Abstract
The chromosomes of ciliates are fragmented at reproducible sites during the development of the polyploid somatic macronucleus, but the mechanisms involved appear to be quite diverse in different species. In Paramecium aurelia, the process is imprecise and results in de novo telomere addition at locally heterogeneous positions. To search for possible determinants of chromosome fragmentation, we have studied an ∼21-kb fragmentation region from the germ line genome of P. primaurelia. The mapping and sequencing of alternative macronuclear versions of the region show that two distinct multicopy elements, a minisatellite and a degenerate transposon copy, are eliminated by an imprecise mechanism leading either to chromosome fragmentation and the formation of new telomeres or to the rejoining of flanking sequences. Heterogeneous internal deletions occur between short direct repeats containing TA dinucleotides. The complex rearrangement patterns produced vary slightly among genetically identical cell lines, show non-Mendelian inheritance during sexual reproduction, and can be experimentally modified by transformation of the maternal macronucleus with homologous sequences. These results suggest that chromosome fragmentation in Paramecium is the consequence of imprecise DNA elimination events that are distinct from the precise excision of single-copy internal eliminated sequences and that target multicopy germ line sequences by homology-dependent epigenetic mechanisms.
In ciliates, the development of the somatic macronucleus involves extensive rearrangements of the germ line genome, including the fragmentation of chromosomes into smaller, acentric molecules healed by de novo telomere addition and the precise excision of numerous internal eliminated sequences (IESs). The rearrangement program can be repeated in each sexual generation because these unicellular eukaryotes separate germ line and somatic functions into two distinct kinds of nuclei, both of which develop after sexual events from mitotic copies of the zygotic nucleus. The diploid micronucleus is transcriptionally silent during vegetative growth and serves only to transmit the unrearranged germ line genome to sexual progeny through the processes of meiosis and karyogamy. The highly polyploid macronucleus (∼800n in Paramecium aurelia species) is responsible for all transcription during vegetative growth but is lost during sexual events and replaced by the newly developed zygotic macronucleus (for general reviews of macronuclear development and associated genome rearrangements, see references 12, 29, 36, 52, and 64).
Developmentally regulated chromosome fragmentation occurs in all ciliates studied, but the processes show important variations between species. The large number of breakage sites (∼40,000) in the haploid genomes of spirotrichs such as Oxytricha, Stylonichia, or Euplotes results in very short macronuclear “chromosomes” that usually contain a single gene (59), while in oligohymenophorans a few hundreds of breakage events give rise to longer macronuclear chromosomes, ranging from <100 to 1,500 kb in Tetrahymena thermophila (11) and from 50 to 800 kb in Paramecium primaurelia (5). Although fragmentation patterns are highly reproducible on a large scale, the addition of telomeric repeats usually does not occur at a precise nucleotide position, thus generating microheterogeneity among the multiple copies of the genome present in each developing macronucleus. Multiple telomere addition sites have been found to be clustered within regions of ≤70 bp in Oxytricha (2, 26, 62), ≤30 bp in Tetrahymena (18), and up to 1 to 2 kb in Paramecium (3, 20, 32). Such microheterogeneity in principle could arise either from multiple cut sites or from variable exonucleolytic trimming of the ends produced by a single cut prior to telomere addition. Euplotes crassus is unique in that it does not exhibit any microheterogeneity: telomeric repeats are reproducibly added at the same nucleotide positions in all macronuclear copies (2).
A second level of heterogeneity can result from alternative processing of the same germ line sequence. In Oxytricha, some fragmentation sites are not cut in all macronuclear copies, thus generating a fraction of macronuclear chromosomes containing several genes (6, 25, 26, 56, 62). Similarly, the ends of some Paramecium macronuclear chromosomes can form at one of several alternative telomere addition regions separated by 2 to 13 kb, each of which shows microheterogeneity in the exact positions of telomeres (1, 4, 20, 55). The whole set of alternative versions is reproducibly generated in each developing macronucleus, indicating that, as in other ciliates (36), chromosome fragmentation and telomere addition occur after substantial endoreplication of the diploid zygotic genome.
Little is known about the mechanisms involved in any species, but cis-acting sequence determinants or reaction intermediates have been characterized in some. A 15-bp chromosome breakage sequence (Cbs) has been shown to be necessary and sufficient for both chromosome breakage and telomere addition in Tetrahymena (19, 65); in the process, the Cbs is eliminated together with 4 to 34 bp of flanking DNA on both sides (18). In E. crassus, a 10-bp consensus sequence directs a staggered double-strand break at a precise distance and in a directional manner, but the Cbs itself is not necessarily eliminated (2, 33, 34). No conserved sequence has been identified so far in the vicinity of Paramecium chromosome fragmentation sites. On the other hand, there is evidence that fragmentation patterns are determined not only by the germ line sequence but also by epigenetic mechanisms. Indeed, cell lines with identical, entirely homozygous micronuclear genomes can reproducibly generate different rearrangement patterns from the same germ line sequence at each sexual generation. These variant patterns show maternal (cytoplasmic) inheritance in breeding analyses (17, 20, 42). Transformation experiments have further revealed that the choice of a particular fragmentation pattern during macronuclear development is governed by the maternal macronucleus, still present in the cytoplasm at that time, through trans-nuclear, homology-dependent effects that are likely to be mediated by RNA molecules (44, 45, 46; O. Garnier, V. Serrano, S. Duharcourt, and E. Meyer, submitted for publication).
To search for possible determinants of chromosome fragmentation in P. primaurelia, we have studied a fragmentation region for which both flanking macronuclear chromosomes are known. As a result of alternative processing, only a fraction of macronuclear copies are fragmented in this region. Previous analyses suggested that an initial break is followed by variable sequence elimination and then healed either by telomere addition, yielding two chromosomes of ∼250 and ∼230 kb, or by religation, resulting in larger chromosomes of ∼480 kb (4). An ∼21-kb segment of micronuclear DNA from this region, overlapping the ends of flanking macronuclear chromosomes, was PCR amplified and entirely sequenced. The germ line sequence is very AT rich and does not appear to contain any functional genes; the only recognizable features are a minisatellite and a degenerate Tc1/mariner transposon, the first such elements described for Paramecium. A detailed analysis of the rearranged chromosomes showed that both elements are removed from all macronuclear copies by imprecise deletions, either independently or together in a single large deletion. Each of these deletions is healed by telomere addition in some copies and by religation in others, suggesting that chromosome fragmentation is only a consequence of imprecise DNA elimination. These deletions resemble those that can be experimentally induced by homology-dependent effects at any locus and may be determined by a similar epigenetic mechanism.
MATERIALS AND METHODS
Paramecium cell lines and cultivation.
P. primaurelia wild-type strains 156 and 168 are well characterized, entirely homozygous laboratory stocks. Cells were grown in a wheat grass powder (Pines International Co.) infusion medium bacterized the day before use with Klebsiella pneumoniae and supplemented with 0.8 mg of beta-sitosterol (E. Merck AG, Darmstadt, Germany)/liter at 18 or 27°C. Basic methods of cell culturing have been described elsewhere (57). Autogamy and conjugation were induced as previously described (42). The ΔPX and ΔPY clones, which carry macronuclear deletions of the PX and PY sequences (see below), were obtained as previously described (43).
Genomic DNA extraction.
Cultures (400 ml) of exponentially growing cells (1,000 cells/ml) were centrifuged. After being washed in 10 mM Tris-HCl (pH 7.0), the pellet was resuspended in a volume of the same buffer equal to the volume of the cell pellet; the suspension was quickly added to 4 volumes of lysis solution (0.44 M EDTA [pH 9.0], 1% sodium dodecyl sulfate [SDS], 0.5% N-laurylsarcosine [Sigma], 1 mg of proteinase K [Merck]/ml) at 55°C. The lysate was incubated at 55°C for at least 5 h, gently extracted once with phenol, and dialyzed twice against TE (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) containing 20% ethanol and once against TE. Micronuclear DNA was prepared as described by Preer et al. (51) and further purified from contaminating low-molecular-weight macronuclear DNA by agarose gel electrophoresis. This DNA was used to amplify some of the Tennessee copies, but its average molecular weight was not high enough to allow efficient long-range amplification of the micronuclear fragmentation region, which was carried out by using total DNA samples (see Results).
DNA restriction, electrophoresis, Southern blotting, and gel purification.
DNA restriction and electrophoresis were carried out according to standard procedures (54). Pulsed-field electrophoresis was carried out with a home-made contour-clamped homogeneous electric field apparatus (10) and 0.25× TBE (1× TBE is 89 mM Tris, 89 mM borate, 2.5 mM EDTA) at 5 V/cm, with cooling to 12°C. Commutation times were 4 s for the best resolution of Asp718-digested macronuclear versions and 50 s for the separation of native macronuclear chromosomes; the migration time was ∼30 h. DNA was transferred from agarose gels to Hybond N+ membranes (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) in 0.4 N NaOH after depurination in 0.25 N HCl. Hybridization was carried out with 7% SDS-0.5 M sodium phosphate-1% bovine serum albumin-1 mM EDTA (pH 7.2) at 61°C. Probes were 32P labeled by random priming to a specific activity of 3 × 109 cpm/μg. Membranes were then washed for 30 min with 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% SDS at 60°C prior to autoradiography or PhosphorImager exposure. DNA fragments were purified from low-melting-temperature agarose gels by treatment with agarase (Sigma) or by using a Qiaquick gel extraction kit (Qiagen). PCR products were purified by using a Qiaquick PCR purification kit (Qiagen) or Microcon centrifugal filter devices (Millipore).
Cloning of probes from the fragmentation region.
Attempts to clone the A5-A6 Asp718 fragment from the different chromosome 2 versions into yeast artificial chromosomes yielded only one 65-kb insert out of 700 screened recombinant clones (4). The YAC insert was gel purified, and different fragments were subcloned in plasmid pUC18 and used as probes on Southern blots. PX is a 1.6-kb HindIII fragment to the right of BamHI site B1, between positions 474 and 2068 of the germ line sequence; PY is a 1.3-kb BglII-SacI fragment to the left of SacI, between positions 18694 and 20029. Attempts to clone the gel-purified ∼13-kb B1-B2 BamHI fragment from version 2a into a lambda phage vector yielded only a 6-kb fragment representing an existing but rare macronuclear version from which one probe was derived to the left of BamHI site B2. A 6-kb fragment containing the SacI site was cloned by screening of an EcoRI library with PY. Similar difficulties were encountered in all cloning strategies and were also observed in the cloning of telomeric PCR products, suggesting that instability is an intrinsic property of many sequences from this AT-rich (79%) region.
PCR amplification.
Classical PCR amplifications were carried out either with the Tfl (Promega) or the Dynazyme (Finnzymes) DNA polymerase according to the manufacturer's specifications. For long-range PCR amplifications, an Expand long-template PCR system (Roche) was used. All reactions were performed with a Techne Progene thermocycler. The primer used for telomeric PCRs was 5′-[(C/A)AACCC]5-3′.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited at GenBank under accession numbers AY325300 (for the 20,816-bp segment containing the entire rearrangement region) and AY326276 to AY326283 (for the Tennessee transposon).
RESULTS
Fragmented and nonfragmented chromosomes are derived from the same germ line sequence.
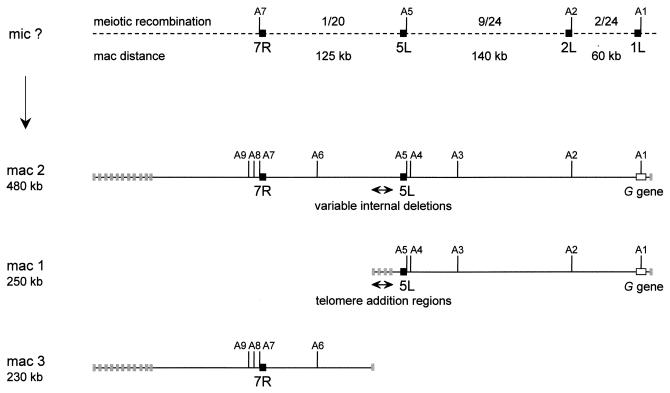
In a previous study, the G surface antigen gene of P. primaurelia (strain 156) was found to be located near the ends of two macronuclear chromosomes of very different sizes (4). Based on restriction mapping (Fig. 1), the smaller chromosome (chromosome 1, ∼250 kb) appeared to be identical to the right part of the larger one (chromosome 2, ∼480 kb). A third chromosome (chromosome 3, ∼230 kb) was found to correspond to the left part of chromosome 2. The same study provided evidence for alternative processing both at the left end of chromosome 1, which has alternative telomere addition regions, and in the corresponding region in the middle of chromosome 2, which has variable internal deletions. These results suggested that facultative breakage of a common micronuclear precursor gives rise to all three macronuclear chromosomes.
FIG. 1.
Restriction maps of macronuclear chromosomes 1, 2, and 3 (mac 1, mac 2, and mac 3, respectively). A1 to A9 are Asp718 sites. The G surface antigen gene is shown as a white box near the right ends of chromosomes 1 and 2. Gray boxes at chromosome ends represent telomere addition regions. Alternative processing at the left ends of chromosomes 2 and 3 results in multiple telomere addition regions distributed over >50 kb. Variable internal deletions were mapped in the middle of chromosome 2, in a region corresponding to the alternative telomere addition regions at the left end of chromosome 1. The broken line at the top represents the unknown micronuclear (mic) sequence. Allelic restriction fragment length polymorphisms were identified in sequences 7R, 5L, 2L, and 1L (black boxes). The numbers of F2 meiotic recombinants and the macronuclear distances between these sequences are indicated.
We first used a genetic test to confirm that chromosomes 1 and 3 are linked in the micronuclear genome. Allelic restriction fragment length polymorphisms that distinguish homozygous strains 156 and 168 were identified for a number of sequences located along these chromosomes. The frequencies of meiotic recombination between these sequences were estimated by counting the number of recombinants among F2 clones obtained by autogamy of F1 156/168 heterozygotes (autogamy is a self-fertilization process resulting in entirely homozygous progeny). As shown in Fig. 1, only 1 F2 clone out of 20 showed meiotic recombination between sequence 5L, which is located in chromosome 1, and sequence 7R, which is located in chromosome 3. This result is clearly different from the 50% of recombinants expected for unlinked loci. Since chromosomes 1 and 2 both contain the G surface antigen gene, which behaves as a single Mendelian locus, we conclude that all three macronuclear chromosomes are produced by alternative processing of the same micronuclear chromosome.
Southern blot mapping of alternative rearrangements.
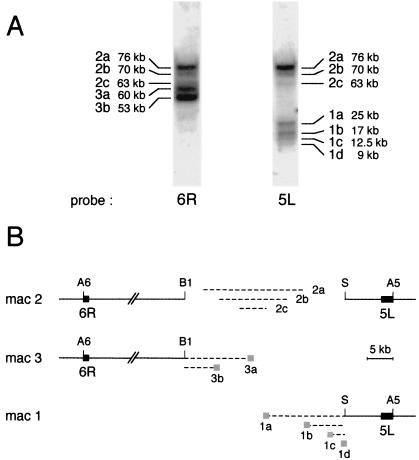
The Asp718 fragment from the middle of chromosome 2, between sites A5 and A6, was previously shown to be heterogeneous in size, appearing as a smeared band on Southern blots of pulsed-field electrophoresis gels (4). The mapping was refined by using DNA samples from the same caryonidal clone as in the previous study, hereafter called the reference clone (a caryonidal clone is a vegetative clone arising from a single event of macronuclear development). Under different pulsed-field conditions (see Materials and Methods), the smear could be resolved into three main bands hybridizing with probes 5L and 6R (Fig. 2A). These fragments, of approximately 76, 70, and 63 kb, identified three main chromosome 2 versions, which were designated 2a, 2b, and 2c, respectively. Better resolution was also obtained for the terminal fragments (between the last Asp718 site and the telomere) of fragmented chromosomes 1 and 3. Probe 5L previously identified three different telomeric fragments from chromosome 1, but under the new conditions, four fragments, of approximately 25, 17, 12.5, and 9.5 kb, were revealed; these represented alternative versions 1a, 1b, 1c, and 1d, respectively. Probe 6R revealed two distinct telomeric fragments from chromosome 3, of ∼60 kb (version 3a) and ∼53 kb (version 3b); only the shorter version was previously characterized.
FIG. 2.
Alternative versions of chromosomes 2, 3, and 1. (A) Southern blot of a pulsed-field electrophoresis gel of Asp718-digested total DNA, hybridized successively with probe 6R to reveal alternative versions of chromosomes 2 and 3 and with probe 5L to reveal alternative versions of chromosomes 2 and 1. (B) Maps of main alternative versions of macronuclear chromosomes 2, 3, and 1 (mac 2, mac 3, and mac 1, respectively) between Asp718 sites A5 and A6. Almost all of the alternative rearrangements occur between BamHI site B1 and SacI site S. The lengths of the B1-S fragments of the different chromosome 2 versions and of the telomeric fragments of the different versions of chromosomes 3 and 1 are represented by broken lines. Gray boxes show the positions of alternative telomere addition regions. Probes 5L and 6R are shown as black boxes.
The breakpoints of chromosome 2 internal deletions, like those of the addition of telomeres in chromosomes 1 and 3, thus occur in a few preferential regions. Using Southern blots of classical electrophoresis gels and several probes derived from the central region (see Materials and Methods), both kinds of alternative rearrangements were shown to occur within a BamHI-SacI fragment (B1-S in Fig. 2B) which contains almost all of the macronuclear heterogeneity. The B1-S fragment was estimated to be approximately 17.5, 12, and 5 kb long in versions 2a, 2b, and 2c, respectively (Fig. 2B). Only a small fraction of macronuclear copies showed deletions extending further to the right and had lost the SacI site.
Germ line sequence of region of alternative rearrangements.
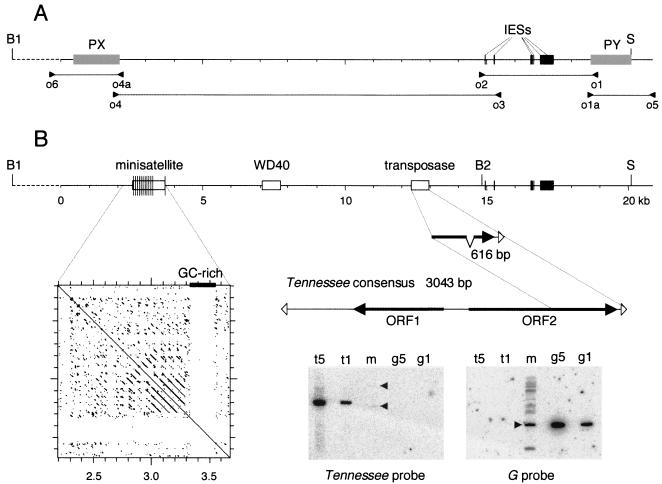
Because many sequences of the macronuclear variants appeared to be unstable after cloning in different vectors (see Materials and Methods), a PCR strategy was chosen to analyze the germ line sequence. To selectively amplify micronuclear DNA from total cellular DNA, in which the micronuclear/macronuclear ploidy ratio is ∼1:200, we took advantage of variant clones in which the sequences located on the left or right sides of the variable region were deleted from all macronuclear copies, so that primers located in these sequences could amplify only micronuclear DNA. These macronuclear deletion variants were obtained by an experimental procedure which does not affect the micronuclear genome (42, 43; see also the last section of Results). The PCR strategy also made use of a micronucleus-specific primer derived from an IES identified in the region (Fig. 3A). Indeed, six short DNA segments were found to be deleted from all macronuclear copies. The unique sequence obtained from mass PCR products showed that these deletions occur at the same positions in all macronuclear molecules and are bounded by two TA dinucleotides, one of which is maintained after deletion (data not shown); thus, they do not differ from previously characterized Paramecium IESs. As shown in Fig. 3A, four micronucleus-specific, long-range amplifications together covered 22.1 kb of the micronuclear sequence. A 20,816-bp segment containing the entire rearrangement region was sequenced directly from mass PCR products (GenBank accession number AY325300). PCR amplification of repeated sequences can lead to trans-PCR artifacts, which could be of some concern here because of the presence of minisatellite repeats (see below). However, the sequences produced from mass PCR products were always unique, suggesting that such artifacts did not occur. This suggestion was later confirmed by other nested, micronucleus-specific amplifications of total DNA, which always yielded products having the expected sizes and restriction maps (data not shown).
FIG. 3.
Germ line sequence of the chromosome fragmentation region. (A) PCR amplification of micronuclear DNA. The positions of primers are indicated by arrowheads. Primers o1 and o1a and primers o4 and o4a are located within the PY and PX sequences, repectively (gray boxes). Micronuclear specificity was achieved by using DNA from ΔPY clones for the o1-o2 and o1a-o5 PCRs and DNA from ΔPX clones for the o3-o4 and o4a-o6 PCRs. In addition, primer o3 was derived from one of the six IESs, 26 to 468 bp long (black boxes), that were identified in the o1-o2 micronuclear product. (B) Features of the germ line sequence. The 20,816-bp sequenced portion (solid line) begins 1.7 kb after BamHI site B1 and ends 0.8 kb after SacI site S. White boxes indicate the positions of the minisatellite and GC-rich sequence, WD40-related sequence, and truncated transposon. Thin vertical lines in the minisatellite represent individual repeats. The dot plot shows a self-comparison matrix of the minisatellite and GC-rich sequence (DNA Strider stringency 15; window 23). The 616-bp sequence of the truncated Tennessee copy is compared with the 3,043-bp consensus sequence derived from the alignment of eight different copies. ORFs are indicated by filled arrows, and terminal 31-bp IRs are indicated by open arrowheads. The Southern blot at the bottom was hybridized successively with a 677-bp probe from the right end of the Tennessee transposon (left panel) and, as a control, with a 1.6-kb EcoRI fragment from the G surface antigen gene (right panel). Lanes t5 and t1 contain 150 and 30 pg of a 3-kb complete Tennessee copy, respectively; these amounts correspond to five copies and one copy per haploid genome in 1 μg of the ∼100-Mb macronuclear genome. Lanes m contain 1 μg of EcoRI-digested DNA from the reference clone. Only two faint bands were revealed by the Tennessee probe in lane m (arrowheads), indicating that two copies of the transposon are partially maintained in the macronucleus, at much less than one copy per haploid genome. Lanes g5 and g1 contain 80 and 16 pg of the 1.6-kb EcoRI fragment from the G surface antigen gene, respectively; these amounts correspond to five copies and one copy per haploid genome. The 1.6-kb fragment revealed by the G surface antigen gene probe in lane m (arrowhead) indicates that the G surface antigen gene is present at one copy per haploid genome in macronuclear DNA; weaker bands arise from paralogous genes that cross-hybridize with the probe under the low-stringency conditions used.
The ∼21-kb germ line sequence has a G+C content of only 21%, which is lower than the 28% average for the macronuclear genome (L. Sperling, personal communication). Internal and database homology searches revealed only a few recognizable features (Fig. 3). One is a series of 11 direct repeats of an AT-rich, ∼69-bp motif with various levels of sequence conservation. This minisatellite is followed by a 224-bp sequence with a relatively high G+C content (36%), which in turn is followed by a single 69-bp repeat in the reverse orientation (Fig. 3B). The only protein coding sequences that could be identified by BLASTX searches are a few degenerate gene remnants, among which is an ∼700-bp sequence containing several repeats with some similarity to the WD40 protein motif (pfam00400). This finding stands in contrast to the high gene density in the macronuclear genome (>68% coding, as determined by analyses of random genome survey sequences) (58) but indicates that functional genes once may have been present in the region. More interestingly, another weak BLASTX hit revealed the presence of a short sequence with homology to those of DDE transposases, a signature of the Tc1-IS630 superfamily of transposons (14).
Characterization of Tennessee, a micronucleus-specific, multicopy transposon.
To determine whether the transposase-related sequence indeed belongs to some transposable element, we used different PCR strategies to search the genome for homologous sequences. Inverse PCRs were performed with restricted and circularized total DNAs and a pair of divergent primers designed to match the conserved transposase motifs so as to maximize the number of different transposon copies that could be amplified. The sequencing of PCR products showed that sequences highly similar to the transposase gene sequence were present at different locations in the genome and allowed us to identify a putative 31-bp inverted repeat (IR) present at both ends of these elements, after which flanking sequences completely differed from one copy to the other. Because the chosen restriction enzymes had sites within the elements, none of the inverse PCRs allowed us to amplify both ends of a single product. However, when several primers that pointed inward and corresponded to different parts of the IR consensus sequence were used alone for PCR amplification of total or micronuclear DNA, additional copies of the same element were obtained. A total of 38 sequences were found to represent at least eight different copies (five full length and three partial), which differed from each other by point mutations and insertions or deletions (GenBank accession numbers AY326276 to AY326283). For most of these copies, the same sequences were obtained with different PCR strategies, ruling out PCR artifacts other than point substitutions.
A multiple alignment of the eight copies allowed us to build a 3,043-bp consensus sequence (see www.biologie.ens.fr/∼lemouel/lemouel1.doc) showing the characteristic features of Tc1/mariner transposons (Fig. 3B). The end sequence of the 31-bp IR (5′-TACAGTCC…-3′) is similar to the consensus sequence of Tc1/mariner IR ends (TACAGTKS…), where the terminal 5′-TA-3′ dinucleotides are the duplicated copies of a TA integration site, and to the consensus sequence of Paramecium IES ends (TAYAGYNR…) (35). The element contains two putative genes, open reading frame (ORF) 1 (ORF1) and ORF2. The conceptual translation of ORF2 yields a 437-amino-acid protein with significant similarity to DDE transposases from prokaryotic and eukaryotic transposons, including the TBE and Tec ciliate elements. The ORF2 product is most similar to the putative transposase of Sardine, a larger transposon recently discovered in the micronuclear genome of P. tetraurelia (O. Garnier, A. Le Mouël, M. Prajer, S. Malinsky, M. Bétermier, and E. Meyer, unpublished data). ORF1 was tentatively identified by TBLASTX comparisons with the Sardine transposon. It appears to contain two short introns and could encode a 248-amino-acid protein with no homolog in the databases. This analysis shows that the transposase sequence initially found in the fragmentation region is a 616-bp truncated copy of a Tc1/mariner transposon, which was named Tennessee.
Like other ciliate transposons, Tennessee appears to be limited to the micronuclear genome of P. primaurelia, as Southern blots showed that the element is almost completely eliminated from the macronuclear genome (Fig. 3B). A comparison of the sequences of the different copies with the consensus sequence shows that most of them are probably not functional, as they have accumulated mutations in the ORFs, including in-frame stop codons and insertions or deletions. The transposase gene of one copy (not the copy from the fragmentation region) is interrupted by a 229-bp insertion that is absent from all other copies. The insertion has a relatively high G+C content (41%) and is flanked by direct repeats formed by the duplication of 19 bp of the transposase coding sequence. Surprisingly, this insertion is significantly similar to the 224-bp GC-rich sequence adjacent to the minisatellite in the fragmentation region (67% identity, with three gaps). The occurrences of this element in different genomic locations could represent mutated copies of an unknown type of transposable element.
PCR mapping of chromosome 2 internal deletions.
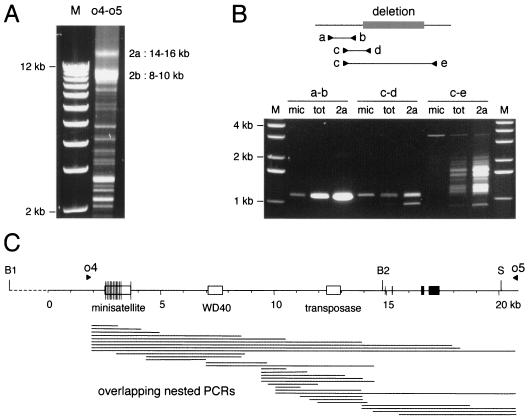
In the germ line genome, the distance between BamHI site B1 and the SacI site should be ∼22 kb if no large insertion occurs in the ∼0.4 kb between B1 and the beginning of the amplified micronuclear sequence. From the measured sizes of the B1-S fragment in the different chromosome 2 versions (∼17.5, ∼12, and ∼5 kb; Fig. 2B), the total lengths of deleted germ line sequences can be calculated to be approximately 4.5 kb for version 2a, 10 kb for 2b, and 17 kb for 2c. To map these deletions in the germ line sequence, we used a two-step PCR procedure. Macronuclear versions 2a and 2b were first amplified by long-range PCR from total DNA of the reference clone with primers o4 and o5, which lie outside the rearrangement region and define a segment about 3 kb shorter than the B1-S fragment (Fig. 4C). Figure 4A shows that abundant products of ∼14 to 16 kb and ∼8 to 10 kb were produced, consistent with the sizes expected for versions 2a (∼14.5 kb) and 2b (∼9 kb). The widths of the bands observed indicate that each of these versions is itself heterogeneous and consists of many slightly different variants. o4-o5 products shorter than 3 kb should correspond to 2c versions, but these were too short and too dispersed for mass analysis and were not studied further. Minor discrete products of 3 to 8 kb are probably amplified from macronuclear versions that are too rare and too dispersed to be detected on Southern blots.
FIG. 4.
PCR mapping of internal deletions. (A) PCR amplification of alternative macronuclear versions. PCR products corresponding to heterogeneous versions 2a and 2b were amplified from total DNA with primers o4 and o5 (see map in panel C) and analyzed on a 0.9% agarose gel. Lane M, markers. The sizes indicated were more precisely measured on different types of gels after the purification of 2a and 2b products. (B) Nested PCRs of o4-o5 products. The principle of the method is illustrated by three primer pairs (a-b, c-d, and c-e) overlapping a deletion observed in version 2a (gray box). For each of the PCRs, the agarose gel shows the amplification products obtained from three different templates: a purified PCR product representing the micronuclear sequence (mic), total cell DNA (tot), and a purified PCR product representing heterogeneous macronuclear version 2a. Lanes M, molecular size markers. (C) Map of the germ line sequence showing the positions of primers o4 and o5 (filled arrowheads). These primers lie outside the rearrangement region, since no macronuclear heterogeneity was detected between B1 and o4 on Southern blots (data not shown). Horizontal lines below the map represent the micronuclear amplification products for the entire set of overlapping nested PCRs.
In a second step, the broad bands corresponding to versions 2a and 2b were gel purified and used as templates for a series of overlapping nested PCRs spanning the entire region. As controls, the same PCRs were performed in parallel with purified PCR products of the micronuclear sequence and with total cell DNA. Figure 4B illustrates the principle of the method with one example of a deletion observed in version 2a. If no deletion occurs between the two primers used (primers a and b), a single product of the same size is obtained with all templates: the gel-purified 2a and micronuclear templates and total DNA. The different amounts reflect the different concentrations of molecules containing this sequence in the three template samples. If at least one primer is located within the deletion (primers c and d), only the micronuclear sequence can be amplified. The single, low-abundance product of micronuclear size that is amplified from total DNA is consistent with the low micronuclear/macronuclear ploidy ratio (compare the relative intensities of the micronuclear and total DNA products in the a-b and c-d amplifications). The product of micronuclear size is also the main product amplified from the 2a template; this result could have been due to contamination of the gel-purified template with the closely migrating micronuclear sequence (amplified from total DNA in the first-step PCR with primers o4 and o5) and/or to rare 2a variants that do not show this deletion. A less abundant, shorter product indeed suggests that one rare 2a variant contains both primers with a smaller deletion between them. Finally, if the two primers used are located on either side of the major deletion (primers c and e), abundant shorter products are made from the 2a template and total DNA. The abundance and size heterogeneity of PCR products obtained with primers c and e indicate that most 2a molecules have an ∼1.6- to 2.8-kb deletion between primers c and e, with boundaries that vary in position over ≥0.6 kb but cluster at a few preferential hot spots, as revealed by prominent bands in the pattern.
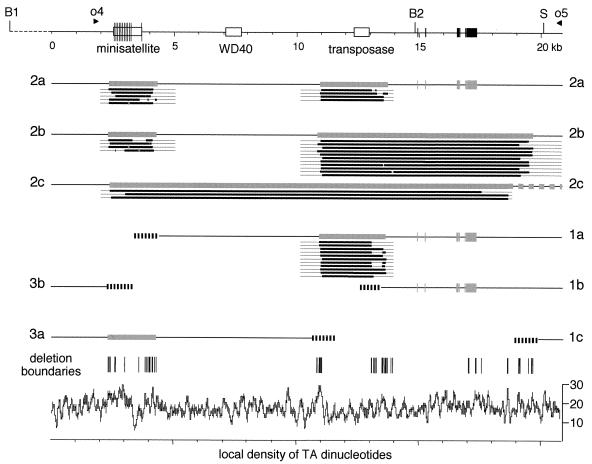
The whole set of overlapping PCRs used is diagrammed in Fig. 4C. A careful examination of the patterns produced allowed us to determine the approximate positions of the major large deletions occurring in versions 2a and 2b (Fig. 5). All of them displayed heterogeneity in the exact positions of boundaries, similar to the example described above. The main deletions were found to be discontinuous: two separate segments are deleted in both version 2a and version 2b, together accounting for most of the measured deletion lengths (the six IESs identified account for 0.7 kb of the deletions in version 2a). An ∼1.0- to 1.7-kb deletion is common to versions 2a and 2b. It removes the minisatellite and GC-rich sequence together with variable lengths of flanking sequences. The second deletion in version 2a (the example described above) is ∼1.6 to 2.8-kb long and removes the truncated Tennessee copy and adjacent sequences. In version 2b, the second deletion starts at the same position on the left side of the transposon but extends much further to the right, removing 8 to 9 kb of DNA.
FIG. 5.
Maps of main alternative rearrangements. The major macronuclear versions are represented by labeled lines (2a, 2b, 2c, 1a, and so forth) below the map of the germ line sequence (top line). Gray boxes in the macronuclear versions show the approximate positions of imprecise internal deletions of the minisatellite and truncated Tennessee copy, as determined by mass PCR mapping. The right end of the large deletion in version 2c is uncertain, as indicated by the disconnected gray boxes. Thick black lines below gray boxes indicate the exact extents of the deletions in individual molecules sequenced from cloned PCR products; thin lines on both sides show the regions covered by the PCRs. The six invariant IESs are shown as black boxes in the germ line sequence; gray boxes in the macronuclear versions indicate the positions of the corresponding precise deletions of the IESs. Hatched regions at the ends of fragmented chromosomes 1 and 3 represent telomere addition regions. The graph at the bottom shows the local density of TA dinucleotides along the germ line sequence (percentage of total dinucleotides computed in a 100-bp window); the short vertical lines above the graph mark the positions of the boundaries of internal deletions for all sequenced molecules (excluding the IESs).
Figure 5 also shows three examples of version 2c deletions that were characterized from cloned molecules in a separate experiment (data not shown). Note that these examples are not representative of version 2c, as the molecules analyzed were among the largest. Most version 2c deletions are larger and must extend further to the right, since there is no evidence for alternative rearrangements to the left of the mapped deletions. The SacI site on the right is indeed deleted in a fraction of version 2c deletions (4). Thus, version 2c deletions are approximately coextensive with version 2b deletions, but the two segments that are deleted separately in version 2b molecules are here bridged in a single large deletion which also removes the ∼6-kb segment of DNA between them.
PCR mapping of regions of telomere additions.
The lengths of the alternative telomeric fragments of chromosomes 1 and 3 could be determined quite precisely by Southern blot mapping of genomic DNA, but whether these fragments were colinear with the germ line sequence up to the telomeric repeats was not known. To characterize their structure and map the telomere addition regions in the germ line sequence, chromosome ends were first amplified from total cell DNA by long-range PCR with one degenerate primer complementary to the G strand of telomeric repeats (see Materials and Methods) and a second primer specific for the region. Different PCRs were used to selectively amplify one or more of the alternative telomeres from each chromosome, which could be identified by their known sizes; all but the 1d telomere were amplified. These telomeric PCR products must be revealed with specific probes on Southern blots, as the telomeric primer by itself is able to amplify a large amount of DNA from many different locations in the genome. The Southern blots were successively hybridized with various oligonucleotide probes chosen from the germ line sequence to scan the expected extent of the telomeric fragments and beyond. With probes located upstream of the telomeres, each of the five amplified fragments appeared as a heterogeneous cluster of bands varying in size by ∼1 kb (data not shown), consistent with the usual microheterogeneity of telomere addition sites. Oligonucleotide probes located beyond the telomeres failed to hybridize, while those located within the telomere addition regions hybridized only to the longest molecules in the heterogeneous patterns, providing direct information about the locations of these regions in the germ line sequence.
Figure 5 shows the positions of the five telomere addition regions analyzed. This analysis revealed the occurrence of internal deletions in the longest versions of chromosomes 1 and 3. Indeed, telomere addition regions 1a and 3a were found to map further in the germ line sequence than was anticipated from the known lengths of the telomeric fragments. Furthermore, some probes located at internal positions in the fragments failed to hybridize to the telomeric PCR products, indicating the positions of internal deletions. Remarkably, these deletions were found to be the same as some of the chromosome 2 internal deletions. Telomeric fragment 3a appeared to carry the same internal deletion of the minisatellite and GC-rich sequence as versions 2a and 2b, and the internal deletion in fragment 1a was the same as the second deletion in version 2a (Fig. 5). The position, extent, and heterogeneity of the internal deletion in fragment 1a were further characterized by the same set of overlapping nested PCRs as those used to analyze the fragment 2a deletion. In particular, the c-e nested amplification produced nearly identical heterogeneous patterns with the 1a telomeric PCR product as a template and with the 2a purified template, confirming that the same 1.6- to 2.8-kb imprecise deletion of the truncated Tennessee copy occurs in versions 1a and 2a.
Several important conclusions can be drawn from the complex pattern of alternative rearrangements presented in Fig. 5. First, the minisatellite and truncated Tennessee copy are removed from all macronuclear copies of the region, including both religated versions (chromosome 2) and fragmented versions (chromosomes 1 and 3). Second, all of the telomere addition regions that were mapped in the region colocalize with the microheterogeneous boundaries of internal deletions, and conversely, all internal deletion boundaries colocalize with telomere addition regions. These data suggest that each of the alternative DNA elimination events can lead either to the ligation of flanking sequences or to the formation of telomeres. Third, an unexpected consequence of alternative processing at two distinct deletion regions is the formation of macronuclear chromosomes with overlapping ends: chromosome 3a implies rejoining at the minisatellite and telomerization at the Tennessee copy, while chromosome 1a implies telomerization at the minisatellite and rejoining at the Tennessee copy.
Sequence analysis of internal deletion junctions and telomere addition sites.
Because of their heterogeneity, the junctions of internal deletions can be studied only in cloned molecules. Although many sequences from the region appear to be unstable in different cloning vectors (see Materials and Methods), some short PCR products could be cloned, and a number of representative molecules were sequenced for each of the main deletions. The sequences confirmed the microheterogeneity evidenced by PCR mapping; Fig. 5 shows the precise extents of the deletions in each of the sequenced molecules. One unexpected finding is that the large deletions of the minisatellite and truncated Tennessee copy that characterize each chromosome 2 version are themselves discontinuous in about one-third of the sequenced molecules, where they actually consist of two or more closely spaced deletions. The distance between adjacent deletions can be as short as 13 bp.
A comparison of the junction sequences with the germ line sequence indicated that in all instances, the deletions occur between two short direct repeats (1 to 8 bp), one of which is maintained in the macronuclear sequence (Fig. 6; the sequences can also be obtained at www.biologie.ens.fr/∼lemouel/lemouel1.doc). The repeats differ in sequence from one deletion event to the next but, strikingly, they all contain almost exclusively thymines and adenines. Furthermore, with one possible exception (which might be due to a PCR artifact), all boundary repeats contain at least one 5′-TA-3′ dinucleotide. The sequencing also confirmed that the boundaries of internal deletions tend to cluster in short segments particularly rich in TA dinucleotides. A plot of the local density of TA dinucleotides, computed in a 100-bp window sliding along the germ line sequence, indeed revealed that most deletion boundaries coincide with peaks in TA density (Fig. 5). There appear to be few other constraints on the use of TA-containing direct repeats as deletion boundaries: in many instances, the same boundary on one side is used in conjunction with different boundaries on the other side; furthermore, the same sequence can be used both as a left boundary and as a right boundary. For instance, the TA in the sequence ATCTAAAATACAATGAAT is the right boundary repeat of one deletion, while the overlapping AATA (ATCTAAAATACAATGAAT) is the left boundary repeat of another.
FIG. 6.
Direct repeats at boundaries of imprecise internal deletions. The top line shows the sequence of one example of a deletion junction; the single repeat maintained in the macronuclear sequence is in bold type. Line 1 shows the germ line sequence around the direct repeats at the right and left boundaries of this deletion; the deleted sequence is shown in lowercase letters. The entire set of sequenced deletions is represented in the same way. Deletions 1 to 8 and 9 to 11 remove the minisatellite in versions 2a and 2b, respectively; deletions 12 to 16, 17 to 29, and 30 to 41 occur around the truncated Tennessee copy in versions 2a, 1a, and 2b, respectively.
For some of the internal deletions, the left boundary was located within the minisatellite, so that a few minisatellite repeats were maintained in the macronuclear sequence. In two instances, one of the macronuclear repeats differed in sequence from the corresponding repeat in the germ line sequence. These variant repeats are unlikely to be simply due to PCR-induced substitutions, since one of them was also present in telomeric PCR products from a different genomic region (see below).
To determine the exact positions of telomere addition sites in a sample of molecules from chromosomes 1 and 3, telomeric PCR products also had to be cloned. However, these sequences appeared to be very unstable in plasmid vectors, and only seven clones corresponding to the 3b telomere could be obtained (data not shown). Their sequencing confirmed the PCR mapping of the 3b telomere addition region; the longest molecules contained some minisatellite repeats before the telomeric repeats. Two clones contained a few variant repeats that were absent from the germ line sequence. Such variant repeats may have been incorporated by trans-PCR artifacts involving a template switch to similar repeats present at the ends of other macronuclear chromosomes. Indeed, one of the primers used for telomeric PCRs had partial homology with minisatellite repeats and yielded clones containing a different set of variant repeats.
Caryonidal variability and experimental modifications of rearrangement patterns.
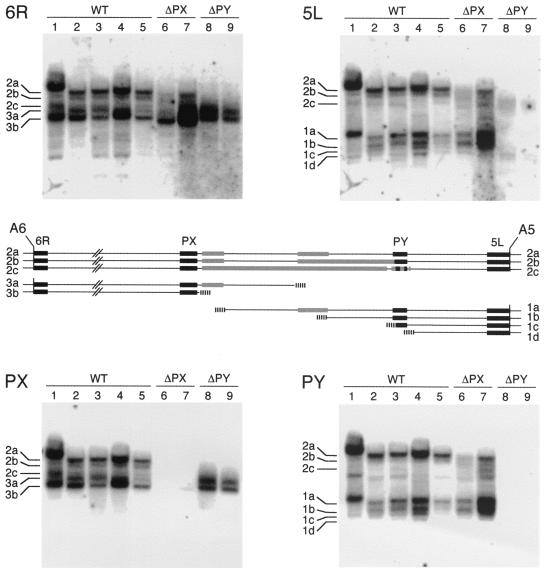
Up to this point, the mapping and sequencing of macronuclear versions were carried out with samples from the reference clone. To examine the caryonidal variability of the complex rearrangement patterns, total DNAs from four independent clones of entirely homozygous strain 156 were digested with Asp718 and analyzed as described in the legend to Fig. 2. A Southern blot of the pulsed-field electrophoresis gel is shown in Fig. 7. After hybridization with probe 6R, which reveals the different versions of chromosomes 2 and 3, or with probe 5L, which reveals the different versions of chromosomes 1 and 2, these four clones (Fig. 7, lanes 1 and 3 to 5) yielded patterns similar to that of the previously studied reference clone (lane 2). However, variations were observed in the numbers, relative intensities, and exact sizes of the different fragments. In particular, clone 1 showed reduced heterogeneity with probe 5L, which revealed a single major version each for chromosomes 1 and 2. Furthermore, these major versions are both slightly larger than their counterparts in the reference clone, versions 1a and 2a. This observation further supports the idea that alternative rearrangements are determined in a coordinated manner in fragmented and nonfragmented versions. Interestingly, the variant pattern of clone 1 was reproduced in its sexual progeny, as shown by an analysis of one postautogamy caryonidal clone (data not shown). Since all clones have identical micronuclear genomes, such inheritance of variant patterns must depend on epigenetic mechanisms, as previously shown for maternally inherited alternative rearrangements of other genomic regions (46).
FIG. 7.
Varibility and experimental modifications of rearrangement patterns. A Southern blot of a pulsed-field electrophoresis gel of Asp718-digested total DNAs from different clones was hybridized successively with probes 6R, 5L, PX, and PY. Lanes 1 to 5, wild-type (WT) caryonidal clones; lanes 6 and 7, ΔPX clones; lanes 8 and 9, ΔPY clones. The different fragments indicated on the sides of the blots are those of the reference clone, which is in lane 2. The map shows the positions of the probes (black boxes) between Asp718 sites A5 and A6 in the main macronuclear versions of the reference clone. Gray boxes represent the average extents of the heterogeneous internal deletions; hatched boxes represent telomere addition regions.
To further test the coordinated regulation of DNA elimination events in fragmented and nonfragmented versions, we took advantage of the fact that targeted deletions of any sequence can be experimentally induced in the developing macronucleus by transformation of the maternal macronucleus with high copy numbers of that sequence prior to the induction of autogamy. Targeting of a sequence located close to the end of a macronuclear chromosome results in a terminal truncation of this chromosome in the new macronucleus, while targeting of an internal sequence leads to imprecise internal deletions slightly larger than the targeted region (42, 43; Garnier et al., submitted). Here we chose to target sequences PX and PY, which are internal sequences in chromosome 2 but which also lie close to the ends of chromosomes 3 and 1, respectively (Fig. 7). Plasmids carrying the PX or PY sequences were microinjected into the macronuclei of vegetative cells of strain 156. Selected high-copy-number transformants (>40,000 copies per macronucleus) were cultured and allowed to undergo autogamy, and total DNA was extracted from sexual progeny to check the structure of the new macronuclear genome.
Figure 7 shows the patterns obtained for two caryonidal clones derived from PX-transformed cells (ΔPX clones; lanes 6 and 7) and two caryonidal clones derived from PY-transformed cells (ΔPY clones; lanes 8 and 9). As expected, hybridization of the Southern blot with probes PX and PY revealed that these sequences are almost entirely deleted from the macronuclear genomes of ΔPX and ΔPY clones, respectively. Hybridization with probe 6R showed that the 3a telomere had a terminal truncation in ΔPX clones, while probe 5L indicated that chromosome 1 telomeres were not affected. Although the reduction in the size of the 3a telomeric fragment is barely noticeable on the pulsed-field Southern blot in Fig. 7, the terminal deletion was more precisely mapped on classical blots with other enzymes and found to be ∼3 kb (data not shown). Similarly, probe 6R revealed that the entire group of bands from chromosome 2 versions was ∼3 to 4 kb shorter in the ΔPX clones, consistent with the PX sequence being deleted as part of an extended deletion of the minisatellite region. Targeting of the PX sequence for deletion appeared to favor fragmentation over religation; quantification of the copy numbers of chromosomes 1, 2, and 3 by pulsed-field electrophoresis of native chromosomes showed that religated chromosome 2 versions represent only 26 to 29% of the total copy number in ΔPX clones, as opposed to 50 to 58% in control clones (data not shown).
In contrast to the PX sequence, which is never deleted in wild-type clones, the PY sequence largely overlaps the 8- to 9-kb extended deletion of the Tennessee copy in version 2b (Fig. 7). In ΔPY clones, the targeted PY deletion appeared to extend over a large distance on the right in both chromosome 1 telomeric fragments and chromosome 2 internal fragments, whereas chromosome 3 remained unaffected, as revealed by probe 6R. The reduced signal intensity obtained with probe 5L indicates that in many copies, the deletion extended into the 5L sequence. We conclude from this experiment that targeting of the sequences located on either side of the variable rearrangement region increases the sizes of the deletions in a coordinated manner in fragmented and rejoined versions.
DISCUSSION
Micronucleus-specific repeated sequences in a chromosome fragmentation region.
In an effort to understand how chromosome fragmentation is determined during macronuclear development in Paramecium, we studied the micronuclear (germ line) sequence of an ∼21-kb fragmentation region for which flanking macronuclear chromosomes were known on both sides. Two distinct sequence elements were found to be eliminated from all macronuclear copies in the region. One is an AT-rich minisatellite flanked by a short GC-rich sequence; the other is a mutated and truncated copy of Tennessee, a novel transposable element of the Tc1/mariner family. The two elements share no common sequence, but they share the characteristic of being multicopy in the germ line genome. In addition to the local repetition of the minisatellite motif, the GC-rich sequence itself appears to be multicopy, since a very similar sequence was found, entirely by chance, within the transposase gene of one Tennessee copy located elsewhere in the genome. Consistent with the general paucity of repeated sequences in the macronuclear genomes of ciliates, the vast majority, if not all, of the copies of these elements are deleted during macronuclear development.
How general is chromosome fragmentation as a consequence of imprecise sequence elimination?
The two germ line-specific elements are eliminated in a variety of alternative ways. The mapping and sequence analysis of the major internal deletions in the macronucleus of one caryonidal clone revealed that each of them has microheterogeneous boundaries, spread over at least 0.5 kb. This clustered distribution of deletion breakpoints is very similar to that of telomere addition sites in fragmented versions. Furthermore, all of the telomere addition regions that were mapped colocalized with heterogeneous boundaries of internal deletions. This finding also appears to be true for clones with slightly different rearrangement patterns as well as for clones where deletions were experimentally lengthened by specific targeting of the PX or PY sequences located at the borders of the fragmentation region. Thus, the same deletion events appear to be healed either by the rejoining of flanking sequences or by telomere addition. The data suggest that rearrangements are initiated by the elimination of specific germ line sequences and that chromosome fragmentation occurs only when flanking sequences fail to be rejoined for at least one of the deletion events in the region. One unexpected consequence of such a mechanism operating on two distinct elements is the formation of macronuclear chromosomes with overlapping ends, similar to those occurring in Oxytricha (62).
Could all fragmentation events result from imprecise elimination of repeated sequences? Facultative rejoining of flanking sequences has not been reported for other fragmentation regions, but many macronuclear chromosomes are known to use alternative telomere addition regions in P. primaurelia and P. tetraurelia. In most instances, the possibility cannot be excluded that some of the alternative forms are in fact rejoined versions, as restriction fragments with heterogeneous internal deletions have the same smeared appearance as telomeric fragments on Southern blots. Strong evidence for facultative religation can be obtained only by physical mapping of entire macronuclear chromosomes, as was done in the present study by using jumping and linking libraries to clone probes from the entire length of chromosome 2 (4). Although such studies are lacking for other chromosomes, pulsed-field Southern blots of native chromosomes have shown that a number of single-copy genes are borne by several macronuclear chromosomes with widely different sizes (50), suggesting that facultative fragmentation may not be uncommon. Finally, we showed that increasing the lengths of deletions in the region studied favors fragmentation over religation. Simple macronuclear telomeres might thus be produced when the deletion of very large germ line regions precludes the rejoining of flanking sequences.
Imprecise DNA elimination mechanism distinct from IES excision.
The heterogeneity of internal deletions occurring in the fragmentation region stands in sharp contrast to the precise excision of IESs, another type of developmentally regulated DNA elimination occurring in Paramecium and other ciliates. Although some IESs have been shown to use alternative excision boundaries a few nucleotides apart (15, 38), a higher level of junction microheterogeneity probably would be lethal, since IESs very frequently are present within coding sequences. Furthermore, IES excision is always followed by the rejoining of flanking sequences; the frequent addition of telomeres to flanking sequences would be disastrous for the same reason.
All precisely excised IESs that have been sequenced so far in Paramecium are short (26 to 882 bp), single-copy DNA segments. However, full-length DNA transposons can be excised precisely in Oxytricha (27, 61), and in Euplotes the same precise mechanism appears to excise both short, single-copy IESs and large transposons (31, 37). Some Tennessee copies in the Paramecium genome may well be excised precisely, but this event would probably require the presence of both terminal IRs. Indeed, the precise excision of Paramecium IESs can be abolished by point mutations in an 8-bp degenerate consensus sequence which is present at both ends in opposite orientations (24, 41) and which is thought to have been conserved from the presumed transposon ancestors of single-copy IESs (35). The truncated Tennessee copy studied here lacks one terminal IR, and the imprecise deletion boundaries lie outside the element, within flanking sequences. Interestingly, the deletions of the truncated Tennessee copy are similar in this respect to the heterogeneous deletions that are promoted by various fragments of the Tlr transposable element in Tetrahymena (63).
These deletions nevertheless show one similarity with the precise excision of IESs. They occur between two short direct repeats, one of which is maintained at the deletion junction; the repeats are 2 to 8 bp long and, with one possible exception, always contain at least one 5′-TA-3′ dinucleotide. This finding is highly significant: in the 7 instances (out of 41) where repeats are limited to two nucleotides, the repeat sequence is TA. IESs are also invariably bounded by two TA dinucleotides, one of which is maintained in the macronuclear sequence after precise excision. A point mutation in one of the TA dinucleotides abolishes developmental excision (40, 53), and the TA dinucleotides were recently shown to be located in the middle of 4-base 5′ overhangs produced by staggered double-strand breaks at both IES ends during excision (23a). Thus, both types of DNA elimination may use the same basic, TA-dependent recombination machinery, with additional mechanisms ensuring the precision of IES excision. It is unclear whether a consensus sequence similar to that of IES ends is present internal to the TA boundaries of imprecise deletions, because the precise TA dinucleotide used cannot be determined when there is more than one in the direct repeats. TA boundaries, however, are used in a much more flexible manner, since the same repeats can be used as right boundaries in some molecules and as left boundaries in others.
Multiple cut sites versus exonuclease action.
A previous model proposed that, after the introduction of a single double-strand break in the fragmentation region, the DNA ends could be degraded by an exonuclease to variable extents and then either rejoined or healed by telomere addition (4). The finding that most chromosome 2 molecules carry at least two distinct deletions does not support such a model. Furthermore, each of the two deletions in versions 2a and 2b itself can be discontinuous. For instance, although the same 8- to 9-kb region around the Tennessee copy is reproducibly deleted in version 2b, the deletion is sometimes achieved by two or more smaller deletions, separated by as little as 80 bp of maintained DNA (13 bp in minisatellite deletions). Thus, the 8- to 9-kb deletion itself is not likely to be initiated by a single cut followed by exonuclease action but could result from repeated rounds of smaller deletions targeting this region. Each deletion event could be initiated by two cuts at TA dinucleotides, which would have to be kept in close spatial proximity to allow the religation of flanking sequences. Alternatively, TA stretches could be exonuclease pause sites or barriers; cuts then could be located elsewhere, but they would still have to be multiple to account for discontinuous deletions.
Could the same cuts be used for telomere addition? The Paramecium telomerase is indeed able to add telomeres to the end of any DNA molecule, without extensive resection (23). However, the sequencing of a few telomeric molecules in this study showed that telomeric repeats are not added at a fixed position relative to the nearest TA dinucleotide in the germ line sequence (data not shown). Thus, if both rearrangement types were initiated by cuts at TA dinucleotides, some trimming of the ends would have to precede telomere addition. Alternatively, random cuts could be substrates for direct telomere addition or for an exonuclease pausing in TA stretches to allow rejoining. The rejoining or telomere addition alternative may involve the nonhomologous end-joining repair pathway, since one of its components, the Ku heterodimer, was also proposed to recruit telomerase to DNA ends in yeasts (49) and mammals (8).
Epigenetic programming of imprecise deletions.
If such low-specificity cleavage is involved, what determines the specificity of the deletions observed? The variability of rearrangement patterns and the inheritance of variant patterns in genetically identical cell lines imply epigenetic mechanisms. Both minisatellites and multicopy transposable elements are often packaged as heterochromatin in eukaryotes, and many lines of evidence support the idea that heterochromatin is specifically deleted during the development of the ciliate macronucleus (13, 16, 22, 28, 30, 39; reviewed in reference 48). For Tetrahymena, it was recently shown that DNA elimination is targeted by the methylation of histone H3 on lysine 9, a modification that is absent from the mature macronucleus (60). The heterogeneous lengths of the deletions occurring in the fragmentation region could reflect the variable spreading of such a heterochromatin mark nucleated in the two germ line-specific, multicopy elements.
What then would determine the formation of heterochromatin over these sequences? The imprecise deletions studied here are strikingly similar to the deletions that can be induced experimentally at any locus by high-copy-number transformation of the maternal macronucleus. The latter also occur between short, TA-containing direct repeats, and their heterogeneous boundaries similarly cluster in TA-rich recombination hot spots located outside the targeted sequence (43). We have shown that such homology-dependent maternal effects can increase the sizes of deletions in the fragmentation region. Transformation of the macronucleus with transgenes at high copy numbers was shown to result in the production of aberrant transcripts from both strands, even when these constructs lacked transcription promoters (21). The formation of double-stranded RNA could explain the silencing of homologous genes observed during the vegetative growth of such transformants, as well as the deletions induced in their sexual progeny, during the development of the new macronuclei. Direct delivery of double-stranded RNA prior to meiosis indeed results in very similar deletions during macronuclear development (Garnier et al., submitted). Thus, the spontaneous deletions of the minisatellite region and truncated Tennessee copy also may be programmed ultimately by homologous RNA molecules, which could be transcribed from these or any other sufficiently similar loci in the germ line genome.
A very similar model has been proposed to explain the capacity of different, nonoverlapping fragments of the Tlr multicopy element of Tetrahymena to determine their own elimination during macronuclear development (63). In this organism, some germ line-specific sequences were indeed shown to be transcribed on both strands before they were deleted (9), and indirect evidence implicates short, 26- to 31-nucleotide RNA molecules that accumulate during macronuclear development in the programmed rearrangements of the genome (47). More generally, there is increasing evidence from diverse eukaryotes that RNA molecules can modify the chromatin structure of homologous genomic sequences (reviewed in reference 7): developmentally regulated chromosome fragmentation in Paramecium may be one manifestation of a basic mechanism which is conserved in eukaryotes and which appears to be essential for the control of transposable elements.
Acknowledgments
This work was initiated by the late François Caron. We thank all members of our laboratory for fruitful discussions and critical reading of the manuscript and Edouard Bray for help with formatting the downloadable data.
This work was supported by the Association pour la Recherche sur le Cancer (grant no. 5733), the Centre National de la Recherche Scientifique (Soutien aux Jeunes Équipes), and the Comité de Paris de la Ligue Nationale contre le Cancer (grant no. 75/01-RS/73).
REFERENCES
- 1.Amar, L. 1994. Chromosome end formation and internal sequence elimination as alternative genomic rearrangements in the ciliate Paramecium. J. Mol. Biol. 236:421-426. [DOI] [PubMed] [Google Scholar]
- 2.Baird, S. E., and L. A. Klobutcher. 1989. Characterization of chromosome fragmentation in two protozoans and identification of a candidate fragmentation sequence in Euplotes crassus. Genes Dev. 3:585-597. [DOI] [PubMed] [Google Scholar]
- 3.Baroin, A., A. Prat, and F. Caron. 1987. Telomeric site position heterogeneity in macronuclear DNA of Paramecium primaurelia. Nucleic Acids Res. 15:1717-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caron, F. 1992. A high degree of macronuclear chromosome polymorphism is generated by variable DNA rearrangements in Paramecium primaurelia during macronuclear differentiation. J. Mol. Biol. 225:661-678. [DOI] [PubMed] [Google Scholar]
- 5.Caron, F., and E. Meyer. 1989. Molecular basis of surface antigen variation in Paramecia. Annu. Rev. Microbiol. 43:23-42. [DOI] [PubMed] [Google Scholar]
- 6.Cartinhour, S. W., and G. A. Herrick. 1984. Three different macronuclear DNAs in Oxytricha fallax share a common sequence block. Mol. Cell. Biol. 4:931-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerutti, H. 2003. RNA interference: traveling in the cell and gaining functions? Trends Genet. 19:39-46. [DOI] [PubMed] [Google Scholar]
- 8.Chai, W., L. P. Ford, L. Lenertz, W. E. Wright, and J. W. Shay. 2002. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J. Biol. Chem. 277:47242-47247. [DOI] [PubMed] [Google Scholar]
- 9.Chalker, D. L., and M. C. Yao. 2001. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 15:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu, G., D. Vollrath, and R. W. Davis. 1986. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234:1582-1585. [DOI] [PubMed] [Google Scholar]
- 11.Conover, R. K., and C. F. Brunk. 1986. Macronuclear DNA molecules of Tetrahymena thermophila. Mol. Cell. Biol. 6:900-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coyne, R. S., D. L. Chalker, and M. C. Yao. 1996. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu. Rev. Genet. 30:557-578. [DOI] [PubMed] [Google Scholar]
- 13.Coyne, R. S., M. A. Nikiforov, J. F. Smothers, C. D. Allis, and M. C. Yao. 1999. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol. Cell 4:865-872. [DOI] [PubMed] [Google Scholar]
- 14.Doak, T. G., F. P. Doerder, C. L. Jahn, and G. Herrick. 1994. A proposed superfamily of transposase genes: transposon-like elements inciliated protozoa and a common “D35E” motif. Proc. Natl. Acad. Sci. USA 91:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubrana, K., A. Le Mouel, and L. Amar. 1997. Deletion endpoint allele-specificity in the developmentally regulated elimination of an internal sequence (IES) in Paramecium. Nucleic Acids Res. 25:2448-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duharcourt, S., and M. C. Yao. 2002. Role of histone deacetylation in developmentally programmed DNA rearrangements in Tetrahymena thermophila. Eukaryot. Cell 1:293-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epstein, L. M., and J. D. Forney. 1984. Mendelian and non-Mendelian mutations affecting surface antigen expression in Paramecium tetraurelia. Mol. Cell. Biol. 4:1583-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan, Q., and M. Yao. 1996. New telomere formation coupled with site-specific chromosome breakage in Tetrahymena thermophila. Mol. Cell. Biol. 16:1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan, Q., and M. C. Yao. 2000. A long stringent sequence signal for programmed chromosome breakage inTetrahymena thermophila. Nucleic Acids Res. 28:895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forney, J. D., and E. H. Blackburn. 1988. Developmentally controlled telomere addition in wild-type and mutant paramecia. Mol. Cell. Biol. 8:251-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galvani, A., and L. Sperling. 2001. Transgene-mediated post-transcriptional gene silencing is inhibited by 3′ non-coding sequences in Paramecium. Nucleic Acids Res. 29:4387-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh, S., and L. A. Klobutcher. 2000. A development-specific histone H3 localizes to the developing macronucleus of Euplotes. Genesis 26:179-188. [DOI] [PubMed] [Google Scholar]
- 23.Gilley, D., J. R. Preer, Jr., K. J. Aufderheide, and B. Polisky. 1988. Autonomous replication and addition of telomere-like sequences to DNA microinjected into Paramecium tetraurelia macronuclei. Mol. Cell. Biol. 8:4765-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Giratias, A., and M. Bétermier. 2003. Processing of double-strand breaks is involved in the precise excision of Paramecium internal eliminated sequences. Mol. Cell. Biol. 23:7152-7162. [DOI] [PMC free article] [PubMed]
- 24.Haynes, W. J., K. Y. Ling, R. R. Preston, Y. Saimi, and C. Kung. 2000. The cloning and molecular analysis of pawn-B in Paramecium tetraurelia. Genetics 155:1105-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrick, G., S. W. Cartinhour, K. R. Williams, and K. P. Kotter. 1987. Multiple sequence versions of the Oxytricha fallax 81-MAC alternate processing family. J. Protozool. 34:429-434. [DOI] [PubMed] [Google Scholar]
- 26.Herrick, G., D. Hunter, K. Williams, and K. Kotter. 1987. Alternative processing during development of a macronuclear chromosome family in Oxytricha fallax. Genes Dev. 1:1047-1058. [DOI] [PubMed] [Google Scholar]
- 27.Hunter, D. J., K. Williams, S. Cartinhour, and G. Herrick. 1989. Precise excision of telomere-bearing transposons during Oxytricha fallax macronuclear development. Genes Dev. 3:2101-2112. [DOI] [PubMed] [Google Scholar]
- 28.Jahn, C. L. 1999. Differentiation of chromatin during DNA elimination in Euplotes crassus. Mol. Biol. Cell 10:4217-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jahn, C. L., and L. A. Klobutcher. 2002. Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 56:489-520. [DOI] [PubMed] [Google Scholar]
- 30.Jahn, C. L., Z. Ling, C. M. Tebeau, and L. A. Klobutcher. 1997. An unusual histone H3 specific for early macronuclear development in Euplotes crassus. Proc. Natl. Acad. Sci. USA 94:1332-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaraczewski, J. W., and C. L. Jahn. 1993. Elimination of Tec elements involves a novel excision process. Genes Dev. 7:95-105. [DOI] [PubMed] [Google Scholar]
- 32.Keller, A. M., A. Le Mouël, F. Caron, M. Katinka, and E. Meyer. 1992. The differential expression of the G surface antigen alleles in Paramecium primaurelia heterozygous cells correlates to macronuclear DNA rearrangement. Dev. Genet. 13:306-317. [DOI] [PubMed] [Google Scholar]
- 33.Klobutcher, L. A. 1999. Characterization of in vivo developmental chromosome fragmentation intermediates in E. crassus. Mol. Cell 4:695-704. [DOI] [PubMed] [Google Scholar]
- 34.Klobutcher, L. A., S. E. Gygax, J. D. Podoloff, J. R. Vermeesch, C. M. Price, C. M. Tebeau, and C. L. Jahn. 1998. Conserved DNA sequences adjacent to chromosome fragmentation and telomere addition sites in Euplotes crassus. Nucleic Acids Res. 26:4230-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klobutcher, L. A., and G. Herrick. 1995. Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res. 23:2006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klobutcher, L. A., and G. Herrick. 1997. Developmental genome reorganization in ciliated protozoa: the transposon link. Prog. Nucleic Acid Res. Mol. Biol. 56:1-62. [DOI] [PubMed] [Google Scholar]
- 37.Klobutcher, L. A., L. R. Turner, and J. LaPlante. 1993. Circular forms of developmentally excised DNA in Euplotes crassus have a heteroduplex junction. Genes Dev. 7:84-94. [DOI] [PubMed] [Google Scholar]
- 38.Ling, K. Y., B. Vaillant, W. J. Haynes, Y. Saimi, and C. Kung. 1998. A comparison of internal eliminated sequences in the genes that encode two K(+)-channel isoforms in Paramecium tetraurelia. J. Eukaryot. Microbiol. 45:459-465. [DOI] [PubMed] [Google Scholar]
- 39.Madireddi, M. T., R. S. Coyne, J. F. Smothers, K. M. Mickey, M. C. Yao, and C. D. Allis. 1996. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination inTetrahymena. Cell 87:75-84. [DOI] [PubMed] [Google Scholar]
- 40.Mayer, K. M., and J. D. Forney. 1999. A mutation in the flanking 5′-TA-3′ dinucleotide prevents excision of an internal eliminated sequence from the Paramecium tetraurelia genome. Genetics 151:597-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayer, K. M., K. Mikami, and J. D. Forney. 1998. A mutation in Paramecium tetraurelia reveals functional and structural features of developmentally excised DNA elements. Genetics 148:139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer, E. 1992. Induction of specific macronuclear developmental mutations by microinjection of a cloned telomeric gene in Paramecium primaurelia. Genes Dev. 6:211-222. [DOI] [PubMed] [Google Scholar]
- 43.Meyer, E., A. Butler, K. Dubrana, S. Duharcourt, and F. Caron. 1997. Sequence-specific epigenetic effects of the maternal somatic genome on developmental rearrangements of the zygotic genome in Paramecium primaurelia. Mol. Cell. Biol. 17:3589-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer, E., and S. Duharcourt. 1996. Epigenetic programming of developmental genome rearrangements in ciliates. Cell 87:9-12. [DOI] [PubMed] [Google Scholar]
- 45.Meyer, E., and S. Duharcourt. 1996. Epigenetic regulation of programmed genomic rearrangements in Paramecium aurelia. J. Eukaryot. Microbiol. 43:453-461. [DOI] [PubMed] [Google Scholar]
- 46.Meyer, E., and O. Garnier. 2002. Non-Mendelian inheritance and homology-dependent effects in ciliates. Adv. Genet. 46:305-337. [DOI] [PubMed] [Google Scholar]
- 47.Mochizuki, K., N. A. Fine, T. Fujisawa, and M. A. Gorovsky. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110:689-699. [DOI] [PubMed] [Google Scholar]
- 48.Nikiforov, M. A., M. A. Gorovsky, and C. D. Allis. 2000. A novel chromodomain protein, pdd3p, associates with internal eliminated sequences during macronuclear development in Tetrahymena thermophila. Mol. Cell. Biol. 20:4128-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peterson, S. E., A. E. Stellwagen, S. J. Diede, M. S. Singer, Z. W. Haimberger, C. O. Johnson, M. Tzoneva, and D. E. Gottschling. 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 27:64-67. [DOI] [PubMed] [Google Scholar]
- 50.Phan, H. L., J. Forney, and E. H. Blackburn. 1989. Analysis of Paramecium macronuclear DNA using pulsed field gel electrophoresis. J. Protozool. 36:402-408. [DOI] [PubMed] [Google Scholar]
- 51.Preer, L. B., G. Hamilton, and J. R. Preer, Jr. 1992. Micronuclear DNA from Paramecium tetraurelia: serotype 51 A gene has internally eliminated sequences. J. Protozool. 39:678-682. [DOI] [PubMed] [Google Scholar]
- 52.Prescott, D. M. 1994. The DNA of ciliated protozoa. Microbiol. Rev. 58:233-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruiz, F., A. Krzywicka, C. Klotz, A. Keller, J. Cohen, F. Koll, G. Balavoine, and J. Beisson. 2000. The SM19 gene, required for duplication of basal bodies in Paramecium, encodes a novel tubulin, eta-tubulin. Curr. Biol. 10:1451-1454. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Scott, J., C. Leeck, and J. Forney. 1993. Molecular and genetic analyses of the B type surface protein gene from Paramecium tetraurelia. Genetics 134:189-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seegmiller, A., K. R. Williams, and G. Herrick. 1997. Two two-gene macronuclear chromosomes of the hypotrichous ciliates Oxytricha fallax and O. trifallax generated by alternative processing of the 81 locus. Dev. Genet. 20:348-357. [DOI] [PubMed] [Google Scholar]
- 57.Sonneborn, T. M. 1970. Methods in Paramecium research. Methods Cell Physiol. 4:241-339. [Google Scholar]
- 58.Sperling, L., P. Dessen, M. Zagulski, R. E. Pearlman, A. Migdalski, R. Gromadka, M. Froissard, A. M. Keller, and J. Cohen. 2002. Random sequencing of Paramecium somatic DNA. Eukaryot. Cell 1:341-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swanton, M. T., J. M. Heumann, and D. M. Prescott. 1980. Gene-sized DNA molecules of the macronuclei in three species of hypotrichs: size distributions and absence of nicks. DNA of ciliated protozoa. VIII. Chromosoma 77:217-227. [DOI] [PubMed] [Google Scholar]
- 60.Taverna, S. D., R. S. Coyne, and C. D. Allis. 2002. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110:701-711. [DOI] [PubMed] [Google Scholar]
- 61.Williams, K., T. G. Doak, and G. Herrick. 1993. Developmental precise excision of Oxytricha trifallax telomere-bearing elements and formation of circles closed by a copy of the flanking target duplication. EMBO J. 12:4593-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams, K. R., T. G. Doak, and G. Herrick. 2002. Telomere formation on macronuclear chromosomes of Oxytricha trifallax and O. fallax: alternatively processed regions have multiple telomere addition sites. BMC Genet. 3:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wuitschick, J. D., and K. M. Karrer. 2003. Diverse sequences within Tlr elements target programmed DNA elimination in Tetrahymena thermophila. Eukaryot. Cell 2:678-689. [DOI] [PMC free article] [PubMed]
- 64.Yao, M. C., S. Duharcourt, and D. L. Chalker. 2002. Genome-wide rearrangements of DNA in ciliates, p. 730-758. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 65.Yao, M. C., and C. H. Yao. 1989. Accurate processing and amplification of cloned germ line copies of ribosomal DNA injected into developing nuclei of Tetrahymena thermophila. Mol. Cell. Biol. 9:1092-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]