Abstract
Colonization of the nasopharynx is the initial step in all infections caused by Streptococcus pneumoniae. The antibody response to carriage was examined in an experimental model of human colonization in healthy adults. Asymptomatic colonization was detected in 6/14 subjects and continued for up to 122 d. Susceptibility to carriage did not correlate with total serum immunoglobulin (Ig)G to the homotypic capsular polysaccharide. All of the colonized subjects, in contrast, developed a serum IgG and secretory IgA response to a 22 kD protein, whereas 7 of 8 subjects who did not become colonized had preexisting antibody to this protein. Analysis of the 22 kD protein identified it as the NH2-terminal region of pneumococcal surface protein A (PspA). Our findings provide evidence for the role of antibody to this protein fragment in preventing pneumococcal carriage by humans.
Keywords: Streptococcus pneumoniae, vaccine, carriage, colonization, pneumococcal surface protein A
Introduction
Streptococcus pneumoniae (the pneumococcus) colonizes the mucosal surface of the human nasopharynx. Infection occurs when the organism spreads beyond this niche into normally sterile parts of the respiratory tract or, in the most serious cases, into the bloodstream. Colonization, therefore, is the crucial first step in the pathogenesis of all pneumococcal disease (1).
The acquisition and widespread dissemination of resistance to β-lactam antibiotics as well as resistance to other classes of antimicrobials has complicated the treatment of pneumococcal infection (2). This problem has emphasized the need for preventive strategies against this common pathogen. The immunodominant antigen of the pneumococcus is the capsular polysaccharide (PnPS) of which there are 90 known types. Antibody to the PnPS of a given type is thought to provide protection only to an isolate of the homologous type (3). Although colonization results in an increase in type-specific serum antibody, it remains unclear whether carriage is an immunizing event for a given type (4). Immunization with a mixture of PnPS of the most prevalent types is protective in adults; but because young children do not respond to T cell–independent polysaccharide antigens, this prophylactic strategy is unsuccessful in the population at highest risk of disease. This finding has led to the recent introduction of a vaccine based on conjugate technology that couples PnPS to an immunogenic carrier protein resulting in a shift to a T cell–dependent immune response (5). To produce an epidemiologically effective PnPS-protein conjugate vaccine, however, multiple types of the 90 known PnPSs must be conjugated to a protein carrier. This requirement results in a complex and expensive multi-component vaccine with restricted potential efficacy because of the limited number of PnPS types that can be included in any single formulation, the possibility of serotype replacement, and the high titer type-specific antibody response to some but not to all types (6, 7).
The focus of this study is the identification of pneumococcal proteins that are immunogenic during carriage. A pneumococcal protein vaccine that would specifically interfere with carriage would avoid many of the problems associated with vaccines based on PnPS and could potentially have the greatest impact on the prevention of disease. A number of pneumococcal proteins that have been shown to induce opsonophagocytic antibodies or to offer some degree of protection in murine models are currently under investigation as novel vaccine candidates (8). Mice, however, are not naturally colonized by S. pneumoniae and, when subjected to infection via artificial routes, are variably susceptible to a limited number of pneumococcal types (9). To avoid the limitations of animal models, this study utilizes experimental human colonization and shows that the presence of serum antibody to a single pneumococcal protein correlates with susceptibility to carriage.
Materials and Methods
Carriage Model.
Pneumococcal carriage was studied at the Respiratory Pathogens Research Unit, Baylor College of Medicine according to an Institutional Review Board approved protocol and written informed consent acknowledging the use of an infectious agent and the availability of treatment for any signs of infection. 14 healthy adults were given a 0.1 ml intranasal inoculum to each naris containing 5,000 CFU (FS92, 123, 162, 177), 7,000 CFU (FS1, 62, 80, 105, 129, 138), or 17,000 CFU (FS11, 31, 150, 163) of a type 23F clinical isolate, P833. This penicillin-sensitive isolate was obtained from a child with otitis media by tympanocentesis and stored at –80°C after minimal passage. Study inclusion criteria included; age between 18 and 40 yr old, HIV seronegative, nonpregnant, nonsmoking, an intact spleen, consume <3 alcoholic beverages per day, no history of pneumococcal vaccination, no penicillin allergy, no history of recurrent respiratory tract infections or chronic illnesses, no close contact with individuals at increased risk for pneumococcal disease, and lack pneumococcal colonization for 4 wk before the inoculation as determined by nasal and throat cultures. A separate study was performed with a clinical type 6B isolate. Serum and nasal washes were obtained before inoculation and every 2 wk after inoculation until pneumococcal carriage was no longer detected in nasal or throat cultures in two consecutive visits.
Pneumococcal Strains and Products.
All pneumococcal isolates and strains were grown in semisynthetic media (C+Y media, pH 6.8) until mid-log phase was reached (OD620 = 0.4) (10). Serotype was confirmed by using the quellung reaction with type specific antisera purchased from the Staten Serum Institut. Culture supernatants were collected after centrifugation for 20 min at 900 g. The bacterial pellet was washed with an equal volume of cold PBS (pH 7.5) and then resuspended in 5% of the original culture volume of cold PBS for lysis by sonication.
Western Blot Analysis.
Bacterial sonicates and concentrated supernatants were used in Western blot analysis by using 10 μg total protein/lane and separated on SDS-PAGE gels (7.5–15%). After electrophoretic transfer onto an Immobilon-P membrane (Millipore), transfer was visualized by staining membranes with Ponceau S (Sigma-Aldrich). Individual lanes were incubated 16 h at 25°C with pre- or postinoculum sera (diluted 1:10,000) or nasal washes (diluted 1:500) from individual volunteers. Protein-specific antibodies were detected with goat anti–human IgG or IgA specific antisera conjugated to alkaline phosphatase (Sigma-Aldrich). 20-fold concentrated pneumococci were treated with PBS containing 2% choline chloride in PBS for 20 min at 4°C to isolate the choline-binding proteins. N-terminal sequencing was determined by Edman degradation.
DNA Sequencing.
Genomic DNA was isolated as described previously (11). The pspA gene was amplified by PCR with primers LSM12 and SKH3, which bind to conserved flanking regions (12). Primers, 5′-AACTGAAGAGAAAGCAAAGC-3′ and 5′-CGTCTAACTCATCAATCTTATCAC-3′, internal to the original primers were used to verify both strands of the sequence of the pspA of strain P833. Sequence analysis was repeated on DNA generated in four independent PCR amplifications.
Enzyme-linked Immunosorbant Assays.
A PCR product containing the nucleotide sequence of pspA from P833 and unique restriction enzyme sites was amplified with the use of primers, 5′-CCGGAATTCTCTCAGCTTCTTCTG-3′ and 5′-GGAATTCCATATGGAAGAAGCTCCC-3′ which contain EcoRI and a NdeI sites respectively. After digestion and ligation of the PCR product to the pET28a vector, His-tagged pneumococcal surface protein A (PspA)31–189 was expressed in the presence of IPTG and purified from E. coli strain BL21 on an activated His-Bind Resin column (Novagen). A similar procedure was followed for generating fragments from the mature PspA from amino acids 31–92, 31–162, and 87–162.
The purified His-tagged PspA31–189 was used to coat 96-well Immulon 2 High Binding plates (DYNEX Technologies) by incubating 0.5 μg/ml in coating buffer (0.015 M Na2CO3, 0.035 M NaHCO3, pH 9.6). After overnight incubation at 4°C, the plates were washed with PBS-Tween 20, blocked with 1% BSA for 1 h at 37°C, and washed again. Twofold serial dilutions of human serum were added and then incubated overnight at 4°C. PspA-specific antibodies were detected by goat anti–human IgG-alkaline phosphatase (Sigma-Aldrich), developed with pNPP, and the absorbance at 415 nm recorded. Titers were determined by calculating the sample dilution at which the absorbance was equal to 0.3. The ELISA to detect antibody to the type 23F capsular polysaccharide followed the protocol established by the Centers for Disease Control.
Lactoferrin Binding Assay.
Fractions containing choline-binding proteins, His-tagged PspA31–189, or purified human lactoferrin (Sigma-Aldrich) were separated on 12.5% SDS-PAGE gels and transferred to Immobilon-P membranes. The ability of PspA to bind human lactoferrin was assessed by using a previously described protocol (13, 14).
Infant Rat Colonization.
The model of pneumococcal carriage in infant rats has been described previously (15). Cells of a streptomycin-resistant transformant of strain P833 were mixed with an equal number of cells of the last isolate cultured from subject FS1 and inoculated into the nares of 2-d-old Sprague-Dawley rats (106 cfu/pup). Nasal washes were examined for 7 d after inoculation by plating on media with and without streptomycin (200 μg/ml) to quantify the carriage of each strain.
Results
Antibody Response to Pneumococcal Proteins.
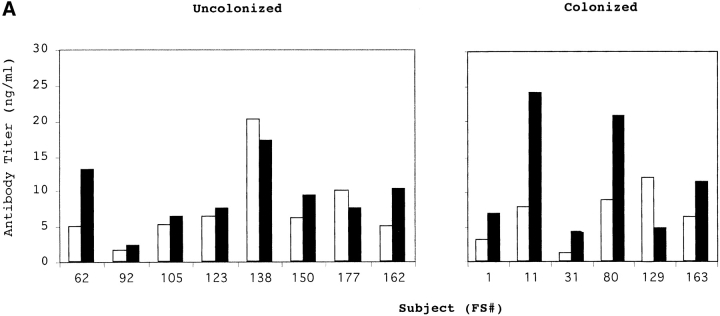
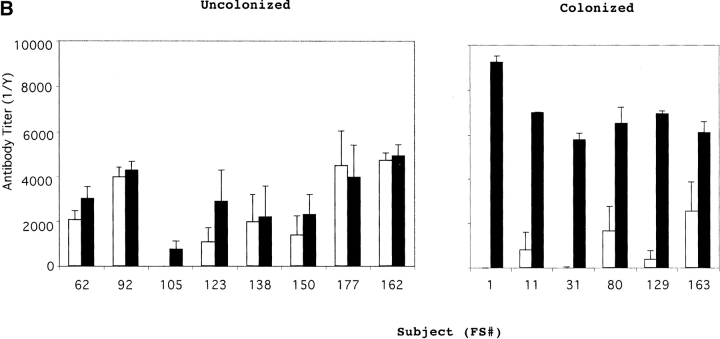
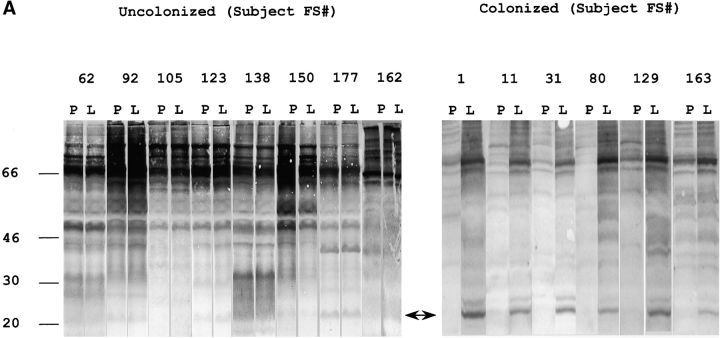
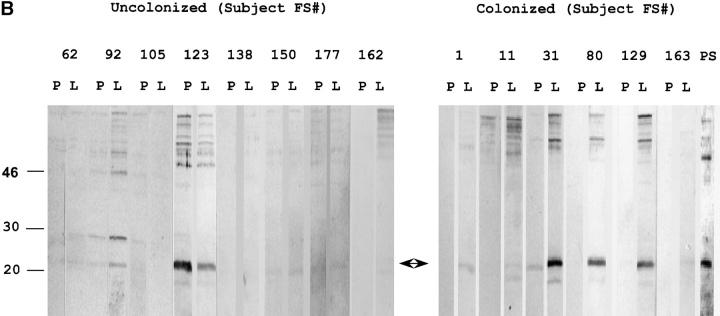
14 healthy adults received an intranasal inoculation of a type 23F clinical isolate. 6 of 14 volunteers carried the type 23F pneumococcus asymptomatically with a duration of carriage ranging from 27–122 d (colonized group); colonization could not be detected in the remaining eight subjects (uncolonized group). Susceptibility to colonization was not predicted by the preinoculation level of serum IgG against the type 23F capsular polysaccharide (PS-23F) as determined by ELISA (Fig. 1 A). On the basis of this unexpected observation, we examined the antibody response to pneumococcal proteins. Western blot analysis of whole cell sonicates of the inoculated strain (P833) were used to compare patterns of reactivity in serum before inoculation (preinoculation) with those of the earliest time point that colonization was not detected (postinoculation) from both colonized and uncolonized subjects. Uncolonized subjects demonstrated no detectable change in the reactivity of serum IgG pre- and postinoculation (Fig. 2 A). In contrast, 6/6 colonized subjects developed an IgG response to a band migrating at 22 kD. Although there were increases in reactivity to other bands after carriage, preinoculation serum IgG from the colonized group did not demonstrate reactivity to the 22 kD band. In addition, 7/8 subjects that did not become colonized had preexisting IgG recognizing this band (P < 0.004 compared with colonized group, Fisher's exact test). A similar pattern of IgG reactivity to this band was seen in immunoblots of nasal washes (unpublished data). Nasal washes were also used to characterize the secretory IgA response (Fig. 2 B). 6/6 colonized versus 1/8 uncolonized subjects acquired or increased sIgA reactivity to the 22 kD band (P < 0.004, Fisher's exact test). These results demonstrate a correlation between the immune response to this band and susceptibility to carriage.
Figure 1.
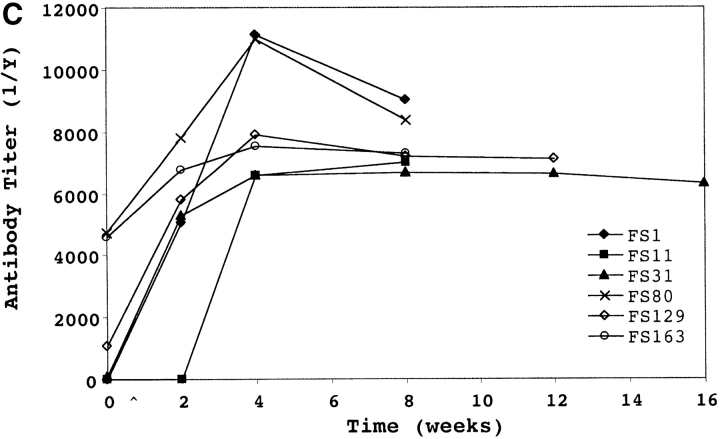
Serum antibody response to (A) type 23F capsular polysaccharide (PS-23F) and (B and C) the recombinant NH2-terminal fragment of PspA (PspA31–189) from strain P833 used as the inoculum in experimental human carriage studies. PS-23F or PspA31–189 was used in ELISAs to determine the PS-23F or PspA31–189 specific IgG titers in uncolonized and colonized subjects (FS#) before (white bars) and at first time point that colonization was no longer detected (black bars). Values shown in panel A are ng/ml amounts of antibody and are from a representative experiment, in B are the mean antibody titers ±SEM for four independent experiments, and in C are a representative determination of the kinetics of the serum IgG response in colonized patients. ^ indicates point of inoculation. The final time point for each subject (FS#) was 2 wk after the last positive throat or nasal culture.
Figure 2.
Western blot analysis demonstrating the antibody response to pneumococcal proteins. Antibodies from individual subjects (indicted by FS#) were used to detect differences in reactivity in immunoblots incubated with specimens obtained preinoculation (P) compared with first time point that colonization was no longer detected (L). Total protein as detected by staining with Ponceau S is shown in lane PS. An arrow indicates the 22-kD band to which the antibody response correlates with susceptibility to carriage. Size markers are in kilodaltons. (A) Whole cell sonicates of the strain used in experimental human carriage studies were used to detect differences in IgG reactivity in serum. (B) Concentrated (100×) culture supernatants from the strain used in experimental human carriage studies were used to detect differences in secretory IgA in nasal washes.
Identification and Characterization of the 22 kD Band.
Further analysis showed that 22 kD band was the most abundant product in culture supernatants (Fig. 2 B, lane PS). After additional purification of culture supernatants by size fractionation, the NH2-terminal sequence, EEAPVAKQFQAERDY, a 14/15 match to a database entry to the NH2 terminus of the mature form of PspA from a type 23F strain, was obtained. This finding was unexpected, as the size of PspA normally ranges from 67–99 kD (12). Sequencing of the pspA gene from P833 revealed a deletion of a single adenosine residue within a group of eight tandem adenosines around position 540 that created a frame-shift leading to premature termination of the mature protein at amino acid 189 (Fig. 3). Thus, the truncated protein, PspA31–189, lacks the choline-binding domain that acts to anchor PspA to the cell surface and prevent its secretion (16).
Figure 3.
Nucleotide sequence and predicted translation product for an internal region of pneumococcal surface protein A (PspA). (A) database entry for a type 23F strain (GenBank/EMBL/DDBJ accession no. AF071811), (B) corresponding sequence for P833 used in nasal inoculations showing a frame-shift mutation within eight adenosines (shown by overlining) causing truncation of PspA as indicated, and (C) corresponding sequence from the last isolate obtained from subject FS1 which contains a compensatory two base pair deletion restoring expression of full-length PspA. Dash indicates a nucleotide deletion. The nucleotide and amino acid position is shown relative to the initiation codon.
Serum IgG Response to PspA31−189.
The immune response to the PspA fragment was quantified by ELISA by using recombinant PspA31–189 from P833 (Fig. 1 B). Comparison of IgG titers to PspA31–189 in pre- and postinoculation serum showed that 7/8 and 2/6 subjects in the uncolonized and colonized groups respectively had preexisting antibody (1/titer > 1,000; P < .09, Fisher's exact test). Serum IgG detected by ELISA correlated with results from Western analysis with the exception of preinoculation IgG to PspA31–189 which was observed in subjects FS80 and FS163 only by ELISA. All of the colonized but none of the uncolonized subjects developed a significant rise in IgG levels to the PspA31–189 (P < 0.0003, Fisher's exact test). When subfragments of PspA31–189 were analyzed, the IgG response was found to be specific to the hypervariable region COOH-terminal to amino acid 92 (unpublished data; reference 12). In contrast to the ELISA results for PspA31–189, the level of preexisting serum IgG specific to the type 23F polysaccharide did not correlate with susceptibility to becoming colonized (mean IgG preinoculation in uncolonized/colonized = 2.75 for PspA31–189 versus 1.17 for PS-23F; Fig. 1). In addition, the colonized group had an average eightfold increase in IgG to PspA31–189, whereas the average increase in IgG titer was only twofold to PS-23F during carriage. The increase in IgG titer to PspA31–189 occurred by 4 wk postinoculation (Fig. 1 C). As carriage was maintained for up to 122 d postinoculation, the rise in the level of IgG to PspA31–189 does not appear to correlate temporally with the loss of carriage. Serum from subjects challenged with an unrelated type 6B pneumococcal isolate (6/7 colonized) had no significant rise by ELISA in IgG titers to PspA31–189 from strain P833 suggesting that the antibody response to this fragment may be strain specific.
Stability of Expression of PspA.
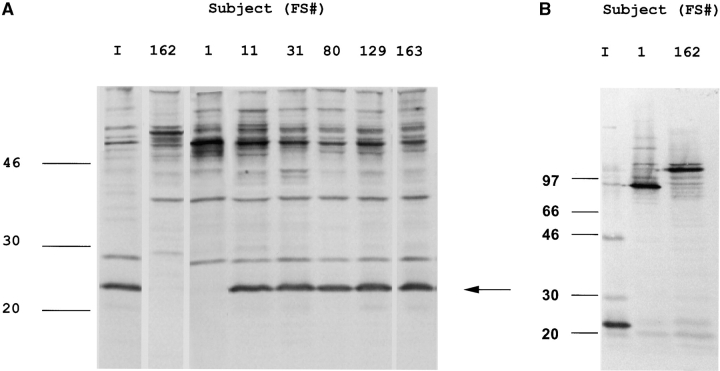
The observation that an isolate expressing a truncated and secreted PspA was still able to colonize the human nasopharynx was unexpected. To determine if this characteristic was maintained throughout the period of carriage, the last isolate from each colonized subject was analyzed. Western blots demonstrated that pneumococci from 5/6 subjects expressed PspA31–189 throughout the entire period of colonization suggesting that the full-length protein is not essential for maintenance of the carrier state (Fig. 4 A). The last isolate from subject FS1 (P833-FS1), in contrast, lost expression of the 22 kD band. Western analysis showed that the loss of the 22 kD band from P833-FS1 was associated with the acquisition of a prominent choline-binding protein of the predicted size of the full-length PspA of P833 (Fig. 4 B). Nucleotide sequence analysis of P833-FS1 showed the loss of two additional nucleotides upstream of the adenosine deletion in P833 restored the in-frame expression of pspA (Fig. 3). As P833-FS1 was the same serotype and the sequence of the entire pspA was otherwise identical to P833, it was concluded that this was a mutant of the same strain and that this mutation most likely occurred in vivo. Thus, P833, a low-passage clinical isolate from a child with otitis media, and P833-FS1 recovered from the nasopharynx after carriage for 40 d both had distinct frameshift mutations in pspA.
Figure 4.
Western blot analysis showing the stability of expression of PspA. Culture supernatants (A) and whole cell sonicates (B) obtained from the last positive nasal or throat culture from each of the colonized subjects (FS#) were compared with the inoculation strain (lane I) by immunoblotting with post-inoculation serum from subject FS31. The arrow indicates the position of the PspA31–189. The isolate recovered from one of the subjects (FS162) at day 3 postinoculation was found to be a type 6A strain expressing a larger full-length PspA. This subject was then considered in the uncolonized group. Size markers are in kilodaltons.
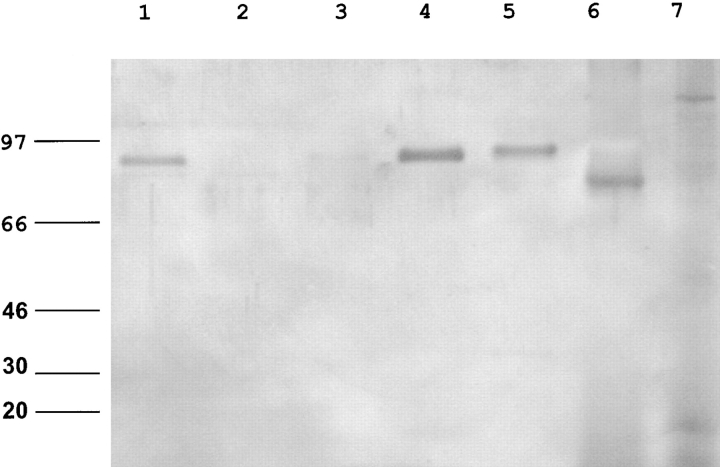
Recent reports suggest that PspA is a lactoferrin binding protein (13). In contrast to the full-length PspA from P833-FS1, neither P833 expressing the truncated protein nor the purified PspA31–189 bound to human lactoferrin, suggesting that human colonization does not require PspA-mediated lactoferrin binding (Fig. 5). In an infant rat model of nasopharyngeal colonization, a mixed inoculum of equal numbers of P833 and P833-FS1 were compared (unpublished data). P833-FS1 out-competed P833 by 48 h after intranasal inoculation, suggesting that there is an advantage to expression of the full-length PspA in carriage in the infant rat, although expression of PspA31–189 is permissive for human colonization.
Figure 5.
Lactoferrin binding by S. pneumoniae. Binding of lactoferrin to extracted choline-binding proteins was detected with an antiserum to human lactoferrin. P282 (PspA+; lane 1), P282::pspA − (lane 2), P833 (inoculation strain; lane 3), last isolate from subject FS1 expressing a full-length PspA (lane 4), a type 6A strain isolated from subject FS162 (lane 5), purified human lactoferrin control (lane 6), and the purified NH2-terminal fragment of PspA (lane 7). Size markers are in kilodaltons.
Discussion
A number of pneumococcal protein vaccine candidates have shown promise in animal studies, although no single immunogen has emerged despite many years of investigation. The current study utilizes an experimental model of colonization by an exclusive human pathogen in the natural host. We focused on the humoral immune response during nasopharyngeal colonization, the first step in the interaction of the pneumococcus and humans. This approach also allowed for the identification of protein targets against which the presence of preexisting antibody may predict susceptibility to carriage.
One of the key features of the experimental model of carriage used in this study was the availability of preinoculation samples. These sera permitted the identification of targets of newly acquired antibody from among the many antigens recognized by preexisting antibody in human serum and secretions. Results of this limited study in 14 adults failed to support the hypothesis that the susceptibility to carriage correlates with diminished levels of type-specific anti-PS antibody (3, 17). Instead, susceptibility to colonization was most closely associated with lack of preexisting systemic (serum IgG) and mucosal (nasopharyngeal sIgA) antibodies against a 22 kD protein subsequently identified as the first 159 amino acids of mature PspA. No other antigens with this characteristic could be detected when serum and nasal washes were screened by Western analysis, a technique limited in its sensitivity by the possibility of antibody against one protein obscuring another. In fact, if the clinical isolate used in the inoculum did not express a truncated form of PspA, allowing for the NH2-terminal portion to be visualized separately from the more conserved COOH-terminal portion, this association may not have been detected (12). The fortuitous use of this natural PspA mutant, however, enabled us to demonstrate a correlation between the presence of preexisting antibody to PspA31–189 and resistance to an intranasal challenge with the homologous strain. Based on this observation, this immunogenic portion of PspA may be an important target of the immune response to the pneumococcus during colonization and would seem to offer promise as a vaccine component to protect specifically against the acquisition of pneumococcal carriage. PspA has previously been shown to be immunogenic and able to generate protection in colonization studies in mice (8, 18). In addition to providing corroborative data in humans, the current study defines a smaller fragment of 96 amino acids (PspA93–189) that may be sufficient as an immunogen against carriage. This fragment of PspA, however, is highly variable in sequence and does not contain amino acids 192–260 previously shown to be important in eliciting cross-protection in murine models (19). As a result of the variability of the NH2-terminal region of PspA, the correlation of preexisting antibody against PspA31–189 and susceptibility to carriage appears to be strain-specific. Our study, therefore, provides evidence to support the immunization strategy proposed by Briles and others based on a mixture of antigenically variable PspA molecules representative of the most common clades but suggests that a different region of PspA may contain the critical epitopes for protection in humans (20).
Although it appears that the NH2-terminal portion of PspA is highly immunogenic and that the immune response to it may block the acquisition of carriage in the natural host, the expression of PspA was noted to be unstable during carriage. Such instability could be a limitation in development of a PspA-based vaccine and we are currently examining the frequency of mutations in PspA in carriage strains. This study shows that S. pneumoniae is able to colonize and persist in the human nasopharynx for as long as 122 d without expression of a full-length form of this major surface protein. A thorough understanding of the contribution of PspA to colonization and disease caused by this organism is still lacking. Two features of PspA in the inoculum strain, however, indicate that it was unlikely to have been fully functional during colonization in 5/6 subjects; PspA31–189 is secreted rather than anchored on the cell surface and it no longer binds to lactoferrin, the only known ligand of PspA. The observation that the challenge strain isolated from an inflamed middle ear space and used in this human model of experimental colonization expressed a truncated protein could be evidence of prior selective immune pressure directed against PspA. This escape mutation resulted in secretion of an immunogenic fragment of PspA and loss of expression of the relatively conserved portions of the protein. The frame-shift mutation in the inoculum strain occurred in a run of eight tandem adenosines. Although not a particularly long repetitive feature in this species with AT rich DNA, tandem repeats are known to be subject to increased rates of mutation by a mechanism involving slip-stranded mispairing (21). In contrast, the last isolate cultured from 1/6 colonized subjects had a second mutation site that restored the expression of a full-length protein. The in vivo instability of expression and frequency of mutations in PspA or other pneumococcal antigens has not been recognized previously. Therefore, the use of experimental carriage studies performed in humans allowed for the identification of a potentially protective immunogen as well as demonstrating a naturally occurring variability in its expression. This study offers a paradigm for using experimental human infection to characterize the immune response to a pathogen and to identify and/or confirm vaccine targets.
Acknowledgments
The authors thank Dr. R. Austrian for critical review of the manuscript and for assistance with quellung experiments.
This work was supported by grants from the U.S. Public Health Service (AI38436 and AI44231 to J.N. Weiser and NO1-AI-65298 to the Respiratory Pathogens Research Unit, Baylor College of Medicine).
References
- 1.Austrian, R. 1986. Some aspects of the pneumococcal carrier state. J. Antimicrob. Chemother. 18:35–45. [DOI] [PubMed] [Google Scholar]
- 2.Tomasz, A. 1995. Pneumococcus at the gates. N. Engl. J. Med. 333:514–515. [DOI] [PubMed] [Google Scholar]
- 3.MacLeod, C.M., R.G. Hodges, M. Heidelberger, and W.G. Bernhard. 1945. Prevention of pneumococcal pneumonia by immunization with specific capsular polysaccharides. J. Exp. Med. 82:445–465. [PMC free article] [PubMed] [Google Scholar]
- 4.Gwaltney, J.J., M. Sande, R. Austrian, and J. Hendley. 1975. Spread of Streptococcus pneumoniae in families. II. Relation of transfer of S pneumoniae to incidence of colds and serum antibody. J. Infect. Dis. 132:62–68. [DOI] [PubMed] [Google Scholar]
- 5.Black, S., H. Shinefield, B. Fireman, E. Lewis, P. Ray, J. Hansen, L. Elvin, K. Ensor, J. Hackell, G. Siber, et al. 2000. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Northern California Kaiser Permanente Vaccine Study Center Group. Pediatr. Infect. Dis. J. 19:187–195. [DOI] [PubMed] [Google Scholar]
- 6.Lipsitch, M., J. Dykes, S. Johnson, E. Ades, J. King, D. Briles, and G. Carlone. 2000. Competition among Streptococcus pneumoniae for intranasal colonization in a mouse model. Vaccine. 18:2895–2901. [DOI] [PubMed] [Google Scholar]
- 7.Obaro, S. 2000. Confronting the pneumococcus: a target shift or bullet change? Vaccine. 19:1211–1217. [DOI] [PubMed] [Google Scholar]
- 8.Ogunniyi, A., R. Folland, D. Briles, S. Hollingshead, and J. Paton. 2000. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. Infect. Immun. 68:3028–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morch, E. 1943. Serological Studies on the Pneumococci. Oxford University Press, London. 193 pp.
- 10.Tomasz, A. 1964. A chemically defined medium for Streptococcus pneumoniae Bacteriol. Proc. 64:29. [Google Scholar]
- 11.Pearce, B.J., Y.B. Yin, and H.R. Masure. 1993. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol. Microbiol. 9:1037–1050. [DOI] [PubMed] [Google Scholar]
- 12.Hollingshead, S., R. Becker, and D. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammerschmidt, S., G. Bethe, P. Remane, and G.S. Chhatwal. 1999. Identification of pneumococcal surface protein A as a lactoferrin-binding protein of Streptococcus pneumoniae. Infect. Immun. 67:1683–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakansson, A., H. Roche, S. Mirza, L.S. McDaniel, A. Brooks-Walters, and D. Briles. 2001. Characterization of binding of human lactoferrin to pneumococcal surface protein A. Infect. Immun. 69:3372–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiser, J.N., R. Austrian, P.K. Sreenivasan, and H.R. Masure. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62:2582–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yother, J., and J.M. White. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagan, R., M. Muallem, R. Melamed, O. Leroy, and P. Yagupsky. 1997. Reduction of pneumococcal nasopharyngeal carriage in early infancy after immunization with tetravalent pneumococcal vaccines conjugated to either tetanus toxoid or diphtheria toxoid. Pediatr. Infect. Dis. J. 16:1060–1064. [DOI] [PubMed] [Google Scholar]
- 18.Briles, D., E. Ades, J. Paton, J. Sampson, G. Carlone, R. Huebner, E. Virolainen, E. Swiatlo, and S. Hollingshead. 2000. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect. Immun. 68:796–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDaniel, L.S., B.A. Ralph, D.O. McDaniel, and D.E. Briles. 1994. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microb. Pathog. 17:323–337. [DOI] [PubMed] [Google Scholar]
- 20.Crain, M.J., W.D. Waltman, J.S. Turner, J. Yother, D.F. Talkington, L.S. McDaniel, B.M. Gray, and D.E. Briles. 1990. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect. Immun. 58:3293–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levinson, G., and G.A. Gutman. 1987. Slipped-stranded mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4:203–221. [DOI] [PubMed] [Google Scholar]