||
| Lhcbm2, 8 | ANGGPLGEGLDPLHPGGAFDPLGLADDP DTFA ELK | 160-194 | 3 | 0.4 | 4.61 | |||
| Lhcbm2, 8 | GPIQNLDDHLANPTAVNAFAYATK | 221-244 | 3 | −0.6 | 1.3 | 5.27 | 6.50 | |
| Lhcbm6 | WAMLGALGCQTPELLAK | 92-108 | 2 | 0.3 | 2.35 | |||
| 22 | Lhcbm1-10 | YRELELIHAR | 3 | −1.2 | 3.68 | |||
| Lhcbm1-10 | ELELIHAR | 2 | 0.8 | −0.1 | 2.27 | 2.39/PICK> | ||
| Lhcbm2, 3, 4, 5, 8, 10 | WAMLGALGCL(I)TPELLAK | 2 | 0.9 | 5.24 | ||||
| Lhcbm3 | QAPASSGIEFYGPNR | 31-45 | 2 | 1.0 | 0.5 | 3.64 | 3.43 | |
| Lhcbm3, 6 | WLGPYSENATPAYLTGEFPGDYGWDTAGL SADPETFKR | 3 | 0.4 | 4.91 | ||||
| Lhcbm3, 7, 10 | GPIQNLDDHLSNPTVNNAFAFATK | 3 | 0.1 | 5.07 | ||||
| Lhcbm4, 6 | FGEAVWFK | 2 | 0.4 | 1.0 | 3.56 | 3.35 | ||
| Lhcbm4, 6 | SSGVEFYGPNR | 2 | 1.0 | −0.7 | 2.49 | 2.43 | ||
| Lhcbm3, 4, 6, 10 | SGTK(Q)FGEAVWFK | 2 | 0.4 | 2.97 | ||||
| Lhcbm4, 6, 7, 9, 10 | VNGGPAGEGLDPLYPGESFDPLGLADDPDT FAELK | 3 | −1.0 | 4.30 | ||||
| Lhcbm6 | GPVQNLDDHLANPTVNNAFAFATK | 222-245 | 3 | 0.0 | 5.41 | |||
| Lhcbm6 | WAMLGALGCQTPELLAK | 92-108 | 2 | 0.1 | 3.01 | |||
| 23 | Lhcbm1-10 | YRELELIHAR | 2 | −0.2 | 3.90 | |||
| Lhcbm1-10 | ELELIHAR | 2 | 0.9 | 0.2 | 2.56 | 2.76 | ||
| Lhcbm2, 3, 4, 5, 8, 10 | WAMLGALGCL(I)TPELLAK | 2 | 0.8 | 4.79 | ||||
| Lhcbm3 | QAPASSGIEFYGPNR | 31-45 | 2 | 0.9 | −0.8 | 3.44 | 3.40 | |
| Lhcbm3 | SSGIEFYGPNR | 35-45 | 2 | 0.9 | 0.3 | 3.07 | 2.28 | |
| Lhcbm3, 6 | WLGPYSENATPAYLTGEFPGDYGWDTAGL SADPETFKR | 3 | 1.3 | 4.02 | ||||
| Lhcbm3, 7, 10 | GPIQNLDDHLSNPTVNNAFAFATK | 3 | 0.0 | −0.2 | 4.67 | 6.34 | ||
| Lhcbm4, 6 | FGEAVWFK | 2 | 0.9 | 0.2 | 3.38 | |||
| Lhcbm4, 6 | SSGVEFYGPNR | 2 | 0.8 | −0.5 | 2.37 | 2.80 | ||
| Lhcbm3, 4, 6, 10 | SGTK(Q)FGEAVWFK | 2 | 0.4 | 3.37 | ||||
| Lhcbm4, 6, 7, 9, 10 | VNGGPAGEGLDPLYPGESFDPLGLADDPD TFAELK | 3 | 0.1 | 3.42 | ||||
| Lhcbm6 | GPVQNLDDHLANPTVNNAFAFATK | 222-245 | 3 | −1.3 | 5.95 | |||
| 24 | Lhcbm1-10 | ELELIHAR | 2 | 1.0 | 0.4 | 2.71 | 2.49 | |
| Lhcbm1 | VNGGPLGEGLDK | 158-169 | 2 | 0.4 | 2.77 | |||
| Lhcbm1-10 | ASTPDSFWYGPER | 2 | 0.5 | 3.27 | ||||
| Lhcbm3, 7, 10 | GPIQNLDDHLSNPTVNNAFAFATK | 3 | 0.7 | 0.9 | 5.38 | 5.68 | ||
| Lhcbm3 | QAPASSGIEFYGPNR | 31-45 | 2 | 1.0 | −0.9 | 3.50 | 3.53 | |
| Lhcbm3 | SSGIEFYGPNR | 35-45 | 2 | 1.2 | 2.26 | |||
| Lhcbm4, 6 | SSGVEFYGPNR | 2 | 0.9 | −0.8 | 2.28 | 2.65 | ||
| Lhcbm3, 4, 6, 10 | SGTK(Q)FGEAVWFK | 2 | 0.6 | 3.20 | ||||
| 30f | CP29 | GSVEAIVQATPDEVSSENR | 102-120 | 2 | −0.7 | 4.67 | ||
| CP29 | FADGLNNGK | 270-278 | 2 | 0.0 | 2.94 | |||
| 31f | CP26 | GWLGGQGGAADLDK | 38-51 | 2 | 0.9 | 4.23 | ||
| CP26 | KLFLPSGLYDR | 58-56 | 2 | 0.9 | 2.91 | |||
| CP26 | YRENELLHAR | 100-109 | 2 | 0.7 | 4.07 | |||
| CP26 | WAMLAAAGILIPEGLQANGANIK | 110-132 | 3 | −0.1 | 4.79 | |||
| CP26 | NGTGPAGYSPGIGK | 185-198 | 2 | −0.2 | 3.04 | |||
| CP26 | HVADPFGYNLLTVLGAEER | 267-285 | 2 | 1.3 | 2.55 | |||
The charge state of the measured ion (z), the calculated deviation of the experimentally determined mass from the theoretical average mass of the peptide (ΔM), and the Xcorr calculated by using the Sequest algorithm are shown. All peptide sequences reported produced Xcorr values equal to or above 1.5, 2.25, or 3.5 for singly, doubly, or triply charged precursor ions, respectively.
The peptide ions identified as lhcb gene products are organized according to the 2-DE protein spots from which they originated (Fig. 1). The gene product and SDS-PAGE mobility identifier are shown. To conserve space in the table, protein designations were shortened; for example, “Lhcbm1, 10” indicates Lhcbm1 and Lhcbm10, and “Lhcbm1-10” indicates Lhcbm1 to Lhcbm10.
The peptide sequence retrieved by database searching is shown.
Positions of identified peptides within the database protein sequence.
Values shown are for iron-sufficient (iron-deficient) conditions.
Present in iron-sufficient and iron-deficient conditions; measured for iron-deficient conditions only.
TABLE 2.
Lhca proteins identified by MS-MS data from protein spots excised from PSI 2-DE mapsa
| 2-DE spot | Proteinb | Peptidec | Positionsd | z | ΔM | Xcorr |
|---|---|---|---|---|---|---|
| 1 | Lhca9 (p22.2) | GALAGDNGFDPLGLGQDEGR | 43-62 | 2 | 0.1 | 4.81 |
| Lhca9 (p22.2) | WYAEAEK | 65-71 | 1 | −0.6 | 1.77 | |
| 2 | Lhca1 (p22.1) | RFTESEVIHGR | 73-83 | 2 | −0.1 | 4.09 |
| Lhca1 (p22.1) | GDAGGVVYPGGAFDPLGFAK | 146-165 | 2 | −0.3 | 4.04 | |
| Lhca1 (p22.1) | FTESEVIHGR | 74-83 | 1 | −0.6 | 3.45 | |
| Lhca9 (p22.2) | PTWLPGLNPPAHLK | 29-42 | 3 | 0.0 | 3.52 | |
| 3 | Lhca5 (p15.1) | GDLAGDYGWDPLGLGADPTALK | 47-68 | 2 | 0.6 | 4.88 |
| Lhca5 (p15.1) | VPNPEMGYPGGIFDPFGFSK | 157-176 | 2 | −0.6 | 3.68 | |
| Lhca5 (p15.1) | NFGSVNEDPIFK | 142-153 | 2 | −0.1 | 3.65 | |
| Lhca5 (p15.1) | LWAPGVVAPEYLK | 34-46 | 2 | −0.1 | 3.02 | |
| Lhca3 (p14.1) | GPFQNLVEHLADPVNNNILTNFGK | 241-264 | 3 | 1.2 | 5.95 | |
| Lhca3 (p14.1) | GSGDAAYPGGPFFNLFNLGK | 186-205 | 2 | −0.6 | 3.95 | |
| Lhca3 (p14.1) | WLQYSEVIHAR | 86-96 | 2 | −0.2 | 3.81 | |
| Lhca7 (p15) | VGLGFPEWYDAGK | 95-107 | 2 | 0.0 | 3.37 | |
| Lhca7 (p15) | GLENGYPGGR | 159-168 | 2 | −0.3 | 3.15 | |
| Lhca7 (p15) | YQEYK | 183-187 | 1 | −0.5 | 1.97 | |
| 4 | Lhca3 (p14.1) | WLQYSEVIHAR | 86-96 | 2 | −0.6 | 3.80 |
| Lhca3 (p14.1) | WLQYSEVIHAR | 86-96 | 3 | 0.0 | 3.56 | |
| Lhca3 (p14.1) | TEAAMK | 206-211 | 1 | −0.4 | 1.78 | |
| Lhca7 (p15) | VGLGFPEWYDAGK | 95-107 | 2 | 0.4 | 3.35 | |
| Lhcbm5 | LSAFYGPDR | 49-57 | 2 | 0.3 | 2.66 | |
| 5 | Lhca3 (p14.1) | WLQYSEVIHAR | 86-96 | 2 | −0.2 | 4.08 |
| Lhca3 (p14.1) | WLQYSEVIHAR | 86-96 | 3 | 0.3 | 3.57 | |
| 6 | Lhca5 (p15.1) | GDLAGDYGWDPLGLGADPTALK | 47-68 | 2 | −1.2 | 4.96 |
| Lhca5 (p15.1) | NFGSVNEDPIFK | 142-153 | 2 | −0.3 | 3.89 | |
| Lhca7 (p15) | VGLGFPEWYDAGK | 95-107 | 2 | −0.2 | 3.47 | |
| Lhca7 (p15) | YQEYK | 183-187 | 1 | −0.4 | 1.72 | |
| Lhca7 (p15) | WYDFK | 136-140 | 1 | −0.4 | 1.55 | |
| 7 | Lhca5 (p15.1) | GDLAGDYGWDPLGLGADPTALK | 47-68 | 2 | −0.8 | 5.06 |
| Lhca5 (p15.1) | NFGSVNEDPIFK | 142-153 | 2 | 0.7 | 4.28 | |
| Lhca5 (p15.1) | VPNPEMGYPGGIFDPFGFSK | 157-176 | 2 | −0.8 | 4.01 | |
| Lhca5 (p15.1) | LWAPGVVAPEYLK | 34-46 | 2 | 0.9 | 3.12 | |
| Lhca6 (p18.1) | GDLPGDFGFDPLGLGANAESLK | 44-65 | 2 | −0.8 | 4.47 | |
| Lhca6 (p18.1) | KPGSVDQDPIFSQYK | 134-148 | 2 | −0.5 | 4.07 | |
| Lhca6 (p18.1) | IPASVSYK | 235-242 | 1 | −0.5 | 1.86 | |
| Lhca7 (p15) | VGLGFPEWYDAGK | 95-107 | 2 | −0.6 | 3.20 | |
| Lhca4 (p14) | WYAQAELMNAR | 95-105 | 2 | −0.5 | 2.82 | |
| 8 | Lhca2 (p19) | ESVPYFPWNEPWNKV | 232-246 | 2 | −0.7 | 3.09 |
| Lhca2 (p19) | LGVNKDNLK | 56-64 | 2 | −0.4 | 2.77 | |
| Lhca2 (p19) | ESVPYFPWNEPWNK | 232-245 | 2 | −0.6 | 2.52 | |
| Lhca2 (p19) or Lhca10 (p18.2) | YEIYKK | 126-131 | 2 | 0.0 | 2.22 | |
| Lhca2 (p19) or Lhca10 (p18.2) | YEIYK | 126-130 | 1 | −0.5 | 1.59 | |
| Lhca2 (p19) | SEEMK | 150-154 | 1 | 0.0 | 1.50 | |
| Lhca2 (p19) or Lhca10 (p18.2) | FWTAGAEK | 95-102 | 1 | −0.6 | 1.70 | |
| Lhca2 (p19) | TLNPGK | 226-231 | 1 | −0.2 | 1.59 | |
| Lhca5 (p15.1) | NFGSVNEDPIFK | 142-153 | 2 | −0.5 | 4.16 | |
| Lhca7 (p15) | VGLGFPEWYDAGK | 95-107 | 2 | −0.2 | 3.61 | |
| 9 | Lhca5 (p15.1) | GDLAGDYGWDPLGLGADPTALK | 47-68 | 2 | −0.4 | 5.69 |
| Lhca5 (p15.1) | NFGSVNEDIFK | 142-153 | 2 | −0.3 | 3.79 | |
| Lhca5 (p15.1) | LWAPGVVAPEYLK | 34-46 | 2 | −0.5 | 3.17 | |
| Lhca6 (p18.1) | KPGSVDQDPIFSQYK | 134-148 | 2 | −0.3 | 5.03 | |
| Lhca6 (p18.1) | GDLPGDFGFDPLGLGANAESLK | 44-65 | 2 | −1.0 | 3.89 | |
| Lhca6 (p18.1) | IPASVSYK | 235-242 | 1 | 0.7 | 1.88 | |
| Lhca2 (p19) | TGETGFLSFAPFDPM[+16]GMK | 132-149 | 2 | 0.0 | 4.07 | |
| Lhca10 (p18.2) | TGETGFLSYAPFDPMGMK | 2 | 0.0 | 2.61 | ||
| Lhca2 (p19) | ESVPYFPWNEPWNKV | 232-246 | 2 | −0.8 | 2.59 | |
| Lhca2 (p19) or Lhca10 (18.2) | FWTAGAEK | 95-102 | 1 | −0.6 | 2.06 | |
| Lhca2 (p19) | LGVNKDNLK | 56-64 | 2 | 0.0 | 3.41 | |
| Lhca2 (p19) | TLNPGK | 226-231 | 1 | −0.4 | 1.59 | |
| 10 | Lhca5 (p15.1) | KLWAPGVVAPEYLK | 33-46 | 2 | −0.1 | 5.00 |
| Lhca5 (p15.1) | VPNPEMGYPGGIFDPFGFSK | 157-176 | 2 | −0.8 | 4.58/PICK> | |
| Lhca5 (p15.1) | GDLAGDYGWDPLGLGADPTALK | 47-68 | 2 | −1.0 | 4.51 | |
| Lhca5 (p15.1) | NFGSVNEDPIFK | 142-153 | 2 | −0.6 | 4.01 | |
| Lhca5 (p15.1) | LWAPGVVAPEYLK | 34-46 | 2 | −0.5 | 3.06 | |
| Lhca6 (p18.1) | KPGSVDQDPIFSQYK | 134-148 | 2 | −0.5 | 4.1 | |
| Lhca6 (p18.1) | AAVVPGQAVAPPCK | 221-234 | 2 | 0.2 | 2.82 | |
| Lhca6 (p18.1) | IPASVSYK | 235-242 | 1 | −0.7 | 1.59 | |
| 11 | Lhca6 (p18.1) | KPGSVDQDPIFSQYK | 134-148 | 2 | −0.3 | 4.42 |
| Lhca6 (p18.1) | GDLPGDFGFDPLGLGANAESLK | 44-65 | 2 | −0.4 | 4.24 | |
| Lhca6 (p18.1) | KPGSVDQDPIFSQYK | 134-148 | 3 | −0.1 | 4.10 | |
| Lhca6 (p18.1) | ESELVHSR | 69-76 | 2 | −0.2 | 2.45 | |
| Lhca6 (p18.1) | IPASVSYK | 235-242 | 1 | −0.6 | 2.06 | |
| 25 | Lhca8 (p18) | WYQQAELIHCR | 68-78 | 2 | −0.4 | 4.24 |
| Lhca8 (p18) | YATGAGPVDNLAAHLK | 211-226 | 2 | −0.2 | 4.04 | |
| Lhca8 (p18) | GTSELGYPGGPFDPLGLSK | 162-180 | 2 | 0.1 | 3.99 | |
| Lhca8 (p18) | AGALNVPEWYDAGK | 96-109 | 2 | −0.2 | 3.36 | |
| Lhca8 (p18) | EADKWADWK | 181-189 | 2 | −0.4 | 2.35 | |
| Lhca8 (p18) | WADWK | 185-189 | 1 | −0.4 | 1.60 | |
| Lhca8 (p18) | KPGSQGEPGSFLGFEASLK | 144-161 | 2 | −0.6 | 5.15 | |
| 26 | Lhca7 (p15) | NPGSQADGSFLGFTEEFK | 141-158 | 2 | −0.5 | 5.12 |
| Lhca7 (p15) | VGLGFPEWYDAGK | 95-107 | 2 | −0.4 | 3.40 | |
| Lhca7 (p15) | GLENGYPGGR | 159-168 | 2 | 0.3 | 2.67 | |
| Lhca7 (p15) | GLENGYPGGR | 159-168 | 2 | −0.1 | 2.51 | |
| Lhca7 (p15) | FFDPMGLSR | 169-177 | 2 | −0.4 | 2.36 | |
| Lhca7 (p15) | YQEYK | 183-187 | 1 | −0.4 | 1.83 | |
| Lhca7 (p15) | VVVEK | 108-112 | 1 | −0.3 | 1.52 | |
| 29 | Lhca1 (p22.1) | GDAGGVVYPGGAFDPLGFAK | 146-165 | 2 | −0.7 | 4.11 |
| Lhca1 (p22.1) | FTESEVIHGR | 74-83 | 2 | −0.3 | 3.57 | |
| 30 | Lhca9 (p22.2) | GALAGDNGFDPLGLGQDEGR | 43-62 | 2 | −0.1 | 4.08 |
| Lhca1 (p22.1) | GDAGGVVYPGGAFDPLGFAK | 146-165 | 2 | −0.6 | 3.95 | |
| 31 | Lhca9 (p22.2) | GALAGDNGFDPLGLGQDEGR | 43-62 | 2 | −0.3 | 6.01 |
| Lhca9 (p22.2) | TQPIEGLTAHLADPFGK | 181-197 | 2 | 0.0 | 3.70 | |
| Lhca9 (p22.2) | WYAEAEK | 65-71 | 1 | −0.6 | 1.60 | |
| 2+ | Lhca1 (p22.1) | GDAGGVVYPGGAFDPLGFAK | 146-165 | 2 | −0.5 | 3.71 |
| Lhca1 (p22.1) | FTESEVIHGR | 74-83 | 2 | −0.3 | 3.33 | |
| Lhca9 (p22.2) | GALAGDNGFDPLGLGQDEGR | 43-62 | 2 | −0.1 | 3.24 | |
| 29+ | Lhca1 (p22.1) | GDAGGVVYPGGAFDPLGFAK | 146-165 | 2 | −0.1 | 4.18 |
| Lhca1 (p22.1) | FTESEVIHGR | 74-83 | 2 | −0.1 | 3.29 |
The charge state of the measured ion (z), the calculated deviation of the experimentally determined mass from the theoretical average mass of the peptide (ΔM), and the Xcorr calculated by using the Sequest algorithm are shown. All peptide sequences reported produced Xcorr values equal to or above 1.5, 2.25, or 3.5 for singly, doubly, or triply charged precursor ions, respectively.
The peptide ions identified as lhca gene products are organized according to the PSI 2-DE protein spots from which they originated. The gene product and SDS-PAGE mobility identifier are shown.
The peptide sequence retrieved by database searching is shown.
Positions of identified peptides within the database protein sequence.
We also specifically searched for N-terminal peptides (see Materials and Methods). From spots 14 to 16, we identified for Lhcbm2 or Lhcbm8 an N-terminal peptide (IEWYGDPR) whose sequence matches perfectly the N-terminal amino acid sequence deduced from Edman amino acid sequencing (19). From spot 23, we identified for Lhcbm3 an N-terminal peptide (SSGIEFYGPNR). An overlapping peptide [(K)QAPASSGIEFYGPNR] was also determined for Lhcbm3 (Table 1), indicating that two N termini exist for Lhcbm3, as suggested for Lhcbm6 (see below). Interestingly, another N-terminal peptide of Lhcbm3 (PASSGIEFYGPNR) was identified by analysis of a tryptic digest derived from LHC proteins separated by native gel electrophoresis (see below). Additionally, these data show that mature Lhcbm3 does not start with the N-terminal amino acids IEF, as does Lhcbm2or Lhcbm8. A corresponding peptide [(K)SSGVEFYGPNR] of Lhcbm4 or Lhcbm6 was also recognized from spots 22 to 24. However, in this instance, it is possible that the peptide originated from a tryptic cut. We also identified two lhca gene products, Lhca3 (p14.1) and Lhca7 (p15), in spots 12 and 13. The numbers in parentheses indicate the relative mobilities of the respective Lhca proteins in SDS-PAGE, in accordance with the nomenclature of Bassi et al. (3). The lhca7 gene product is defined by an assembled EST contig (Grossman et al., unpublished) and has been identified for the first time at the protein level (see below).
To test the significance of our data gathering and processing procedures, in an independent experiment we analyzed protein spots 12 to 24 derived from 2-DE-separated thylakoid membrane proteins isolated from iron-deficient cells (grown for 5 days in iron-deficient medium). It was shown recently that in the process of adaptation to iron deficiency, Lhcb proteins remain stable whereas, the Lhca3 protein and other Lhca proteins disappear (33). While the Lhcbm proteins identified from protein spots 12 to 24 were almost identical to those found under iron-sufficient conditions, no lhca-related gene products could be identified from protein spots 12 and 13 derived from iron-deficient thylakoid membrane proteins (Table 1). In contrast to the findings under iron-sufficient conditions, one Lhcbm1 peptide was detected from spot 24 under iron-deficient conditions. Although this result might suggest differential protein processing for this polypeptide, we cannot exclude the possibility that it was found because of horizontal streaking of the protein due to electrophoresis in the second dimension. Overall, these data indicate that the peptides identified by the MS-MS and subsequent Sequest analyses are significant and specific for the single spots processed. Interestingly, we identified from spots 23 and 24 under iron-sufficient as well as iron-deficient conditions several peptides from PsbO, the oxgen-evolving enhancer protein 1 (30). Although PSII polypeptides are also degraded in the process of adaptation to iron deficiency, it appears that Lhca3 (p14.1) and Lhca7 (p15) are more severely affected and thus represent primary targets of iron deficiency, as already demonstrated (33).
Beside PsbO, several other peptides deduced from the EST data could be identified from the MS-MS data. From spot 23, two peptides corresponding to a Lon protease homologue (AV397248) could be recognized. However, no other significant hits (more than one peptide for the same putative protein) were obtained.
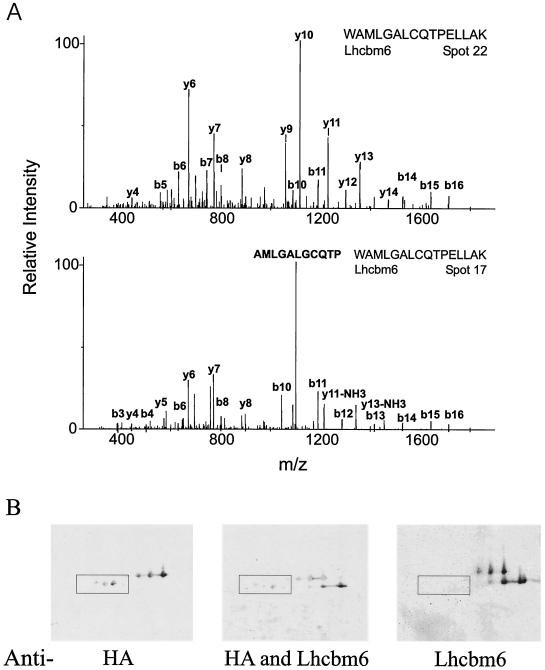
Interestingly, lhcbm6-related gene products with different molecular masses and different isoelectric points were found. The MS-MS data identified peptide WAMLGALGCQTPELLAK, which is specific for Lhcbm6, in spots 22 and 23 as well as in spots 16 and 17 with a high level of significance (Fig. 2A). A previous MS analysis of 2-DE-separated Lhcbm6 had already indicated that this LHCII protein might contain two alternative N-terminal protein cleavage sites (20). An HA tag was introduced into the lhcbm6 gene in such a way that it was localized behind the second suggested N-terminal processing site [28APKS(HA tag)SGVE35] in the expressed gene product (23). Anti-HA monoclonal antibodies recognized two protein bands with different molecular masses (mass difference, 1 to 2 kDa) after SDS-PAGE fractionation of thylakoid membranes from the Lhcbm6-HA-tagged algal strain. Immunoblot experiments with 2-DE-separated Lhcbm6-HA-tagged thylakoid membranes, anti-Lhcbm6 peptide antibodies (directed against a sequence upstream of the second processing site), and anti-HA antibodies revealed that the lower-molecular-mass spots recognized by the anti-HA antibodies are not recognized by the anti-Lhcbm6 antibodies; these data demonstrate that processing occurs at the N terminus (Fig. 2B). Thus, the Lhcbm 2-DE mapping results and the tagging experiment independently confirmed that two differentially N-terminally processed Lhcbm6 forms exist in vivo. It should be noted that the lower-molecular-mass protein spots detected by the anti-HA antibodies are considerably shifted in their isoelectric points toward more acidic values compared to the higher-molecular-mass HA-tagged protein spots. This finding is also indicative of N-terminal protein processing, since the N-terminal part of the Lhcbm6 protein is enriched with positively charged amino acids. Fractionation of whole HA-tagged cells by one-dimensional SDS-PAGE and subsequent immunoblot analyses with anti-HA antibodies revealed two protein bands with different molecular masses, indicating that N-terminal processing also occurs in whole cells and is not an artifact due to the membrane preparation.
FIG. 2.
Two differentially N-terminally processed forms of Lhcbm6. (A) MS-MS spectra of peptide WAMLGALGCQTPELLAK from spot 22 (without Fe) and spot 17 (with Fe). The CID mass spectra of the doubly protonated precursor ion peaks measured at m/z 930.5 and 930.4 for spots 22 and 17, respectively, are shown. (B) To identify differentially processed Lhcbm6 forms, an immunoblot of 2-DE-separated thylakoid membranes of Lhcbm6-HA-tagged strain Lep2 was probed with an anti-Lhcbm6 peptide antibody (recognizes the N terminus) (3) and an antibody recognizing the HA epitope. Boxed areas indicate the positions of the processed forms of HA-tagged Lhcbm6.
Phosphorylation of Lhcbm3.
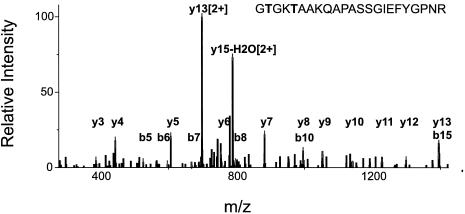
Currently, it is not known for Chlamydomonas which of the major Lhcbm proteins is phosphorylated, although it is known that these proteins become phosphorylated during the process of state transitions (46). In order to identify phosphorylated Lhcbm proteins, thylakoid membranes were in vitro phosphorylated and immediately subjected to native green gel electrophoresis. The green monomeric LHC band was excised, proteins were digested in the gel with trypsin, and peptides were analyzed by MS-MS. The MS-MS data were specifically matched with the Lhcbm database that contains a set of different N termini (see Materials and Methods). Thus, the Lhcbm3 peptide [(K)GTGKTAAKQAPASSGIEFYGPNR] was reproducibly found to exhibit a molecular mass shift of 80 Da, representative of phosphorylation at a Thr or Ser residue. The peptide was found as a triply charged ion. Sequest identifies this peptide with a correlation factor of 3.2 or 3.3 when the second or first N-terminal Thr residue becomes phosphorylated, respectively, whereas no significant scores for phosphorylated Ser residues are found. Our data do not allow us to determine which of the two Thr residues is phosphorylated. We also cannot exclude the possibility that this peptide represents a mixed population, where either one of the two Thr residues is phosphorylated.
A closer analysis of the fragmentation spectrum (Fig. 3) revealed a series of y- and b-type ions, including those that exhibit a shift in molecular mass of 80 Da; these data indicate that the fragmentation spectrum represents the CID pattern of the respective phosphorylated peptide. It is known that phoshorylated peptides do not fragment well in MS-MS analyses; therefore, we interpret the Sequest results as being significant, although the correlation factor is slightly under the cutoff value used throughout this study. In this analysis, we identified several peptides already found in the 2-DE analysis as well as three Lhcbm5-specific peptides and two Lhcbm4- or Lhcbm9-specific peptides (two triply charged ions with correlation factors of 5.2 and 3.8, respectively: AKWLGPYSENSTPAYLTGEFPGDYGWDTAGLSADPETFK and WLGPYSENSTPAYLTGEFPGDYGWDTAGLSADPETFKR). The latter finding indicates that Lhcbm4 or Lhcbm9 is expressed at the protein level; thus, another Lhcbm protein is added to the total number of Lhcb proteins found in this study. These large peptides were not found in the 2-DE analysis, probably because of their size. Thus, we identified eight distinct Lhcb proteins. We also detected a singly charged ion with a molecular mass of 1,394.7 Da (deviation from theoretical mass, 0.2 Da). Sequest analysis of the MS-MS data revealed a significant match, at a correlation factor of 2.00, with an Lhcbm3-specific peptide (33PASSGIEFYGPNR45). This peptide represents another putative N terminus for Lhcmb3 (see above). It should be noted that the phosphorylation site that we determined would be removed by this protein processing. Interestingly, such N-terminal protein processing can be also described for Lhcbm6 (see above).
FIG. 3.
MS-MS spectrum of phosphorylated Lhcbm3 peptide GTGKTAAKQAPASSGIEFYGPNR. The data indicate that either the first or the second Thr (in bold type) becomes phosphorylated. The CID mass spectrum of the triply protonated precursor ion peak measured at m/z 797.65 was identified from a tryptic digest of a green LHC band obtained after fractionation of in vitro-phosphorylated thylakoid membranes by native gel electrophoresis. The b-type ions b5, b6, b7, b8, and b10 represent phophorylated ions, indicating phosphorylation of either of the two Thr residues.
Lhca protein map.
For high-resolution 2-DE protein mapping of Lhca proteins, we separated enriched PSI particles by 2-DE (Fig. 4). The putative Lhca protein spots (20) were excised, digested in the gel with trypsin, and analyzed by LC-MS-MS. A total of 20 protein spots were investigated. The minor LHCII antenna protein CP26 (Lhcb5) was found in spots 16 and 18, indicating copurification with enriched PSI particles (data not shown). However, none of the other major Lhcb proteins could be identified in any of the other spots, demonstrating a reasonable enrichment of LHCI (PSI) proteins versus LHCII proteins. Interestingly, a peptide specific to Lhcbm5 was recognized in spot 4. Lhcbm5 is 1 of 10 putative lhcbm gene products expressed in C. reinhardtii (11). Lhcbm5 is not among the major Lhcbm proteins identified in 2-DE-separated thylakoid membranes (see above), indicating that this Lhcb protein is a rather minor component of the Lhcbm proteins which can be copurified with PSI particles.
Peptides related to lhca gene products could be identified in 18 protein spots. In eight protein spots (1, 5, 11, 25, 29, 29+, 30, and 31), peptides that could be correlated only with one lhca gene product were found. In the other spots, the MS-MS data revealed peptides deduced from more than one lhca gene product. At least nine different lhca-related gene products were predicted by searching the Chlamydomonas EST database with MS data. For nine Lhca proteins, the respective genes were found as assembled EST contigs (Table 2), and seven are represented completely in the first release of the genomic database assembly (Table 3). Of these nine, three are identified for the first time at the protein level; Fig. 5 shows selected MS-MS spectra. In accordance with the nomenclature of Bassi et al. (3), the numbers in parentheses in Fig. 4 represent the relative mobility identifiers of the respective Lhca proteins in SDS-PAGE (see also Table 2). The N-terminal amino acid sequences of six Lhca proteins (p14, p14.1, p15.1, p18, p18.1, and p22.1) were reported by Bassi et al. (3). The respective contigs of the newly identified lhca gene products are 20020630.7235.1, 20020630.8317.1, and 200206304096.2. For the last contig sequence, a corresponding gene has been already annotated (Merchan and Fernandez, 2000).
TABLE 3.
C. reinhardtii lhca and lhcb genes and gene products as determined by EST, cDNA, or genomic sequences in comparison with tryptic peptide sequences obtained by MS
| Chlamydomonas LHC proteins | Related gene in vascular plants | Method(s) of identification | Annotation or EST contiga | Position of identity to sequences in respective scaffolds of genomic database (version 1.0 assembly) |
|---|---|---|---|---|
| Lhcbm1 | lhcII-4b | 2-DE-LC | AB051210 | 1839 |
| AB051206 | ||||
| AF495473 | ||||
| Lhcbm2 | lhcII-3b | (Mature sequence identical to Lhcbm8) | AB051209 | 385 (transit peptide sequences are found in scaffold 87) |
| AB051205 | ||||
| Lhcbm3 | lhcII-1.3b | 2-DE-LC | AB051208 | 354 |
| AB051204 | ||||
| Lhcbm4c | lhcb2b | Native PAGE-LC | AF104630 | 688d |
| Lhcbm5 | lhcb3b | 2-DE-LC | AF104631 | 168 |
| Lhcbm6 | cabII-1b | 2-DE-LC | AF95472 | 1191 (Not complete) |
| M24072.1 | ||||
| Lhcbm7 | lhcb2.2 | (Not unequivocally) | AF479779 | Not found |
| Lhcbm8 | cabII-2b | 2-DE-LC | AF330793 | 385 |
| Lhcbm9c | lhcb2.2 | Native PAGE-LC | AF479778 | 688 |
| Lhcbm10 | lhcb1.5 | (Not unequivocally) | AF479777 | Not found |
| Lhcb5 (CP26) | lhcb5 | 2-DE | AB050007 | 800 |
| Lhcb4 (CP29) | lhcb4.3 | 2-DE | 20021010.7231.1 | 538 (first 37 N-terminal amino acids not found) |
| Lhca1 (p22.1) | lhca1 | 2-DE-LC | AF104633 | 497 |
| Lhca2 (p19) | lhca2 | 2-DE-LC | 20020630.8317.1 | 218e |
| Lhca3 (p14.1) | lhca3 | 2-DE-LC | 20020630.1214.1 | 1152 (first seven N-terminal amino acids not found) |
| Lhca4 (p14) | lhca2 or lhca4 | 2-DE-LC | 20020630.608.1 | 106 |
| Lhca5 (p15.1) | lhca2 or lhca4 | 2-DE-LC | 20020630.154.1 | 3122 |
| Lhca6 (p18.1) | lhca2 or lhca4 | 2-DE-LC | 20020630.6026.1 | 653 |
| Lhca7 (p15) | lhca5 | 2-DE-LC | 20020630.7235.1 | 870 |
| Lhca8 (p18) | lhca5 | 2-DE-LC | 20021010.4209.1 | 299 |
| Lhca9 (p22.2) | lhca2 | 2-DE-LC | AF244524 | 419 |
| Lhca10 (p18.2) | 2-DE-LC | av391905 | Not found |
C. reinhardtii EST contig or EMBL-EBI accession numbers of previously reported genes for Lhcbm1 (AB051210, AB051206, and AF495473 [11, 42]), Lhcbm2 (AB051209 and AB051205 [42]), Lhcbm3 (AB051208 and AB051204 [42]), lhcbm4 (AF104630 [O'Connor et al., 1999]); Lhcbm5 (AF104631 [O'Connor et al., 1999]), Lhcbm6 (M24072.1 and AF495472 [11, 23]), Lhcbm7 (AF479779 [11]), Lhcbm8 (AF330793. [38]), Lhcbm9 (AF479778 [11]), Lhcbm10 (AF479777 [11]), Lhcb5 (CP26 and AB050007 [32]), Lhca1 (AF104633 [O'Connor et al., 1998]), and Lhca9 (AF244524 [Merchan and Fernandez, 2000]).
Homology to this vascular plant Lhcb gene was reported by Elrad et al. (11).
Peptides identical to those deduced from lhcbm4 and lhcbm9 gene products were found.
Ala207 is replaced by Arg in the genomic sequence.
Ile198 is replaced by Val in the genomic sequence.
FIG. 5.
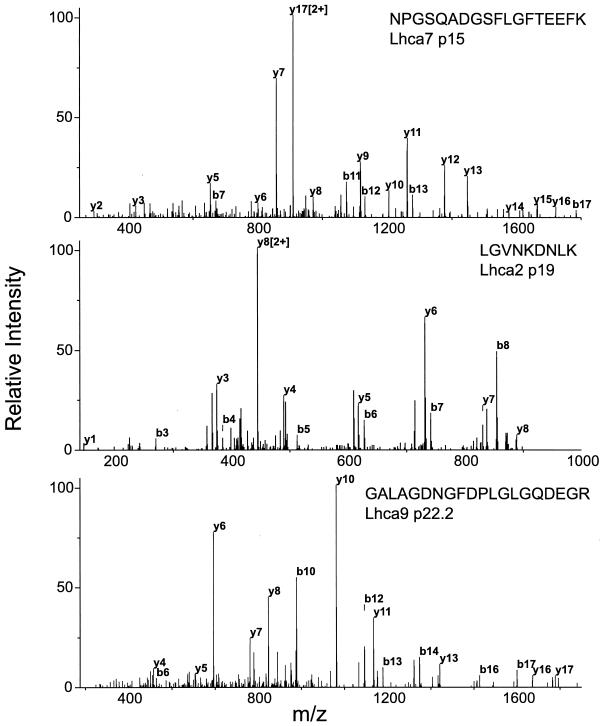
MS-MS spectra of newly identified lhca gene products. The CID mass spectra of the doubly protonated precursor ion peaks measured at m/z 966.3, 500.9, and 980.6 for Lhca7 (p15), Lhca2 (p19), and Lhca9 (p22.2), respectively, are shown. The amino acid sequence retrieved by the Sequest program for the MS-MS data is shown in the upper right-hand corner of each spectrum. The ion peaks that corresponded to theoretical b-type (N-terminal) or y-type (C-terminal) fragments of the precursors are labeled. Peaks identified as doubly charged are labeled only for high intensities. The mass-to-charge ratios of the measured fragment ion peaks are shown below the x axis, and the relative intensities of the total ion current measured are shown along the y axis.
The MS-MS data obtained for the novel Lhca proteins p19 or p15 and p22.2 resulted in sequence coverages of more than 15 and 25%, respectively (Table 2). For the other lhca gene products, the MS-MS data gathered resulted in sequence coverages of more than 20%. One of the peptides identified from spot 9 (TGETGFLSYAPFDPMGMK) corresponds to a sequence deduced from an open reading frame of a single EST clone (AV391905). This peptide is highly similar to a peptide derived from Lhca2 (p19) (TGETGFLSFAPFDPMGMK) and differs by a mass of 16 Da due to a change of Tyr to Phe. Unfortunately, this EST is not found in an assembled EST contig. However, the corresponding open reading frame codes for an almost complete lhca-related gene product that could represent a novel Lhca subunit. It was designated Lhca10 (p18.2) and included in further Lhca protein analyses. It should be noted that the mass difference of 16 Da could also be explained by oxidation of the first Met, as predicted by Sequest analysis. The Xcorr for this possibility is 4.07; in comparison, the Xcorr is 2.61 when the sequence from the EST clone is used. These data indicate that this MS-MS spectrum more likely represents the modified peptide derived from Lhca2 (p19) and not the novel peptide. However, our data cannot exclude the possibility that Lhca10 exists.
From spots excised from the PSI 2-DE map, we also identified peptides from proteins other than Lhc. However, in none of these instances could more than one peptide corresponding to a respective polypeptide sequence be recognized (data not shown); these data thus were not regarded as significant.
To verify the different lhca gene products by MS-MS, in an independent experiment we digested the entire isolated PSI complex with trypsin, separated the resulting peptides by reversed-phase LC, and analyzed them by MS-MS. Analysis of the MS-MS data with Sequest confirmed the presence of nine distinct lhca gene products, which were also identified by the 2-DE approach (data not shown). We could identify several peptides for each of the nine Lhca proteins. In contrast, no peptide for putative Lhca10 (p18.2) could be identified. Interestingly, we identified by database searches an assembled EST contig (20020630.8279.1) that is highly homologous to the p18 protein sequence deduced from contig 20021010.4209.1. This sequence differs at only one position, which would alter a putative tryptic peptide, KPGSQDEPGSFLGFEASLK, deduced from the sequence of contig 20021010.4209.1 (Table 2). This peptide has an Asp instead of a Gly at position 6. This exchange is not supported by the genomic database.
Phylogenetic analysis of Lhca proteins.
Phylogenetic analysis of C. reinhardtii Lhca precursor proteins Lhca1 (p22.1), Lhca3 (p14.1), Lhca4 (p14), Lhca5 (p15.1), Lhca6 (p18.1), and Lhca9 (p22.2) has already indicated that p14.1 and p22.1 are related to Lhca3 and Lhca1 proteins from vascular plants, respectively (42) (Table 3), whereas the other proteins could not be assigned to a distinct vascular plant homologue. Proteins p14, p15.1, p18, and p18.1 were therefore assigned to vascular plant Lhca2 or Lhca4 protein (42) (Table 3). Reanalysis of these proteins by BLAST similarity searches showed that p18 was most similar to Lhca5 from Arabidopsis (24) (Table 3). When we investigated the novel precursor proteins p15, p19, and p22.2 by BLAST similarity searches, it appeared that p15 was highly similar to Lhca5, while p19 and p22.2 were most similar to Lhca2 from Arabidopsis (24) (Table 3).
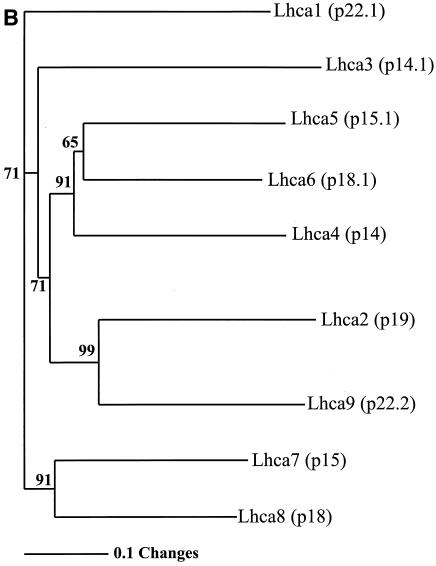
To cluster the different lhca gene products, we performed phylogenetic analysis of Lhca precursor protein sequences (aligned in Fig. 6A by the CLUSTAL W program) (43) by using the neighbor-joining method (38). This analysis resulted in three major clusters, which represent the Lhca1 homologue p22.1; the Lhca3, Lhca2 or Lhca4, and Lhca2-like homologues p14.1, p15.1, p18.1, p14, p19, p22.2 and p18.2 (p18.2 is not included in Fig. 6); and the Lhca5-like homologues p15 and p18. The second cluster can be further differentiated. The Lhca3 protein seems to be distinct from the Lhca2 or Lhca4 and Lhca2-like proteins. It appears that the Lhca2 or Lhca4 homologues p15.1 and p18.1 are more closely related to each other than to p14. It is likely that polypeptides p19 and p18.2 as well as polypeptides p15.1 and p18.1 were generated by more recent gene duplications. In summary, phylogenetic analysis indicates that Lhca proteins from C. reinhardtii can be divided into five distinct classes, where the Lhca1 and Lhca3 homologues are represented by only one gene product and the Lhca2 or Lhca4, Lhca2-like, and Lhca5-like homologues are represented by more than one gene product (Fig. 6B).
FIG. 6.
Phylogenetic analyses. (A) CLUSTAL W alignment of Lhca protein sequences. The CLUSTAL W alignment of full-length protein sequences was performed by using the European Molecular Biology Laboratory-European Bioinformatics Institute computer server with default values. Sequences for the tryptic peptides identified by LC-MS and database searching are shown in bold type; overlapping sequences are indicated in grey. The identities of the protein sequences are as follows: Lhca1, p22.1; Lhca3, p14.1; Lhca7, p15; Lhca8, p18; Lhca5, p15.1; Lhca6, p18.1; Lhca4, p14; Lhca2, p19; and Lhca9, p22.2. The putative Lhca10 (p18.2) sequence is not shown, since a complete contig assembly is not available in the database. (B) Neighbor-joining tree of the CLUSTAL W alignment of Lhca protein sequences. The C. reinhardtii Lhca protein sequences group into three clusters. One of the clusters is split into three groups. The tree supports the assignment of Lhca8 (p18) as an Lhca5-like protein, in contrast to the previously suggested annotation as a vascular plant Lhca2- or Lhca4-like protein.
Stoichiometry of Lhca proteins.
To determine the relative stoichiometry of the LHCI protein subunits, Hippler et al. recently (20) evaluated “spot volumes” (by using Phoretix 2-DE software) of the individual LHCI proteins after staining of 2-DE-separated PSI particles with colloidal Coomassie blue. According to this study, the Lhca1 protein (spot 2 in Fig. 4) appeared to be the most abundant Lhca protein, followed by the Lhca3 protein and the p18 protein represented in spot 25. The use of LC-MS-MS in the present study allowed us to identify peptides with a very high sensitivity. Thus, it appears that in spot 2, which represents Lhca1 (20), a peptide of another Lhca protein (Lhca9; p22.2) is identified. This protein is also present in spots 1, 30, and 31. The fact that two and three peptides of p22.2 are identified in spots 1 and 31—although these spots are rather faint—makes it very likely that p22.2 is represented in spot 2 only in very small amounts. From these data we can conclude that Lhca1 is the most abundant protein in spot 2 and therefore the most abundant Lhca protein. It is followed in abundance by Lhca8 (p18), a vascular plant Lhca5-like protein, which is present in relative amounts that are about 10-fold smaller. Peptides identified in spot 8 are derived almost exclusively from Lhca2 (p19) (a vascular plant Lhca2-like protein). Thus, spot 8 represents mainly p19 and is, according to its staining intensity, also about 10-fold less abundant than Lhca1. The relative amounts of Lhca3 (p14.1), Lhca5 (p15.1), Lhca6 (p18.1), and Lhca7 (p15) (a vascular plant Lhca5-like protein) are difficult to determine from our 2-DE map, since these proteins are always found together with other Lhca proteins. However, from the number of peptides found, it is likely that their abundance is at least comparable to that of p18 and p19. In contrast, the Lhca9 protein (p22.2) has a very low abundance and is more then 50-fold less abundant than the Lhca1 protein.
Modifications of Lhcb and Lhca proteins.
For Lhcb proteins as well as for Lhca proteins, it is apparent that bands of spots that are represented by individual gene products appear across the 2-DE maps. A possible explanation for this finding could be modification of the peptide amino terminus or the side chains of lysine or arginine by isocyanic acid. Such protein carbamylation would result in so-called carbamylation trains. For Lhcbm6, such trains are evident on the 2-DE maps (Fig. 1 and 2). Therefore, we specifically searched the MS-MS data of all Lhcbm proteins for protein carbamylation, but the results were negative. These data indicate that the Lhcbm polypeptides produce natural trains which could be caused by posttranslational modifications, such as phosphorylation (see above), or charge heterogeneity. Most of the Lhca proteins (p22.2, p22.1, p19, p18.1, p15.1, p15, and p14.1) can be found at different positions on the 2-DE maps. The exact reason for these data is unknown. We could not identify differentially N-terminally processed Lhca proteins by MS-MS, although we specifically searched the MS-MS data for such processing events (see Materials and Methods). However, it is tempting to speculate that besides posttranslational modification, protein processing may account for the fact that particular Lhca proteins are found at different positions on the 2-DE maps.
DISCUSSION
In this study, we took advantage of a recently developed procedure to separate transmembrane thylakoid proteins by 2-DE. We thereby established detailed two-dimensional protein maps of light-harvesting proteins from C. reinhardtii by analyzing excised trypsin-digested protein spots with MS. In addition, we examined tryptic digests of isolated PSI (LHCI) particles by coupling LC and MS. We also took advantage of the sequence information provided by the Chlamydomonas genome project, which allowed us to retrieve peptide sequences from a large Chlamydomonas EST database as well as from a genomic database by using the MS data. With this information, we predicted eight distinct Lhcb proteins and nine different Lhca proteins. Of these 17 different gene products, 14 were completely represented in the first C. reinhardtii genomic database assembly (version 1.0) (Table 3). For other gene products, complete genes could not be reconstructed, but the genomic assembly did represent partial sequences. For the putative lhca10 gene product, no 100% matching sequences could be found in the genomic database. Since lhcbm6 (formerly lhcb1) (23), which was the first LHC-encoding cDNA that was cloned, is not completely represented, further assemblies of the genomic data are needed before final conclusions can be drawn. Therefore, the putative lhca10 gene product could represent a 10th Lhca subunit.
The MS and succeeding bioinformative analyses revealed a high level of complexity of the light-harvesting proteins in C. reinhardtii. In addition to the large number of different LHC proteins, we also identified N-terminal protein processing of the lhcbm3 and lhcbm6 gene products. This finding, which was evident from the MS data as well as from the Lhcbm6-HA tagging experiment (Fig. 2), demonstrates that two differentially N-terminally processed forms of Lhcbm3 and Lhcbm6 exist in thylakoid membranes. What could be the cause for the differential processing? Differential processing of pre-LHC proteins has already been reported in in vitro studies for wheat, pea, tobacco, tomato, and corn (5, 6, 8, 27, 35). A secondary processing site in the wheat sequence is believed to be within the mature protein (7) and is, interestingly, beyond Thr3, which has been found to be phosphorylated in thylakoid membranes from spinach and Arabidopsis (31, 45). It has been shown that the recombinant stromal processing peptidase can process pre-Lhcb1, which would be homologous to Chlamydomonas Lhcbm3 or Lhcbm6, at the secondary site (37). Thus, different stromal processing peptidases that process the transit sequence at different positions could be the cause for the different N-terminal protein products observed for Lhcbm3 or Lhcbm6. What could be the function of such processing? Such a processing event would remove the phosphorylation site at Thr3 in Lhcb1 of vascular plants and would thereby inhibit the ability of the respective Lhcb1 protein to migrate to the stromal membranes and participate in the process of state transitions. Our data indicate that Lhcbm3 is phosphorylated in Chlamydomonas. In the 2-DE map, Lhcbm3 is found in the upper row of Lhcbm protein spots, corresponding to the position of protein P11, which is one of the major phosphorylated protein bands seen in SDS-PAGE analysis under state II conditions in Chlamydomonas (46). Interestingly, phosphorylation of the second Thr residue in the respective Lhcbm3 peptide [(K)GT*GKT*AAKQAPASSGIEFYGPNR] corresponds exactly to the phosphorylation of Thr3 in vascular plants (RKT*AAKAKQ…) (asterisks indicate phosphorylated residues) (31, 45). The MS data identify two N termini of Lhcbm3 in which, due to N-terminal protein processing, both phosphorylation sites are removed. Therefore, we suggest that such N-terminal protein processing could be a regulatory event in the process of state transitions in Chlamydomonas as well as vascular plants.
Our proteomic study reveals that among the 10 distinct lhca gene products (including the putative lhca10 gene product), 5 different phylogenetic types of Lhca proteins are expressed in Chlamydomonas. Chlamydomonas is the first plant where the expression of Lhca5-like proteins can be demonstrated. The lhca gene family in Arabidopsis is composed of six nuclear genes (lhca1 to lhca6); of these, genes lhca5 and lhca6 are expressed only at very low RNA levels (24). The Lhca proteins identified in 2-DE of enriched PSI particles are copurified with the PSI complex and therefore belong to the antenna that is functionally connected to PSI. The PSI (LHCI) protein complex in Chlamydomonas may contain as many as 14 light-harvesting proteins per reaction center, as revealed by electron microscopy (15). Such structural data indicate eight Lhca (Lhca1 to Lhca4) proteins per PSI complex for PSI (LHCI) proteins in vascular plants (4). These data suggest that the LHCI antenna in Chlamydomomas is larger than that in vascular plants. Our proteomic data show, as already suggested (3, 20), that the variability of Lhca proteins also is greater in Chlamydomonas than in vascular plants. These data suggest that the variability in light harvesting due to the high level of diversity of Lhca proteins could be an additional important regulatory parameter in response to changing physiological conditions. Such a dynamic adaptation of the Lhca antenna has been shown for Chlamydomonas in response to iron deficiency (33), which induces the remodeling of the Lhca antenna, including subunit degradation and replacement.
The fact that several Lhca proteins are found at different positions in the 2-DE map makes an exact determination of the stoichiometry of individual Lhca polypeptides difficult. However, an intriguing question is how and in which stoichiometry these subunits are organized within the complex. It appears that Lhca protein expression is realized on a rather dynamic scale in Chlamydomonas. In future experiments, it will be important to determine the exact stoichiometry of individual Lhca proteins. This goal could be achieved by immunotitrations with specific antibodies directed against distinct Lhca proteins or by MS with synthetic peptides for the quantification of respective tryptic Lhca peptides. Additionally, it will be interesting to monitor how the relative abundances of individual Lhca and Lhcb proteins change under different physiological conditions and in different genetic backgrounds.
Acknowledgments
E. J. Stauber and A. Fink contributed equally to this work.
We are grateful to M. Stäger for excellent technical assistance. We thank A. Romualdi for bioinformative assistance.
This work was supported by grant Hi739/1-3 from the Deutsche Forschungsgemeinschaft and Freistaat Thüringen (Nachwuchsgruppe: die Plastidäre Proteinausstattung bei Differenzierungs- und Regulationsvorgängen) to M.H.
REFERENCES
- 1.Asamizu, E., K. Miura, K. Kucho, Y. Inoue, H. Fukuzawa, K. Ohyama, Y. Nakamura, and S. Tabata. 2000. Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Res. 7:305-307. [DOI] [PubMed] [Google Scholar]
- 2.Asamizu, E., Y. Nakamura, S. Sato, H. Fukuzawa, and S. Tabata. 1999. A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. DNA Res. 6:369-373. [DOI] [PubMed] [Google Scholar]
- 3.Bassi, R., S. Y. Soen, G. Frank, H. Zuber, and J. D. Rochaix. 1992. Characterization of chlorophyll a/b proteins of photosystem I from Chlamydomonas reinhardtii. J. Biol. Chem. 267:25714-25721. [PubMed] [Google Scholar]
- 4.Boekema, E. J., P. E. Jensen, E. Schlodder, J. F. van Breemen, H. van Roon, H. V. Scheller, and J. P. Dekker. 2001. Green plant photosystem I binds light-harvesting complex I on one side of the complex. Biochemistry 40:1029-1036. [DOI] [PubMed] [Google Scholar]
- 5.Clark, S. E., M. S. Abad, and G. K. Lamppa. 1989. Mutations at the transit peptide-mature protein junction separate two cleavage events during chloroplast import of the chlorophyll a/b-binding protein. J. Biol. Chem. 264:17544-17550. [PubMed] [Google Scholar]
- 6.Clark, S. E., J. E. Oblong, and G. K. Lamppa. 1990. Loss of efficient import and thylakoid insertion due to N- and C-terminal deletions in the light-harvesting chlorophyll a/b binding protein. Plant Cell 2:173-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, S. E., and G. K. Lamppa. 1991. Determinants for cleavage of the chlorophyll a/b binding protein precursor: a requirement for a basic residue that is not universal for chloroplast imported proteins. J. Cell Biol. 114:681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cline, K. 1988. Membrane insertion, proteolytic processing assembly into LHCII and localisation to appressed membranes occur in chloroplast lysates. Plant Physiol. 86:1120-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corradini, D., C. G. Huber, A. M. Timperio, and L. Zolla. 2000. Resolution and identification of the protein components of the photosystem II antenna system of higher plants by reversed-phase liquid chromatography with electrospray-mass spectrometric detection. J. Chromatogr. A 886:111-121. [DOI] [PubMed] [Google Scholar]
- 10.Depege, N., S. Bellafiore, and J. D. Rochaix. 2003. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299:1572-1575. [DOI] [PubMed] [Google Scholar]
- 11.Elrad, D., K. K. Niyogi, and A. R. Grossman. 2002. A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14:1801-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eng, J., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez, E., R. Schnell, L. P. Ranum, S. C. Hussey, C. D. Silflow, and P. A. Lefebvre. 1989. Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 86:6449-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleischmann, M. M., S. Ravanel, R. Delosme, J. Olive, F. Zito, F. A. Wollman, and J. D. Rochaix. 1999. Isolation and characterization of photoautotrophic mutants of Chlamydomonas reinhardtii deficient in state transition. J. Biol. Chem. 274:30987-30994. [DOI] [PubMed] [Google Scholar]
- 15.Germano, M., A. E. Yakushevska, W. Keegstra, H. J. van Gorkom, J. P. Dekker, and E. J. Boekema. 2002. Supramolecular organization of photosystem I and light-harvesting complex I in Chlamydomonas reinhardtii. FEBS Lett. 525:121-125. [DOI] [PubMed] [Google Scholar]
- 16.Gomez, S. M., J. N. Nishio, K. F. Faull, and J. P. Whitelegge. 2002. The chloroplast grana proteome defined by intact mass measurements from liquid chromatography mass spectrometry. Mol. Cell. Proteomics 1:46-59. [DOI] [PubMed] [Google Scholar]
- 17.Harris, E. H. 1989. The Chlamydomonas sourcebook. A comprehensive guide to biology and laboratory use. Academic Press, Inc., San Diego, Calif. [DOI] [PubMed]
- 18.Hippler, M., F. Drepper, J. Farah, and J. D. Rochaix. 1997. Fast electron transfer from cytochrome c6 and plastocyanin to photosystem I of Chlamydomonas reinhardtii requires PsaF. Biochemistry 36:6343-6349. [DOI] [PubMed] [Google Scholar]
- 19.Hippler, M., K. Biehler, A. Krieger-Liszkay, J. van Dillewjin, and J. D. Rochaix. 2000. Limitation in electron transfer in photosystem I donor side mutants of Chlamydomonas reinhardtii. Lethal photo-oxidative damage in high light is overcome in a suppressor strain deficient in the assembly of the light harvesting complex. J. Biol. Chem. 275:5852-5859. [DOI] [PubMed] [Google Scholar]
- 20.Hippler, M., J. Klein, A. Fink, T. Allinger, and P. Hoerth. 2001. Towards functional proteomics of membrane protein complexes: analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J. 28:595-606. [DOI] [PubMed] [Google Scholar]
- 21.Hippler, M., B. Rimbault, and Y. Takahashi. 2002. Photosynthetic complex assembly in Chlamydomonas reinhardtii. Protist 153:197-220. [DOI] [PubMed] [Google Scholar]
- 22.Huber, C. G., A. M. Timperio, and L. Zolla. 2001. Isoforms of photosystem II antenna proteins in different plant species revealed by liquid chromatography-electrospray ionization mass spectrometry. J. Biol. Chem. 276:45755-45761. [DOI] [PubMed] [Google Scholar]
- 23.Imbault, P., C. Wittemer, U. Johanningmeier, J. D. Jacobs, and S. H. Howell. 1988. Structure of the Chlamydomonas reinhardtii cabII-1 gene encoding a chlorophyll-a/b-binding protein. Gene 73:397-407. [DOI] [PubMed] [Google Scholar]
- 24.Jansson, S. 1999. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4:236-240. [DOI] [PubMed] [Google Scholar]
- 25.Jasper, F., B. Quednau, M. Kortenjan, and U. Johanningmeier. 1991. Control of cab gene expression in synchronized Chlamydomonas reinhardtii cells. J. Photochem. Photobiol. 11:139-150. [DOI] [PubMed] [Google Scholar]
- 26.Kindle, K., R. Schnell, E. Fernandez, and P. Lefebvre. 1989. Stable nuclear transformation of Chlamydomonas reinhardtii. J. Cell Biol. 109:2589-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohorn, B. D., and D. Yakir. 1990. Movement of newly imported light-harvesting chlorophyll-binding protein from unstacked to stacked thylakoid membranes is not affected by light treatment or absence of amino-terminal threonines. J. Biol. Chem. 265:2118-2123. [PubMed] [Google Scholar]
- 28.Kuhlbrandt, W., D. N. Wang, and Y. Fujiyoshi. 1994. Atomic model of plant light-harvesting complex by electron crystallography. Nature 367:614-621. [DOI] [PubMed] [Google Scholar]
- 29.Lamppa, G. K., and M. S. Abad. 1987. Processing of a wheat light-harvesting chlorophyll a/b protein precursor by a soluble enzyme from higher plant chloroplasts. J. Cell Biol. 105:2641-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayfield, S. P., G. Schirmer-Rahire, H. Frank, H. Zuber, H., and J. D. Rochaix. 1989. Analysis of the genes of the OEE1 and OEE3 proteins of the photosystem II complex from Chlamydomonas reinhardtii. Plant Mol. Biol. 12:683-693. [DOI] [PubMed] [Google Scholar]
- 31.Michel, H., P. R. Griffin, J. Shabanowitz, D. F. Hunt, and J. Bennett. 1991. Tandem mass spectrometry identifies sites of three post-translational modifications of spinach light-harvesting chlorophyll protein II. Proteolytic cleavage, acetylation, and phosphorylation. J. Biol. Chem. 266:17584-17591. [PubMed] [Google Scholar]
- 32.Minagawa, J., K. C. Han, N. Dohmae, K. Takio, and Y. Inoue. 2001. Molecular characterization and gene expression of lhcb5 gene encoding CP26 in the light-harvesting complex II of Chlamydomonas reinhardtii. Plant Mol. Biol. 46:277-287. [DOI] [PubMed] [Google Scholar]
- 33.Moseley, J. L., T. Allinger, S. Herzog, P. Hoerth, E. Wehinger, S. Merchant, and M. Hippler. 2002. Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21:6709-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peter, G. F., and J. P. Thornber. 1991. Biochemical composition and organization of higher plant photosystem II light-harvesting pigment proteins. J. Biol. Chem. 266:16745-16754. [PubMed] [Google Scholar]
- 35.Pichersky, E., N. E. Hoffman, V. S. Malik, R. Bernatzky, L. Szabo, and A. R. Cashmore. 1987. The tomato Cab-4 and Cab-5 genes encode a second type of CAB polypeptides localized in photosystem II. Plant Mol. Biol. 9:109-120. [DOI] [PubMed] [Google Scholar]
- 36.Porra, R. J., W. A. Thompson, and P. E. Kriedemann. 1989. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975:384-394. [Google Scholar]
- 37.Richter, S., and G. K. Lamppa. 1998. A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc. Natl. Acad. Sci. USA 95:7463-7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Santoni, V., M. Molloy, and T. Rabilloud. 2000. Membrane proteins and proteomics: un amour impossible? Electrophoresis 21:1054-1070. [DOI] [PubMed] [Google Scholar]
- 41.Strauss, C., Mussgang, J. H., and O. Kruse. 2001. Ligation-mediated suppression-PCR as a powerful tool to analyse nuclear gene sequences in the green alga Chlamydomonas reinhardtii. Photosynth. Res. 70:311-320. [DOI] [PubMed] [Google Scholar]
- 42.Teramoto, H., T. Ono, and J. Minagawa. 2001. Identification of Lhcb gene family encoding the light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Cell Physiol. 42:849-856. [DOI] [PubMed] [Google Scholar]
- 43.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Wijk, K. J. 2001. Challenges and prospects of plant proteomics. Plant Physiol. 126:501-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vener, A. V., A. Harms, M. R. Sussman, and R. D. Vierstra. 2001. Mass spectrometric resolution of reversible protein phosphorylation in photosynthetic membranes of Arabidopsis thaliana. J. Biol. Chem. 276:6959-6966. [DOI] [PubMed] [Google Scholar]
- 46.Wollman, F. A., and P. Delepelaire. 1984. Correlation between changes in light energy distribution and changes in thylakoid membrane polypeptide phosphorylation in Chlamydomonas reinhardtii. J. Cell Biol. 98:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zolla, L., S. Rinalducci, A. M. Timperio, and C. G. Huber. 2002. Proteomics of light-harvesting proteins in different plant species. Analysis and comparison by liquid chromatography-electrospray ionization mass spectrometry. Photosystem I. Plant Physiol. 130:1938-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]