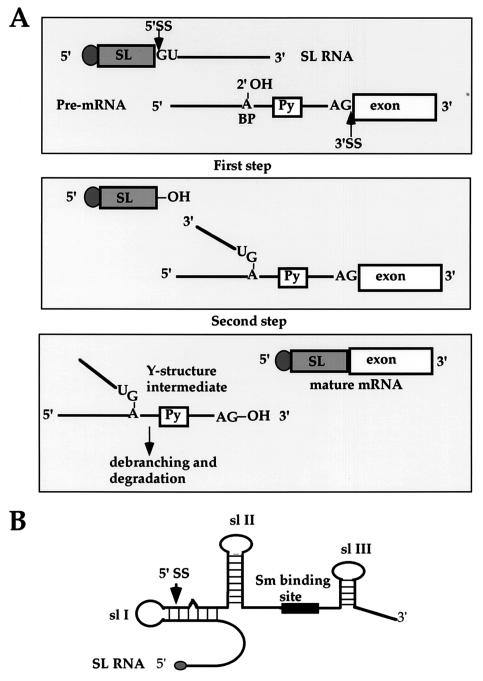
mRNA maturation in trypanosomes differs from the process in most eukaryotes mainly because protein-coding genes are transcribed into polycistronic RNAs in this organism (78). Studies from the ongoing genome project suggest that the entire chromosome may be transcribed as large transcripts, but thus far there is no evidence to support the existence of conventional polymerase II promoters (62). The process of trans splicing was discovered 20 years ago when it was found that the different variant surface glycoprotein mRNAs in Trypanosoma brucei carry a common 39-nucleotide (nt) sequence, namely, the spliced leader (SL) sequence (9). It was later established that all trypanosome mRNAs undergo trans splicing (2). The source of the SL sequence was found to be a small capped RNA, the SL RNA (14, 58). Thus, the SL addition serves two purposes: it functions together with polyadenylation in dissecting the polycistronic transcripts, and it provides the cap to mRNAs (2). trans splicing proceeds through a two-step transesterification reaction, analogous to cis splicing but forming a Y structure instead of a lariat intermediate (illustrated in Fig. 1A) (61, 88). Although first discovered in trypanosomes, the process was later found in nematodes (41), euglenoids (91), trematodes (73), and recently in chordates (97). Surprisingly, after almost a decade of searching for cis splicing, a single gene carrying a cis-spliced intron was discovered, suggesting that these two splicing processes coexist in trypanosomes, as in all other organisms capable of trans splicing (51).
FIG. 1.
Mechanism of trans splicing. (A) Schematic representation of trans splicing. The 5′ splice site GU on the SL RNA and the 3′ splice site AG on the pre-mRNA are indicated. BP, branch point; Py, polypyrimidine tract. (B) Secondary structure of SL RNA. The three stem-loop structures (sl I, II, and III), the 5′ splice site, and the Sm-binding site are indicated. The shadowed dot at the 5′ end of SL RNA indicates the cap 4 structure.
In the last decade, studies have focused on elucidating the mechanism and machinery of pre-mRNA processing in these organisms. The major findings included (i) the identification of the first cis-spliced intron and U1 snRNA that may function exclusively in this process, (ii) the finding and unraveling the function of U5 and SLA1, and (iii) the existence of coupling between trans splicing and polyadenylation. In this review we summarize studies performed mainly in vivo to elucidate structure-function aspects of the SL RNA and the snRNAs with which it interacts. The unique modifications on the SL RNA, including capping and pseudouridylation and their role in SL RNA function and biogenesis, are described. The regulation of splicing and its linkage to polyadenylation is discussed, and data are provided for the existence of splicing factors whose function is well characterized in other eukaryotes.
STRUCTURE-FUNCTION ANALYSIS OF THE SL RNA
In the absence of an in vitro system for trans splicing, most of the structure-function studies were performed in vivo in Leptomonas seymouri, Leptomonas collosoma, and Leishmania tarentolae by using tagged SL RNAs (49, 53, 85, 86, 108). For some unknown reason, this approach does not work for T. brucei (Elisabetta Ullu, unpublished data). The SL RNA secondary structure of all organisms carrying out trans splicing is similar; it is composed of three stem-loops. In trypanosomatids, The SL sequence is 39 to 41 nt, followed by an intron of variable length that can be folded into two stem-loops separated by a single-stranded region (Fig. 1B). This region is analogous to the region in snRNAs that binds the core proteins and is known as the Sm-like site, since in several trypanosomatid species it deviates from the canonical Sm binding sequence (11). Despite the evolutionary conservation of the secondary structure, the SL RNA sequence is not conserved among different organisms capable of trans splicing. However, within the trypanosomatids the exon sequence shows a considerable degree of conservation at the primary sequence level. One characteristic feature of the SL RNA is the presence of a hypermodified cap structure referred to as cap 4 because the 4 nt after the 7-methylguanosine (m7G) are modified (71, 23, 6).
In trypanosomatids SL RNA genes do not contain internal promoter elements (13), and the conservation of the exon sequence may stem from constraints imposed on SL-snRNA interactions and protein(s) binding. Several studies in recent years have addressed the function of various domains in the trypanosomatid SL RNA with regard to transcription, trans-splicing activity, assembly into the SL RNP, and 3′-end formation, but no unifying view has emerged with regard to the role in trans splicing and cap modification, most probably because of differences inherent in the various trypanosomatid systems analyzed and/or the assays used to monitor trans splicing (49, 53, 85, 86, 108). The most recent study attempted to reconcile the discrepancy that had emerged from these different studies (49, 53). The major difference regarding the function of the exon emerged from studies on the 5′ splice site region and its vicinity, especially stem I. In L. tarentolae and L. collosoma changing the entire stem I sequence did not affect trans splicing. However, mutations in L. seymouri in the region comprising the intramolecular base pairing severely inhibited trans splicing and cap modification, suggesting that this domain may provide a binding site for SL-specific methyltransferase(s) (49). This discrepancy can be reconciled considering that only in L. seymouri did the mutations completely disrupt the structure of domain I, whereas mutations in the other systems still afforded the formation of a stem, suggesting that the sequence of stem I but not its structure can tolerate changes (53).
The function of the SL RNA intron sequence in trans splicing is puzzling. For instance, in the L. collosoma and L. tarentolae systems, changing the sequence and structure of stem-loop II and III severely affected trans splicing (53, 86) and in L. seymouri the intron domain was tolerant to mutations; only stem-loop II was indispensable for function (49). In L. tarentolae and L. collosoma, mutations in the Sm site and its length affected not only trans splicing but also modification at the fourth cap nucleotide (53, 86). Interestingly, in L. tarentolae, mutations in the Sm site and stem-loop III affected 3′-end formation of the SL RNA, resulting in the accumulation of SL RNA with longer 3′ tails (86, 87).
The differences in the results obtained in the three experimental systems may stem from the different assays used; a quantitative poison primer extension assay used in L. seymouri and L. collosoma, as opposed to reverse transcription-PCR and Northern analysis in L. tarentolae. Only in L. collosoma was the utilization in splicing monitored separately for both steps of splicing and was the differential expression of the SL RNA mutants taken in account (108). One major difference between the systems, which can affect the results, is the level of expression of the tagged SL RNA compared to the wild-type transcript, which was as high as the wild-type RNA in L. collosoma and L. seymouri (53, 108) and much lower in L. tarentolae (85-87). In addition, the different tags introduced in the SL RNA may have different effects on the function of the tagged molecule.
Note that in the nematode in vitro system the exon was found to be dispensable for the reaction, indicating that conservation in the exon region among the nematode SL RNA does not stem from its role in the trans-splicing reaction. However, the Sm site and 3 nt downstream were shown to be essential for the reaction because of their role in interacting with U6 snRNA, suggesting that the Sm site and its vicinity has a special and specific role in nematode trans splicing which seems to be different in trypanosomes (see discussion of SL-snRNA interactions below) (31).
snRNAs INVOLVED IN trans AND cis SPLICING
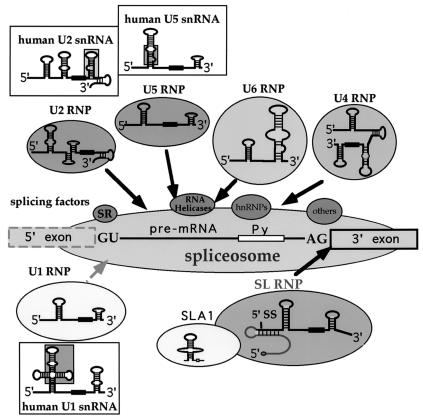
In this section we will discuss the structure and function of snRNAs, which seem to have unique features in trypanosomes and function in trans and cis splicing, such as U2, U4, U5, U6, SLA1, which is most probably specific to trans splicing, and U1, which is specific to cis splicing (Fig. 2). Because of space limitation, we chose to discuss snRNAs that have unique features in trypanosomes. Detailed information on U6 and U4 from different trypanosomatid species is not covered in this review but can be found in (8, 26, 44, 105).
FIG. 2.
Trypanosome snRNA repertoire. The snRNAs are organized around a cis-splicing substrate that can potentially bind all of the depicted trypanosome snRNAs. On the pre-mRNA, the 5′ and 3′ splice sites and polypyrimidine track (Py) are indicated. The trypanosome snRNAs are circled. The mammalian snRNA counterparts are boxed (30). The missing domains from the trypanosome snRNAs are in shaded boxes placed on the mammalian counterparts. The Sm-binding site is indicated by a black heavy line. Putative splicing factors are indicated.
U2.
The U2 homologue was identified in T. brucei by immunoprecipitation with anti-trimethyl-guanosine (TMG) antibodies (60). Phylogenetic studies of the U2 snRNAs from different species suggest that the U2 snRNA is shorter than its metazoan counterparts and is missing stem-loop III (Fig. 2). The trypanosomatid U2 secondary structure suggests the presence of only two helices in the region of stem-loop II (32). More importantly, the putative branch point recognition region does not contain the highly conserved GUAGUA sequence, which interacts by base-pairing with the branch point sequences in yeast and mammals, suggesting that additional factor(s) may be involved in bringing the trypanosome U2 RNP into the spliceosome (32, 60). U2 RNP cleavage with antisense oligonucleotide inhibited trans splicing in permeable cells, demonstrating its direct role in trans splicing (92).
U5 snRNA.
The failure to identify the U5 snRNAs was puzzling for many years because it was not identified in the initial screen for TMG-capped U snRNAs (60). Initially, SLA1 (see below) was proposed to be the homologue of U5 based on the presence of a sequence that seems to be related to the invariant U5 loop (102). The identification of SLA1 as a U5 homologue was discarded first because the L. seymouri homologue did not possess this “invariant” sequence (67) and because later the “true” U5 was identified (7, 22, 107). U5 has several unique properties: it is the smallest in nature, and it lacks several conserved structural domains. U5 lacks a TMG cap, m7G, and has a 5′-end phosphate terminus. The invariant loop contains an A instead of C in the second position and it lacks 2′-O-methylation characteristic of the invariant loop (22, 107). As opposed to the U5 of T. brucei, the L. collosoma and the L. seymouri U5 is longer and possesses a second stem-loop structure. A tri-snRNP complex composed of U4/U6 · U5 was detected (22, 107). Analyses of mutations introduced in the Sm site of the L. seymouri U5 suggest that U5 snRNA binds common proteins during a transient cytoplasmic phase before the U5 is translocated to the nucleus as in mammals but in contrast to yeast (7).
SLA1.
SLA1 was first proposed to be the trypanosome U5 homologue based on the presence sequence related to the U5 invariant sequence. After the discovery of U5 (see above), the function of SLA1 still remained unresolved. It was later suggested that SLA1 may possess some U1 function, since all trypanosomatid U1 snRNAs lack the stem-loop II, the binding site of protein U1A, which plays an essential role in linking splicing and polyadenylation; this structure is present in SLA1 (80). However, it was recently shown that SLA1 belongs to a group of RNAs that guide modification on other RNAs, such as rRNA and snRNAs (46). The presence of SLA1 in a cluster carrying C/D snoRNAs (76, 77) and our recent finding that such clusters also carry H/ACA RNA (45) prompted us to examine whether SLA1 belongs to this class of RNAs. Indeed, it was suggested that SLA1 may function in guiding pseudouridylation on the SL RNA. First, the pseudouridine at position −12 (relative to the 5′ splice site) is conserved in all trypanosomatids tested. Second, a potential for base-pair interaction between SLA1 and SL RNA is in agreement with the canonical rules for guiding the modification. Third, mutations in the SL RNA proposed interaction domain with SLA1 abolished pseudouridylation.
U1.
The last U snRNA to be identified in trypanosomes is U1. U1 was identified in Crithidia fasciculata (79) and T. brucei (21, 65) and is smaller than other U1 RNAs, since stem-loops II and III in prototype U1 are missing (Fig. 2). U1 most probably functions in cis splicing but not in trans splicing as in nematodes (19), since there is no potential for interaction of U1 with the 5′ splice site of SL RNA. The U1 has the potential to interact by base pairing with the 5′ splice site of the poly(A) polymerase (PAP) gene that undergoes cis splicing (51). Point mutations in the PAP 5′ splice site region, which are thought to disrupt the U1-5′ splice site interaction, abolished cis splicing of PAP in vivo (51). Two proteins associated with the U1 small nuclear ribonucleoprotein (snRNP) were identified (40 and 65 kDa). The 65-kDa protein is the T. brucei poly(A)-binding protein I. This association is intriguing because it may suggest that the U1 snRNP mediates the interaction between the cis splicing and polyadenylation machinery in trypanosomes, which is similar to the coupling that was observed between trans splicing and polyadenylation (see section discussing this topic below). The 40-kDa protein is related to U1-70K. The U1A may be missing from the particle because its binding domain is absent from the trypanosome RNA. Note that additional proteins may bind the U1 snRNP and that these proteins may have dislodged during the stringent affinity selection protocol (65). U1 snRNP/5′-splice site binding assays were developed, confirming the recognition of the 5′ splice site by U1 snRNP (65). Since only a single gene containing a cis-spliced intron has been identified in trypanosomes (51), it is not yet possible to conclude whether the extensive base pairing between U1 and the 5′ splice site of PAP is a common feature to trypanosomes cis-spliced introns.
SL-snRNA INTERACTIONS
Several approaches were used to reveal and validate potential base-pair interactions of snRNA-SL RNA essential to form the trans-spliceosome. In vivo UV cross-linking was used to identify snRNA that interact with the SL RNA by base pairing (22, 101, 102). A genetic approach was utilized to identify splicing defects in SL RNA due to its failure to interact with snRNAs. This analysis was followed by compensatory mutation in the snRNA interacting domain to suppress the splicing defect (108).
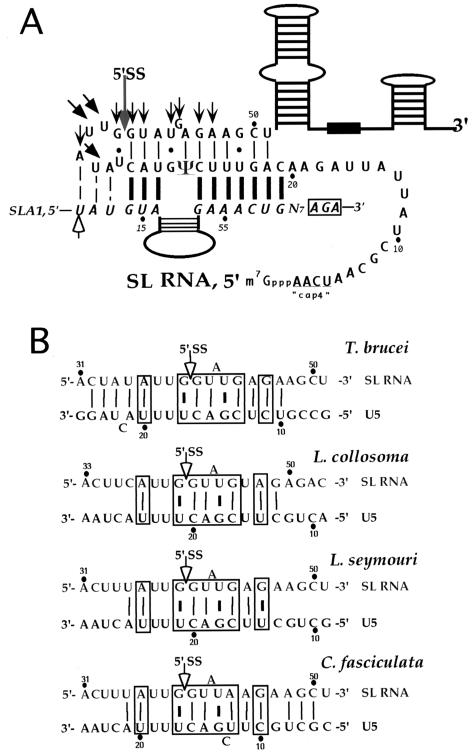
In vivo cross-linking with the bifunctional reagent psoralen led to the identification of two small RNAs, termed SLA1 and SLA2, the U5 homologue (see previous sections). The base pairing suggested by the psoralen cross-linking of the T. brucei U5 RNA with the SL RNA enables alignment of these sequences in a manner exactly like that proposed for cis splicing (22, 107) (Fig. 3A). However, in L. collosoma, L. seymouri, and Crithidia spp., as opposed to T. brucei, the potential for base pairing is less extensive (7, 107) (Fig. 3B). To validate the interaction of the SL RNA with the U5 and U6 snRNA, mutations were introduced at positions −4, −1, +1, +4, +5, and +7/+8 of SL RNA. All mutants exhibited altered splicing phenotypes compared to the parental strain (108). Compensatory mutations were introduced in U5 and U6 and suppression was observed only for positions +5 and +7/+8 with U5 mutations and for position +5 also with a U6 mutation, supporting the existence of a base-pair interaction of U5 and U6 with the SL RNA (108). The failure of the other mutations to compensate for the defects suggests that other protein factor(s) interact with these key positions or that the mutations disrupted other essential inter or intramolecular base pairing. This genetic study supported a unique interaction between the U5 and the SL RNA intron region that resembles the interaction of U1 with the 5′ splice site. The trypanosome U5 may therefore function in trans splicing in locating the 5′ splice site, and this interaction is mediated by extensive base pairing with the intron region, a finding analogous to the interaction of U1 with the same domain. The striking difference in invariant loops between U5 snRNAs in the trypanosome and all other organisms is the presence of an A instead of a C at the second position of the loop. This change may reflect the need to preserve the base-pair interaction between position +2 of the intron and the second position of the U5 invariant loop, especially since this position is located exactly at the middle of the SL-U5 duplex.
FIG. 3.
Intra- and intermolecular base pairing across the SL RNA 5′ splice site. (A) Potential base-pairing interaction between U5-SL RNA and SLA1-SL RNA. The intramolecular base-pair interactions across the T. brucei 5′ splice site are indicated by thin bars. The interaction between the SL RNA and SLA1 is indicated by heavy bars, and the interactions with U5 are indicated by arrows (22). The cross-linked sites on SL RNA to either U5 or SLA1 are indicated by filled arrows (22, 102), and the cross-linked sites of SL RNA on SLA1 is indicated by an open arrow (102). The 5′ splice site is indicated by a gray arrow. (B) Potential for base-pair interactions of U5 with 5′ splice site of SL RNA from different trypanosomatid species. The nucleotides on SL and U5 snRNAs are numbered. The 5′ splice site is indicated by an open arrow. The most conserved interactions are boxed. Accession numbers (where available) and references for the SL and U5 snRNAs are as follows: T. brucei SL RNA (58) and the U5 RNA (22); L. collosoma SL RNA (58) and the U5 RNA (107); L. seymouri SL RNA X07487 and the U5 RNA (5); and C. fasciculata SL RNA J03470 and the U5 RNA AF182356.
The compensatory genetic approach was also utilized to study the SL-U6 snRNA interaction across the 5′ splice site, and support was gained for interaction of U6 RNA with the intron at position +5 (108), analogous to that of U6 in cis splicing (50). Interestingly, Crithidia U6 carries a C-to-U change in the second position of the ACAGAG sequence and a compensatory base change of the corresponding SL RNA at position +5 (105). These phylogenetic data also support the 5′ splice site-U6 interaction. Surprisingly, mutations in L. seymouri (49) and in L. tarentolae (86) in positions +4 and +5 did not affect trans splicing. However, these mutations did not change a single nucleotide but several positions, and therefore the large sequence and structural change may circumvent the need for base pairing with these positions. In nematodes, U6 seems to have a unique function since it interacts with the SL RNA in the Sm site and 3 nt downstream from it, serving as a bridge between the SL RNA and the pre-mRNA (31). Although an analogous interaction can be envisioned in different trypanosomatid species (26), no functional proof of such interaction was obtained, since mutations in 3 nt downstream from the Sm site that affected trans splicing in the nematodes had no severe trans-splicing defect in L. collosoma (53). These data suggest that trypanosomes and nematodes differ in the mechanism by which the SL RNA is brought into the spliceosome. In nematodes, the U6 has a dominant role (31), whereas in trypanosomes it is the interactions of the SL RNA with the U5 and U6 across the 5′ splice site that seem to be essential for the reaction (108). We currently do not know of any proteins that may function to bring the SL RNP into the spliceosome.
Of special interest is the role of the SLA1 and SLA1-SL RNA interactions for the trans splicing. It is currently unknown whether the modified base is essential for the trans-splicing reaction itself or whether this pseudouridylation is a by-product of the need of SLA1 to base-pair with SL RNA. Mutations that changed the region of the exon, including a change in U28 (the modified nucleotide) to A, did not affect trans splicing in L. tarentolae (85, 86). However, no study had yet addressed the role of only this nucleotide for trans splicing. However, the presence of modified bases on snRNAs around functionally important regions was demonstrated for the cis splicing (28), so the pseuodouridine may stabilize certain important interactions within the trans-spliceosome catalytic core. Note that the 5′ splice site selection of SL RNA may involve the formation of multiple base-pair interactions that require melting the intramolecular SL RNA duplex in stem I and forming base-pair interactions with U5 and U6 (46). The presence of the pseudouridine at position −12 may therefore contribute to the fine-tuning of the intramolecular versus intermolecular interactions during the recognition of the 5′ splice site or it may stabilize an SL RNA structure that is needed to facilitate the intermolecular base pairing in the intron-exon junction (46). Another possibility is that SLA1 functions as a chaperonic RNA and is needed to maintain a certain structure that is essential for subsequent interactions of the SL RNA with the capping machinery and the SL RNA-binding proteins.
snRNP BIOGENESIS, Sm, AND snRNP PROTEINS
SL RNA biogenesis.
SL RNA is transcribed by RNA polymerase II (13, 29, 25). The promoter of the SL RNA is extragenic and does not contain internal promoter elements (3, 13, 29, 36). A poly(T) tract that varies in length exists downstream to the coding region of the SL RNA and was postulated to serve as the termination element for SL RNA transcription (87). Using permeable cells, it was demonstrated that SL RNA cap 4 was added cotranscriptionally in a 5′-to-3′ direction. Transcripts of between 56 and 67 nt were partially modified and carried methyl groups on the first two adenosine residues, whereas a fully modified cap 4 structure was present on transcripts of 117 nt carrying the Sm site (52). Surprisingly, accurate modification of the entire cap structure required SL RNA being in its SL RNP complex (95). Because modification of the first 2 nt took place before the Sm-like site was synthesized, it was proposed that the SL RNA capping takes place in the nucleus (52). More recently, evidence obtained by in situ hybridization suggests that SL RNA can also be found in the cytoplasm (109). Treatment with the karyopherin-specific inhibitor leptomycin B eliminated the cytoplasmic SL RNA and affected the modification on the cap +4 position and 3′-end formation, suggesting that the cytoplasmic stage is necessary for SL RNA biogenesis. This finding resembles the finding on the maturation of snRNPs in trypanosomatids demonstrating that mutation in the Sm site resulted in cytoplasmic accumulation of U4 and U5 in L. seymouri (7, 8). In mammals, capping (formation of trimethylguanosine) is coupled to Sm assembly, and these two serve as nuclear import signals (55). Since the SL RNP, like other snRNPs, binds Sm proteins (27, 66), its maturation pathway resembles the pathway of snRNPs in trypanosomes, as well as in metazoa. Indeed, mutation in the SL RNA Sm site affected capping at the +4 position (53, 86). The discrepancy regarding the cellular localization of cap 4 formation and which of the steps takes place cotranscriptionally will be resolved by the purification and characterization of the methylase(s) that carry out these specific modifications. Note that cap modification includes base and sugar methylations and that these modifications may be carried out by different methylases localized in different compartments. In addition to cap 4 modification, the SL RNA undergoes pseudouridylation at position −12 (46). It is currently unknown where and when during SL RNA biogenesis pseudouridylation takes place. One can envision a model for SL RNA biogenesis where the m7G and the first two cap modification serves as an export signal of SL RNA to the cytoplasm where assembly with Sm proteins and capping at position +4 takes place. The fully modified cap 4 and the Sm proteins may serve as an import signal of SL RNP to the nucleus.
Another unique property related to SL RNA biogenesis is the 3′-end formation of the RNA, which seems to be different from the termination of snRNAs (72). In vitro and in vivo data on mutants in the Sm site and stem-loop III suggest that transcription termination in the T tract is followed by trimming that generates the mature 3′ end (87). It is currently unknown whether the nucleolytic function that generates the 3′ end of the SL RNA involves the concerted action of the endonucleolytic and/or exosome function and in which compartment in the cell it takes place. The trimming is coupled to the Sm assembly, since mutations in the Sm site do not undergo proper 3′-end processing (87). This trimming process may serve as a quality control mechanism for the SL RNP that directs only the SL RNA that is properly assembled and trimmed to the spliceosome. Longer heterogeneous SL RNAs (at the 3′ end) were accumulated in permeable T. brucei cells that were incubated with an oligonucleotide complementary to the Sm site (94). Another interesting phenomenon related to SL RNA maturation is the finding of stage-specific heterogeneity at the 3′ end of the Leishmania donovani amastigote SL RNA that carries an 80-nt poly(A) tail (42). This polyadenylated RNA is present only in amastigotes and may represent another regulatory process that SL RNA undergoes. This process may be related to the adenylation and deadenylation control that take place in the maturation of several snRNAs and 7SL RNA (72).
Sm and snRNP proteins.
The identification of common core proteins that bind SL and U2 RNPs as classical Sm proteins was surprising (27, 63, 64, 66) since core proteins of the snRNPs were not recognized by the anti-Sm sera that recognize cognate proteins from yeast to humans (57, 63, 64). The binding sites for the Sm proteins in trypanosome snRNAs are relatively degenerate in comparison to the well-conserved sites of other eukaryotes (60). In contrast to the high correlation between Sm proteins trimethylation and nuclear localization in other eukaryotes, there is no such correlation in trypanosomes. The Sm proteins were shown to bind U1, U2, and U4 that harbor a TMG cap; to SL RNA that possesses a special cap 4 structure; and to U5, which has no cap. The trypanosome Sm core proteins were identified by affinity selection of SL and the U2 snRNPs (27, 66). Seven Sm proteins were identified and cloned from T. brucei. The trypanosome Sm proteins are a minimal version of the canonical Sm proteins, because three of the seven, in particular SmB, are smaller than homologues from other organisms (66). There are several significant deviations in the conserved Sm motifs 1 and 2 present in all known Sm proteins. In contrast to the many changes in charged amino acids, there are almost 11 conserved hydrophobic amino acids in these motifs (66). It is of particular interest that the SmD1 and D3 are lacking the C-terminal RG dipeptide repeats of the human homologues. The arginines in this RG domain are usually methylated, constituting an important determinant of the Sm epitope. The lack of the RG-rich domain may explain why the trypanosome Sm proteins are not recognized by Sm antibodies (66). The arginine methylation is the signal for recognition of SMN in metazoa, which is a key player in Sm assembly and serves to selectively assure the binding of Sm proteins to snRNAs (24). Thus far, there is no evidence for the existence of SMN in trypanosomes.
Only two snRNP-specific proteins were identified biochemically and their coding genes analyzed. The first snRNP protein to be cloned and sequenced was a U2-specific protein of T. brucei homologous to human U2A′ (31% identity). The trypanosome protein has unique domains missing from its homologous protein; it has a C-terminal extension and two insertions, one of which is a leucine repeat. Since the C-terminal domain of this protein shares no significant homology with U2A′; this domain may contribute a function specific for trypanosomes (17).
The U5-specific protein PRP8 is one of the most crucial factors in cis splicing, since it is involved in splice site selection (82). The T. brucei protein p277 shares 40% identity and 61% similarity with the yeast PRP8 homologue. However, the positions believed to interact with the polypyrimidine tract are not conserved in the trypanosome protein, suggesting that the U-rich region is recognized in trypanosomes in a manner different from that in yeast (48).
RECOGNITION OF THE 3′ SPLICE SITE, POLYPYRIMIDINE TRACT, AND BRANCH POINT IN trans SPLICING
The AG dinucleotide at the 3′ splice site and its associated upstream polypyrimidine tracts are the most prominent and highly conserved cis-acting sequences in the pre-mRNA substrate. The elimination of the native splice acceptor site led to splicing at the next AG downstream and the accumulation of Y-branched intermediate, supporting a scanning model by splicing factors that represent a cross talk between factors that bind to the polypyrimidine tract and the 3′ splice site (37). Studies in mammalian cells demonstrate that the splicing factor U2AF65 recognizes and binds to the polypyrimidine tract and branch point (96), and the factor U2AF35 binds to the 3′ splice site (104). Splicing at the first AG downstream of the branch point may be a consequence of combined interactions between these two factors. A U2AF35 homologue bearing an identity of 34% was identified in the T. brucei genome. Recently, U2AF35 was cloned and analyzed from Trypanosoma cruzi, and this protein shares 38% identity with the human homologue (99). Interestingly, the central RNA-binding domain in U2AF35 is surrounded by two zinc finger motifs and instead of the C-terminal SR domain, the trypanosome protein has a different Zn finger motif which is reminiscent of the ones characterized for kinetoplastid DNA/RNA single-stranded binding proteins (99). The U2AF65 homologue was not yet identified in the trypanosomatid genome databases. However, significant homology was identified to hnRNP I (Table 1), which is known to bind to the polypyrimidine tract and affects the binding of U2AF65 (5). The PRP8 protein may also assist in locating the 3′ splice site and, indeed, the region of PRP8 that was shown in yeast to be involved in binding to the 3′ splice site is conserved in the trypanosome protein (48).
TABLE 1.
Putative splicing factors present in the T. brucei genome databasea
| Splicing factor | Accession no. | %
|
Function | ||
|---|---|---|---|---|---|
| P | I | S | |||
| Helicases | |||||
| PRP02-S.c | TRYP10.0.000033 | 78 | 36 | 56 | Branch point recognition |
| PRP22-S.c | TRYP10.0.001375 | 60 | 48 | 66 | Release of mRNA from spliceosome |
| PRP28-S.c | TRYP10.0.000031 | 64 | 36 | 59 | Replacing U1 by U6 |
| PRP43-S.c | CONTIG10724* | 75 | 60 | 71 | Release of lariat intermediate from spliceosome |
| hnRNPs | |||||
| hnRNP C-h | tryp_Ixb-62d11.plc | 48 | 30 | 53 | Heterogeneous nuclear RNP |
| hnRNP F-h | EMBL:AC007864 | 45 | 32 | 54 | Heterogeneous nuclear RNP |
| hnRNP I-h | TRYP9.0.002804 | 43 | 26 | 46 | Binding to polypyrimidine tract |
| SR protein | |||||
| U2AF35-h | TRYP10.0.000037 | 65 | 34 | 50 | Binding to 3′ splice site |
| Others | |||||
| PRP04-S.c | TRYP10.0.001185 | 62 | 29 | 44 | U4.U6-U5 interaction |
| PRP11-S.c | EMBL:AC008031 | 33 | 28 | 45 | U2 addition |
| PRP31-S.p | TRYP10.0.000313 | 65 | 27 | 44 | U4.U6-U5 assembly |
| PRP17-h | EMBL:AC105379 | 59 | 33 | 54 | Second catalytic step |
| DSK1-S.p | EMBL:AC008146 | 85 | 40 | 58 | SR protein kinase |
The sequences and accession numbers are from either http://www.sanger.ac.uk/projects/T brucei or GenBank. The identities and similarities are derived from a comparison with protein homologues from yeast or human sources as indicated. P, portion of the T. brucei gene currently present in the genome project. I and S, identity and similarity, respectively, to the sequence presented in the left column. Factor suffixes: h, Homo sapiens; S.c, Saccharomyces cerevisiae; S.p, Schizosaccharomyces pombe.
In mammalian introns the polypyrimidine tract serves a dual function: first in recognizing the branch site and then for 3′ splice site selection (75). In yeast, the polypyrimidine is needed for effective identification of the 3′ splice sites that are distant from the branch site (69). In T. brucei the polypyrimidine tract was identified as a major determinant for accurate trans splicing (34, 56, 81). In addition, in Leishmania the minimal requirement for efficient trans splicing is an AG 3′ splice site and even a synthetic polypyrimidine tract (18). In many trypanosome genes the polypyrimidine occurs most frequently in the region between positions −20 and −10, but differences exist in the length of the tract and its location relative to the 3′ splice site. These differences may account for the different trans-splicing efficacies of various genes (34, 56).
Branch point recognition in trypanosomes seems to be completely different from that of other eukaryotes since the region has no potential to interact with the U2 RNA by base-pairing (32). In yeast (Saccharomyces cerevisiae) the branch is mostly adenosine and is found within a highly conserved YUACUAAC sequence (68). In mammals, the branch point consensus is less stringent and the seqeunce is YNCURAY (74). The open question is how the branch point is recognized in trypanosomes. Branch points were mapped initially only for the T. brucei tubulin genes and more recently for a T. cruzi gene and is usually the first adenosine upstream of the polypyrimidine tract (37, 70). More recently, the Bindereif group mapped additional branch point sites. In 13 of 15 cases, the identified branch site is an adenosine. In one case, as well as in the cis-spliced intron of the poly(A) polymerase gene, an unusual branch was mapped to a cytidine. Comparison of the branch point sequences led to a relatively degenerate consensus that are poorly matched to the U2 interaction region (positions 33 to 38 of T. brucei U2 RNA). These data suggest that, in contrast to yeast and higher eukaryotes, additional sequences and/or signals might be used for branch point selection in trypanosomes, and extended protein-RNA interactions (not RNA-RNA interactions) most probably play a vital role in this recognition (S. Lücke, K. Meissner, and A. Bindereif, unpublished data).
SPLICING FACTORS IDENTIFIED IN TRYPANOSOMATIDS
RNA-binding proteins were cloned and sequenced in several trypanosomatid species, but their exact role in splicing was not elucidated further. TSR1IP was identified in T. brucei and shows homology to the U1 70-kDa domain. The protein contains an SR domain, as well as an acidic/arginine domain, homologous to the U1 70-kDa domain that interacts with the poly(A) polymerase to inhibit polyadenylation (39). However, its identity as a 70-kDa homologue is currently puzzling, since an U1-specific protein whose homology to the U1 70-kDa domain was recently identified by Palfi et al. (65). A protein (TSR1) was identified by using a yeast two-hybrid screen that interacts with TSR1IP (38). TSR1 carries two RNA recognition motifs and can interact with the splicing factors U2AF65/U2AF35 in the yeast two-hybrid system. It is tempting to speculate that these two proteins may be involved in the recruitment of SL RNA or U1 RNP into the spliceosome or in the coupling of the splicing and polyadenylation. Another interesting splicing factor is the T. cruzi XB1 protein (106), which was shown by a yeast three-hybrid screen to bind to the stem-loop II of the SL RNA (106). However, no direct role of any of these factors in trans or cis splicing was demonstrated. The use of RNAi offers an elegant and quick way to investigate the role of such splicing factors. As a first step, we searched the genome database for the presence of splicing factors whose function has been clearly demonstrated in yeast and mammals. A summary of these factors is listed in Table 1. We concentrated on certain helicases that participate in distinct steps in splicing to unwind transient interactions during spliceosome assembly. The search identified homologues to prp2 that function in the RNA rearrangements before the first step of splicing (90); homologues to prp22 that function in the release of the spliced mRNA from the spliceosome (15); homologues to prp28, which functions in the unwinding of U1 to allow the interaction of U6 with 5′ splice site (84); and homologues to prp43 that release the lariat intermediate from the spliceosome (4). It is currently unknown whether these helicases function in both cis and trans splicing. It is anticipated that all of the above-mentioned helicases will be shared between cis and trans splicing except prp28, which should function only in cis splicing. Indeed, RNAi silencing of prp43 in T. brucei was shown to result in the lethal phenotype, accumulation of cis- and trans-splicing precursors, reduction in the mature RNAs, and accumulation of the SL RNA (X.-h. Liang and S. Michaeli, unpublished results). Interestingly, numerous factors associated with U2 binding were revealed. These include factors such as prp16, which functions in the fidelity of branch point recognition (12), and prp11 and prp 9, which function in U2 recruitment (103). Of special interest are splicing factors such as an homologue to DSK1, a protein kinase whose targets are SR proteins. This finding suggests that alternative splicing exists in trypanosomes and is regulated by phosphorylation (89). The presence of hnRNP proteins such as C, F, and I homologues in trypanosomes is intriguing, since these proteins are present in metazoa but not in yeast (40). hnRNP proteins, among their multiple functions, are known to regulate splicing by serving as splicing repressors or activators (1, 83). For instance, hnRNP I serves as a splicing repressor in the brain (5).
LINKAGE BETWEEN trans SPLICING AND POLYADENYLATION
In cis splicing, factors involved in capping, splicing, and polyadenylation interact with the carboxy-terminal domain of RNA polymerase II at an early stage of mRNA production, and these processes are therefore coupled to transcription (59). In trypansomes, poly(A) addition is coupled to trans splicing of the downstream gene (35, 43, 56, 98). No specific sequence, such as the signal for polyadenylation (AAUAAA) in higher eukaryotes, seems to be present in trypanosomatid mRNAs (2). The polypyrimidine tract plays a major role in the coupling of these processes, since mutating the polypyrimidine tract led to aberrant poly(A) site choice (35). Inhibition of trans splicing in permeabilized cells abolished mRNA 3′-end formation (93). It has been suggested that the factors required for 3′-end cleavage and polyadenylation is associated with the pre-mRNA after the 3′ splice site region has been marked by the machinery of the spliceosome (56). The finding that the location of poly(A) site moves in concert with the 3′ splice site AG in Leishmania supports a model in which the factor(s) responsible for cleaving the pre-mRNA at the poly(A) site can bind only a certain short distance away from the 3′ splice site (43). After cleavage at the 3′ splice site, the same factor could direct polyadenylation upstream. This putative factor can be shared by the 3′-end formation and trans-splicing machineries, thus effectively coupling the two processes (56). U1 snRNP is essential for the coupling of cis splicing and polyadenylation (100). Analogously, an SL RNP specific protein may coordinate the interaction between trans-splicing and polyadenylation machineries.
REGULATION OF trans SPLICING
It is currently unknown how the efficiency of trans splicing of different genes is determined. Although the polypyrimidine tract definitely contributes to the selection of the 3′ splice site, other determinants that affect splicing efficacy were not identified. For instance, how sequences at the branch point influence the process and what is the optimal distance between the polypyrimidine tract and the first 3′ splice site are currently unknown.
To be able to modulate trans splicing, one should envision the presence of regulatory sequences that bind factors that can enhance or repress splicing. Indeed, it was demonstrated that the efficient use of α-tubulin 3′ splice site is dependent upon the presence of exon sequences (47). Exonic sequences that positively regulate trans splicing might be similar to the cis-splicing enhancers described in other systems. The best-characterized splicing enhancers known in mammals consist of short, purine-rich sequences that bind to specific members of the SR protein family (33). The presence of SR and hnRNP proteins known to function as splicing activators or repressors suggests that the machinery for carrying out alternative splicing exists in trypanosomes (Table 1). Interestingly, the splicing of lysosomal cysteine proteases genes in Leishmania is regulated most probably via binding of stage-specific factor that interacts with the splicing-polyadenylation machinery and binds to a regulatory element in the intergenic region (10). More recently, alternative trans splicing was demonstrated in T. cruzi for the LYT1 gene that functions in the lytic pathway (54).
cis SPLICING AND COORDINATION WITH trans SPLICING
A twist in our understanding of pre-mRNA splicing in trypanosomes occurred when the first cis-spliced intron was discovered. The PAP gene was found to carry an intron that obeys the GU/AG rule of cis-spliced introns (51). Interestingly, PAP mRNAs are trans spliced at the very 5′ end, as well as at the internal 3′ splice site, generating two alternatively trans-spliced products. The main question is how the two processes of cis and trans splicing are coordinated. In nematodes, it was found that the absence of a proper 5′ splice site at a certain distance from the 3′ splice site forces the substrates to undergo trans splicing (16). In trypanosomes, however, it seems that competition exists between these two processes, since the same AG 3′ splice site can be used once in trans splicing or can serve as the 3′ splice site for the removal of the intron. The decision as to whether to utilize a 3′ splice site for cis or trans splicing may depend on the presence of canonical 5′ splice site and may be modulated by factors such as U1 snRNP and certain SR proteins. It is currently unknown how widespread cis splicing is in trypanosomatids. From a phylogenetic point of view, the discovery of cis splicing in trypanosomes unifies the theme that every organism bearing trans splicing also carries out cis splicing.
CONCLUDING REMARKS AND PERSPECTIVES
As described above, the complete subset of snRNAs involved in trans splicing was determined, including U5. The finding of U1 snRNA occurred just before the discovery of cis splicing. However, U1 most probably functions only in cis splicing, as in nematodes. The domains of the SL RNA important for trans splicing were identified, and the role of the exon and intron sequences for trans splicing was established. However, no evidence was found for an SL-U6 interaction analogous to the interaction in nematodes. In contrast, a unique interaction between the SL RNA intron regions adjacent to the 5′ splice site with U5 and U6 snRNA seems to be important for trans splicing. SLA1, which interacts with the exon sequence, was found to be a small RNA that guides pseudouridylation on SL RNA. Faithful SL RNA transcription systems were established, as well as an in vitro system for the unique capping of the SL RNA. The coupling between the trans splicing and capping of the SL RNA was found, as well as a linkage between trans splicing and polyadenylation. The role of the polypyrimidine tract for trans splicing was also established, and the exon enhancers were described. The SL and U2 RNPs were purified to homogeneity, and Sm proteins were shown to bind to SL and U snRNPs. Several splicing factors were identified, but their direct functional role in trans splicing has not yet been demonstrated.
However, the trypanosome trans-splicing research suffers from the lack of an in vitro system. We now have a better understanding of how a trypanosome pre-mRNA substrate is structured, and it is time to reattempt to reconstitute the reaction in vitro. Perhaps the first step toward achieving this goal would be to reconstitute the efficient binding of SL RNP and U2 RNP to the synthetic pre-mRNA substrate. Other challenging questions are how the 5′ splice site of the SL RNA associates efficiently with the 3′ splice site present in the target pre-mRNA to be trans spliced and also how the decision is made whether a certain 3′ splice site is marked for trans or cis splicing. Recent studies in nematodes identified two SL RNP-specific proteins, one of which makes contact with the SF1/BBP (branch point binding protein). The association of the SL RNP-specific protein with the BBP that interacts with the branch point and with U2AF65 explains how the bridge between the SL RNA and the pre-mRNA is being formed in nematodes (20). At present, no SL RNP-specific proteins have been identified in trypanosomes analogous to the proteins in nematodes. However, it should be noted that major differences exist between the trypanosome and the nematode systems regarding the role of the SL RNA intron and exon sequences for trans splicing and the role of the U6 and U5 snRNAs for the reaction. One can envision a single spliceosome carrying both U1 and SL RNP, with the competition between U1 and SL RNP binding being the major determinant for selecting cis or trans splicing. The binding of SL RNP or U1 snRNP may depend on perfect binding of U1 to the 5′ splice site, as well as its association with SR proteins. Several SR proteins have been determined in trypanosomes, and their role in trans or cis splicing needs to be established. Finally, unraveling the machinery of trans splicing that does not exist in the host of these medically and economically important parasites still gives hope for a therapeutic intervention directed toward the trans-splicing-specific components.
Acknowledgments
We thank Elisabetta Ullu, Albrecht Bindereif, and Zofia Palfi from their many helpful discussions and valuable comments regarding the manuscript.
The research was supported in part by the German Israel Foundation. S.M. is an International Research Scholar of the Howard Hughes Medical Institute.
REFERENCES
- 1.Adams, M. D., D. Z. Rudner, and D. C. Rio. 1996. Biochemistry and regulation of pre-mRNA splicing. Curr. Opin. Cell Biol. 8:331-339. [DOI] [PubMed] [Google Scholar]
- 2.Agabian, N. 1990. trans splicing of nuclear pre-mRNAs. Cell 61:1157-1160. [DOI] [PubMed] [Google Scholar]
- 3.Agami, R., R. Aly, S. Halman, and M. Shapira. 1994. Functional analysis of cis-acting DNA elements required for expression of the SL RNA gene in the parasitic protozoan Leishmania amazonensis. Nucleic Acids Res. 22:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arenas, J. E., and J. N. Abelson. 1997. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. USA 94:11798-11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashiya, M., and P. J. Grabowski. 1997. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 3:996-1015. [PMC free article] [PubMed] [Google Scholar]
- 6.Bangs, J. D., P. F. Crain, T. Hashizume, J. A. McCloskey, and J. C. Boothroyd. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267:9805-9815. [PubMed] [Google Scholar]
- 7.Bell, M., and A. Bindereif. 1999. Cloning and mutational analysis of the Leptomonas seymouri U5 snRNA gene: function of the Sm site in core RNP formation and nuclear localization. Nucleic Acids Res. 27:3986-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell, M., R. Wohner, and A. Bindereif. 2000. U4 small nuclear RNA genes of trypanosomes: cloning of the Leptomonas seymouri gene and mutational analysis of core snRNP assembly. Gene 247:77-86. [DOI] [PubMed] [Google Scholar]
- 9.Boothroyd, J. C., and G. A. Cross. 1982. Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5′ end. Gene 20:281-289. [DOI] [PubMed] [Google Scholar]
- 10.Brooks, D. R., H. Denise, G. D. Westrop, G. H. Coombs, and J. C. Mottram. 2001. The stage-regulated expression of Leishmania mexicana CPB cysteine proteases is mediated by an intercistronic sequence element. J. Biol. Chem. 276:47061-47069. [DOI] [PubMed] [Google Scholar]
- 11.Bruzik, J. P., K. Van Doren, D. Hirsh, and J. A. Steitz. 1988. Trans splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature 335:559-562. [DOI] [PubMed] [Google Scholar]
- 12.Burgess, S. M., and C. Guthrie. 1993. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73:1377-1391. [DOI] [PubMed] [Google Scholar]
- 13.Campbell, D. A., N. R. Sturm, and M. C. Yu. 2000. Transcription of the kinetoplastid spliced leader RNA gene. Parasitol. Today 16:78-82. [DOI] [PubMed] [Google Scholar]
- 14.Campbell, D. A., D. A. Thornton, and J. C. Boothroyd. 1984. Apparent discontinuous transcription of Trypanosoma brucei variant surface antigen genes. Nature 311:350-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Company, M., J. Arenas, and J. Abelson. 1991. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349:487-493. [DOI] [PubMed] [Google Scholar]
- 16.Conrad, R., R. F. Liou, and T. Blumenthal. 1993. Conversion of a trans-spliced C. elegans gene into a conventional gene by introduction of a splice donor site. EMBO J. 12:1249-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cross, M., B. Wieland, Z. Palfi, A. Gunzl, U. Rothlisberger, H. W. Lahm, and A. Bindereif. 1993. The trans-spliceosomal U2 snRNP protein 40K of Trypanosoma brucei: cloning and analysis of functional domains reveals homology to a mammalian snRNP protein. EMBO J. 12:1239-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curotto de Lafaille, M. A., A. Laban, and D. F. Wirth. 1992. Gene expression in Leishmania: analysis of essential 5′ DNA sequences. Proc. Natl. Acad. Sci. USA 89:2703-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denker, J. A., P. A. Maroney, Y. T. Yu, R. A. Kanost, and T. W. Nilsen. 1996. Multiple requirements for nematode spliced leader RNP function in trans-splicing. RNA 2:746-755. [PMC free article] [PubMed] [Google Scholar]
- 20.Denker, J. A., D. M. Zuckerman, P. A. Maroney, and T. W. Nilsen. 2002. New components of the spliced leader RNP required for nematode trans-splicing. Nature 417:667-670. [DOI] [PubMed] [Google Scholar]
- 21.Djikeng, A., L. Ferreira, M. D'Angelo, P. Dolezal, T. Lamb, S. Murta, V. Triggs, S. Ulbert, A. Villarino, S. Renzi, E. Ullu, and C. Tschudi. 2001. Characterization of a candidate Trypanosoma brucei U1 small nuclear RNA gene. Mol. Biochem. Parasitol. 113:109-115. [DOI] [PubMed] [Google Scholar]
- 22.Dungan, J. M., K. P. Watkins, and N. Agabian. 1996. Evidence for the presence of a small U5-like RNA in active trans-spliceosomes of Trypanosoma brucei. EMBO J. 15:4016-4029. [PMC free article] [PubMed] [Google Scholar]
- 23.Freistadt, M. S., G. A. Cross, and H. D. Robertson. 1988. Discontinuously synthesized mRNA from Trypanosoma brucei contains the highly methylated 5′ cap structure, m7GpppA*A*C(2′-O)mU*A. J. Biol. Chem. 263:15071-15075. [PubMed] [Google Scholar]
- 24.Friesen, W. J., S. Massenet, S. Paushkin, A. Wyce, and G. Dreyfuss. 2001. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell 7:1111-1117. [DOI] [PubMed] [Google Scholar]
- 25.Gilinger, G., and V. Bellofatto. 2001. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 29:1556-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldring, A., and S. Michaeli. 1995. The U6 snRNA-encoding gene of the monogenetic trypanosomatid Leptomonas collosoma. Gene 156:139-144. [DOI] [PubMed] [Google Scholar]
- 27.Goncharov, I., Z. Palfi, A. Bindereif, and S. Michaeli. 1999. Purification of the spliced leader ribonucleoprotein particle from Leptomonas collosoma revealed the existence of an Sm protein in trypanosomes: cloning the SmE homologue. J. Biol. Chem. 274:12217-12221. [DOI] [PubMed] [Google Scholar]
- 28.Gu, J., J. R. Patton, S. Shimba, and R. Reddy. 1996. Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions. RNA 2:909-918. [PMC free article] [PubMed] [Google Scholar]
- 29.Günzl, A., E. Ullu, M. Dorner, S. P. Fragoso, K. F. Hoffmann, J. D. Milner, Y. Morita, E. K. Nguu, S. Vanacova, S. Wunsch, A. O. Dare, H. Kwon, and C. Tschudi. 1997. Transcription of the Trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol. Biochem. Parasitol. 85:67-76. [DOI] [PubMed] [Google Scholar]
- 30.Guthrie, C., and B. Patterson. 1988. Spliceosomal snRNAs. Annu. Rev. Genet. 22:387-419. [DOI] [PubMed] [Google Scholar]
- 31.Hannon, G. J., P. A. Maroney, Y. T. Yu, G. E. Hannon, and T. W. Nilsen. 1992. Interaction of U6 snRNA with a sequence required for function of the nematode SL RNA in trans-splicing. Science 258:1775-1780. [DOI] [PubMed] [Google Scholar]
- 32.Hartshorne, T., and N. Agabian. 1990. A new U2 RNA secondary structure provided by phylogenetic analysis of trypanosomatid U2 RNAs. Genes Dev. 4:2121-2131. [DOI] [PubMed] [Google Scholar]
- 33.Hertel, K. J., K. W. Lynch, and T. Maniatis. 1997. Common themes in the function of transcription and splicing enhancers. Curr. Opin. Cell Biol. 9:350-357. [DOI] [PubMed] [Google Scholar]
- 34.Huang, J., and L. H. Van der Ploeg. 1991. Requirement of a polypyrimidine tract for trans-splicing in trypanosomes: discriminating the PARP promoter from the immediately adjacent 3′ splice acceptor site. EMBO J. 10:3877-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hug, M., H. R. Hotz, C. Hartmann, and C. Clayton. 1994. Hierarchies of RNA-processing signals in a trypanosome surface antigen mRNA precursor. Mol. Cell. Biol. 14:7428-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huie, J. L., P. He, and V. Bellofatto. 1997. In vitro transcription of the Leptomonas seymouri SL RNA and U2 snRNA genes using homologous cell extracts. Mol. Biochem. Parasitol. 90:183-192. [DOI] [PubMed] [Google Scholar]
- 37.Hummel, H. S., R. D. Gillespie, and J. Swindle. 2000. Mutational analysis of 3′ splice site selection during trans-splicing. J. Biol. Chem. 275:35522-35531. [DOI] [PubMed] [Google Scholar]
- 38.Ismaili, N., D. Perez-Morga, P. Walsh, A. Mayeda, A. Pays, P. Tebabi, A. R. Krainer, and E. Pays. 1999. Characterization of a SR protein from Trypanosoma brucei with homology to RNA-binding cis-splicing proteins. Mol. Biochem. Parasitol. 102:103-115. [DOI] [PubMed] [Google Scholar]
- 39.Ismaili, N., D. Perez-Morga, P. Walsh, M. Cadogan, A. Pays, P. Tebabi, and E. Pays. 2000. Characterization of a Trypanosoma brucei SR domain-containing protein bearing homology to cis-spliceosomal U1 70-kDa proteins. Mol. Biochem. Parasitol. 106:109-112. [DOI] [PubMed] [Google Scholar]
- 40.Kaufer, N. F., and J. Potashkin. 2000. Analysis of the splicing machinery in fission yeast: a comparison with budding yeast and mammals. Nucleic Acids Res. 28:3003-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krause, M., and D. Hirsh. 1987. A trans-spliced leader sequence on actin mRNA in Caenorhabditis elegans. Cell 49:753-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamontagne, J., and B. Papadopoulou. 1999. Developmental regulation of spliced leader RNA gene in Leishmania donovani amastigotes is mediated by specific polyadenylation. J. Biol. Chem. 274:6602-6609. [DOI] [PubMed] [Google Scholar]
- 43.LeBowitz, J. H., H. Q. Smith, L. Rusche, and S. M. Beverley. 1993. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 7:996-1007. [DOI] [PubMed] [Google Scholar]
- 44.Li, L., L. R. Otake, Y. X. Xu, and S. Michaeli. 2000. The trans-spliceosomal U4 RNA from the monogenetic trypanosomatid Leptomonas collosoma: cloning and identification of a transcribed tRNA-like element that controls its expression. J. Biol. Chem. 275:2259-2264. [DOI] [PubMed] [Google Scholar]
- 45.Liang, X. H., L. Liu, and S. Michaeli. 2001. Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA. J. Biol. Chem. 276:40313-40318. [DOI] [PubMed] [Google Scholar]
- 46.Liang, X. H., Y. X. Xu, and S. Michaeli. 2002. The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the SL RNA. RNA 8:237-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez-Estrano, C., C. Tschudi, and E. Ullu. 1998. Exonic sequences in the 5′ untranslated region of alpha-tubulin mRNA modulate trans splicing in Trypanosoma brucei. Mol. Cell. Biol. 18:4620-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lücke, S., T. Klockner, Z. Palfi, M. Boshart, and A. Bindereif. 1997. Trans mRNA splicing in trypanosomes: cloning and analysis of a PRP8-homologous gene from Trypanosoma brucei provides evidence for a U5-analogous RNP. EMBO J. 16:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lücke, S., G. L. Xu, Z. Palfi, M. Cross, V. Bellofatto, and A. Bindereif. 1996. Spliced leader RNA of trypanosomes: in vivo mutational analysis reveals extensive and distinct requirements for trans splicing and cap 4 formation. EMBO J. 15:4380-4391. [PMC free article] [PubMed] [Google Scholar]
- 50.Madhani, H. D., R. Bordonne, and C. Guthrie. 1990. Multiple roles for U6 snRNA in the splicing pathway. Genes Dev. 4:2264-2277. [DOI] [PubMed] [Google Scholar]
- 51.Mair, G., H. Shi, H. Li, A. Djikeng, H. O. Aviles, J. R. Bishop, F. H. Falcone, C. Gavrilescu, J. L. Montgomery, M. I. Santori, L. S. Stern, Z. Wang, E. Ullu, and C. Tschudi. 2000. A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA 6:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mair, G., E. Ullu, and C. Tschudi. 2000. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem. 275:28994-28999. [DOI] [PubMed] [Google Scholar]
- 53.Mandelboim, M., C. L. Estraño, C. Tschudi, E. Ullu, and S. Michaeli. 2002. On the role of exon and intron sequences in trans-splicing utilization and cap 4 modification of the trypanosomatid Leptomonas collosoma SL RNA. J. Biol. Chem. 277:35210-35218. [DOI] [PubMed] [Google Scholar]
- 54.Manning-Cela, R., A. González, and J. Swindle. 2002. Alternative splicing of LYT1 transcripts in Trypanosoma cruzi. Infect. Immun. 70:4726-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mattaj, I. W. 1986. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 46:905-911. [DOI] [PubMed] [Google Scholar]
- 56.Matthews, K. R., C. Tschudi, and E. Ullu. 1994. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 8:491-501. [DOI] [PubMed] [Google Scholar]
- 57.Michaeli, S., T. G. Roberts, K. P. Watkins, and N. Agabian. 1990. Isolation of distinct small ribonucleoprotein particles containing the spliced leader and U2 RNAs of Trypanosoma brucei. J. Biol. Chem. 265:10582-10588. [PubMed] [Google Scholar]
- 58.Milhausen, M., R. G. Nelson, S. Sather, M. Selkirk, and N. Agabian. 1984. Identification of a small RNA containing the trypanosome spliced leader: a donor of shared 5′ sequences of trypanosomatid mRNAs? Cell 38:721-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minvielle-Sebastia, L., and W. Keller. 1999. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 11:352-357. [DOI] [PubMed] [Google Scholar]
- 60.Mottram, J., K. L. Perry, P. M. Lizardi, R. Lührmann, N. Agabian, and R. G. Nelson. 1989. Isolation and sequence of four small nuclear U RNA genes of Trypanosoma brucei subsp. brucei: identification of the U2, U4, and U6 RNA analogs. Mol. Cell. Biol. 9:1212-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murphy, W. J., K. P. Watkins, and N. Agabian. 1986. Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: evidence for trans splicing. Cell 47:517-525. [DOI] [PubMed] [Google Scholar]
- 62.Myler, P. J., and K. D. Stuart. 2000. Recent developments from the Leishmania genome project. Curr. Opin. Microbiol. 3:412-416. [DOI] [PubMed] [Google Scholar]
- 63.Palfi, Z., and A. Bindereif. 1992. Immunological characterization and intracellular localization of trans-spliceosomal small nuclear ribonucleoproteins in Trypanosoma brucei. J. Biol. Chem. 267:20159-20163. [PubMed] [Google Scholar]
- 64.Palfi, Z., A. Gunzl, M. Cross, and A. Bindereif. 1991. Affinity purification of Trypanosoma brucei small nuclear ribonucleoproteins reveals common and specific protein components. Proc. Natl. Acad. Sci. USA 88:9097-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palfi, Z., W. S. Lane, and A. Bindereif. 2002. Biochemical and functional characterization of the cis-spliceosomal U1 small nuclear RNP from Trypanosoma brucei. Mol. Biochem. Parasitol. 121:233-243. [DOI] [PubMed] [Google Scholar]
- 66.Palfi, Z., S. Lucke, H. W. Lahm, W. S. Lane, V. Kruft, E. Bragado-Nilsson, B. Seraphin, and A. Bindereif. 2000. The spliceosomal snRNP core complex of Trypanosoma brucei: cloning and functional analysis reveals seven Sm protein constituents. Proc. Natl. Acad. Sci. USA 97:8967-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palfi, Z., G. L. Xu, and A. Bindereif. 1994. Spliced leader-associated RNA of trypanosomes. Sequence conservation and association with protein components common to trans-spliceosomal ribonucleoproteins. J. Biol. Chem. 269:30620-30625. [PubMed] [Google Scholar]
- 68.Parker, R., P. G. Siliciano, and C. Guthrie. 1987. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 49:229-239. [DOI] [PubMed] [Google Scholar]
- 69.Patterson, B., and C. Guthrie. 1991. A U-rich tract enhances usage of an alternative 3′ splice site in yeast. Cell 64:181-187. [DOI] [PubMed] [Google Scholar]
- 70.Patzelt, E., K. L. Perry, and N. Agabian. 1989. Mapping of branch sites in trans-spliced pre-mRNAs of Trypanosoma brucei. Mol. Cell. Biol. 9:4291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perry, K., K. P. Watkins, and N. Agabian. 1987. Trypanosome mRNAs have unusual “cap 4” structures acquired by addition of a spliced leader. Proc. Natl. Acad. Sci. USA 84:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perumal, K., and R. Reddy. 2002. The 3′ end formation in small RNAs. Gene Expr. 10:59-78. [PMC free article] [PubMed] [Google Scholar]
- 73.Rajkovic, A., R. E. Davis, J. N. Simonsen, and F. M. Rottman. 1990. A spliced leader is present on a subset of mRNAs from the human parasite Schistosoma mansoni. Proc. Natl. Acad. Sci. USA 87:8879-8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rautmann, G., and R. Breathnach. 1985. A role for branchpoints in splicing in vivo. Nature 315:430-432. [DOI] [PubMed] [Google Scholar]
- 75.Reed, R. 1989. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 3:2113-2123. [DOI] [PubMed] [Google Scholar]
- 76.Roberts, T. G., J. M. Dungan, K. P. Watkins, and N. Agabian. 1996. The SLA RNA gene of Trypanosoma brucei is organized in a tandem array which encodes several small RNAs. Mol. Biochem. Parasitol. 83:163-174. [DOI] [PubMed] [Google Scholar]
- 77.Roberts, T. G., N. R. Sturm, B. K. Yee, M. C. Yu, T. Hartshorne, N. Agabian, and D. A. Campbell. 1998. Three small nucleolar RNAs identified from the spliced leader-associated RNA locus in kinetoplastid protozoans. Mol. Cell. Biol. 18:4409-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sather, S., and N. Agabian. 1985. A 5′ spliced leader is added in trans to both alpha- and beta-tubulin transcripts in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 82:5695-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schnare, M. N., and M. W. Gray. 1999. A candidate U1 small nuclear RNA for trypanosomatid protozoa. J. Biol. Chem. 274:23691-23694. [DOI] [PubMed] [Google Scholar]
- 80.Schnare, M. N., and M. W. Gray. 1999. Spliced leader-associated RNA from Crithidia fasciculata contains a structure resembling stem/loop II of U1 snRNA. FEBS Lett. 459:215-217. [DOI] [PubMed] [Google Scholar]
- 81.Schurch, N., A. Hehl, E. Vassella, R. Braun, and I. Roditi. 1994. Accurate polyadenylation of procyclin mRNA in Trypanosoma brucei is determined by pyrimidine-rich elements in the intergenic regions. Mol. Cell. Biol. 14:3668-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Siatecka, M., J. L. Reyes, and M. M. Konarska. 1999. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 13:1983-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith, C. W., and J. Valcarcel. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25:381-388. [DOI] [PubMed] [Google Scholar]
- 84.Staley, J. P., and C. Guthrie. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell 3:55-64. [DOI] [PubMed] [Google Scholar]
- 85.Sturm, N. R., J. Fleischmann, and D. A. Campbell. 1998. Efficient trans-splicing of mutated spliced leader exons in Leishmania tarentolae. J. Biol. Chem. 273:18689-18692. [DOI] [PubMed] [Google Scholar]
- 86.Sturm, N. R., and D. A. Campbell. 1999. The role of intron structures in trans-splicing and cap 4 formation for the Leishmania spliced leader RNA. J. Biol. Chem. 274:19361-19367. [DOI] [PubMed] [Google Scholar]
- 87.Sturm, N. R., M. C. Yu, and D. A. Campbell. 1999. Transcription termination and 3′-end processing of the spliced leader RNA in kinetoplastids. Mol. Cell. Biol. 19:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sutton, R. E., and J. C. Boothroyd. 1986. Evidence for trans-splicing in trypanosomes. Cell 47:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tang, Z., T. Kuo, J. Shen, and R. J. Lin. 2000. Biochemical and genetic conservation of fission yeast Dsk1 and human SR protein-specific kinase 1. Mol. Cell. Biol. 20:816-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teigelkamp, S., M. McGarvey, M. Plumpton, and J. D. Beggs. 1994. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 13:888-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tessier, L. H., M. Keller, R. L. Chan, R. Fournier, J. H. Weil, and P. Imbault. 1991. Short leader sequences may be transferred from small RNAs to pre-mature mRNAs by trans-splicing in Euglena. EMBO J. 10:2621-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tschudi, C., and E. Ullu. 1990. Destruction of U2, U4, or U6 small nuclear RNA blocks trans-splicing in trypanosome cells. Cell 61:459-466. [DOI] [PubMed] [Google Scholar]
- 93.Ullu, E., K. R. Matthews, and C. Tschudi. 1993. Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol. Cell. Biol. 13:720-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ullu, E., and C. Tschudi. 1993. 2′-O-methyl RNA oligonucleotides identify two functional elements in the trypanosome spliced leader ribonucleoprotein particle. J. Biol. Chem. 268:13068-13073. [PubMed] [Google Scholar]
- 95.Ullu, E., and C. Tschudi. 1995. Accurate modification of the trypanosome spliced leader cap structure in a homologous cell-free system. J. Biol. Chem. 270:20365-20369. [DOI] [PubMed] [Google Scholar]
- 96.Valcarcel, J., R. K. Gaur, R. Singh, and M. R. Green. 1996. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science 273:1706-1709. [DOI] [PubMed] [Google Scholar]
- 97.Vandenberghe, A. E., T. H. Meedel, and K. E. Hastings. 2001. mRNA 5′-leader trans-splicing in the chordates. Genes Dev. 15:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vassella, E., R. Braun, and I. Roditi. 1994. Control of polyadenylation and alternative splicing of transcripts from adjacent genes in a procyclin expression site: a dual role for polypyrimidine tracts in trypanosomes? Nucleic Acids Res. 22:1359-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vázquez, M., C. Atorrasagasti, N. Bercovich, R. Volcovish, and M. J. Levin. 2003. Unique festures of the Trypanosoma cruzi U2AF35 splicing factor. Mol. Biochem. Parasitol. 128:77-81. [DOI] [PubMed] [Google Scholar]
- 100.Wassarman, K. M., and J. A. Steitz. 1993. Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev. 7:647-659. [DOI] [PubMed] [Google Scholar]
- 101.Watkins, K. P., and N. Agabian. 1991. In vivo UV cross-linking of U snRNAs that participate in trypanosome trans-splicing. Genes Dev. 5:1859-1869. [DOI] [PubMed] [Google Scholar]
- 102.Watkins, K. P., J. M. Dungan, and N. Agabian. 1994. Identification of a small RNA that interacts with the 5′ splice site of the Trypanosoma brucei spliced leader RNA in vivo. Cell T. 6:171-182. [DOI] [PubMed] [Google Scholar]
- 103.Wiest, D. K., C. L. O'Day, and J. Abelson. 1996. In vitro studies of the Prp9 · Prp11 · Prp21 complex indicate a pathway for U2 small nuclear ribonucleoprotein activation. J. Biol. Chem. 271:33268-33276. [DOI] [PubMed] [Google Scholar]
- 104.Wu, S., C. M. Romfo, T. W. Nilsen, and M. R. Green. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402:832-835. [DOI] [PubMed] [Google Scholar]
- 105.Xu, G. L., B. Wieland, and A. Bindereif. 1994. Trans-spliceosomal U6 RNAs of Crithidia fasciculata and Leptomonas seymouri: deviation from the conserved ACAGAG sequence and potential base pairing with spliced leader RNA. Mol. Cell. Biol. 14:4565-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu, P., L. Wen, G. Benegal, X. Wang, and G. A. Buck. 2001. Identification of a spliced leader RNA binding protein from Trypanosoma cruzi. Mol. Biochem. Parasitol. 112:39-49. [DOI] [PubMed] [Google Scholar]
- 107.Xu, Y. X., H. Ben-Shlomo, and S. Michaeli. 1997. The U5 RNA of trypanosomes deviates from the canonical U5 RNA: the Leptomonas collosoma U5 RNA and its coding gene. Proc. Natl. Acad. Sci. USA 94:8473-8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu, Y. X., L. Liu, and S. Michaeli. 2000. Functional analyses of positions across the 5′ splice site of the trypanosomatid spliced leader RNA: implications for base-pair interaction with U5 and U6 snRNAs. J. Biol. Chem. 275:27883-27892. [DOI] [PubMed] [Google Scholar]
- 109.Zeiner, G. M., N. R. Sturm, and D. A. Campbell. 2003. Exportin 1 mediates nuclear export of the kinetoplastid spliced leader RNA. Eukaryot. Cell 2:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]