Abstract
Antigen receptor loci are composed of numerous variable (V), diversity (D), and joining (J) gene segments, each flanked by recombination signal sequences (RSSs). The V(D)J recombination reaction proceeds through RSS recognition and DNA cleavage steps making it possible for multiple DNA double strand breaks (DSBs) to be introduced at a single locus. Here we use ligation-mediated PCR to analyze DNA cleavage intermediates in thymocytes from mice with targeted RSS mutations at the endogenous TCRβ locus. We show that DNA cleavage does not occur at individual RSSs but rather must be coordinated between RSS pairs flanking gene segments that ultimately form coding joins. Coordination of the DNA cleavage step occurs over great distances in the chromosome and favors intra- over interchromosomal recombination. Furthermore, through several restrictions imposed on the generation of both nonpaired and paired DNA DSBs, this requirement promotes antigen receptor gene integrity and genomic stability in developing lymphocytes undergoing V(D)J recombination.
Keywords: lymphocyte, antigen receptor, RAG, DNA cleavage, DNA repair
Introduction
The precise specificity and vast diversity of the adaptive immune response is achieved through the expression of lymphocyte antigen receptors with polymorphic variable regions. Genes encoding variable regions are assembled during lymphocyte development from V, D, and J gene segments by a site specific DNA recombination process, termed V(D)J recombination (1). Variable gene segments (V, D, and J) are bordered by recombination signal sequences (RSSs)* composed of conserved heptamers and nonamers flanking nonconserved 12 or 23 base pair spacers (hereafter referred to as 12-RSSs and 23-RSSs, respectively) (1). Recombination only occurs between variable gene segments flanked by 12- and 23- RSSs, a restriction known as the 12/23 rule (1). The V(D)J recombination reaction proceeds through DNA cleavage and joining steps and is performed by an enzymatic complex collectively referred to as the V(D)J recombinase. This complex includes the lymphoid specific recombinase activating gene (RAG)–1 and –2 proteins which introduce DNA double strand breaks (DSBs) precisely at the RSS/coding segment border (2–4). The resulting hairpin closed coding ends and blunt phosphorylated signal ends are then joined by generally expressed proteins involved in the nonhomologous end joining pathway of DNA DSB repair to form coding and signal joins, respectively (5–8).
There are six antigen receptor loci that encode the TCRα, β, γ, and δ chains and the Ig H and L (κ and λ) chains. These loci are composed of many variable gene segments with flanking RSSs. DNA cleavage by RAG-1/2 at isolated RSSs could result in numerous DNA DSBs at multiple loci in developing lymphocytes. Although some of these DSBs would be resolved through standard coding and signal join formation, many would require resolution by other means such as open-and-shut or hybrid join formation before cell cycle progression is permitted (9). Furthermore, these DSBs could be resolved through interchromosomal V(D)J recombination or serve as substrates for RAG-1/2 mediated transposition resulting in chromosomal translocations (10, 11). Accordingly, mechanisms that limit DNA DSB generation to gene segments that ultimately participate in coding join formation would promote efficient variable region gene assembly and genomic stability.
The V(D)J recombination reaction is regulated in tissue, lineage, and developmental stage specific manners (12, 13). Although in some cases this regulation may occur at postcleavage steps, it is primarily effected at the DNA cleavage step of the reaction by limiting accessibility of antigen receptor genes to the V(D)J recombinase (12–14). This occurs through, among other things, changes in chromatin structure, methylation, and/or transcription that restricts DNA cleavage by the V(D)J recombinase (12, 13). For example, DNA DSB generation in Ig genes is restricted to developing B cells with further restriction of IgH and IgL chain gene DSBs to pro-B and pre-B cells, respectively (15). Similar considerations apply to DSBs generated in TCR genes during T cell development (15). However, the extent of DNA DSB generation in an accessible locus will be determined primarily by regulatory features of the V(D)J recombination reaction. In this regard, accumulation of DSBs can be limited by restrictions placed on the DNA cleavage step and/or by promoting rapid resolution of DSBs (i.e., as open-and-shut joins) introduced at gene segments that do not ultimately participate in coding join formation.
Analysis of the DNA cleavage step in vitro and on extrachromosomal and transgenic recombination substrates has demonstrated that it is most efficient when 12/23 RSS pairing is permitted (16–19). This restriction, which provides the basis for the 12/23 rule, reflects a requirement for 12/23 RSS synapsis before DNA cleavage (20, 21). However, cleavage at isolated RSSs is observed in these analyses, and the extent to which this restriction limits DNA DSB generation at the numerous variable gene segments in endogenous antigen receptor loci is not known. Furthermore, RSSs impose significant constraints on the V(D)J recombination reaction beyond enforcing 12/23 compatibility (22, 23). Whether these restrictions occur at the DNA cleavage step, further limiting DNA DSB generation, is also unknown.
TCRβ variable region genes are assembled from Vβ, Dβ, and Jβ gene segments with Dβ to Jβ rearrangement preceding Vβ to Dβ rearrangement. We have analyzed the DNA cleavage step of the V(D)J recombination reaction in the context of endogenous TCRβ gene rearrangements during normal thymocyte development. These experiments used mice with targeted mutations of RSSs in the endogenous TCRβ locus and, importantly, were designed to evaluate the effects of these mutations on DNA cleavage not only at mutant RSSs but also at their normal wild-type RSS partners. Our findings provide important insights into the regulation of the DNA cleavage step of the V(D)J recombination reaction in vivo within the context of the chromosome. In addition, they have implications for the manner in which DNA DSBs are limited during chromosomal V(D)J recombination and are discussed in the context of maintaining genomic stability during antigen receptor variable region gene assembly in developing lymphocytes.
Materials and Methods
Flow Cytometric Analyses.
Thymocytes were stained with FITC-conjugated anti-CD8, PE-conjugated anti-CD4, or PE-conjugated as described previously (22). Stained samples were analyzed on a Becton Dickinson FACScan™.
Isolation of Thymocyte DNA.
Whole thymocytes were treated with anti-CD4 (RL172.4), anti-CD8 (3.155), and complement as described previously (22). The resulting cells were lysed and DNA isolated as described previously (24).
Oligonucleotide Primers.
The BW-1, BW-2, and BW-1H oligonucleotide primers have been described previously (25). The sequences of other primers used are as follows. 5A: 5′-TGACGCACAGCCTTAGGG-3′; 5B: 5′-GGGCTGTTACTTCTTCATAGGGTGG-3′ P2: 5′-GCTTATCTGGTGGTTTC-3′; 3A: 5′-TTGACGATAGACTTGTGGC-3′; 3B: 5′-CAGTGGAACTTACATCCCTTTTGTG-3′; P1:5′-CCTCTCTCAAGGTCCATCAA-3′; BWJ: 5′-CCGGGAGATCTGAATTCCACATCA-3′; JA: 5′-CTGTCTGTCCCAAGGC-3′; JB: 5′-TCTC-CTGGGTCGCAAGCC-3′; V2: 5′-TCCTGGGGACAAAGA-GGTCAAATC-3′. VA2: 5′-TTCAGTCAGTGTTTATATG-AGC-3′; VB2: 5′-GATGGTTTCAATGGTGCTCTG-3′; VP2: 5′-GAACCAAAGTCTATGCTCTGAG-3′; VA11: 5′-TAGCCTGGATCTGTTCTG-3′; VB11: 5′-AGGTCTCTGGGCTATCTA-3′; VP11: 5′-TCCCAGCCTATTTAGTCTTCAG-3′; VA8.3: 5′-GATGACACCAATGAGTGAAG-3′; VB8.3: 5′-CTGAAACTATGGCTGTGAGA-3′; VP8.3: 5′-TTTCAGAATGACCAGGACTG-3′. V15: 5′-CCCATCAGTCATCCCAACTTATCC-3′. VA15: 5′-GACTCTTGAGACCTGACTATC-3′. VB15: 5′-GCAATGAACCTGTCTGAGTTG-3′. VP15: 5′-GGAGTGAGTAGACTTGTGTTG-3′. VA12: 5′- GGACCATTAGTAGAGGACAAAC-3′. VB12: 5′-CCTGATATTCCCGTACTGGGACTGGC-3′; VP12: 5′-GAAATCTTGGGTGGAAAATGAC-3′; R2U: 5′-TCACTCAAATCCTC-TTCAGA-3′R2D: 5′-CCCAGAGAACCACAGAAA-3′; R2P: 5′-GCTGACTGCCTACCCCAT-3′.
Ligation-mediated PCR and PCR.
The BW linker was generated by annealing the BW-1 and BW-2 oligonucleotides. Purified thymocyte genomic DNA (2–3 μg) was ligated to 100 pmoles of the BW linker in a volume of 50–60 μl with 1–3 units T4 DNA Ligase (Promega) at 16°C for 12–14 h. Ligated samples were extracted with phenol and chloroform before PCR analysis. Heminested PCRs were performed using the BW-1H or BWJ primers and A series primers (i.e., VA12, VA11, JA) in the primary reaction and BW-1H or BWJ primers and B series primers (i.e., VB12, VB11, JB) in the secondary reaction. Primary ligation-mediated PCRs (LMPCRs) were performed on 30–40 ng of ligated genomic DNA with 10 pmoles of each primer, 50 mM KCl, 10 mM Tris (pH 8.0), 1–2 mM MgCl2 and Taq in 50 μl reaction volume. The conditions for primary LMPCR reactions were 92°C, 1 min; 50–60°C, 1 min; and 72°C, 1 min for 18 cycles. Secondary LMPCRs were performed on 10 μl of the primary LMPCR using primary LMPCR conditions. Cycling conditions for secondary LMPCR reactions were 92°C, 1 min; 55–65°C, 1 min; and 72°C, 1min for 28 cycles. The RAG-2 gene was amplified using the R2U and R2D primers using the primary LMPCR reaction conditions and the secondary LMPCR cycling conditions. Signal joins were amplified by a heminested PCR of nonligated genomic DNA using primary and secondary LMPCRs conditions with V primers and the 5A (primary PCR) and 5B (secondary PCR) primers.
Southern Blotting.
PCR products were size fractionated on 1–1.5% agarose gels and transferred to Zeta-probe membranes (Bio-Rad Laboratories) followed by hybridization with oligonucleotide probes as described previously (22).
Results
Experimental Approach.
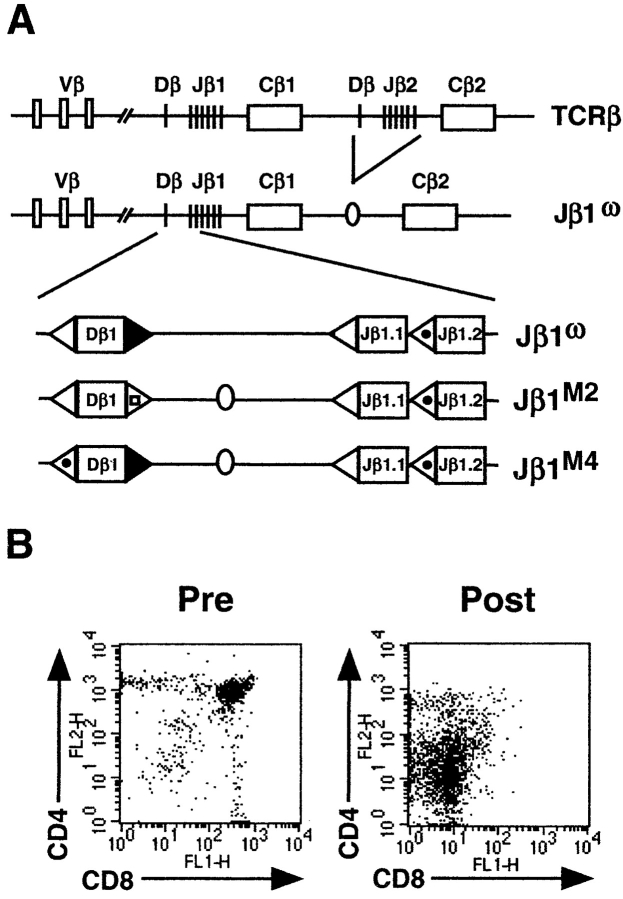
The mouse TCRβ locus consists of 21 functional Vβ gene segments spanning ∼650 kb upstream of two DJβ gene clusters (DJβ1 and DJβ2), each with a single Dβ gene segment, six functional Jβ gene segments, and associated constant region genes (Cβ1 and Cβ2; Fig. 1 A). The Jβ1ω allele has a targeted deletion of the DJβ2 gene cluster limiting rearrangements on this allele to the DJβ1 gene cluster (22). Vβ and Jβ gene segments are flanked by 23- and 12-RSSs, respectively, and the Dβ1 gene segment by a 5′ 12-RSS and a 3′ 23-RSS. Because direct Vβ to Jβ rearrangement is prohibited, despite 12/23 compatibility, all rearrangements (Dβ to Jβ and Vβ to Dβ) on the Jβ1ω allele involve the Dβ1 gene segment (22, 23).
Figure 1.
Schematic of the wild-type and mutant TCRβ alleles. (A) Wild-type and modified Jβ1ω, Jβ1M2 Jβ1M4 TCRβ alleles. Shown are the 5′ Dβ1 and Jβ1.1 12-RSSs (open triangles), Jβ1.2 12-RSS (dotted open triangle), the 3′ Dβ1 23-RSS (closed triangle), and the 3′ Dβ1 23-RSS with the heptamer mutation (open triangle with open square). The 70 base pair fragment containing a loxP site is also shown (oval). (B) Flow cytometric analysis of thymocytes before (Pre) and after (Post) treatment with anti-CD4, anti-CD8, and complement.
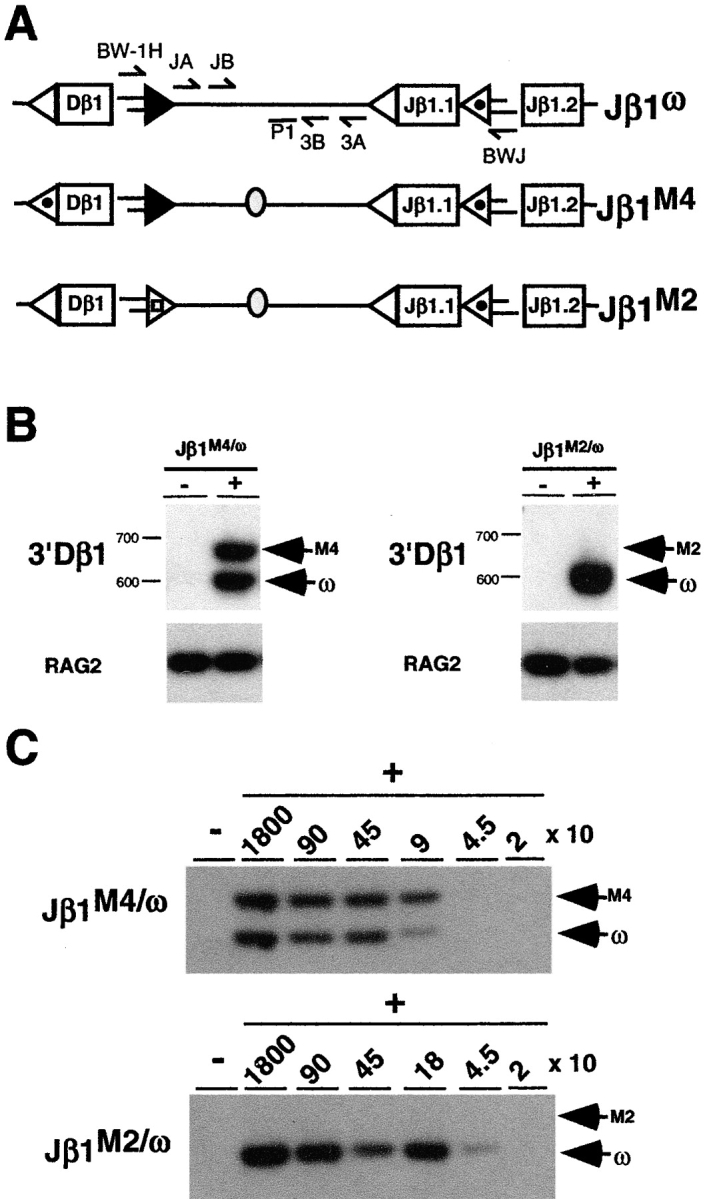
DNA cleavage intermediates were assayed in thymocytes from mice heterozygous for 5′ or 3′ Dβ1 RSS mutations on the Jβ1ω allele (Jβ1M4/ω and Jβ1M2/ω, respectively, see below; Fig. 1 A). Importantly, as all rearrangements must use the Dβ1 gene segment, the effects of these mutations could be assayed not only at the mutant Dβ1 RSSs but also at their native Vβ or Jβ1 RSS partners. To enrich for cells actively undergoing TCRβ gene rearrangements, thymocytes were treated with anti-CD4, anti-CD8, and complement with the resulting cells being >90% CD4−CD8− (double negative, DN; Fig. 1 B). Signal end DNA cleavage intermediates were assayed in genomic DNA isolated from DN thymocytes by LMPCR using a nested pair of TCRβ locus-specific primers and a linker-specific primer. Polymorphisms on the mutant (Jβ1M4 and Jβ1M2) and wild-type (Jβ1ω) alleles enabled direct quantitative comparison of signal end DNA cleavage intermediates derived from each allele in samples from thymocytes heterozygous for RSS mutations (Jβ1M4/ω and Jβ1M2/ω).
DNA Cleavage Intermediates from Dβ to Jβ Rearrangement.
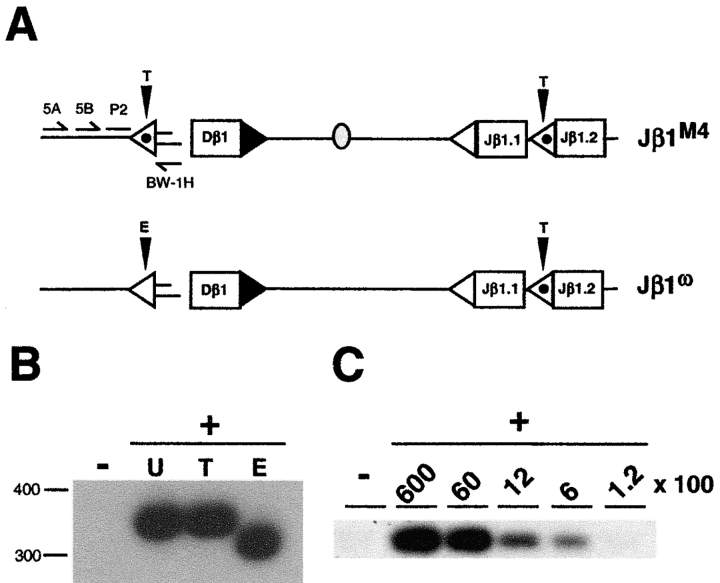
The Jβ1M4 allele was generated by precisely replacing the 5′Dβ1 12-RSS on the Jβ1ω allele with the Jβ1.2 12-RSS without alterations in the nucleotides flanking the RSS (22; Fig. 1 A). Dβ to Jβ rearrangement occurs on both alleles in Jβ1M4/ω thymocytes (22). Accordingly, equivalent levels of DNA cleavage intermediates would be expected from Dβ to Jβ rearrangement at each allele in Jβ1M4/ω DN thymocytes. Assay of linker ligated genomic DNA from Jβ1M4/ω DN thymocytes revealed equivalent levels of 3′Dβ1 signal ends derived from the Jβ1M4 and Jβ1ω alleles (Fig. 2, A and B) . Similarly, equivalent levels of Jβ1.1 and Jβ1.2 signal ends were detected from the Jβ1M4 and Jβ1ω alleles (Fig. 2, A and C, and data not shown). Products from linker ligated Jβ1.2 signal ends could be detected from as little as 90 cell equivalents of ligated genomic DNA demonstrating the assay is sensitive at low template levels (Fig. 2 C). LMPCR products derived from Jβ1M4 and Jβ1ω alleles can be distinguished due to size differences generated by the introduction of a loxP site between Dβ1 and Jβ1.1 on the Jβ1M4 allele (Fig. 2, A–C). All products were ligase dependent, and sequence analysis of these and subsequent products revealed that they were generated by blunt end ligation of the linker to precisely cut signal ends (data not shown). These findings demonstrate that the experimental approach used here permits the quantitative assessment of DNA cleavage intermediates generated during rearrangement at the TCRβ locus in DN thymocytes. This is evidenced by the detection of equivalent levels of DNA cleavage intermediates from Dβ to Jβ rearrangement at the Jβ1M4 and Jβ1ω alleles in Jβ1M4/ω thymocytes.
Figure 2.
DNA cleavage is coordinated at 3′Dβ and Jβ RSSs. (A) Schematic of the Jβ1ω, Jβ1M2, and Jβ1M4 alleles as described in the legend to Fig. 1 with the BW linker ligated to cleaved 3′ Dβ1 and Jβ1.2 signal ends. The position of the oligonucleotide primers used to amplify linker ligated to the 3′ Dβ1 (BW-1H, 3A, and 3B) and Jβ1.2 (BWJ, JA, and JB) signal ends are shown as is the position of the oligonucleotide (P1) used to probe these PCR products. Genomic DNA from Jβ1M4/ω or Jβ1M2/ω DN thymocytes was incubated with the BW linker in the presence (+) or absence (−) or T4 DNA ligase and heminested LMPCRs performed to detect 3′ Dβ1 (B) and Jβ1.2 (C) signal ends as described in the Materials and Methods section. Analyses of Jβ1.2 signal ends was performed on ligated thymocyte DNA that was serially diluted into nonligated DNA keeping the total amount of DNA constant. Cell equivalents of ligated template DNA are indicated. Products from ligation of the BW linker to signal ends from the Jβ1ω (ω), Jβ1M4 (M4), and Jβ1M2 (M2) alleles are indicated. Molecular weight markers are shown. Also shown is a RAG-2 gene PCR performed on ligated and nonligated template DNA and probed with the R2P oligonucleotide.
DNA Cleavage Is Coordinated at Dβ and Jβ RSSs.
The Jβ1M2 allele exhibits a block in Dβ to Jβ rearrangement due to a mutation of the 3′Dβ1 RSS heptamer (CACAGTG to ATTTTAA; reference 23; Fig. 1 A). As expected, 3′Dβ1 signal ends from the Jβ1ω, but not the Jβ1M2, allele were readily detected by LMPCR of genomic DNA from Jβ1M2/ω DN thymocytes (Fig. 2, A and B). The Jβ1 RSSs on the Jβ1M2 allele are unaltered and as such are identical to those on the Jβ1ω allele. Strikingly, although Jβ1.1 and Jβ1.2 signal ends can be readily detected from the Jβ1ω allele, these signal ends are not detected from the Jβ1M2 allele in Jβ1M2/ω DN thymocytes (Fig. 2 C, and data not shown). Low levels of cleavage at the Jβ1.2 RSS on the Jβ1M2 allele should be detected, if present, as cleavage at the Jβ1.2 RSS the Jβ1ω allele can be detected from as little as 45 cell equivalents of Jβ1M2/ω DN thymocyte DNA (Fig. 2 C). Together, these findings demonstrate that DNA DSB generation at Dβ and Jβ RSSs is coordinated during Dβ to Jβ rearrangement, as mutation of the 3′Dβ 23-RSS prevents cleavage at native Jβ 12-RSSs on the same allele. This occurs within the context of the 12/23 rule as DNA cleavage is not coordinated between Jβ1 12-RSSs on the Jβ1M2 allele. However, it is notable that cleavage at Jβ1 12-RSSs is not promoted by Vβ 23-RSSs suggesting that coordinated cleavage is regulated by constraints beyond 12/23 compatibility (see below).
Requirements for DNA Cleavage at the 5′ Dβ RSS.
Replacing the 5′Dβ1 12-RSS with the Jβ1.2 12-RSS on the Jβ1M4 allele causes a block in Vβ to Dβ rearrangement due to a requirement for the 5′Dβ1 12-RSS to target Vβ rearrangement beyond simply enforcing the 12/23 rule (22; Fig. 1 A). To determine whether this restriction occurs at the DNA cleavage step of the V(D)J recombination reaction, 5′Dβ1 signal end cleavage intermediates were assayed in Jβ1M4/ω DN thymocytes (Fig. 3 A). These analyses yielded a 350 base pair LMPCR product expected from ligation of the linker to 5′Dβ1 signal ends from either the Jβ1M4 or Jβ1ω allele (Fig. 3 B). This product digested completely with Eco0109 (present only in the 5′Dβ1 RSS) and not Taq I (present only in the Jβ1.2 12-RSS), demonstrating that it is derived entirely from Jβ1ω 5′Dβ1 signal ends (Fig. 3, A and B). Signal ends from cleavage at the Jβ1ω 5′Dβ1 RSS can be detected from less than 600 cell equivalents of Jβ1M4/ω DN thymocyte DNA (Fig. 3 C).
Figure 3.
Cleavage at the 5′Dβ RSS. (A) Schematic of the Jβ1ω and Jβ1M4 alleles as described in the legend to Fig. 1 and the position of the oligonucleotide primers (5A, 5B, and BW-1H) used to amplify linker ligated to 5′ Dβ1 signal ends and the oligonucleotide probe (P2) for these products. The Taq1 (T) and Eco0109 (E) sites present in the native Jβ1.2 and 5′ Dβ1 RSSs are indicated. (B) Linker ligated (+) and nonligated (−) Jβ1M4/ω DN thymocyte DNA assayed for 5′Dβ RSS signal ends by hemi-nested LMPCR using the above primer sets as described in Materials and Methods section. Undigested (U) LMPCR products or products digested with TaqI (T) or Eco0109 (E) are indicated. (C) 5′Dβ signal ends were assayed in ligated Jβ1M4/ω DN thymocyte DNA that was serially diluted into nonligated DNA keeping the total amount of DNA constant. The cell equivalents of ligated DNA are indicated. Molecular weight markers are shown.
The inability to detect Jβ1M4 5′Dβ1 signal ends could result from efficient cleavage at this RSS followed by rapid resolution of signal ends through signal join formation or through hybrid and open-and-shut join formation which may occur in settings where standard joining of RAG-1/2 cleavage products is prohibited (26–29). Analysis of Jβ1M4/ω DN thymocyte DNA by PCR revealed that Vβ/Dβ signal and hybrid joins were derived entirely from the Jβ1ω allele as evidenced by the digestion of PCR products with Eco0109 and not Taq I (Fig. 4) . PCR analysis of Jβ1M4 DJβ rearrangements in 20 previously described Jβ1M4/ω T cell hybridomas demonstrated that none had undergone Vβ/Dβ hybrid join formation (22; data not shown). Furthermore, sequence analyses of these rearrangements revealed only one with sequence heterogeneity at the 5′Dβ1 heptamer-coding junction consistent with possible open-and-shut joining (data not shown). Together these data demonstrate that the absence of detectable Jβ1M4 5′Dβ1 signal ends in Jβ1M4/ω DN thymocytes does not result from efficient cleavage at this RSS with rapid resolution of signal ends as signal, open-and-shut or hybrid joins. Rather the inability to detect these ends reflects a primary defect at the DNA cleavage step due to constraints imposed by the 5′Dβ1 RSS beyond simply having an appropriate spacer length.
Figure 4.
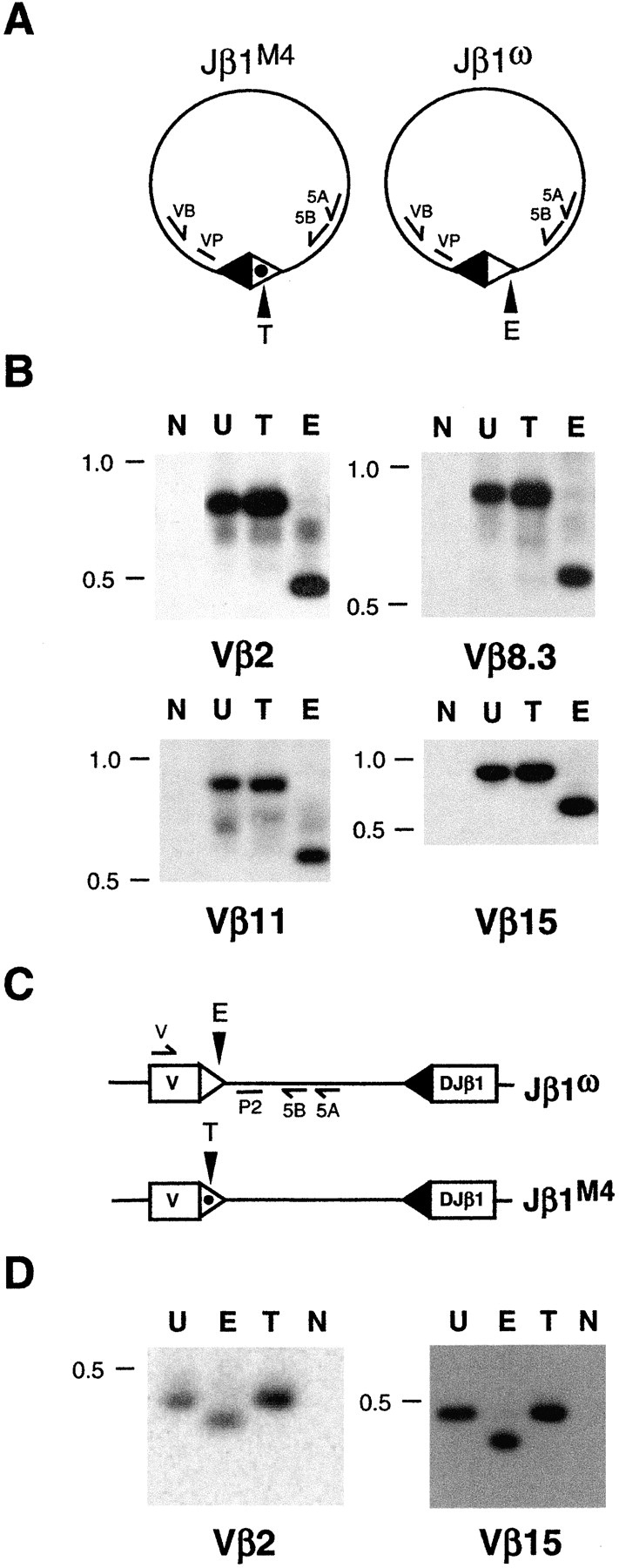
Vβ/Dβ signal and hybrid joins. Shown are schematics of Vβ/Dβ signal joins (A) and Vβ/Dβ hybrid joins (C) on the Jβ1M4 and Jβ1ω alleles. The Vβ 23-RSS (filled triangle) and the Jβ1M4 and Jβ1ω 5′Dβ 12-RSSs (open and open dotted triangles, respectively), the Eco0109 (E) and Taq I (T) sites in the 5′Dβ and Jβ1.2 RSSs are indicated. (B) Vβ/Dβ signal joins were amplified from Jβ1M4/ω DN thymocyte DNA by heminested PCR using the 5A/5B oligonucleotide primers and VB2 (Vβ2), VB8.3 (Vβ8.3), VB11 (Vβ11), or VB15 (Vβ15) oligonucleotide primers. PCR products were probed with VP2, VP8.3, VP11, and VP15 oligonucleotides, respectively. (D) Vβ/Dβ hybrid joins were amplified by heminested PCR using the 5A/5B oligonucleotide primers and V2 (Vβ2) or V15 (Vβ15) oligonucleotide primers. PCR products were probed with the P2 oligonucleotide. VP8.3, VP11, and VP15, respectively. Shown are PCR products from reactions without template DNA (N) or those from reactions with template DNA after the PCR products were undigested (U) or TaqI (T) or Eco0109 (E) digested. Molecular weight markers are shown.
Unique Restrictions Coordinate DNA Cleavage during Vβ to Dβ Rearrangement.
To determine whether the novel restriction on DNA cleavage at the 5′ Dβ1 RSS is coordinated with Vβ RSSs, Jβ1M4/M4, and Jβ1ω/ω mice were bred to SJL mice to generate SJLβ/Jβ1M4 and SJLβ/Jβ1ω mice, respectively. The SJL TCRβ locus has a 100 kb deletion that deletes nine functional Vβ gene segments, including Vβ8.3, Vβ11, and Vβ12 (Fig. 5 A). Thus, DNA DSBs at RSSs flanking these Vβ gene segments in SJLβ/Jβ1M4 and SJLβ/Jβ1ω mice must be derived from the Jβ1M4 and Jβ1ω alleles, respectively. Signal ends from DNA cleavage at Vβ8.3, Vβ11, and Vβ12 RSSs on the Jβ1ω allele were detected over a 100-fold dilution of linker ligated DNA from SJLβ/Jβ1ω DN thymocytes (Fig. 5 B). Strikingly, Vβ8.3, Vβ11, and Vβ12 RSS signal ends from the Jβ1M4 allele were not detected over similar dilutions of linker ligated DNA from SJLβ/Jβ1M4 DN thymocytes (Fig. 5 B). The observed differences were not due to differential LMPCR efficiencies between the two samples as signal ends from cleavage at Vβ2, Vβ15, and 5′Dβ1 RSSs (present on the Jβ1M4, Jβ1ω, and SJLβ alleles) were detected at similar levels in SJLβ/Jβ1M4 and SJLβ/Jβ1ω thymocyte DNA (Fig. 5 B). As expected, 5′Dβ1 RSS cleavage intermediates in SJLβ/Jβ1M4 thymocytes were derived entirely from the SJLβ allele (data not shown). Together these findings demonstrate that Vβ to Dβ rearrangement proceeds through a coordinate DNA cleavage step that is constrained by RSSs beyond simple 12/23 compatibility, as replacing the 5′Dβ1 12-RSS with the Jβ1.2 12-RSS prevents cleavage at native Vβ 23-RSSs on the Jβ1M4 allele.
Figure 5.
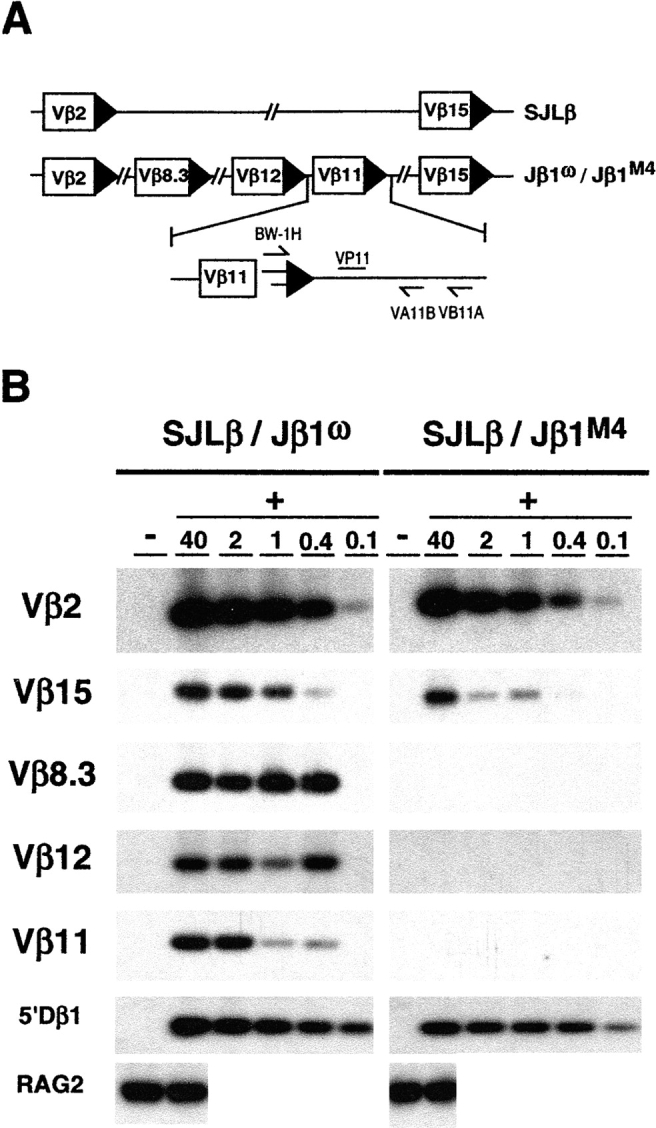
Coordination of DNA cleavage during Vβ to Dβ rearrangement. (A) Schematic of Vβ 2, 8.3, 12, 11, and 15 gene segments on the SJLβ, Jβ1M2, and Jβ1M4 alleles. Also shown are the oligonucleotides used to amplify BW linker ligated Vβ11 signal ends (BW-1H, VA11, and VB11). (B) Ligated (+) DNA from SJLβ:Jβ1M2 and SJLβ:Jβ1M4 DN thymocytes was diluted into nonligated (−) DNA (relative dilutions are indicated) and assayed by a hemi-nested LMPCR for cleavage at the Vβ 2, 15, 8.3, 12, and 11 RSSs. PCR products were probed with the VP2, VP15, VP8.3, VP12, and VP11 oligonucleotides, respectively. Assay of cleavage at the 5′Dβ RSS was performed as described in the legend to Fig. 3. RAG-2 gene PCR products on ligated and nonligated DNA samples are also shown.
Discussion
By analyzing mice with RSS mutations in the endogenous TCRβ locus, we demonstrate that DNA cleavage is coordinated between appropriate RSS pairs in the context of chromosomal V(D)J recombination. Importantly, this requirement restricts the introduction of DNA DSBs primarily to pairs of RSSs flanking gene segments that subsequently participate in coding join formation. Cleavage at Jβ RSSs fails to occur in the absence of a functional 3′ Dβ RSS (Jβ1M2 allele). Similarly, cleavage at Vβ RSSs fails to occur in the absence of a functional 5′ Dβ RSS (Jβ1M4 allele) demonstrating that coordination of DNA cleavage occurs over chromosomal distances in excess of 400 kb.
These findings reveal several important constraints imposed on the generation of paired DNA DSBs during chromosomal V(D)J recombination. As was observed from in vitro analyses, we find that DNA cleavage in vivo is also regulated within the context of the 12/23 rule (20, 21, 30). However, pairing and cleavage between the 5′Dβ1 12-RSS and Vβ 23-RSSs on the Jβ1M4 allele did not occur, demonstrating the existence of restrictions on RSS synapsis beyond 12/23 compatibility. This may be due to the functional synapsis of Vβ/Dβ and Dβ/Jβ, but not Vβ/Jβ, 12/23 RSS pairs. Alternatively, expression of developmental stage and/or lineage specific factors may alter the ability of the V(D)J recombinase to use specific RSSs or RSS pairs to generate a synaptic complex. Such factors may function, for example, by promoting RSS synapsis over great distances in the chromosome. Importantly, these findings suggest that some aspects of the developmental regulation of variable region gene assembly may occur at the level of the V(D)J recombination reaction itself.
Cleavage at Vβ or Jβ RSSs lacking appropriate RSS partners (Jβ1M4 or Jβ1M2 alleles, respectively) was not coordinated through pairing with cryptic RSSs which are predicted to occur at significant frequencies within the genome. This suggests that mechanisms are in place that disfavor synapsis involving cryptic RSSs. Such mechanisms would limit rearrangements involving cryptic RSS within the TCRβ locus which would be nonfunctional and possibly preclude subsequent rearrangements. Furthermore, rearrangements involving cryptic RSSs outside of the locus could result in chromosomal deletions, inversions, or translocations (see below).
Coordination of the DNA cleavage step also promotes efficient assembly of variable region genes by limiting the generation of nonpaired DNA DSBs. For example, cleavage at isolated RSSs during Dβ to Jβ rearrangement could lead to the introduction of as many as 15 DSBs at a single TCRβ allele (all 3′Dβ and Jβ RSSs). DNA DSBs not ultimately resolved as coding and signal joins would have to be resolved as open-and-shut or hybrid joins. Open-and-shut joins accompanied by signal end nucleotide loss would preclude subsequent rearrangement of the gene segment. Hybrid join formation would limit further rearrangements by changing the RSS spacer lengths of the involved gene segments. In addition, hybrid join formation may prevent expression of fully assembled variable region genes. For example, hybrid join resolution of DNA DSBs at Vβ and Jβ gene segments flanking a fully assembled (VDJβ) variable region gene would invert the orientation of the variable region gene with respect to the constant region gene. In this regard the requirement for cleavage at Vβ 23-RSSs to be coordinated with 5′Dβ 12-RSSs, and not Jβ 12-RSSs, prevents both nonpaired and paired introduction of DNA DSBs at Vβ 23-RSSs and Jβ 12-RSSs.
The generation of DNA DSB intermediates during V(D)J recombination poses a risk for genomic instability through their resolution as chromosomal translocations. This potential risk is underscored by the high incidence of chromosomal translocations and lymphoid tumors in mice deficient in proteins involved in the response to and repair of DNA DSBs (31–34). The requirement for coordinated DNA cleavage reduces the risk of chromosomal translocations by limiting the generation of nonpaired signal ends which could be joined to DNA DSBs generated elsewhere in the genome or serve as substrates for RAG-1/2–mediated transposition. This requirement also promotes chromosomal stability within the context of paired DNA DSB generation. Variable region genes generated through interchromosomal V(D)J recombination, resulting in chromosomal translocations, are found at very low frequencies in mature T cells (35). Analyses of extrachromosomal recombination substrates have suggested that interchromosomal V(D)J recombination may be prohibited at the joining step with persisting DNA DSBs signaling apoptosis (36). These findings, although controversial (37), suggest that developing lymphocytes may attempt interchromosomal recombination at a higher frequency than would be predicted from analyses of completed rearrangements in mature lymphocytes.
Our findings demonstrate that coordination of the DNA cleavage step occurs preferentially in an intrachromosomal manner. In this regard the 3′Dβ1 RSS on the Jβ1ω allele coordinates cleavage with Jβ RSSs on the Jβ1ω allele (intrachromosomal) and not the Jβ1M2 allele (interchromosomal) in Jβ1M2/ω thymocytes. Similarly, the 5′Dβ1 RSS on the SJLβ allele coordinates cleavage only with the Vβ RSSs on the SJLβ allele in SJLβ/Jβ1M4 thymocytes. These findings demonstrate that interchromosomal V(D)J recombination is prohibited primarily at the DNA cleavage step but do not exclude the possibility that mechanisms may also exist at the joining step of the reaction. V(D)J recombination between two alleles of an antigen receptor locus would result in a chromosomal translocation that would not likely promote genomic instability. Importantly, though, this restriction would also be expected to limit recombination between different loci and to cryptic RSSs in nonreceptor loci which could lead to pathologic chromosomal translocations. The bias for coordinating cleavage between RSSs on the same allele may be due to the probability of synapsis between RSSs linked on the same versus separate chromosomes or to features of synaptic complex assembly. Alternatively, it may be inherent to the manner in which the V(D)J recombination reaction is regulated if, for example, mechanisms are in place to ensure that a single allele rearranges at a time. Such mechanisms may be important to allow for allelic exclusion, which requires that a newly assembled variable region gene be “tested” to determine whether it encodes a functional antigen receptor chain before rearrangement proceeds on the other allele (38).
Acknowledgments
We thank O. Kanagawa for helpful advice and reagents and E. Unanue, K. Murphy, M. Krangel, and B. Khor for critical review of the manuscript.
This work is supported in part by the National Institutes of Health grant 1 R01 AI47829-01 (B.P. Sleckman). B.P. Sleckman is a recipient of an Investigator Award in General Immunology and Cancer Immunology from the Cancer Research Institute and a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund. R.E. Tillman is supported by a predoctoral training grant in tumor immunology from the Cancer Research Institute. M.M. Hughes is supported by a National Institutes of Health graduate training grant.
Footnotes
Abbreviations used in this paper: DSB, double strand break; LMPCR, ligation-mediated PCR; RAG, recombinase activating gene; RSS, recombination signal sequence.
References
- 1.Tonegawa, S. 1983. Somatic generation of antibody diversity. Nature. 302:575–581. [DOI] [PubMed] [Google Scholar]
- 2.Schatz, D.G., M.A. Oettinger, and M.S. Schlissel. 1992. V(D)J recombination: molecular biology and regulation. Annu. Rev. Immunol. 10:359–383. [DOI] [PubMed] [Google Scholar]
- 3.van Gent, D.C., J.F. McBlane, D.A. Ramsden, M.J. Sadofsky, J.E. Hesse, and M. Gellert. 1995. Initiation of V(D)J recombination in a cell-free system. Cell. 81:925–934. [DOI] [PubMed] [Google Scholar]
- 4.McBlane, J.F., D.C. van Gent, D.A. Ramsden, C. Romeo, C.A. Cuomo, M. Gellert, and M.A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 83:387–395. [DOI] [PubMed] [Google Scholar]
- 5.Gellert, M. 1997. Recent advances in understanding V(D)J recombination. Adv. Immunol. 64:39–64. [DOI] [PubMed] [Google Scholar]
- 6.Weaver, D.T. 1995. V(D)J recombination and double-strand break repair. Adv. Immunol. 58:29–85. [DOI] [PubMed] [Google Scholar]
- 7.Moshous, D., I. Callebaut, R. de Chasseval, B. Corneo, M. Cavazzana-Calvo, F. Le Deist, I. Tezcan, O. Sanal, Y. Bertrand, N. Philippe, et al. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 105:177–186. [DOI] [PubMed] [Google Scholar]
- 8.Jeggo, P.A. 1998. Identification of genes involved in repair of DNA double-strand breaks in mammalian cells. Radiat. Res. 150:S80–91. [PubMed] [Google Scholar]
- 9.Lewis, S.M., J.E. Hesse, K. Mizuuchi, and M. Gellert. 1988. Novel strand exchanges in V(D)J recombination. Cell. 55:1099–1107. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal, A., Q.M. Eastman, and D.G. Schatz. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 394:744–751. [DOI] [PubMed] [Google Scholar]
- 11.Hiom, K., M. Melek, and M. Gellert. 1998. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 94:463–470. [DOI] [PubMed] [Google Scholar]
- 12.Sleckman, B.P., J.R. Gorman, and F.W. Alt. 1996. Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu. Rev. Immunol. 14:459–481. [DOI] [PubMed] [Google Scholar]
- 13.Hesslein, D.G., and D.G. Schatz. 2001. Factors and forces controlling V(D)J recombination. Adv. Immunol. 78:169–232. [DOI] [PubMed] [Google Scholar]
- 14.Hempel, W.M., P. Stanhope-Baker, N. Mathieu, F. Huang, M.S. Schlissel, and P. Ferrier. 1998. Enhancer control of V(D)J recombination at the TCRbeta locus: differential effects on DNA cleavage and joining. Genes Dev. 12:2305–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanhope-Baker, P., K.M. Hudson, A.L. Shaffer, A. Constantinescu, and M.S. Schlissel. 1996. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 85:887–897. [DOI] [PubMed] [Google Scholar]
- 16.McMurry, M.T., C. Hernandez-Munain, P. Lauzurica, and M.S. Krangel. 1997. Enhancer control of local accessibility to V(D)J recombinase. Mol. Cell. Biol. 17:4553–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steen, S.B., L. Gomelsky, and D.B. Roth. 1996. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells. 1:543–553. [DOI] [PubMed] [Google Scholar]
- 18.Eastman, Q.M., T.M. Leu, and D.G. Schatz. 1996. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 380:85–88. [DOI] [PubMed] [Google Scholar]
- 19.van Gent, D.C., D.A. Ramsden, and M. Gellert. 1996. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 85:107–113. [DOI] [PubMed] [Google Scholar]
- 20.Hiom, K., and M. Gellert. 1998. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell. 1:1011–1019. [DOI] [PubMed] [Google Scholar]
- 21.Agrawal, A., and D.G. Schatz. 1997. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 89:43–53. [DOI] [PubMed] [Google Scholar]
- 22.Bassing, C.H., F.W. Alt, M.M. Hughes, M. D'Auteuil, T.D. Wehrly, B.B. Woodman, F. Gartner, J.M. White, L. Davidson, and B.P. Sleckman. 2000. Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature. 405:583–586. [DOI] [PubMed] [Google Scholar]
- 23.Sleckman, B.P., C.H. Bassing, M.M. Hughes, A. Okada, M. D'Auteuil, T.D. Wehrly, B.B. Woodman, L. Davidson, J. Chen, and F.W. Alt. 2000. Mechanisms that direct ordered assembly of T cell receptor beta locus V, D, and J gene segments. Proc. Natl. Acad. Sci. USA. 97:7975–7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sleckman, B.P., C.G. Bardon, R. Ferrini, L. Davidson, and F.W. Alt. 1997. Function of the TCRα enhancer in αβ and γδ T cells. Immunity. 7:505–515. [DOI] [PubMed] [Google Scholar]
- 25.Schlissel, M., A. Constantinescu, T. Morrow, M. Baxter, and A. Peng. 1993. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 7:2520–2532. [DOI] [PubMed] [Google Scholar]
- 26.Melek, M., M. Gellert, and D.C. van Gent. 1998. Rejoining of DNA by the RAG1 and RAG2 proteins. Science. 280:301–303. [DOI] [PubMed] [Google Scholar]
- 27.Bogue, M.A., C. Wang, C. Zhu, and D.B. Roth. 1997. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 7:37–47. [DOI] [PubMed] [Google Scholar]
- 28.Han, J.O., S.B. Steen, and D.B. Roth. 1997. Ku86 is not required for protection of signal ends or for formation of nonstandard V(D)J recombination products. Mol. Cell. Biol. 17:2226–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis, S.M., and J.E. Hesse. 1991. Cutting and closing without recombination in V(D)J joining. EMBO J. 10:3631–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiom, K., and M. Gellert. 1997. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 88:65–72. [DOI] [PubMed] [Google Scholar]
- 31.Liao, M.J., and T. Van Dyke. 1999. Critical role for Atm in suppressing V(D)J recombination-driven thymic lymphoma. Genes Dev. 13:1246–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao, Y., D.O. Ferguson, W. Xie, J.P. Manis, J. Sekiguchi, K.M. Frank, J. Chaudhuri, J. Horner, R.A. DePinho, and F.W. Alt. 2000. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 404:897–900. [DOI] [PubMed] [Google Scholar]
- 33.Xu, Y., T. Ashley, E.E. Brainerd, R.T. Bronson, M.S. Meyn, and D. Baltimore. 1996. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10:2411–2422. [DOI] [PubMed] [Google Scholar]
- 34.Elson, A., Y. Wang, C.J. Daugherty, C.C. Morton, F. Zhou, J. Campos-Torres, and P. Leder. 1996. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. USA. 93:13084–13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aster, J.C., and J. Sklar. 1992. Interallelic V(D)J trans-rearrangement within the beta T cell receptor gene is infrequent and occurs preferentially during attempted D beta to J beta joining. J. Exp. Med. 175:1773–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han, J.O., S.B. Steen, and D.B. Roth. 1999. Intermolecular V(D)J recombination is prohibited specifically at the joining step. Mol. Cell. 3:331–338. [DOI] [PubMed] [Google Scholar]
- 37.Tevelev, A., and D.G. Schatz. 2000. Intermolecular V(D)J recombination. J. Biol. Chem. 275:8341–8348. [DOI] [PubMed] [Google Scholar]
- 38.Alt, F.W., G.D. Yancopoulos, T.K. Blackwell, C. Wood, E. Thomas, M. Boss, R. Coffman, N. Rosenberg, S. Tonegawa, and D. Baltimore. 1984. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 3:1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]