More than a century after eosinophil granulocytes were baptized by Paul Ehrlich their role in defense mechanisms against parasitic infections and in allergic reactions has been firmly established (1). In contrast to the wealth of functional knowledge about these cells, their developmental origin, relationship to other blood cell types, and the critical transcription factors that determine their fate have long been elusive. Work in the 1990's showed a key role of both GATA and C/EBP transcription factors through experiments in which their expression was enforced in transformed chicken and murine cell lines. In addition, eosinophil-specific promoters were found to be regulated by an interplay between these factors. In several new papers this concept has been confirmed and applied to the reprogramming of normal human and murine cells. In addition, two studies show that GATA-1–deficient mice lack eosinophils, nicely complementing earlier work that demonstrated a lack of eosinophils in C/EBPα-deficient mice. In the following these papers will be discussed and interpreted in the light of the earlier results obtained with transformed cell systems.
GATA-1 As an Inducer of Eosinophil Formation.
In a striking example of cellular engineering, in this issue, Hirasawa et al. (2) have succeeded in efficiently generating eosinophils from human fetal blood cells. To do this, they isolated hematopoietic progenitors from human cord blood (CD34+ fraction), infected them with a retrovirus that encodes GATA-1 (together with GFP as an indicator), and sorted GFP-positive cells 60 h later. The cells were then grown in SCF and GM-CSF for 5 d and subsequently for an additional 8 d under either myeloid conditions (with the same factors) or eosinophil conditions (with IL-5). Surprisingly, perhaps, the GATA-1–transduced cells cultured under myeloid conditions contained eosinophilic granules and expressed eosinophil peroxidase as well as major basic protein but lacked myeloid markers. In contrast, the vector-only transduced cells were predominantly myelomonocytic and lacked eosinophil markers. GATA-2 exerted a similar effect, while dominant negative forms of the factors (produced by fusion to the Drosophila engrailed protein as a transcriptional repressor) completely prevented eosinophil formation in cells grown with IL-5. They also showed that the COOH-terminal zinc finger of GATA-1 (needed for DNA-binding) is required for this effect, while the NH2-terminal zinc finger (needed for interaction with FOG-1, see below) is not.
Using a related approach Heyworth et al. obtained similar results with mouse bone marrow cells (Tarik Enver, personal communication). Hematopoietic progenitors were isolated from bone marrow (CD34+/c-kit+ fraction of cyclophosphamide-treated mice), infected for 2 d with a GATA-1/GFP-encoding retrovirus, sorted for GFP expression, and cultured in soft agar. Here again, a dramatic phenotypic shift was observed away from myelomonocytic colonies toward colonies that consisted predominantly of erythrocytes, eosinophils, and basophils. To ensure that this was not due to selection of preexisting eosinophil progenitors, the experiments were repeated with a hormone inducible form of GATA-1 (GATA-1/ERT) and similar results were obtained. The generation of erythroid cells in addition to eosinophils in this study cannot simply be explained by the presence of erythropoietin in the culture medium as erythroid differentiation was also seen in IL-3 alone. Whether this apparent discrepancy with the GATA-1–transduced human cord blood experiments (where only eosinophils were observed) reflects differences between species or the tissues or vectors used for infection remains to be seen.
Isolate CD34+ Cells, Express C/EBPα and … Also Get Eosinophils.
Using the same protocol as in the GATA-1 experiments described above, the Tsukuba group recently described the formation of eosinophil and neutrophil granulocytes by enforced expression of C/EBPα in cord blood progenitors, although at lower efficiencies (3). The observation that eosinophils can be obtained from CD34+ cord blood cells with both a zinc finger-type and a leucine zipper-type transcription factor may at first seem paradoxical. Assuming that eosinophils are specified by the coexpression of both GATA-1 and C/EBPα factors (see also below), the observations can be explained by the following two models (Fig. 1) . In model A, the two transcription factors act on a homogeneous population of cells. This could be a common myeloid precursor that is both GATA-1, C/EBPα low/negative, and in this scenario expression of either (GATA-1 or C/EBPα) would lead to the up-regulation of the other factor (another target could be a population of committed immature eosinophils but this is a possibility that we consider less likely; see below). Alternatively, (model B) each of the two factors could affect a distinct cell type within the CD34+ population: one consisting of a precursor that is GATA-1− but C/EBPα+, possibly representing a granulocyte/macrophage progenitor (GMP); and another that is C/EBP− but GATA-1+, possibly representing a megakaryocytic/erythroid progenitor (MEP). Transduction with GATA-1 would complement C/EBPα in the myeloid cells and C/EBPα would complement GATA-1 in the MEP cells, in each case leading to the outgrowth of eosinophils. A combination of models A and B is also possible. In support of the second model is the fact that CD34 is expressed not only by multipotent progenitors, but also by committed myeloid and erythroid precursors (for a review, see reference 4). In the following we will discuss experiments, obtained with a transformed cell system, that additionally favor model B.
Figure 1.
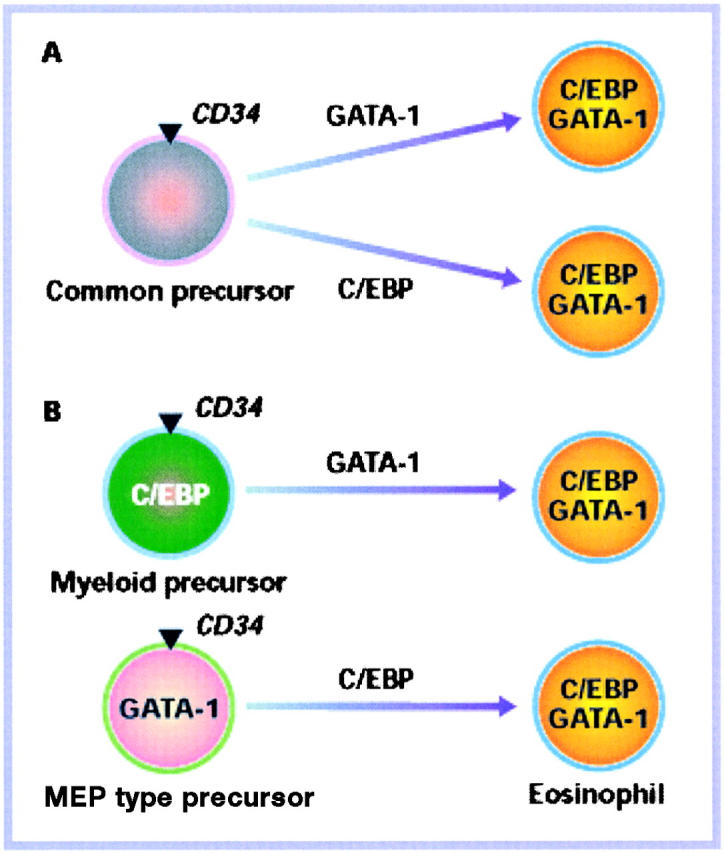
Two models explaining how GATA-1 and C/EBP might induce eosinophil formation in CD34+ cells.
GMP-type Myeloid Progenitors and MEP-type Progenitors Can Be Converted Into Eosinophils by GATA-1 and C/EBP, Respectively.
The first clue as to the origins of eosinophils came from the serendipitous finding that phorbolester treatment of E26 leukemia virus-transformed chicken cells converts them into eosinophils and myeloblasts in a concentration-dependent manner (5, 6; Fig. 2 A). These cells, called E26-“MEPs” for “Myb-Ets transformed progenitors,” resemble normal MEP cells in that they can be induced to differentiate into megakaryocytes and erythrocytes by the inactivation of either the Myb or Ets domains of the Myb-Ets oncoprotein (7–9). Enforced C/EBPα or β expression mimics the phorbolester effect, leading to the formation of eosinophils through a partial down-regulation of GATA-1 (10, 11). In contrast, PU.1 induces these cells to mature into myeloblasts through a complete down-regulation of GATA-1 (12). This process is reversible as enforced expression of GATA-1 in myeloid cells leads to the formation of eosinophils when expressed at low levels and of MEP cells, when expressed at high levels (13; Fig. 2 B).
Figure 2.
Generation of eosinophils in transformed cells. (A) E26 leukemia virus transformed MEPs can be induced to differentiate into either eosinophils or myeloblasts by phorbolester treatment, depending on whether protein kinase C (PKC) activity is high or low. (B) The three types of E26 transformed cells can be converted into one another (arrows) by enforced expression of the transcription factors indicated.
There is yet another player in this process: FOG-1. This “Friend of GATA-1” protein, which was first identified as a GATA-1 binding protein, is essential for megakaryocyte formation (14). It is expressed in normal cells as well as in E26-transformed MEPs but not in eosinophils and myeloid cells. Importantly however, its enforced expression in eosinophils induces them to “de-differentiate” into MEPs, while the factor has no effect on myeloblasts (15). As is the case in the induction of eosinophils from human cord blood cells (2) the NH2-terminal zinc finger, which binds FOG-1, is not required for the switch (14). Thus, in this transformed cell system, lineage transitions can be induced according to a simple binary code that defines cell identity: intermediate levels of GATA-1 plus C/EBPα/β specifies eosinophils; high GATA-1 plus FOG-1 specifies MEPs; and PU.1 plus C/EBPβ specifies myeloblasts (Fig. 2). In addition to these synergisms, two types of cross-antagonisms play an essential role in the process of determining whether a cell differentiates along the eosinophil or myeloid pathway: C/EBP and FOG-1 on the one hand and PU.1 and GATA-1 on the other. These antagonisms which are discussed in more detail elsewhere (9, 16, 17) but both ensure that active commitment to one cell fate coincides with a repression of other cell fates.
GATA and C/EBP Cooperate on Eosinophil-specific Promoters.
Key to deciphering the process that governs eosinophil lineage commitment at the molecular level has been the dissection of transcriptional control elements that regulate eosinophil-specific genes (18–20). A well studied example is the eos47 gene (encoding EOS47, the avian ortholog of the mammalian melanotransferrin gene) which in bone marrow is specific to early eosinophils (21). The elements governing its lineage-specific expression reside within a 309 bp promoter region and consist of binding sites for Myb-, Ets-, C/EBP-, and GATA-type transcription factors. C/EBPα and Ets-1 were found to cooperate in both the binding to and activation of the eos47 promoter, mediated through an interaction between the DNA-binding domains of the factors (18). Interestingly, while low levels of GATA-1 enhanced promoter activation by C/EBPα and Ets-1, high levels led to a repression (which was further enhanced by FOG-1), with GATA-1 on its own having little effect (15). Thus, positive and negative regulation of the EOS47 promoter by GATA, C/EBP, and FOG-1 closely mimics their effects on cell phenotype: here also, moderate levels of GATA-1–induced eosinophil formation while high levels of GATA-1, as well as FOG-1, led instead to the formation of MEP cells (11, 13, 15, 18). Unlike C/EBP and GATA factors, Ets-1 and c-Myb, because of their widespread distribution across the hematopoietic system, probably contribute to basal promoter activity and do not appear to directly participate in lineage specification of eosinophils.
Studies of the mammalian granule major basic protein (MBP) promoter, likewise, showed a cooperative effect of C/EBPβ and GATA-1 (19, 20). Here also FOG-1 was found to lead to a repression of the promoter (15, 19). Taken together, these results strongly support the eosinophil-specific combinatorial code that has emerged from the enforced expression experiments discussed before.
Which of the GATA and C/EBP Family Members Are the Key Players In Vivo?
As GATA-1, GATA-2, C/EBPα, and C/EBPβ are all known to be expressed in eosinophils and to have the capacity to induce eosinophil formation, do they all play a role in eosinophil formation in vivo? Studies with knockout mice suggest that this is not so and that the key players are in fact GATA-1 and C/EBPα. Thus, mice lacking C/EBPα show a profound absence of neutrophils and eosinophils (22). Although GATA-1 is essential for the formation of erythroid and megakaryocytic cells, more recent data suggest that it is also essential for the formation of eosinophils. In this issue, Hirasawa et al. show that GATA-1–deficient fetal liver cells lack the ability to form eosinophils unless GATA-1 (or GATA-2) is reintroduced into these cells (2). Similarly, in another article in this issue, Yu et al. (23) describe a regulatory element in the GATA-1 promoter that selectively governs expression of this gene in eosinophils. Disruption of this element leads to the formation of mice that completely lack eosinophils. Interestingly, these mice still have (immature) mast cells (23). In contrast, earlier work of the same group had shown that GATA-2 knockout mice display a general reduction of hematopoiesis, but a complete lack of mast cells (24). A role of GATA-2 in mast cell formation is also supported by the observation that GATA-2, but not GATA-1, induces the formation of mast cells in multipotent progenitors, in conjunction with moderate levels of PU.1 (Singh, H., personal communication). In aggregate, these experiments indicate that GATA-1 together with C/EBPα is the key player for the formation of eosinophils in vivo while GATA-2 is crucial for the formation of mast cells.
Normal Origins of Eosinophils.
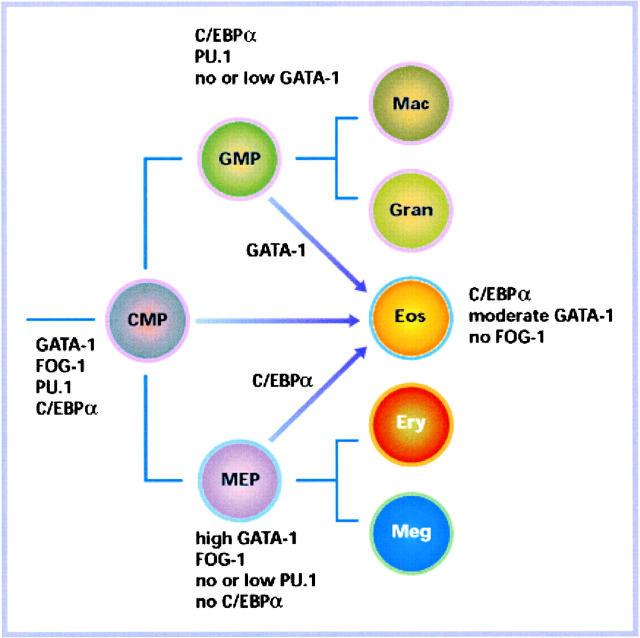
Eosinophils developed relatively late during vertebrate evolution, perhaps by the “recycling” of transcription factors that had been “invented” earlier. This may have occurred through the introduction of thresholds that play out on novel, eosinophil-specific promoters, such as that described in the study by Yu et al. (23) see also Nerlov et al. [25]). A possible scenario of how transcription factors specify myeloid and erythroid lineages and how eosinophils might fit into this scheme is shown in Fig. 3 , based on the evidence discussed above as well as the known expression patterns of these factors in normal hematopoietic progenitors (26). In this scenario CMPs coexpress a number of lineage-restricted factors such as GATA-1/FOG-1 on the one hand and PU.1/C/EBP on the other, reflecting the concept of “priming” that has been suggested by Enver and colleagues (27). Lineage decisions would then ensue by the up-regulation of one program (PU.1/C/EBP for myeloid cells and GATA-1/FOG-1 for erythroid cells) and would be stabilized by the GATA-1/PU.1 and FOG-1/C/EBP cross-antagonisms. In this model eosinophils represent a cellular intermediate which is specified by moderate levels of GATA-1 together with C/EBPα and the absence of FOG-1 (for reviews, see references 9, 16, and 17). It is less clear how eosinophils originate during normal hematopoietic differentiation. One speculation, akin to model B in Fig. 1, is that they form as a separate branch from either GMPs or MEPs (indicated by the top and bottom arrows in Fig. 3). Alternatively, they may form from CMPs (middle arrow in Fig. 3). Recent work by Akashi and colleagues has shown that both CMPs and GMPs can generate eosinophil colonies in the presence of IL-3, SCF, and IL-5, while MEPs do not (Koichi Akashi, personal communication). This would suggest that the third route, conversion from MEPs into eosinophils via up-regulation of C/EBPα is not normally used.
Figure 3.
Roads to eosinophil formation. The scheme, which is based on work by Akashi et al. (reference 26) shows combinatorial transcription factor codes that specify GMPs and MEPs on the one hand, and eosinophils on the other. It also depicts alternative pathways of eosinophil formation during normal hematopoiesis. CMP, common myeloid progenitor; GMP, granulocyte/macrophage progenitor; MEP, megakaryocyte/erythroid progenitor; Mac, macrophage; Gran, neutrophil granulocyte; Eos, eosinophil; Ery, erythrocyte; Meg, megakaryocyte.
Along with the understanding of how eosinophils are generated comes the potential for new therapies of allergic diseases and asthma. Eosinophils are known to be major mediators of lung tissue damage in chronic allergy. Preventing their influx and ability to degranulate has been explored as a potential point for therapeutic intervention. Tissue-specific inhibition of either C/EBPα or GATA-1 function, such as by expression of antisense/dominant negative forms in erythroid/myeloid progenitors or of FOG-1 in eosinophils, may offer a means of blocking eosinophil formation.
Acknowledgments
We would like to thank Koichi Akashi, Stuart Orkin, Tarik Enver, and Harinder Singh for sharing unpublished data and for discussions.
References
- 1.Rothenberg, M.E. 1998. Eosinophilia. N. Engl. J. Med. 338:1592–1600. [DOI] [PubMed] [Google Scholar]
- 2.Hirasawa, R., R. Shimizu, S. Takahashi, M. Osawa, S. Takayanagi, Y. Kato, M. Onodera, N. Minegishi, M. Yamamoto, K. Fukao, H. Taniguchi, H. Nakauchi, and A. Iwama. 2002. Essential and instructive roles of GATA factors in eosinophil development. J. Exp. Med. 195:1473–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwama, A., M. Osawa, R. Hirasawa, N. Uchiyama, S. Kaneko, M. Onodera, K. Shibuya, A. Shibuya, C. Vinson, D.G. Tenen, and H. Nakauchi. 2002. Reciprocal roles for CCAAT/enhancer binding protein (C/EBP) and PU.1 transcription factors in Langerhans cell commitment. J. Exp. Med. 195:547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krause, D.S., M.J. Fackler, C.I. Civin, and W.S. May. 1996. CD34: structure, biology, and clinical utility. Blood. 87:1–13. [PubMed] [Google Scholar]
- 5.Rossi, F., M. McNagny, G. Smith, J. Frampton, and T. Graf. 1996. Lineage commitment of transformed haematopoietic progenitors is determined by the level of PKC activity. EMBO J. 15:1894–1901. [PMC free article] [PubMed] [Google Scholar]
- 6.Graf, T., K. McNagny, G. Brady, and J. Frampton. 1992. Chicken “erythroid” cells transformed by the Gag-Myb-Ets-encoding E26 leukemia virus are multipotent. Cell. 70:201–213. [DOI] [PubMed] [Google Scholar]
- 7.McNagny, K.M., and T. Graf. 1996. Acute avian leukemia viruses as tools to study hematopoietic cell differentiation. Curr. Top. Microbiol. Immunol. 212:143–162. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen, J.S., R. Doyonnas, and K.M. McNagny. 2002. Avian models to study the transcriptional control of hematopoietic lineage-commitment and identify lineage-specific genes. Cells, Tissues, Organs. In press. [DOI] [PubMed] [Google Scholar]
- 9.Graf, T. 2000. Transcription Factors that Induce the Commitment of Multipotent Progenitors: Lessons from the MEP System. Oxford University Press, Oxford. 355–363.
- 10.Muller, C., E. Kowenz-Leutz, S. Grieser-Ade, T. Graf, and A. Leutz. 1995. NF-M (chicken C/EBP beta) induces eosinophilic differentiation and apoptosis in a hematopoietic progenitor cell line. EMBO J. 14:6127–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nerlov, C., K.M. McNagny, G. Doderlein, E. Kowenz-Leutz, and T. Graf. 1998. Distinct C/EBP functions are required for eosinophil lineage commitment and maturation. Genes Dev. 12:2413–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nerlov, C., and T. Graf. 1998. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 12:2403–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulessa, H., J. Frampton, and T. Graf. 1995. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9:1250–1262. [DOI] [PubMed] [Google Scholar]
- 14.Tsang, A.P., Y. Fujiwara, D.B. Hom, and S.H. Orkin. 1998. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 12:1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Querfurth, E., M. Schuster, H. Kulessa, J.D. Crispino, G. Doderlein, S.H. Orkin, T. Graf, and C. Nerlov. 2000. Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev. 14:2515–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graf, T. 2002. Differentiation plasticity of hematopoietic cells. Blood. 99:3089–3101. [DOI] [PubMed] [Google Scholar]
- 17.Cantor, A.B., and S.H. Orkin. 2001. Hematopoietic development: a balancing act. Curr. Opin. Genet. Dev. 11:513–519. [DOI] [PubMed] [Google Scholar]
- 18.McNagny, K.M., M.H. Sieweke, G. Doderlein, T. Graf, and C. Nerlov. 1998. Regulation of eosinophil-specific gene expression by a C/EBP-Ets complex and GATA-1. EMBO J. 17:3669–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi, Y., H. Nishio, K. Kishi, S.J. Ackerman, and T. Suda. 1999. C/EBPbeta and GATA-1 synergistically regulate activity of the eosinophil granule major basic protein promoter: implication for C/EBPbeta activity in eosinophil gene expression. Blood. 94:1429–1439. [PubMed] [Google Scholar]
- 20.Yamaguchi, Y., S.J. Ackerman, N. Minegishi, M. Takiguchi, M. Yamamoto, and T. Suda. 1998. Mechanisms of transcription in eosinophils: GATA-1, but not GATA-2, transactivates the promoter of the eosinophil granule major basic protein gene. Blood. 91:3447–3458. [PubMed] [Google Scholar]
- 21.McNagny, K.M., F. Rossi, G. Smith, and T. Graf. 1996. The eosinophil-specific cell surface antigen, EOS47, is a chicken homologue of the oncofetal antigen melanotransferrin. Blood. 87:1343–1352. [PubMed] [Google Scholar]
- 22.Zhang, D.E., P. Zhang, N.D. Wang, C.J. Hetherington, G.J. Darlington, and D.G. Tenen. 1997. Absence of granulocyte colony-stiumulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. USA. 94:569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu, C., A.B. Cantor, H. Yang, C. Browne, R.A. Wells, Y. Fujiwara, and S.H. Orkin. 2002. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J. Exp. Med. 195:1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai, F.Y., and S.H. Orkin. 1997. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 89:3636–3643. [PubMed] [Google Scholar]
- 25.Nerlov, C., D.G. Tenen, and T. Graf. 2000. Regulatory Interactions Between Transcription Factors and Their Role in Hematopoietic Lineage Determination. Oxford University Press, Oxford. 363–367.
- 26.Akashi, K., D. Traver, T. Miyamoto, and I.L. Weissman. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. [DOI] [PubMed] [Google Scholar]
- 27.Enver, T., and M. Greaves. 1998. Loops, lineage, and leukemia. Cell. 94:9–12. [DOI] [PubMed] [Google Scholar]