Abstract
1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) is a neurotoxin that causes parkinsonism in humans and nonhuman animals, and its use has led to greater understanding of the pathogenesis of Parkinson’s disease. However, its molecular targets have not been defined. We show that mice lacking the gene for poly(ADP-ribose) polymerase (PARP), which catalyzes the attachment of ADP ribose units from NAD to nuclear proteins after DNA damage, are dramatically spared from MPTP neurotoxicity. MPTP potently activates PARP exclusively in vulnerable dopamine containing neurons of the substantia nigra. MPTP elicits a novel pattern of poly(ADP-ribosyl)ation of nuclear proteins that completely depends on neuronally derived nitric oxide. Thus, NO, DNA damage, and PARP activation play a critical role in MPTP-induced parkinsonism and suggest that inhibitors of PARP may have protective benefit in the treatment of Parkinson’s disease.
Parkinson’s disease (PD) is a common and disabling idiopathic neurodegenerative disorder characterized by tremor, bradykinesia, rigidity, and balance difficulties. These motor abnormalities are attributed to depletion of brain dopamine (DA) that results from the dramatic loss of dopaminergic neurons in the substantia nigra pars compacta (SNpc) (1–3). Although there are therapies available for PD that help alleviate symptoms, they may produce major side effects and lose efficacy over time as they do not modify the progressive neurodegeneration in PD (4). Insight into the neurodegenerative process in PD comes from the use of the selective neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), which replicates parkinsonian motor signs in human and nonhuman animals (5–11). As in PD, MPTP can produce loss of dopaminergic neurons within the SNpc, Lewy body-like intraneuronal eosinophilic inclusions, markers of increased oxidative stress, and decrements in mitochondrial complex I activity (5–8). Recent studies suggest that nitric oxide (NO) and superoxide anion (O2−) may play a role in MPTP neurotoxicity through mechanisms that are not known (12–16). Peroxynitrite is thought to play a prominent role in O2− and NO-mediated neurotoxicity, which can result in cell death with both apoptotic and nonapoptotic morphologies (17–19). NO, O2−, and peroxynitrite have vast potential targets, but share at least one common downstream target in that they damage DNA (17–20). DNA damage is a prime activator of poly(ADP-ribose) polymerase (PARP, EC 2.4.4.30), which uses NAD as a substrate to transfer ADP ribose groups to a variety of nuclear proteins. PARP is activated by binding to DNA ends or strand breaks, and its activity is strictly proportional to the number of DNA breaks, whereas it is totally inactive in the absence of DNA breaks (20–25). One of the earliest nuclear events that follows DNA strand breakage in response to exposure to free radicals is the poly(ADP-ribosyl)ation of nuclear proteins that are localized predominantly adjacent to the DNA strand breaks. Although PARP is a prominent caspase cleavage target during apoptosis, it is unlikely to play a prominent role in apoptotic cell death as cells lacking the gene for PARP are equally susceptible to apoptosis induced by tumor necrosis factor α, Fas ligand, or γ-irradiation (26, 27). Furthermore, PARP is activated primarily by single-strand nicks of DNA that typically occur after free radical damage, but it is insensitive to double-strand DNA ends that typically occur during apoptosis (28). The exact function of PARP is not clear, but it is thought to play accessory roles in DNA replication, genomic stability, recombination, and DNA repair (20–23, 29, 30). Although massive activation of PARP in acute injury of neurons during stroke leads to cell death, most likely through energy depletion (31–34), its potential role in a chronic neurodegenerative process is not known. In the present study we show that PARP activation is instrumental in MPTP-induced parkinsonism and dopaminergic neuronal loss.
METHODS
Animals.
All experiments were approved and conformed to the guidelines set by the Institutional Animal Care Committee. To avoid differences caused from strain effect or divergent genetic lines, PARP−/− mice used in this study were on a pure 129 Sv/Ev background (26) with the colony maintained by outbreeding with purebred 129 Sv/Ev wild-type (WT) controls (Taconic Farms). Thus the PARP−/− mice are of the same strain as controls, and inbreeding effects are minimized. All mice were also age- and gender-matched (male, 60–90 days old) to avoid known effects of age and estrogen on MPTP-induced neurotoxicity.
MPTP Treatment.
PARP−/− and WT mice were housed three to a cage in a temperature-controlled room with 12-hr dark/light cycle and free access to food and water for the duration of the experiment. Each mouse received four i.p. injections of MPTP-HCl (20 mg/kg free base, Research Biochemicals) in saline or saline alone at 2-hr intervals.
Measurement of Striatal DA, Dihydroxyphenylacetic Acid (DOPAC), and Homovanillic Acid (HVA) Levels.
HPLC with electrochemical detection was used to measure striatal levels of DA, DOPAC, and HVA (14, 15). Seven days after the last MPTP injection, mice (4–6 per group) were sacrificed, brains were quickly removed, and striata were dissected out on an ice-cold glass Petri dish (35). Samples were immediately frozen on dry ice and stored at −80°C until analysis. On the day of the assay, tissue samples were sonicated in 50 volumes of 0.1 M perchloric acid containing 25 μg/ml of dihydrobenzylamine (Sigma) as internal standard. After centrifugation (15,000 × g, 10 min, 4°C), 20 μl of supernatant was injected onto a C18-reversed phase RP-80 catecholamine column (ESA, Bedford, MA). The mobile phase consisted of 90% of a solution of 50 mM sodium phosphate, 0.2 mM EDTA, and 1.2 mM heptanesulfonic acid (pH = 3.5) and 10% methanol. Flow rate was 1.0 ml/min. Peaks were detected by a Coulochem 5100A detector (E1 = −0.04 V, E2 = + 0.35 V) (ESA). Data were collected and processed on a computerized Dynamax data manager (Rainin Instruments).
Measurement of Striatal l-Methyl-4-Phenylpyridimium (MPP+) Levels.
HPLC with UV detection (wavelength 295 nm) was used to measure striatal MPP+ levels (14). PARP−/− and WT mice (three per time point) were sacrificed 90, 120, and 240 min after the fourth MPTP injection. Striata were dissected as above, immediately frozen, and stored at −80°C until analysis. On the day of the assay, tissue samples were sonicated in 5 vol of 5% tricholoracetic acid containing 5 μg/ml of 4-phenylpyridine (Sigma) as internal standard. After centrifugation (as for catecholamines), 50–100 μl of supernatant was injected onto a cation-exchange Ultracyl-CS column (Beckman, San Ramon, CA). The mobile phase consisted of 90% of a solution of 0.1 M acetic acid and 75 mM triethylamine-HCl (pH 2.35 adjusted with formic acid), and 10% acetonitrile. The flow rate was 1.5 ml. Data were collected and processed as above.
Immunohistochemistry.
For tyrosine hydroxylase (TH) immunohistochemistry and Nissl staining, at 7 days after the last dose of MPTP, mice (five per group) were perfusion-fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (PB), pH 7.4 as described (15). After postfixation in the same fixative solution, and cryoprotection in 20% sucrose/PB, brains were frozen and serially sectioned (30 μm for TH) through the entire midbrain. Alternate sections were stained for Nissl or TH. Neurons containing TH were demonstrated by incubating the tissue sections successively with a rabbit polyclonal anti-TH antibody (1:1,000, Eugene Tech, Ridgefield Park, NJ), a biotinylated–conjugated polyclonal goat anti-rabbit antibody (1:200; Vector Laboratories), and a horseradish-peroxidase-conjugated avidin/biotin complex (Vector) as described (15, 36).
Poly(ADP-Ribose) Polymer Western Blots and Immunohistochemistry.
Ventrolateral midbrain and striata were dissected from mice treated with MPTP and immediately frozen. Samples were homogenized in buffer (sucrose/DTT) and centrifuged (5 min, 14,000 × g), and the pellet was resuspended in buffer. Protein concentrations were determined by the Bradford assay, and equal samples were loaded on a gradient SDS/PAGE (30 μg per lane). The gels were transferred to a nitrocellulose membrane and incubated with anti-poly(ADP- ribose) mAb. Membranes were stained with Ponceau S (0.1%) to confirm equal loading and transfer. After blocking of nonspecific sites, membranes were incubated with antibodies to poly(ADP-ribose) (1:250). Bands were visualized via chemiluminesence. For immunohistochemistry mice were perfusion-fixed in 4% paraformaldehyde in 0.1 M PB (pH 7.4) first by deeply anesthetizing the animal with i.p. pentobarbital. Chilled 1× PBS was infused into the left ventricle as blood was allowed to escape the right atrium. Once blood was replaced by PBS, the PB-paraformaldehyde was infused. After infusion, the brain was removed and allowed to postfix in PB-paraformaldehyde for 4 hr and then transferred to 20% glycerol/PB for cryoprotection. The brains were blocked and frozen for sliding microtome sectioning. Serial sections (40 μm) through the midbrain and striatum were taken and incubated with a polyclonal guinea pig antibody to poly(ADP-ribose) (1:400) in PBS-12% BSA overnight at 4°C. Sections were washed for 10 min × 3 in PBS-12% BSA and incubated with secondary anti-guinea pig antibodies (biotin-conjugated, Jackson ImmunoResearch) in PBS-12% BSA at room temperature for 45 min. Texas-red (Vector Laboratories) was used to visualize the immunostaining. Sections were washed for 10 min × 2 in PBS-12% and then for 10 min × 2 in PBS. Slides were mounted with PBS in 80% glycerol and visualized by fluorescence microscopy through a green filter. A black and white camera digitally captured the image to a computer, which then was pseudocolored red (IP Scanalytics, Fairfax, VA).
Stereology.
The total number of TH- and Nissl-stained SNpc neurons were counted from five mice per group by using the optical fractionator (37), an unbiased method of cell counting that is not affected by either the volume of reference (i.e., SNpc) or the size of the counted elements (i.e., neurons). Neuronal counts were performed by using a computer-assisted image analysis system consisting of a Zeiss Axiophot photomicroscope equipped with a MC-XYZ-LC (Applied Scientific Instrumentation, Eugene, OR) computer-controlled motorized stage, a DAGE-MTA (Michigan, IN) video camera, a Macintosh 9600 workstation, and NeuroZoom morphometry software (Scripps Research Institute, La Jolla, CA) (38). In agreement with this method, TH- and Nissl-stained neurons were counted in the right and left SNpc of every fourth section throughout the entire extent of the SNpc. Each midbrain section was viewed at low power (×10 objective), and the SNpc was outlined by using the set of anatomical landmarks defined previously (15, 36). Then at a random start, the number of TH- and Nissl-stained cells were counted at high power (×100 oil; numerical aperture 1.4). To avoid double counting of neurons with unusual shapes, TH- and Nissl-stained cells were counted only when their nuclei were optimally visualized, which occurred only in one focal plane. In addition, neurons were differentiated from nonneuronal cells, including glia, in the Nissl stain by the exclusion of cells that did not have a clearly defined nucleus, cytoplasm, and a prominent nucleolus. Although some small neurons may be excluded, these criteria should reliably exclude all non-neuronal cells. After all of the TH- and Nissl-stained neurons were counted, the total numbers of TH- and Nissl-stained neurons in the SNpc were calculated by using the formula described by West et al. (37).
Statistical Analysis.
Throughout the experiments, the investigators were blinded to the genotype of the mice (i.e., PARP−/− or WT) and the treatment received (i.e., MPTP or saline). All values are expressed as the mean ± SEM. Differences among means were analyzed by using one-way or two-way ANOVA. When two-way ANOVA was appropriate, the different genotypes and treatments were used as the independent factors. When ANOVA showed significant differences, pair-wise comparisons between means were tested by Fisher or Newman-Keuls post hoc tests. In all analyses, the null hypothesis was rejected at the 0.05 level. All statistical analyses were performed by using statview (Abacus Software, San Francisco).
RESULTS
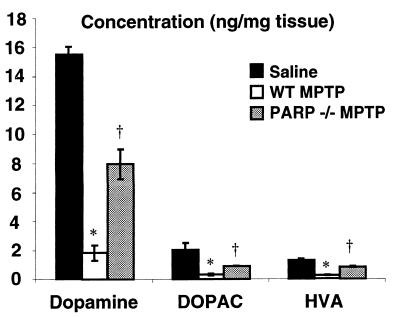
Most pharmacological PARP inhibitors lack specificity and have poor central nervous system bioavailability, raising serious questions about the validity of in vivo studies (20–22). To overcome these problems and to ascertain whether PARP activation participates in MPTP neurotoxicity, we examined the effects of MPTP in mice with targeted disruption of the gene that encodes for PARP (PARP−/−) (26, 39) compared with strain-, age-, and gender-matched WT controls (Fig. 1). It is important to mention that the PARP−/− mice used in this study are congenic and outbred with the 129 Sv/Ev strain (26), thus any observed phenotype is most likely caused by the absence of the PARP gene and is not the result of genetic strain effects. One week after four injections of MPTP at 20 mg/kg, an 80–90% reduction in striatal DA, DOPAC, and HVA is observed in WT mice (Fig. 1). PARP−/− mice are resistant to the toxic effects of MPTP and show significantly lower reductions in DA, DOPAC, and HVA levels than WT mice (Fig. 1).
Figure 1.
PARP−/− mice are resistant to the toxic effects of MPTP. HPLC with electrochemical detection of DA and metabolites, HVA and DOPAC, 1 week after MPTP administration. All animals are on a pure genetic background of 129 SvEv. No differences in striatal DA or metabolite content was observed in WT or PARP−/− animals after saline injection, and these values are graphically combined (n = 10). WT animals (n = 8) injected with MPTP (20 mg/kg ×4) have significant reductions in DA and metabolites compared with saline controls and PARP−/− (n = 5). Two-way ANOVA, P < 0.004 for DA, HVA, and DOPAC. Student-Newman-Keuls post hoc analysis, ∗, P < 0.05 WT MPTP vs. saline, †, P < 0.05 WT MPTP vs. PARP−/− MPTP.
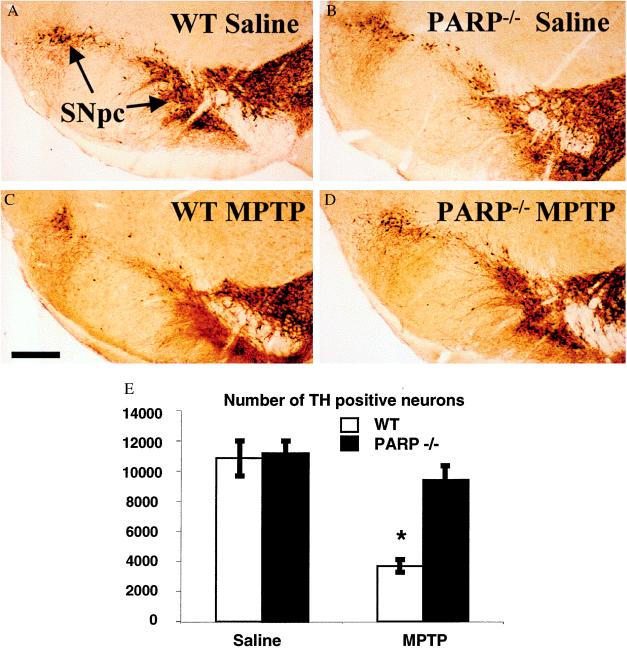
In addition to the destruction of DA nerve terminals in the striatum, there is an accompanying loss of DA cell bodies in the SNpc in PD. Furthermore, MPTP-induced destruction of DA nerve terminals does not necessarily equate with loss of cell bodies (40, 41). Thus, it is important to determine whether the absence of PARP protects against the actual loss of DA neurons in the MPTP mouse model. By using stereological techniques, we counted the number of nigral TH-positive neurons in saline-injected WT and PARP−/− mice 1 week after four injections of MPTP at 20 mg/kg (Fig. 2). In WT and PARP−/− mice there is a large number of TH-positive cell bodies intermingled with a dense network of TH-positive nerve fibers within the SNpc, and there is no significant difference in the number of TH-positive cells between the two groups of saline-injected animals (data not shown). MPTP causes a 60% reduction in nigral TH-positive neurons in WT mice compared with saline-injected controls. The loss of SNpc TH-positive neurons after MPTP is virtually abolished in PARP−/− mice (Fig. 2). MPTP also causes a 60% reduction in Nissl-stained SNpc neurons after MPTP in WT mice, whereas Nissl-stained SNpc neurons are completely spared in PARP−/− mice (Fig. 2). The preservation of both TH and Nissl-stained neurons in the PARP−/− mice indicates that the absence of PARP prevents the MPTP-induced death of SNpc neurons.
Figure 2.
DA neurons from PARP−/− mice are resistant to MPTP neurotoxicity. TH immunostaining of representative midbrain sections 7 days after MPTP adminstration from (A) saline-injected WT, (B) saline-injected PARP−/−, (C) MPTP-injected WT, and (D) MPTP-injected PARP−/− mice. (E) A significant reduction of TH-immunopositive neurons is seen in the WT mice receiving MPTP (n = 5) compared with saline controls (n = 8) (∗, ANOVA with Fisher post hoc, P < 0.0001 WT MPTP vs. saline). No statistical difference is seen between saline controls and PARP−/− (n = 4) 1 week after MPTP administration (ANOVA). Counts of Nissl-stained neurons in midbrain yielded similar results (data not shown).
MPTP requires conversion to MPP+ by monoamine oxidase B (MAO-B) to elicit neurotoxicity (42). MPP+ then is transported into DA neurons where it concentrates in the mitochondria and inhibits complex I (43–46). To ensure that the protection afforded by the disruption of the PARP gene is not caused by alterations in MPP+ concentrations in the brain we monitored MAO-B activity and striatal MPP+ levels in WT and PARP−/− mice. Brain MAO-B activity is not significantly different between WT and PARP−/− mice (Table 1). Because striatal MPP+ levels correlate significantly with the degree of DA neurotoxicity, we also monitored striatal MPP+ levels in WT versus PARP−/− mice. Previously we had shown that striatal MPP+ content reaches a peak level approximately 90 min after MPTP injection (15). Accordingly, we determined striatal MPP+ content at 90 min after four injections of MPTP as well as 2 and 4 hr after the fourth MPTP injection (Table 2). At no time point was the striatal MPP+ level significantly different between WT and PARP−/− mice. Thus, the absence of the functional PARP protein by genetic knockout accounts for MPTP resistance in PARP−/− mice.
Table 1.
MAO-B catalytic activity in drug naive WT and PARP−/− mice
| MAO-B activity
|
||
|---|---|---|
| Km, mM | Vmax, pmol/mg per min | |
| WT | 5.21 ± 1.36 | 148.2 ± 11.9 |
| PARP−/− | 5.46 ± 1.71 | 169.7 ± 16.4 |
Data are the means ± SEM, n = 3 for each value. No significant difference exists in MAO-B catalytic activity (ANOVA) between PARP−/− or WT.
Table 2.
Striatal MPP+ levels in WT and PARP−/− mice
| MPP+ levels, μg/gm striatum
|
|||
|---|---|---|---|
| 90 min | 120 min | 240 min | |
| WT | 3.33 ± 0.35 | 1.95 ± 0.38 | 0.44 ± 0.06 |
| PARP−/− | 3.03 ± 0.35 | 2.12 ± 0.28 | 0.40 ± 0.13 |
MPP+ levels were determined at the times indicated after treatment with MPTP. Data are the means ± SEM, n = 3 for each value. No significant difference exists in MPP+ levels (two-way ANOVA) between PARP−/− or WT.
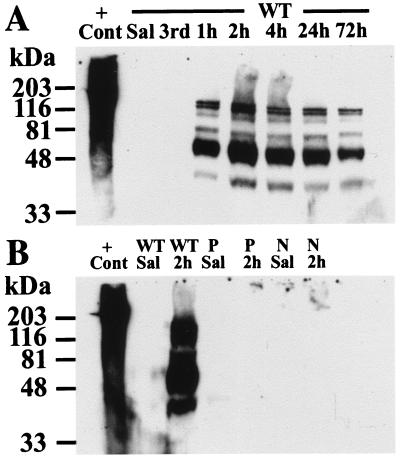
Poly(ADP-ribose) formation via nuclear protein modification is a marker of PARP catalytic activity (20–22, 24, 25, 29, 30, 47–49). In preliminary experiments we examined several time points after MPTP injection to determine when PARP is activated (data not shown). We monitored poly(ADP-ribosyl)ation by using a highly selective and specific mAb to poly(ADP-ribose). We failed to observe any PARP activity in the mouse ventral midbrain, which contains the SNpc until after the fourth injection of MPTP (Fig. 3). Poly(ADP-ribosyl)ation of nuclear proteins is maximal at 2 hr after the fourth injection, and it is still present at 72 hr after the fourth injection. No poly(ADP-ribose) is detected in the striatum of WT animals after MPTP administration, confirming the specificity of the detection of poly(ADP-ribose) (data not shown). In addition to undergoing automodification, PARP catalyzes the poly(ADP-ribosyl)ation of several nuclear proteins, including histones, topoisomerase I and II, DNA polymerase a, proliferating cell nuclear antigen, and p53, all of which play a role in reactions involving DNA strand breaks. PARP is usually the major protein that is poly(ADP-ribosyl)ated; however, unexpectedly we observe that several of these potential alternative acceptors are strongly poly(ADP-ribosyl)ated after MPTP. The precise identities of the ADP-ribosylated proteins under these conditions have not been established; however, based on prior observations, the bands at 116 kDa, 100 kDa, and 53 kDa may represent PARP, topoisomerase I and p53, respectively (49–51). In striking contrast there is no poly(ADP-ribose) formation in the PARP−/− mice after MPTP administration (Fig. 3).
Figure 3.
MPTP induces marked levels of poly(ADP-ribosyl)ation of nuclear proteins. Immunoblot analysis with mAb to poly(ADP-ribose). (A) No evidence of poly(ADP-ribosyl)ation is seen until after the fourth injection of MPTP. Polymer formation peaked at 2 hr after the fourth dose and is still detectable at 72 hr. (B) No polymer formation is seen in PARP−/− or nNOS−/− mice. No polymer formation is seen in the striata of WT animals after MPTP (not shown). P, PARP−/−; N, nNOS−/−; 3rd, time at the third injection of MPTP; h, hours after the fourth dose of MPTP; +Cont, sonicated Hela cells plus NAD as a positive control. This experiment has been replicated three times with similar results.
NO is thought to play a major role in activating PARP through its ability to promote nonapoptotic DNA damage (32, 33). To determine whether there is a link between NO and PARP in MPTP-induced parkinsonism, we monitored poly(ADP-ribosyl)ation after MPTP administration in mice lacking the gene for neuronal NO synthase (nNOS−/−) (52). Remarkably, we do not detect any poly(ADP-ribose) formation in nNOS−/− mice (Fig. 3). These data coupled with the observation that NO plays a role in MPTP-induced cell death (12, 13, 15, 16) indicate that NO-induced DNA damage is necessary for PARP activation in MPTP neurotoxicity.
To ascertain whether poly(ADP-ribose) formation was specific to MPTP-injured DA neurons in the SNpc, we monitored the cellular localization of poly(ADP-ribose) formation via immunocytochemistry in the SNpc of WT versus PARP−/− mice and nNOS−/− mice (Fig. 4). We observe intense nuclear poly(ADP-ribosyl)ation in TH-positive SNpc neurons from WT mice but fail to observe any poly(ADP-ribose) formation in nuclei of PARP−/− mice and nNOS−/− mice (Fig. 4).
Figure 4.
PARP is activated in DA neurons after MPTP intoxication. Immunohistochemical staining with an anti-poly(ADP-ribose) antibody (pseudocolored in red) in the ventral midbrain. (A) WT after MPTP delivery demonstrates intense and specific staining of DA neurons. (B) PARP−/− midbrains are devoid of immunostaining. (C) nNOS−/− mice lack poly(ADP-ribose) formation after MPTP. Poly(ADP-ribose) is not detectable in saline-injected animals (data not shown). These images were obtained from animals 4 hr after the fourth injection of MPTP. These experiments have been replicated three times with similar results.
DISCUSSION
The major finding of this study is that PARP activation is a principal determinant of MPTP-induced dopaminergic cell death. The profound protection against MPTP neurotoxicity in the PARP−/− mice coupled with the observation that DNA is fragmented after MPTP both in vivo and in vitro (53, 54) implicates DNA damage in MPTP-induced neuronal killing. Previous studies indicate that neuronally derived NO and O2− play an important role in MPTP neurotoxicity. Protection against MPTP neurotoxicity is provided by selective nNOS inhibitors or the absence of the nNOS gene (12, 13, 15). MPP+ directly inhibits complex I of the mitochondria, leading to the generation of superoxide anion (55, 56). The combination of NO and O2−, which forms peroxynitrite, is probably the force behind DNA damage in MPTP-induced neurotoxicity as the footprints of peroxynitrite-, nitrotyrosine- (12), and nitrotyrosine-modified proteins (57), are readily detected after MPTP administration. Our observations that PARP is not activated in nNOS−/− mice after MPTP links NO, peroxynitrite, and PARP activation in cell death mediated by MPTP. PARP activation is not seen until the fourth injection of MPTP in our paradigm. It is likely that a critical threshold of peroxynitrite formation is required to initiate the cascade of DNA damage and PARP activation. Consistent with this notion are the observations that neuronal death begins after the fourth dose of MPTP (36), coinciding with the peak of peroxynitrite-mediated nitration of TH (57), which parallels the peak of poly(ADP-ribose) formation.
The TH-positive and Nissl-stained neurons of the SNpc are completely spared from the neurotoxic effects of MPTP in the PARP−/− mice. This finding contrasts with only a partial, but significant, sparing of striatal DA content in the PARP−/− mice. It is well known that the destruction of striatal DA terminals does not necessarily equate with cell loss (58). The complete sparing of dopaminergic neurons, but the loss of striatal DA content may be caused by peroxynitrite-mediated nitration and inactivation of TH (57), the rate-limiting enzyme of DA biosynthesis.
After MPTP neurotoxicity, the following sequence of events presumably takes place. MPTP is converted by MAO-B to MPP+, which then is taken up into DA neurons via the DA transporter into cell bodies and projections, followed by transport into the mitochondria (42–46). Once there, MPP+ potently inhibits complex I, which poisons the mitochondrial electron transport chain, leading to decrements in cellular ATP and formation of O2−. NO combines with O2− to form peroxynitrite, which leads to DNA damage that activates PARP. PARP activation depletes NAD via poly(ADP-ribosyl)ation of nuclear proteins, and ATP is further depleted in an effort to resynthesize NAD, leading to cell death by energy depletion. Consistent with this notion is the observation that replacement of cellular energy stores provides protection against MPTP neurotoxicity (59–61). Although energy depletion is thought to play a prominent role in PARP-mediated cell death (20–22), the pattern of poly(ADP-ribosyl)ation of several nuclear proteins raises the interesting possibility that PARP activation also could contribute to cell death through alternative pathways (20–22, 50, 51). These alternative pathways might account for the profound neuroprotection afforded by deletion of the PARP gene, but only partial protection with replacement of energy stores (59–61).
The role of PARP activation in other forms of cell death, such as cerebral ischemia, glutamate excitotoxicity, cytokine, and free radical-mediated damage to pancreatic islet cells as well as cardiac damage after occlusion of coronary arteries, suggests that PARP may be a critical choke point in a variety of important pathologic conditions (20–22). The failure to detect PARP activation in nNOS−/− mice after MPTP administration suggests that NO is critical in this process. Although prior studies indicate that the absence of PARP leads to short-term protection against acute injury (20–22), we demonstrate that neurons are alive and functioning for an extended period after a toxic insult. Further understanding of these events may inspire novel therapeutics in the treatment of neurodegenerative disorders.
Acknowledgments
We thank Ann Schmidt for secretarial assistance. A.S.M. is supported by U.S. Public Health Service/Clinical Investigation Development Award Grant NS 1KO8NS02035–01, the Herbert Friedburg Fellowship, The Parkinson Disease Foundation Lillian-Schorr Award, and an American Parkinson Disease Association Special Research Grant. S.P. is a recipient of the Cotzias Award of the American Parkinson Disease Association and is supported by U.S. Public Health Service Grant NS37345, U.S. Department of Defense Grant NS38586, The Parkinson’s Disease Foundation, The Smart Foundation, and The Lowenstein Foundation. V.J.L. is supported by the Parkinson’s Disease Foundation. M.E.S. and C.M.S.-R. are supported in part by National Cancer Institute Grants CA2544 and CA74175, the U.S. Air Force Office of Scientific Research Grant AFOSR-89–0053, and the U.S. Army Medical Research and Development Command (Contract DAMD17–90-C-0053). V.L.D is supported by U.S. Public Health Service Grant NS33142 and is a National Alliance for Research on Schizophrenia and Depression Staglin Music Festival Investigator. T.M.D. is supported by U.S. Public Health Service Grants NS 33277 and NS 38377, the Paul Beeson Faculty Scholar Award in Aging Research, the National Parkinson’s Foundation, and the Edward D. and Anna Mitchell Family Foundation. Under an agreement between the Johns Hopkins University and Guilford Pharmaceuticals, T.M.D. and V.L.D. are entitled to a share of sales royalty received by the university from Guilford. T.M.D. and the university also own Guilford stock, and the university stock is subject to certain restrictions under university policy. The terms of this arrangement are being managed by the university in accordance with its conflict-of-interest policies.
ABBREVIATIONS
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- PARP
poly(ADP-ribose) polymerase
- PD
Parkinson’s disease
- DA
dopamine
- SNpc
substantia nigra pars compacta
- DOPAC
dihydroxyphenylacetic acid
- HVA
homovanillic acid
- TH
tyrosine hydroxylase
- PB
phosphate buffer
- WT
wild type
- MPP+
l-methyl-4-phenyl-pyridimium
- MAO-B
monoamine oxidase B
- nNOS
neuronal NO synthase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Hornykiewicz O. Clin Neurol Neurosurg. 1992;94:S9–S11. doi: 10.1016/0303-8467(92)90008-q. [DOI] [PubMed] [Google Scholar]
- 2.Fahn S. Cecil’s Textbook of Medicine. Philadelphia: Saunders; 1988. [Google Scholar]
- 3.Youdim M B, Riederer P. Sci Am. 1997;276:52–59. doi: 10.1038/scientificamerican0197-52. [DOI] [PubMed] [Google Scholar]
- 4.Olanow C W. Trends Neurosci. 1993;16:439–444. doi: 10.1016/0166-2236(93)90070-3. [DOI] [PubMed] [Google Scholar]
- 5.Forno L S, DeLanney L E, Irwin I, Langston J W. Adv Neurol. 1993;60:600–608. [PubMed] [Google Scholar]
- 6.Mizuno Y, Ikebe S, Hattori N, Kondo T, Tanaka M, Ozawa T. Adv Neurol. 1993;60:282–287. [PubMed] [Google Scholar]
- 7.Dexter D T, Sian J, Rose S, Hindmarsh J G, Mann V M, Cooper J M, Wells F R, Daniel S E, Lees A J, Schapira A H, et al. Ann Neurol. 1994;35:38–44. doi: 10.1002/ana.410350107. [DOI] [PubMed] [Google Scholar]
- 8.Schapira A H. Adv Neurol. 1996;69:161–165. [PubMed] [Google Scholar]
- 9.Kopin I J, Markey S P. Annu Rev Neurosci. 1988;11:81–96. doi: 10.1146/annurev.ne.11.030188.000501. [DOI] [PubMed] [Google Scholar]
- 10.Heikkila R E, Sieber B A, Manzino L, Sonsalla P K. Mol Chem Neuropathol. 1989;10:171–183. doi: 10.1007/BF03159727. [DOI] [PubMed] [Google Scholar]
- 11.Langston J W. Neurology. 1996;47:S153–S160. doi: 10.1212/wnl.47.6_suppl_3.153s. [DOI] [PubMed] [Google Scholar]
- 12.Matthews R T, Beal M F, Fallon J, Fedorchak K, Huang P L, Fishman M C, Hyman B T. Neurobiol Dis. 1997;4:114–121. doi: 10.1006/nbdi.1997.0141. [DOI] [PubMed] [Google Scholar]
- 13.Hantraye P, Brouillet E, Ferrante R, Palfi S, Dolan R, Matthews R T, Beal M F. Nat Med. 1996;2:1017–1021. doi: 10.1038/nm0996-1017. [DOI] [PubMed] [Google Scholar]
- 14.Przedborski S, Kostic V, Jackson-Lewis V, Naini A B, Simonetti S, Fahn S, Carlson E, Epstein C J, Cadet J L. J Neurosci. 1992;12:1658–1667. doi: 10.1523/JNEUROSCI.12-05-01658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Przedborski S, Jackson-Lewis V, Yokoyama R, Shibata T, Dawson V L, Dawson T M. Proc Natl Acad Sci USA. 1996;93:4565–4571. doi: 10.1073/pnas.93.10.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz J B, Matthews R T, Muqit M M, Browne S E, Beal M F. J Neurochem. 1995;64:936–939. doi: 10.1046/j.1471-4159.1995.64020936.x. [DOI] [PubMed] [Google Scholar]
- 17.Beckman J S. J Dev Physiol. 1991;15:53–59. [PubMed] [Google Scholar]
- 18.Iadecola C. Trends Neurosci. 1997;20:132–139. doi: 10.1016/s0166-2236(96)10074-6. [DOI] [PubMed] [Google Scholar]
- 19.Samdani A F, Dawson T M, Dawson V L. Stroke. 1997;28:1283–1288. doi: 10.1161/01.str.28.6.1283. [DOI] [PubMed] [Google Scholar]
- 20.Szabo C, Dawson V L. Trends Pharmacol Sci. 1998;19:287–298. doi: 10.1016/s0165-6147(98)01193-6. [DOI] [PubMed] [Google Scholar]
- 21.Berger N A. Radiat Res. 1985;101:4–15. [PubMed] [Google Scholar]
- 22.Milam K M, Cleaver J E. Science. 1984;223:589–591. doi: 10.1126/science.6420886. [DOI] [PubMed] [Google Scholar]
- 23.Thraves P J, Kasid U, Smulson M E. Cancer Res. 1985;45:386–391. [PubMed] [Google Scholar]
- 24.Juarez-Salinas H, Sims J L, Jacobson M K. Nature (London) 1979;282:740–741. doi: 10.1038/282740a0. [DOI] [PubMed] [Google Scholar]
- 25.Benjamin R C, Gill D M. J Biol Chem. 1980;255:10502–10508. [PubMed] [Google Scholar]
- 26.Wang Z Q, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner E F. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leist M, Single B, Kunstle G, Volbracht C, Hentze H, Nicotera P. Biochem Biophys Res Commun. 1997;233:518–522. doi: 10.1006/bbrc.1997.6491. [DOI] [PubMed] [Google Scholar]
- 28.Shall S. Mol Cell Biochem. 1994;138:71–75. doi: 10.1007/BF00928445. [DOI] [PubMed] [Google Scholar]
- 29.de Murcia G, Menissier de Murcia J. Trends Biochem Sci. 1994;19:172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 30.de Murcia J M, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver F J, Masson M, Dierich A, LeMeur M, et al. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endres M, Wang Z Q, Namura S, Waeber C, Moskowitz M A. J Cereb Blood Flow Metab. 1997;17:1143–1151. doi: 10.1097/00004647-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Eliasson M J, Sampei K, Mandir A S, Hurn P D, Traystman R J, Bao J, Pieper A, Wang Z Q, Dawson T M, Snyder S H, Dawson V L. Nat Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Dawson V L, Dawson T M, Snyder S H. Science. 1994;263:687–689. doi: 10.1126/science.8080500. [DOI] [PubMed] [Google Scholar]
- 34.Cosi C, Suzuki H, Milani D, Facci L, Menegazzi M, Vantini G, Kanai Y, Skaper S D. J Neurosci Res. 1994;39:38–46. doi: 10.1002/jnr.490390106. [DOI] [PubMed] [Google Scholar]
- 35.Reches A, Wagner H R, Jiang D, Jackson V, Fahn S. Life Sci. 1982;31:37–44. doi: 10.1016/0024-3205(82)90398-8. [DOI] [PubMed] [Google Scholar]
- 36.Jackson-Lewis V, Jakowec M, Burke R E, Przedborski S. Neurodegeneration. 1995;4:257–269. doi: 10.1016/1055-8330(95)90015-2. [DOI] [PubMed] [Google Scholar]
- 37.West M J. Neurobiol Aging. 1993;14:275–285. doi: 10.1016/0197-4580(93)90112-o. [DOI] [PubMed] [Google Scholar]
- 38.Bloom F E, Young W G, Nimchinsky E A, Hof P R, Morrision J H. Neuronal Vulnerability and Informatics in Human Disease. Mahwah, NJ: Lawrence Erlbaum; 1997. [Google Scholar]
- 39.Wang Z Q, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner E F. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 40.Hallman H, Olson L, Jonsson G. Eur J Pharmacol. 1984;97:133–136. doi: 10.1016/0014-2999(84)90521-1. [DOI] [PubMed] [Google Scholar]
- 41.Hallman H, Lange J, Olson L, Stromberg I, Jonsson G. J Neurochem. 1985;44:117–127. doi: 10.1111/j.1471-4159.1985.tb07120.x. [DOI] [PubMed] [Google Scholar]
- 42.Heikkila R E, Manzino L, Cabbat F S, Duvoisin R C. Nature (London) 1984;311:467–469. doi: 10.1038/311467a0. [DOI] [PubMed] [Google Scholar]
- 43.Javitch J A, D’Amato R J, Strittmatter S M, Snyder S H. Proc Natl Acad Sci USA. 1985;82:2173–2177. doi: 10.1073/pnas.82.7.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicklas W J, Youngster S K, Kindt M V, Heikkila R E. Life Sci. 1987;40:721–729. doi: 10.1016/0024-3205(87)90299-2. [DOI] [PubMed] [Google Scholar]
- 45.Ramsay R R, Dadgar J, Trevor A, Singer T P. Life Sci. 1986;39:581–588. doi: 10.1016/0024-3205(86)90037-8. [DOI] [PubMed] [Google Scholar]
- 46.Vyas I, Heikkila R E, Nicklas W J. J Neurochem. 1986;46:1501–1507. doi: 10.1111/j.1471-4159.1986.tb01768.x. [DOI] [PubMed] [Google Scholar]
- 47.Lindahl T, Satoh M S, Poirier G G, Klungland A. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 48.Lautier D, Lagueux J, Thibodeau J, Menard L, Poirier G G. Mol Cell Biochem. 1993;122:171–193. doi: 10.1007/BF01076101. [DOI] [PubMed] [Google Scholar]
- 49.Simbulan-Rosenthal C M, Rosenthal D S, Iyer S, Boulares A H, Smulson M E. J Biol Chem. 1998;273:13703–13712. doi: 10.1074/jbc.273.22.13703. [DOI] [PubMed] [Google Scholar]
- 50.Ferro A M, Higgins N P, Olivera B M. J Biol Chem. 1983;258:6000–6003. [PubMed] [Google Scholar]
- 51.Whitacre C M, Hashimoto H, Tsai M L, Chatterjee S, Berger S J, Berger N A. Cancer Res. 1995;55:3697–3701. [PubMed] [Google Scholar]
- 52.Huang P L, Dawson T M, Bredt D S, Snyder S H, Fishman M C. Cell. 1993;75:1273–1286. doi: 10.1016/0092-8674(93)90615-w. [DOI] [PubMed] [Google Scholar]
- 53.Tatton N A, Kish S J. Neuroscience. 1997;77:1037–1048. doi: 10.1016/s0306-4522(96)00545-3. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J, Pieper A, Snyder S H. J Neurochem. 1995;65:1411–1414. doi: 10.1046/j.1471-4159.1995.65031411.x. [DOI] [PubMed] [Google Scholar]
- 55.Ramsay R R, Singer T P. Biochem Biophys Res Commun. 1992;189:47–52. doi: 10.1016/0006-291x(92)91523-s. [DOI] [PubMed] [Google Scholar]
- 56.Hasegawa E, Kang D, Sakamoto K, Mitsumoto A, Nagano T, Minakami S, Takeshige K. Arch Biochem Biophys. 1997;337:69–74. doi: 10.1006/abbi.1996.9726. [DOI] [PubMed] [Google Scholar]
- 57.Ara J, Przedborski S, Naini A B, Jackson-Lewis V, Trifiletti R R, Horwitz J, Ischiropoulos H. Proc Natl Acad Sci USA. 1998;95:7659–7663. doi: 10.1073/pnas.95.13.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.German D C, Nelson E L, Liang C L, Speciale S G, Sinton C M, Sonsalla P K. Neurodegeneration. 1996;5:299–312. doi: 10.1006/neur.1996.0041. [DOI] [PubMed] [Google Scholar]
- 59.Beal M F, Matthews R T, Tieleman A, Shults C W. Brain Res. 1998;783:109–114. doi: 10.1016/s0006-8993(97)01192-x. [DOI] [PubMed] [Google Scholar]
- 60.Mukherjee S K, Klaidman L K, Yasharel R, Adams J D., Jr Eur J Pharmacol. 1997;330:27–34. doi: 10.1016/s0014-2999(97)00171-4. [DOI] [PubMed] [Google Scholar]
- 61.Chan P, DeLanney L E, Irwin I, Langston J W, Di Monte D. Ann NY Acad Sci. 1992;648:306–308. doi: 10.1111/j.1749-6632.1992.tb24564.x. [DOI] [PubMed] [Google Scholar]