Abstract
Over the course of several million years, the eukaryotic gut symbionts of lower termites have become adapted to a cellulolytic environment. Up to now it has been believed that they produce nutriments using their own cellulolytic enzymes for the benefit of their termite host. However, we have now isolated two endoglucanases with similar apparent molecular masses of approximately 36 kDa from the not yet culturable symbiotic Archaezoa living in the hindgut of the most primitive Australian termite, Mastotermes darwiniensis. The N-terminal sequences of these cellulases exhibited significant homology to cellulases of termite origin, which belong to glycosyl hydrolase family 9. The corresponding genes were detected not in the mRNA pool of the flagellates but in the salivary glands of M. darwiniensis. This showed that cellulases isolated from the flagellate cells originated from the termite host. By use of a PCR-based approach, DNAs encoding cellulases belonging to glycosyl hydrolase family 45 were obtained from micromanipulated nuclei of the flagellates Koruga bonita and Deltotrichonympha nana. These results indicated that the intestinal flagellates of M. darwiniensis take up the termite's cellulases from gut contents. K. bonita and D. nana possess at least their own endoglucanase genes, which are still expressed, but without significant enzyme activity in the nutritive vacuole. These findings give the impression that the gut Archaezoa are heading toward a secondary loss of their own endoglucanases and that they use exclusively termite cellulases.
Cellulose is the major polysaccharide component of plant cell walls and the most abundant renewable energy source on earth. In the biological conversion of cellulose to glucose, at least three distinct types of glycolytic enzymes are involved. Endoglucanases (endo-1,4-β-glucanase, EC 3.2.1.4) randomly hydrolyze 1,4-β bonds of the cellulose chains. Cellobiohydrolases (exo-1,4-β-glucanase, EC 3.2.1.91) cleave cellobiosyl units from nonreducing ends of the cellullose chains. β-Glucosidases (EC 3.2.1.21) cleave glucosyl units from nonreducing ends of cello-oligosaccharides. The diverse spectra of cellulases are classified into 12 of the 57 glycosyl hydrolase families based on amino acid sequence similarities (5).
Termites are among the most important lignocellulose-digesting insects and possess a great variety of symbiotic microorganisms in their hindguts, including Bacteria, Archaea and Eukarya, i.e., protozoa and yeasts (7). In the digestive tracts of lower termites, cellulose seems to be synergistically degraded by flagellates, bacteria, and yeasts (3, 7, 15) as well as by the termite′s own cellulases (14, 17). Cellulases of termite origin belong to glycosyl hydrolase family 9 (GHF9). Flagellated protozoa belonging to the Archaezoa are unique symbionts in the phylogenetically lower termites. They are cellulolytic and produce acetate from cellulose for the benefit of their hosts (10). Only three species of the flagellate flora have been obtained in culture: Trichomitopsis termopsidis (19, 20) and Trichomympha sphaerica (21) from a Zootermopsis sp. and Trichomitus trypanoides from Reticulitermes santonensis (1) and from Reticulitermes flavipes (6). The problem of pure culture restricted the identification and characterization of cellulases of protozoan origin until Ohtoko et al. (11) obtained diverse genes of protist cellulases of glycosyl hydrolase family 45 (GHF45) from the termite Reticulitermes speratus by means of a culture-independent PCR approach.
The termite Mastotermes darwiniensis is the only species of the most primitive termite family, Mastotermitidae. It harbors four large flagellate species in the hindgut: the hypermastigotes Koruga bonita, Deltotrichonympha nana, and Deltotrichonympha operculata and the trichomonad Mixotricha paradoxa. Two small flagellate species also thrive in the intestine: the trichomonads Metadevescovina extranea and Pentatrichomonoides scroa (7). The flagellates themselves harbor extracellular and intracellular prokaryotes (4, 18). None of the flagellates of M. darwiniensis have been cultivated to date. The present study reports on the isolation and distribution of cellulases and cellulase genes from the host and its symbiotic flagellates.
MATERIALS AND METHODS
Organisms and cultures.
The termite Mastotermes darwiniensis Froggatt was obtained from the Bundesanstalt für Materialforschung und Materialprüfung (Berlin, Germany). The animals were fed with pine wood and cultured at 28°C in metallic vessels containing humid vermiculite.
Isolation of the flagellates.
Termites were surface sterilized in 70% ethanol and then washed in sterile phosphate-buffered saline (PBS) buffer (5 mmol of Na2HPO4/liter, 5 mmol of NaH2PO4/liter, and 130 mmol of NaCl/liter [pH 7.4]). The hindgut was removed from the abdomen and dissected from the midgut. The contents of the hindgut were suspended in sterile PBS buffer. The suspended contents were centrifuged for 1 min at 10 × g for sedimentation of the large flagellates as described by Berchtold and König (2). The pellet was washed twice and finally suspended in PBS buffer. The prepared mixture of the flagellate cells was used for mRNA extraction and cellulase isolation.
Isolation of nuclei from the flagellates.
For identification of the origin of the sequences amplified from the flagellate cells, nuclei were isolated from single cells of the hypermastigotes with the aid of a micromanipulator as described by Fröhlich and König (4). The isolated nuclei were directly used for nested PCR.
Isolation of cellulases from the flagellates.
The flagellate cells prepared from 50 termites were suspended in 8 ml of PBS buffer and vortexed vigorously for 10 min. Cellular debris was removed by centrifugation (at 15,000 × g for 4 min at 4°C). The supernatant (crude extract) was purified by anion-exchange chromatography using a Mono Q column (type HR5/5; Pharmacia, Erlangen, Germany) in 20 mM N-methyl-piperazine buffer (pH 4.5). Proteins were eluted with an NaCl gradient (0.0 to 1.0 M) in the same buffer. Fractions containing cellulase activity against carboxymethyl cellulose (CMC) were pooled and concentrated (15 μl) by using ultrafiltration (UFV5BGC25; molecular weight cutoff, 10,000; Millipore, Schwalbach, Germany). After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the proteins on the gel were transferred to polyvinylidene fluoride membranes (IPVH 10100; Millipore) with a tank blot apparatus (Mini Trans-Blot electrophoretic transfer cell; Bio-Rad, Munich, Germany) and stained with Coomassie brilliant blue. Protein bands were sequenced from the N termini by the method of Edman degradation by ZMMK Servicelabor (Cologne, Germany).
Enzyme assays.
Cellulase activity assays were performed by determination of reducing sugars released from CMC (8). For detection of cellulase activity by SDS-PAGE (0.1% SDS), the protein bands were separated in a gel containing 0.1% CMC. Following SDS-PAGE, the gel was equilibrated in 50 mM sodium phosphate buffer (pH 7.2) for 1 h and incubated for 30 min at 37°C. Enzyme bands were visualized by staining the gel with 0.1% Congo red for 30 min and subsequently washing with a 1 M NaCl solution. Clear zones around the protein bands indicated cellulase activity.
PCR with degenerate primers.
Based on the N-terminal sequences of the isolated cellulases, two degenerate oligonucleotide forward primers, MdK and MdN (Table 1), were designed. Another degenerate oligonucleotide forward primer, F45F (Table 1), corresponding to the consensus amino acid sequences of most members of GHF45 (11), was synthesized. As a template for PCR, mRNA was extracted from the mixture of the whole-flagellate cells, as well as from the salivary glands of the termite M. darwiniensis, and purified by using an RNeasy Mini Kit (Qiagen, Hilden, Germany). Reverse transcription was performed with universal adaptors (Table 1) in order to prepare cDNA strands (SuperScript First-Strand synthesis system for reverse transcription-PCR; Invitrogen, Karlsruhe, Germany). Rapid amplification of 3′ cDNA ends (3′-RACE) was performed to amplify the cellulase genes by using the forward primers and adaptor primers (adap1 or adap2 [Table 1]).
TABLE 1.
Oligonucleotide primers
| Primer | Target gene (positiona) | Sequence (5′-3′) |
|---|---|---|
| F45F | Flagellate cellulase (61-80) | ACNMGNTAYTGGGAYTGYTG |
| 183R | Flagellate cellulase (243-268) | CYGCTGCYGCWGTSCCCATGAARTAG |
| 68R | Flagellate cellulase (128-151) | GCTGGYYRTTYGCATTRCACGAGTTCA |
| FM01F1 | Flagellate cellulase (−3-22) | ACTATGATTGTTGTCTTTGTGATTG |
| FM01F2 | Flagellate cellulase (−3-22) | ACTATGTTTGTTGCCTTTGTAATTG |
| MdK | Termite cellulase (49-71) | GCNTAYGAYTAYAARGAYGTNYT |
| MdN | Termite cellulase (49-71) | GCNTAYGAYTAYAAYGAYGTNYT |
| 231R | Termite cellulase (272-298) | ATGAGTATCCTGCTGGATGGTCCACAA |
| 163R | Termite cellulase (204-230) | CCGAACTTGACATAGTCACCAGCATCA |
| MD01F | Termite cellulase (−2-17) | CAATGAGGGTCCTCCTTTG |
| AdapdT | Universal adaptor | TCCCAACACAATGGAATTGC(T)17 |
| Adap1 | Adaptor primer | TCCCAACACAATGGAATTG |
| AdapdT2 | Universal adaptor | CGTAAGATGAACGTTCGAGTACTCA(T)17 |
| Adap2 | Adaptor primer | CGTAAGATGAACGTTCGAG |
| FM1R1 | Clones 1-7 and 1-12 (541-561) | TCGGAACTCACAACCTGCCTT |
| FM2R1 | Clones 2-2 and 2-10 (538-558) | CCKGAACTGGMATCCTGWTTG |
| FM1F2 | Clones 1-7 and 1-12 (319-337) | ACTTTCACGAGTGGCCAG |
| FM1R2 | Clones 1-7 and 1-12 (519-537) | CTCAGCTGGAATCTTGGCA |
| FM2F2 | Clones 2-2 and 2-10 (318-335) | CACGTTCACGAGTGGTACT |
| FM2R2 | Clones 2-2 and 2-10 (516-533) | GYGGACGGAAGCTTACTG |
Relative to ATG start codons.
Amplification of full-length cDNAs.
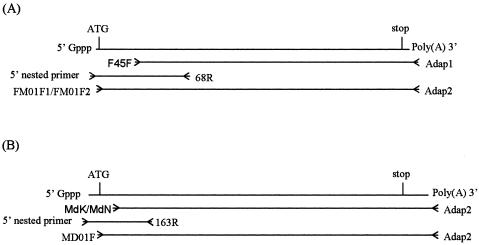
The PCR strategy for the amplification of the full-length cDNAs of cellulase genes is shown in Fig. 1. Gene-specific primers and nested primers were designed from the internal sequences of the DNA fragments amplified by degenerate PCR in order to amplify the 5′-end cDNAs. The 5′-end cDNAs of cellulase genes in the termite M. darwinienis were obtained by 5′-RACE (GeneRacer kit; Invitrogen) using primer 231R and nested primer 163R (Table 1) in combination with adaptor primers (GeneRacer 5′ primer and GeneRacer 5′ nested primer; Invitrogen). The 5′-end cDNAs of cellulase genes in flagellates were amplified by using the same method with primer 183R and nested primer 68R (Table 1). Based on the 5′-end cDNA sequences of cellulase genes, the specific primer MD01F (Table 1) was designed to amplify the full-length cDNAs of termite cellulases, and two specific primers, FM01F1 and FM01F2 (Table 1), were constructed to amplify the full-length cDNAs of flagellate cellulases by 3′-RACE with adaptor primer adap2. The PCR products were purified and cloned into pCR2.1 (Original TA Cloning kit; Invitrogen) for sequencing. DNA sequencing was carried out by Genterprise (Mainz, Germany).
FIG. 1.
RACE-PCR strategies for amplification of full-length cDNAs of cellulase genes of the flagellates (A) and of the termite M. darwiniensis (B) according to the Invitrogen RACE-PCR instruction manual.
Gene sequence analysis.
The following software and databases were used for the alignment and phylogenetic analysis of nucleotide and amino acid sequences: (i) BLASTN/BLASTP (http://www.ncbi.nlm.gov/BLAST) and CLUSTAL W (alignment of nucleotide and amino acid sequences) (13), (ii) ExPasy-ProtParam Tool and SignalP (prediction of physicochemical parameters of a protein sequence and signal peptide cleavage site; available at http://www.expasy.org/tools/), and (iii) PHYLIP (version 3.5c; phylogenetic tree and bootstrap analysis).
The following sequences were used for phylogenetic analysis of members of GHF45 (with EMBL accession numbers given in parentheses): clones 1-12, 1-7, 2-10, and 2-2 from the flagellates of the termite M. darwiniensis (AJ494857, AJ494858, AJ494859, and AJ494860, respectively), representative clones 2-6, 7-50, 7-10, and 45-6 from the protists of the termite R. speratus (BAA98036, BAA98041, BAA98043, and BAA98048, respectively), EGV of the fungus Humicola insolens (P43316), egl4 of the fungus Humicola grisea (BAA74957), Kfam1 of the fungus Fusarium oxysporum (P45699), EGI of the fungus Ustilago maydis (P54424), EGB of the bacterium Pseudomonas fluorescens (P18126), and endoglucanases of the fungus Alternaria alternata (AAF05700) and the beetle Phaedon cochleariae (CAA76931). For phylogenetic analysis, only those alignment positions which could be unambiguously aligned were used. A total of 207 amino acid characters were used for construction of phylogenetic trees.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper are available at the EMBL nucleotide sequence database under accession numbers AJ494857 to AJ494860 for the four full-length cDNA clones derived from the hindgut flagellates and AJ511339 to AJ511343 for the five full-length cDNA clones derived from the salivary glands of the termite M. darwiniensis.
RESULTS AND DISCUSSION
Isolation of cellulases from the flagellates.
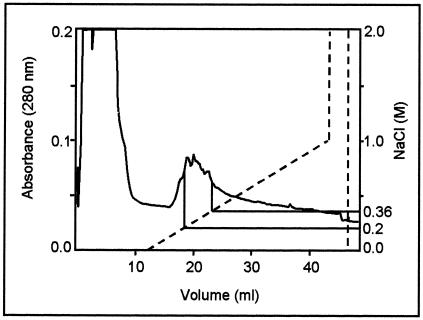
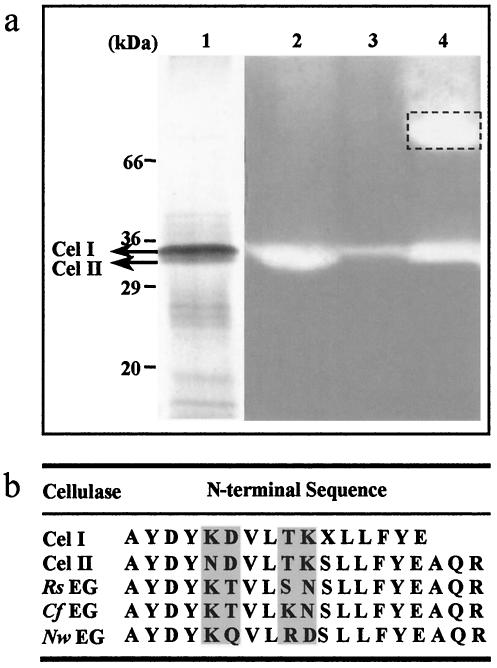
The cellulases of the large flagellates were isolated by ion-exchange chromatography. The crude extract of the flagellate cells from 50 termites suspended in 8 ml of PBS buffer was subjected to chromatography on a Mono-Q column. Figure 2 shows the elution profile of the flagellate extract after separation by column chromatography. Most proteins were found in the exclusion volume. Fractions with cellulase activity appeared between NaCl concentrations of 0.20 to 0.36 M. These fractions (4 ml) were pooled and concentrated to 15 μl. SDS-PAGE of the concentrated fractions revealed two close protein bands (Cel I and Cel II; molecular masses, ∼36 kDa) with cellulolytic activity (Fig. 3a). It was also found that the extract of the salivary glands of the termite M. darwiniensis had a cellulase activity with a similar molecular mass (Fig. 3a). The N′ termini of the two bands, Cel I and Cel II, were sequenced. At the N′ termini of Cel I and Cel II, respectively, 15 and 19 amino acid residues were identified. BLAST analysis of the partial sequences indicated significant homology among Cel I, Cel II, and termite endoglucanases, which belong to GHF9 (Fig. 3b).
FIG. 2.
Elution profile of the separated cell extract from flagellates on a Mono-Q column. Fractions containing cellulase activity against CMC appeared between NaCl concentrations of 0.20 and 0.36 M. The dashed line represents the NaCl gradient used to elute the column. Protein was determined by UV absorbance of the fractions at 280 nm.
FIG. 3.
SDS-PAGE and amino acid sequences of cellulases. (a) Zymogram of enriched cellulases. Lane 1, enriched cellulases from flagellate cell lysate obtained by anion-exchange chromatography; lane 2, activity test of enriched cellulases from flagellate cell lysate obtained by anion-exchange chromatography; lane 3, activity test of cellulases from flagellate cell lysate; lane 4, activity test of cellulases from the salivary glands of M. darwiniensis. Dashed box indicates a not yet characterized additional cellulolytic activity of the salivary gland extract. (b) Comparison of the N-terminal partial sequences of isolated cellulases from flagellate cells (Cel I and Cel II) with the known termite endoglucanases of GHF9. Rs EG, cellulase from R. speratus (accession number BAA31326); Cf EG, cellulase from Coptotermes formosanus (accession number BAB40696); Nw EG, cellulase from Nasutitermes walkeri (accession number BAA33709); X, unidentified amino acid. Variable amino acid positions are shaded.
Cloning of cellulase cDNAs of the flagellates.
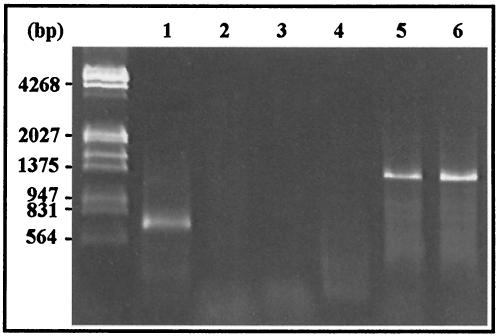
Two forward degenerate primers, MdK and MdN, were designed from the N′-terminal amino acid sequences obtained by protein sequencing of Cel I and Cel II, respectively, in order to amplify the corresponding genes of GHF9 from archaezoal DNA. In addition, a PCR-based approach using conserved cellulase sequences (family specific) was also applied to amplify protozoal cellulase genes. According to the hydrophobic cluster analysis-based cellulase family classification of Henrissat and Bairoch (5), it was possible to design degenerate oligodeoxyribonucleotides that encoded amino acid sequences conserved in an intrafamily (but not interfamily) manner for PCR amplification of the genes coding for specific cellulase family members (12). Since cultivation of the symbiotic protists in the hindguts of termites is difficult, the cloning strategy could be an efficient way to identify genes responsible for lignocellulose degradation in symbiotic protists (11). By using 3′-RACE with the degenerate primer F45F, specific for GHF45, and the universal adaptor primer adap1, ca. 600-bp fragments (3′-cDNA sequences excluding adaptor regions) were amplified from the cDNA derived from the mRNA of the large flagellate cell fraction (from 50 termites). With the degenerate primers MdK and MdN, no PCR products were obtained from the same flagellate templates (Fig. 4).
FIG. 4.
Amplification of cellulase genes by 3′-RACE. Lanes 1 to 3, mRNAs extracted from whole flagellate cells with primers F45F, MdK, and MdN, respectively; lanes 4 to 6, mRNAs extracted from the salivary glands of M. darwiniensis with primers F45F, MdK, and MdN, respectively.
The five identified cDNA clones with inserts of 600 bp amplified from the whole flagellate cells were sequenced. Sequencing revealed that the inserts contained the coding sequences for cellulases belonging to GHF45. These five clones showed 89 to 99% and 95 to 100% identities at the nucleotide and amino acid levels, respectively. Based on the cDNA sequences, the gene-specific reverse primer 183R and the nested primer 68R were synthesized (Table 1) for the amplification of 5′-cDNA sequences by using nested 5′-RACE. A 190-bp PCR fragment amplified from the whole flagellate cells was cloned and sequenced. Each of five identified clones contained the putative ATG start codon at the 5′ end. The five clones were divided into two groups on the basis of amino acid sequence identity (100% identity). Two gene-specific forward primers, FM01F1 and FM01F2 (containing the ATG start codon), corresponding to the two groups of N′-terminal amino acid sequences, were designed for amplification of the full-length cDNAs of cellulase genes of the flagellates by using 3′-RACE.
With primer FM01F1 or FM01F2, a PCR product of ca. 670 bp (excluding the adaptor region) was amplified from cDNA derived from the whole flagellate cells. The four identified cDNA clones (clones 1-7 and 1-12 with primer FM01F1; clones 2-2 and 2-10 with primer FM01F2) were sequenced. The clones shared 79.7 to 93.6% identity at the nucleotide levels. Each of the clones revealed a complete open reading frame (ORF) of 657 bp, or 660 bp when a stop codon was observed about 15 bp upstream of the poly(A) tail. The ORFs were translated into proteins encoding 219 to 220 amino acids, with remarkable similarity to known cellulases from GHF45. The conserved sequence motif (T-R-Y-W-D-C-C) of most members of GHF45 was present in the translated amino acid sequences of these clones. The deduced cellulase proteins had molecular masses of 23.3 ± 0.1 kDa and contained putative signal peptides of 11 amino acids. Similarly, the cellulases of the protists from R. speratus have putative signal peptides of 12 to 15 amino acids (11).
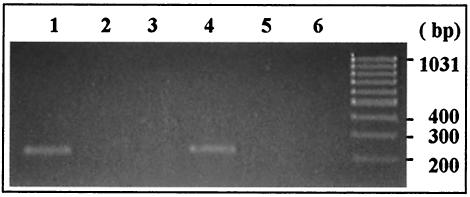
In order to identify the origin of the sequences, two sets of primers, set I, corresponding to the conserved sequences of clones 1-7 and 1-12 (primers FM01F1 and FM1R1 and nested primers FM1F2 and FM1R2 [Table 1]), and set II, corresponding to the conserved sequences of clones 2-2 and 2-10 (primers FM01F2 and FM2R1 and nested primers FM2F2 and FM2R2 [Table 1]), were used for nested PCR with nuclei isolated from the hypermastigotes (the whole product amplified from the first PCR was purified as the template for nested PCR). The PCR products should be 219 and 216 bp long, respectively. Set I resulted in a PCR product of 219 bp from the nuclei of K. bonita. No PCR product was amplified from the nuclei of D. nana or D. operculata (Fig. 5). With primers of set II, a PCR product of 216 bp was obtained from the nuclei of D. nana, and no product was obtained from K. bonita or D. operculata. The results suggested that clones 1-7 and 1-12 originated from the hypermastigote K. bonita and that clones 2-2 and 2-10 originated from the hypermastigote D. nana. Furthermore, the cellulase genes of the hypermastigotes could be transcribed and expressed in vivo according to Fig. 4. The 219-bp PCR product from K. bonita showed sequence similarities of 97% to clone 1-12 and 94% to clone 1-7. The 216-bp sequence amplified from D. nana shared 97% identity with clone 2-10 and 96% identity with clone 2-2. By using nuclei for PCR, we did not obtain any DNA product from D. operculata. In view of the similarity between Deltotrichonympha spp., we propose that the cellulase gene of D. operculata was present, but this gene could not be cloned with the clone-specific primers of set I or set II.
FIG. 5.
Nested PCR with the nuclei of the flagellates. Lane 1, primer set I and five nuclei of K. bonita; lane 2, primer set II and five nuclei of K. bonita; lane 3, primer set I and five nuclei of D. nana; lane 4, primer set II and five nuclei of D. nana; lane 5, primer set I and five nuclei of D. operculata; lane 6, primer set II and five nuclei of D. operculata.
Amplification of cellulase genes of M. darwiniensis.
Due to the significant homology of the N′-terminal sequences of Cel I, Cel II, and termite endoglucanases, the degenerate forward primers MdK and MdN were used for amplification of cellulase genes with cDNAs derived from the salivary glands of M. darwiniensis. It is known that cDNAs of the endoglucanases from M. darwiniensis have been amplified only from salivary gland tissue (14). With the forward primer MdK or MdN and the universal adaptor primer adap2, a PCR fragment of ca. 1,400 bp (3′ cDNA sequences excluding the adaptor region) was amplified by 3′-RACE. The cDNA sequences of five isolated clones showed a nucleotide sequence similarity of more than 99% in 811 bp with the known partial cDNA sequences of salivary cellulases from M. darwiniensis (EMBL accession numbers AAF63724 and AAF63725). The PCR strategy for full-length cDNAs of the cellulases of M. darwiniensis was the same as that used for the intestinal flagellates. The gene-specific reverse primer 231R and the nested primer 163R were designed for amplification of the 5′ cDNA by 5′ nested RACE. The 5′-RACE PCR fragment of 270 bp was cloned and sequenced. The cDNA clones represented two groups of N′-terminal sequences of salivary cellulases (excluding the signal peptides), with a difference of 1 in 77 deduced amino acids; these were similar to the N′-terminal sequences of the isolated cellulases Cel I and Cel II, even in variable regions. In subsequent 3′-RACE, the gene-specific forward primer Md01F, constructed on the basis of the 5′ cDNA sequences (containing the ATG start codon), resulted in a PCR product of ca. 1,450 bp (excluding the adaptor region). The five clones identified were sequenced and divided into three groups based on nucleotide sequence similarity. Clones Md13 and Md5, containing a coding region of 1,344 bp, formed one group with more than 99% amino acid sequence identity. In the second group, clones Md6 and Md11 had ORFs of 1,341 and 1,347 bp, respectively, that shared more than 99% amino acid sequence identity and showed an average of 87% amino acid sequence similarity to clones Md13 and Md5. Clone Md1, with a coding region of 1,341 bp, fell into a third group; it showed a mean amino acid sequence similarity of 98% to clones Md6 and Md11, with a sequence difference only at the C′ terminus, and a mean amino acid sequence identity of 89% with clones Md13 and Md5. The predicted amino acid sequence indicated that the molecular masses were 49.1 ± 0.5 kDa. Each of the cellulase proteins contained a leader peptide consisting of 16 amino acids.
Comparisons of amino acid sequences.
Database searches revealed that the full-length cDNAs cloned from the hindgut flagellates of the termite M. darwiniensis exhibited a high degree of amino acid sequence similarity to cellulases belonging to GHF45. The alignment with homologous sequences is shown in Fig. 6. The four deduced amino acid sequences share more than 84% identity with each other and display 50 to 58% identity to four representative sequences from the symbiotic protists in the hindgut of the termite R. speratus, 47 to 57% identity to two sequences (EGV and egl4) from Humicola spp., 46 to 51% identity to Kfam1 of F. oxysporum, 44 to 45% identity to the endoglucanase of P. cochleariae, 38 to 42% identity to the endoglucanase of A. alternata, 37 to 40% identity to EGI of U. maydis, and 34 to 38% identity to EGB of P. fluorescens. It is also noted that the endoglucanases of flagellate origin reported in this study appear to consist only of a single catalytic domain and contain neither a cellulose-binding domain nor a spacer sequence, like the amino acid sequences from the hindgut protists of R. speratus (11).
FIG. 6.
Alignment of amino acid sequences of endoglucanases from the symbiotic flagellates of the termite M. darwiniensis with homologous cellulases of GHF45. Only the region of the catalytic domain is shown for the seven reference sequences. Reference sequences (with the corresponding amino acid positions in the alignment given in parentheses) are as follows: Fo, F. oxysporum (amino acids 21 to 225); Hi, H. insolens (amino acids 1 to 205); Hg, H. grisea (amino acids 22 to 219); Aa, A. alternata (amino acids 21 to 239); Pc, P. cochleariae (amino acids 37 to 242); Um, U. maydis (amino acids 26 to 240); Pf, P. fluorescens (amino acids 268 to 504). For optimal alignment, three short segments in the amino acid sequences of P. fluorescens were deleted and are indicated by a triangle between two underlined amino acid residues (SSYNAPGDP, AACKQQLGYNASLSQYKSCVL, and FGSRGLT). The conserved region for the PCR primer design is boxed. The putative 11-amino-acid signal peptide of the flagellate cellulases is italicized. Amino acid identity is indicated by asterisks. Strongly or weakly conserved changes are indicated by colons and dots, respectively.
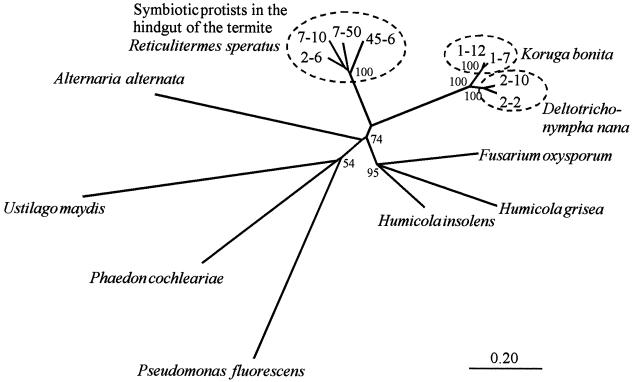
Figure 7 shows an unrooted phylogenetic tree based on the amino acid sequences of members of GHF45. In the tree, the four sequences from the hindgut flagellates of the termite M. darwiniensis form a monophyletic clade, suggesting that the hindgut protists containing the sequences share the latest common ancestor with each other. This is in agreement with the phylogenetic analysis of these flagellates using SSU ribosomal DNA sequences that showed that the large hindgut flagellates have a close evolutionary relationship (4). The four sequences are divided into two subgroups with a bootstrap value of 100%. In one group, clones 1-7 and 1-12 show 96.9% amino acid identity. In another group, clones 2-2 and 2-10 share 93.2% amino acid identity. The tree shows the close phylogenetic relationship of cellulases from the hindgut flagellates in M. darwiniensis and the hindgut protists in R. speratus. However, the bootstrap value is not high in this region.
FIG. 7.
Unrooted phylogenetic tree of GHF45 members based on the amino acid sequence alignment of the catalytic domains (shown in Fig. 6) without putative signal sequences. All gaps in the alignment were omitted, leaving 207 amino acid positions for phylogenetic analysis. Bootstrap values (100 replicates) are indicated at their respective branching points. Bar, 0.20 amino acid substitution.
Origin of the cellulases isolated from the intestinal flagellates.
Due to the lack of pure cultures, it is difficult to separate and identify the cellulases of the hindgut flagellates. By isoelectric focusing, the cellulase activity in the cell extract of the flagellates was mainly found in a band at a pH value of about 3.5, while most proteins focused in a pH range between 4.5 and 6.0 (data not shown). Thus, by using ion-exchange chromatography, two endoglucanases were isolated from the flagellate cell extract prepared from 50 termites, and these were then subjected to protein sequencing. BLAST analysis of the 15 to 19 amino acid residues at the N′ termini indicated that the isolated cellulases have a high similarity to GHF9. With the degenerate primer MdK or MdN, based on the N-terminal sequences of the cellulases isolated from flagellate cells, no corresponding gene was found in the flagellate cells. However, by using these primers, ca. 1,300 bp of cellulase genes (excluding the adaptor region and 3′ untranslated region) was amplified from the salivary glands of M. darwiniensis. The cloned full-length cDNAs showed derived N-terminal sequences similar to those of the isolated cellulases Cel I and Cel II, even in variable regions. Therefore, we confidently propose that the cellulases isolated from the nutritive vacuole of the flagellates originated from the termite M. darwiniensis. No corresponding gene could be found in the flagellate cells. Probably the cellulases are secreted from the salivary glands of M. darwiniensis. During the mechanical grinding of wood particles by the termites, the cellulases are attached to the wood particles or mixed with them; then the attached cellulases or the mixture moves to the hindgut, where it is endocytosed by the flagellates.
It has been shown that the endoglucanases of the termite Coptotermes formosanus are restricted to the salivary glands, the foregut, and the midgut (9). But according to our work, the main endoglucanase activity found in cells of the hindgut flagellates of M. darwiniensis is likely to originate from the termite cellulases. It has also been found that 40% of the endoglucanase activity of M. darwiniensis is present in the hindgut and that most (ca. 84%) of the cellulase activity of the whole hindgut is present in the flagellate extract (16). This implies that a certain amount of termite cellulases, secreted from the salivary glands, moves into the hindgut and enters the flagellate cells. They may be involved in the digestion of cellulose in the flagellate cells. The flagellates' own cellulases could not be detected by an enzyme essay on SDS-PAGE gels. Thus, we assume that the endoglucanases of termite origin play an important role in cellulose degradation in the flagellates.
It is conceivable that in the course of about 300 million years, the symbiosis between the termite and initially free-living Archaezoa affected the enzymatic equipment of both organisms, such that the excess of termite cellulases led to a disuse of the flagellates' own enzymes. The lack of selection pressure in the hindgut possibly directed the mutation and inactivation of cellulolytic enzymes, from which the corresponding genes are still expressed. The production of an inactive enzyme may result in a complete loss of the corresponding genes. This means that the symbiotic Archaezoa are progressing to a state devoid of their own cellulolytic activities, as was probably the case before the existence of cellulose-containing plants. Presently, the symbiotic gut Archaezoa of the primitive Australian termite M. darwiniensis owe their endocellulolytic activity to their host.
REFERENCES
- 1.Berchtold, M., A. Breunig, and H. König. 1995. Culture and phylogenetic characterization of Trichomitus trypanoides Duboscq & Grassè 1924, n. comb.: a trichomonad flagellate isolated from the hindgut of the termite Reticulitermes santonensis Feytaud. J. Eukaryot. Microbiol. 42:388-391. [DOI] [PubMed] [Google Scholar]
- 2.Berchtold, M., and H. König. 1995. Phylogenetic position of the two uncultivated trichomonads Pentatrichomonoides scroa Kirby and Metadevescovina extranea Kirby from the hindgut of the termite Mastotermes darwiniensis Froggatt. Syst. Appl. Microbiol. 18:567-573. [Google Scholar]
- 3.Breznak, J. A., and A. Brune. 1994. Role of microorganisms in the digestion of lignocellulose by termites. Annu. Rev. Entomol. 39:453-487. [Google Scholar]
- 4.Fröhlich, J., and H. König. 1999. Rapid isolation of single microbial cells from mixed natural and laboratory populations with the aid of a micromanipulator. Syst. Appl. Microbiol. 22:249-257. [DOI] [PubMed] [Google Scholar]
- 5.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huntenburg, W., L. Stockert, H. E. Smith-Somerville, and H. E. Buhse. 1986. Trichomitus trypanoides (Trichomonadida) from the termite Reticulitermes flavipes. 1. In vitro cultivation and cloning. Trans. Am. Microsc. Soc. 105:211-222. [Google Scholar]
- 7.König, H., J. Fröhlich, M. Berchtold, and M. Wenzel. 2002. Diversity and microhabitats of the hindgut flora of termites. Recent Res. Microbiol. 6:125-156. [Google Scholar]
- 8.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 9.Nakashima, K., H. Watanabe, H. Saitoh, G. Tokuda, and J.-I. Azuma. 2002. Dual cellulose-digesting system of the wood-feeding termite, Coptotermes formosanus Shiraki. Insect Biochem. Mol. Biol. 32:777-784. [DOI] [PubMed] [Google Scholar]
- 10.Odelson, D. A., and J. A. Breznak. 1985. Nutrition and growth characteristics of Trichomitopsis termopsidis, a cellulolytic protozoan from termites. Appl. Environ. Microbiol. 49:614-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohtoko, K., M. Ohkuma, S. Moriya, T. Inoue, R. Usami, and T. Kudo. 2000. Diverse genes of cellulase homologues of glycosyl hydrolase family 45 from the symbiotic protists in the hindgut of the termite Reticulitermes speratus. Extremophiles 4:343-349. [DOI] [PubMed] [Google Scholar]
- 12.Sheppard, P. O., F. J. Grant, P. J. Oort, C. A. Sprecher, D. C. Foster, F. S. Hagen, A. Upshall, G. L. McKnight, and P. J. O'Hara. 1994. The use of conserved cellulase family-specific sequences to clone cellulase homologue cDNAs from Fusarium oxysporum. Gene 150:163-167. [DOI] [PubMed] [Google Scholar]
- 13.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokuda, G., N. Lo, H. Watanabe, M. Slaytor, T. Matsumoto, and H. Noda. 1999. Metazoan cellulase genes from termites: intron/exon structures and sites of expression. Biochim. Biophys. Acta 1447:146-159. [DOI] [PubMed] [Google Scholar]
- 15.Varma, A., B. K. Kolli, J. Paul, S. Saxena, and H. König. 1994. Lignocellulose degradation by microorganisms from termite hills and termite guts: a survey on the present state of art. FEMS Microbiol. Rev. 15:9-28. [Google Scholar]
- 16.Veivers, P. C., A. M. Musca, R. W. O'Brien, and M. Slaytor. 1982. Digestive enzymes of the salivary glands and gut of Mastotermes darwiniensis. Insect Biochem. 12:35-40. [Google Scholar]
- 17.Watanabe, H., H. Noda, G. Tokuda, and N. Lo. 1998. A cellulase gene of termite origin. Nature 394:330-331. [DOI] [PubMed] [Google Scholar]
- 18.Wenzel, M., R. Radek, G. Brugerolle, and H. König. 2003. Identification of the ectosymbiotic bacteria of Mixotricha paradoxa involved in movement symbiosis. Eur. J. Protistol. 39:11-23.
- 19.Yamin, M. A. 1978. Axenic cultivation of the cellulolytic flagellate Trichomitopsis termopsidis (Cleveland) from the termite, Zootermopsis. J. Protozool. 25:535-538. [Google Scholar]
- 20.Yamin, M. A. 1980. Cellulose metabolism by the termite flagellate Trichomitopsis termopsidis. Appl. Environ. Microbiol. 39:859-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamin, M. A. 1981. Cellulose metabolism by the flagellate Trichonympha from a termite is independent of endosymbiotic bacteria. Science 211:58-59. [DOI] [PubMed] [Google Scholar]