Abstract
Gut intraepithelial CD8 T lymphocytes (T-IEL) are distinct from thymus-derived cells and are thought to derive locally from cryptopatch (CP) precursors. The intermediate stages of differentiation between CP and mature T-IEL were not identified, and the local differentiation process was not characterized. We identified and characterized six phenotypically distinct lineage-negative populations in the CP and the gut epithelium: (a) we determined the kinetics of their generation from bone marrow precursors; (b) we quantified CD3-ε, recombination activating gene (Rag)-1, and pre-Tα mRNAs expression at single cell level; (c) we characterized TCR-β, -γ, and -α locus rearrangements; and (d) we studied the impact of different mutations on the local differentiation. These data allowed us to establish a sequence of T cell precursor differentiation in the gut. We also observed that the gut differentiation varied from that of the thymus by a very low frequency of pre-Tα chain mRNA expression, a different kinetics of Rag-1 mRNA expression, and a much higher impact of CD3 ε/δ and pre-Tα deficiencies. Finally, only 3% of CP cells were clearly involved in T cell differentiation, suggesting that these structures may have additional physiological roles in the gut.
Keywords: T cells, precursors, differentiation, pre-TCRα, CD3-ε
Introduction
The gut epithelium is in continuous contact with food antigens and the intestinal flora, and harbors a major pool of effector T cells. These gut intraepithelial T lymphocytes (T-IEL)* secrete IFN-γ and kill target cells ex vivo (1, 2). They are as numerous as all T cells present in both the spleen and lymph nodes (3).
T-IEL have different origins in normal mice (4, 5). About half derive from activated T cells migrating from the peripheral lymphoid organs to the gut and have a memory phenotype (TCR-αβ+ CD5+ CD28+ LFA-1+ CD44+ CD8αβ+ or CD4+). The other half (CD8αα T-IEL) differs from peripheral T cells. These have a different phenotype (CD5− CD28− LFA-1− B220+, most expressing CD8αα homodimers) and most are TCR-γδ+. Their CD3 complex also has a different composition: the CD3-ζ chain is associated with the ζ analogue FcεRIγ chain (6).
These T cells are thought to develop locally (7). Structures at the base of some gut crypts named cryptopatches (CP) contain lineage markers negative (Lin−) Thy1+ c-kit+ IL-7R+ CD25+ cells (8, 9) that harbor gut T cell precursors. Indeed, CP cells isolated from nude mice reconstitute gut T cells (9). Moreover, after injection of bone marrow (BM) cells into athymic mice, CP reconstitution preceded gut T cell generation, further indicating that CP cells are precursors of mature intraepithelial lymphocytes (IEL) (10). Molecular characterization of sorted Lin− CP cells, however, showed no evidence of advanced T cell differentiation. These cells expressed only rare CD3-ε messages and germline TCR-β transcripts, but no TCR rearrangements or pre-Tα expression was detected (10). These results demonstrate that CP must undergo additional steps of differentiation before they generate mature T cells. It is likely such differentiation occurs in the epithelium since recombinase (4, 11) and pre-Tα mRNAs (12, 13) are present in CD3− IEL. The sequence of the developmental process between CP and mature IEL, however, is yet unknown. Gut T cell differentiation may also differ from thymus T cell generation. As CD3− gut precursors do not express CD3-ζ chains, other CD3 proteins may have a unique role in extrathymic differentiation (6, 13). The local development may favor the production of TCR-γδ+ cells. In particular, the gut environment may have a reduced capacity to induce TCR-αβ rearrangements or pre-Tα chain expression. This latter chain is an essential component of the pre-TCR that drives the expansion of CD44− CD25+ thymocytes and their differentiation toward the TCR-αβ lineage. Expression of this pre-TCR is responsible for the predominance of TCR-αβ production by the thymus (14, 15).
In the present article, we identified by phenotype and characterized all Lin− cell types recovered from the mouse gut, including CP and IEL. To identify pathways of differentiation, we established the kinetics of generation of these Lin− populations from BM precursors, in the absence of a thymus. To determine checkpoints in this differentiation process, we studied the impact of mutations of genes involved in T cell differentiation. To characterize the gut differentiation process, we detailed molecular characterization, including PCR studies of gene expression at single-cell (16) and TCR rearrangements. These results allowed the determination of the sequence of precursor development in the gut. They also demonstrated striking differences in this differentiation process, when compared with T cell differentiation in the thymus.
Materials and Methods
Mice.
C57BL/6 mice were 4–6-wk-old. Normal mice were obtained from Charles River Laboratories. C57BL/6, nu/nu (nude) (Ly5. 2), CD3-εΔ5/Δ5–deficient (17), recombination activating gene (Rag)-2–deficient (18), and TCR-α–deficient mice (19) were obtained from the Center for Development of Advanced Experimentation Techniques. TCR-δ–deficient mice (20) were a gift from P. Pereira (Institut Pasteur, Paris, France). Pre-Tα–deficient mice and Ly5.1 Rag-2, and IL-2Rγ double–deficient mice (Rag/γc−) (21) were obtained from our breeding facilities.
Cell Suspensions and Immunofluorescence Analysis.
The small intestine (gut) was washed. The Peyer's patches were visualized under 20× magnification and removed. The gut was opened lengthwise and cut into 3–4-cm pieces. The gut epithelium was detached from the gut wall as described previously (5). Intraepithelial lymphocytes were recovered from supernatants. These were treated to remove mucins and passed through glass wool columns to remove epithelial cells. The remaining gut pieces were digested with 100 U/ml of collagenase and 5 U/ml of DNase (5). Cells recovered from both IEL and wall digests were centrifuged in a Ficoll-Hypaque gradient (density = 1.077).
Both IEL and wall lymphocytes (WL) were incubated with an anti-CD16/CD32 Fc–receptor mAb (2.4G2) to block Fc-mediated antibody binding, previous to cell surface staining. To identify Lin− populations from the gut, we initially studied which Abs (recognizing BM-derived mature cells) would label our preparations. Based on these results we devised a cocktail of mAbs including anti-CD3 (145-2C11), anti-CD19 (1D3), anti-TCRαβ (H57-597), anti–TCR-δ (GL3), anti-erythroid cells (TER-119), anti-GR1 (8C5), anti–Mac-1 (M1-70), and anti-IgM (Southern Biotechnology Associates, Inc.) to separate Lin+ cells. This cocktail excluded gut dendritic cells that coexpress Mac-1. The Lin− gut cells express low levels of NK1.1, similar to those of NK1.1 T cells (unpublished data). Since mature CD8αα T-IEL are a subpopulation of NK T cells, and express low levels of NK1.1 (22), we could not include anti-NK1.1 mAb in our Lin+ mixture.
The antibodies used for surface staining of Lin− cells were from PharMingen: anti-Ly5. 2 (104-2.1), anti-CD8α (53-6.7), anti-CD25 (PC61), anti-CD44 (IM7), anti-B220 (RA3-6B2; Caltag), anti-Thy1.2 (53-2.1), anti–c-kit (3C1). Anti–IL-7R (A7R34) were a gift from D. Guy-Grand (Institut Pasteur, Paris, France). Biotinylated antibodies were revealed with allophycocyanin-streptavidin (Molecular Probes) or Red613-streptavidin (GIBCO BRL).
Four-color immunofluorescence analysis was performed using a FACS® Calibur flow cytometer (Becton Dickinson) and CellQuest software (Becton Dickinson) for data acquisition and analysis.
Immunohistochemistry.
Small intestine (5 μm), acetone-fixed frozen sections were air-dried and pretreated with avidin/biotin blocking kit (Vector Laboratories), and labeled with anti–c-kit followed by biotinylated anti–rat IgG (Jackson ImmunoResearch Laboratories). Detection was according to Vectorstain ABC kit followed by DAB substrate kit for peroxidase (Vector Laboratories). Slides were counterstained with hematoxylin. Control stainings without the primary Ab or with an isotype control were run in parallel.
Thymectomy and Reconstitutions.
To allow complete removal of the very small thymus rudiment of Rag2/γc− mice, both thymus lobes were carefully dissociated from surrounding structures. A surgical thread was placed in between each thymus lobe and the sternum, and a knot around the thymus pediculum was used to isolate each lobe, which was removed with a forceps. Thymectomized mice were irradiated at 300 rads and injected with 5 × 106 BM cells from nude mice, or with 2.5 to 5 × 105 Lin− WL from normal mice.
PCR Analysis.
Lin− cells were purified from bulk populations by two rounds of cell sorting. Cell contaminants, if present, were below our detection limit (0.1%). Various numbers of cells were plated per well using a FACS® Vantage equipped with an automatic cell deposition unit (Becton Dickinson). RT–PCR analysis was done as previously described (16). Briefly, cells present in each well were lysed, the mRNA coding for different genes were reversed transcribed using specific 3′ primers, and cDNAs were amplified in a modified nested two-step PCR. In the first PCR round, primers amplifying multiple genes were present. The first PCR products were then split and each gene was amplified separately in a second PCR round. This assay is able to identify 10 molecules of mRNA per cell (16). The following primers were used: HPRT (23); CD3-ε 5′ primers: 5′-ACCAGTGTAGAGTTGACGTG-3′, 5′-TTTCTCGGAAGTCGAGGACA-3′ (nested), and 3′ primers 5′-GATGGCTCATAGTCTGGGTT-3′; Rag-1 5′ primers: 5′-CCAAGCTGCAGACATTCTAGCACTC-3′ and 5′-CAGACATTCTAGCA-CTCTGG-3′ (nested); 3′ primers: 5′-CAACATCTGCCTT-CACGTCGATCC-3′ and 5′-GCTTGACTTCCCATCAGC-ATGGA-3′ (nested); pre-Tα 5′ primers: 5′-AACAGGTAGC-TCCTGGCTGCA-3′ and 5′-GCGTCAGGTGTCAGGCT-CTA-3′ (nested), or 5′-CACACTGCTGGTAGATGGAA-3′ (nested for the long form only); and 3′ primers: 5′-CCTG-GGCTTTGTTCTTCCTG-3′ and 5′-GAGCAGTAGTGTCCAGCATC-3′ (nested). None of the primer combinations amplify genomic DNA. TCR-β chain DJ and V(D)J rearrangements (24, 25), and Vα–Jα rearrangements (26) in majority populations were studied as described elsewhere. Depending on the rearrangement the expected PCR products are: DJ, seven bands ranging from 1,279 to 224 bp; and V(D)J ranging in size from 850 to 150 bp. VJγ rearrangements were amplified using two 5′ primers recognizing the Vγ2 gene and the Vγ5 gene, respectively, and one 3′ primer recognizing the Jγ1 gene. Primers and probes were a gift from P. Pereira. The amplified products were analyzed on 1.5% agarose gel, transferred to nitrocellulose membranes, and hybridized with 33P-labeled specific probes. To allow quantification of rearrangements, all PCRs were done with equalized samples (as determined by germline Cβ quantification in nonsaturating conditions with 30 amplification cycles) on an average of 104 cells. The intensity of different bands was quantified in a phosphoimager (Amersham Pharmacia Biotech). For comparison, mature TCR-αβ cells were studied simultaneously. For the study of TCR rearrangements in minority populations, 500 or 200 cells from each population were sorted using an automatic cell deposition unit, and DNA was extracted and amplified as we have previously described (27).
Results
Identification of Lin− Populations in CP and Epithelium.
Characterization of T cell differentiation in the gut is hindered by the difficulty to isolate Lin− CP cells. Isolation procedures require micromanipulation, and nearly two-thirds of the recovered cells are mature adjacent cells (10). We therefore devised a novel purification method. Because CP are subepithelial, they should remain attached to the gut wall after removal of the epithelium. In this case, they should be recovered by collagenase treatment of the gut wall.
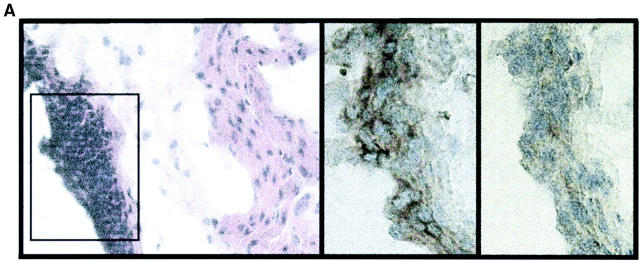
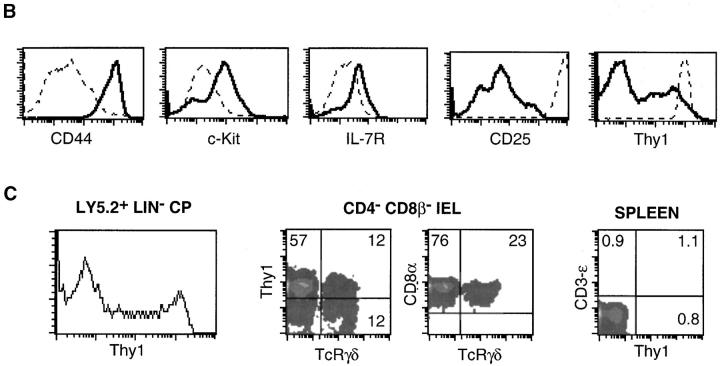
To test this hypothesis, we studied tissue sections of the gut wall after epithelium removal. We could clearly visualize that CPs remained attached to the gut wall (Fig. 1 A). The wall was then digested and the mature hematopoietic-derived cells present in these isolates were identified. Based on these results, a mixture of mAbs was prepared to exclude mature cell types and isolate Lin− WL (see Materials and Methods). Characterization of these Lin− WL showed that these cells were CD45+ CD8− CD44+ c-kit+ IL-7R+ CD25+, and about half expressed Thy1 (Thy1–CP) (Fig. 1 B). This phenotype was previously described as unique to CP cells (9, 10). Indeed, direct isolates from inside or outside CP zones demonstrated that CD44+ c-kit+ IL-7R+ CD25+ cells were only present in CP isolates and not in other parts of the gut (9, 10). The Lin− CP were also compared with Rag−/− thymocytes (10) to characterize the relative expression intensity of these markers. We also compared the Lin− WL with the Rag−/− thymus. The intensity of CD44, c-kit, and IL-7R expression was higher, and that of CD25 lower in WL than in the Rag−/− thymus (Fig. 1 B). The same relative intensity of expression was found between CP cells isolated by micromanipulation and the Rag−/− thymus (10). Based on the previously defined unique CP characteristics (9, 10) we conclude that the Lin− WL cells we isolated are in fact CP cells.
Figure 1.
Isolation of Lin− CP cells. (A) Tissue sections of the gut wall, after epithelium removal, colored with hematoxylin-eosin (left), anti–c-kit (middle), and an isotype control (right). The CP structure outlined in the left was magnified in the middle and right panels. (B) Cell suspensions from collagenase digests of the gut wall were labeled with a mixture of mAbs recognizing mature hematopoietic cells to gate Lin− populations. Histograms show the expression levels of different markers in Lin− WL (full lines) and in the Rag−/− thymocytes (dotted lines). Please note that the distribution of these markers was similar in Thy1+ and Thy1− WL. (C) 250,000–500,000 Ly 5.2+ Lin− WL were injected into thymectomized Ly 5.1+ Rag2/γc− hosts. Ly 5.2+ donor cells were identified in gut wall and IEL, 5 wk after transfer. The left graph shows Lin− populations from the gut wall. The absolute number of these cells was as in normal mice. The middle graphs show T-IEL. The right graph shows T cell labeling in the spleen. Results are from one out of two experiments.
CP cells were previously demonstrated to reconstitute T-IEL in athymic mice (9). To directly demonstrate that Lin− WL were CP cells, Ly5.2+ Lin− WL were isolated and injected into Ly5.1+ thymectomized Rag2/γc–deficient mice. These hosts are devoid of both CP and IEL (21). As shown in Fig. 1 C, Lin− WL cells reconstitute Lin− populations from the gut wall (Fig. 1 C, left) and mature CD8 T-IEL (Fig. 1 C, middle), but they do not reconstitute peripheral T cells (Fig. 1 C, right). These results show that microdissection is not the only way to isolate CP cells. They demonstrate a simple method to isolate functional CP cells from the murine gut.
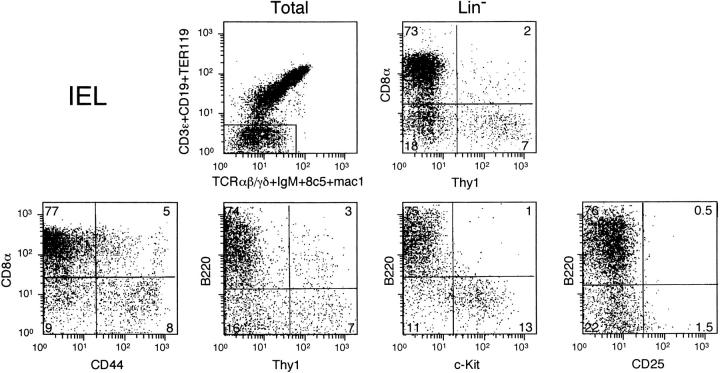
Next, we characterized Lin− cells from IEL. When defining Lin+ cells in IEL, we found cells expressing low levels of Lin markers (Fig. 2, upper left). Characterization of this population showed that they were Mac-1 low but negative for all other lineage markers including MHC class II and CD11c (unpublished data). Mac-1–low cells coexpressed high levels of CD8 and B220, like most Lin− IEL (see below). We included this Mac-1–low population in Lin− IEL, as it is well known that thymic T cell precursors are also Mac-1 low (28).
Figure 2.
The phenotype of Lin− IEL. The upper-left graph shows the total lymphoid population and the window used to select Lin− cells. The upper-right graph shows Thy1/CD8α expression in Lin− IEL. The lower graphs show Lin− cells labeled with anti-CD8 and/or anti-Thy1 Abs, together with other markers.
Lin− IEL could be subdivided by their expression of Thy-1 and CD8 into four cell types (Fig. 2, upper right). 6–10% were CD8− Thy1+ (Thy-1 single positive [SP] cells). Approximately 50–75% expressed CD8 only (CD8-SP). The other two populations were CD8− Thy1− populations (double negative [DN]) and CD8+ Thy1+ cells (double positive [DP]). As only four colors are available for analysis, direct and simultaneous characterization of other markers expressed by these populations is not always possible. Comparison of cells present in the different quadrants using different marker combinations allowed us to deduce their phenotype in more detail. Some of these combinations are shown in the lower graphs. Thy1-SP-IEL shared several markers with Thy1-CP cells. They were (Fig. 2, lower left to right) CD44+, B220− (B220+ Thy1+ cells coexpressing CD8), and c-kit+. Contrary to Thy1-CP, however, the Thy1-SP-IEL were CD25− (Fig. 2, right) and IL-7R− (unpublished data). CD8-SP-IEL lacked CD44 expression (Fig. 2, left), contrary to mature CD8 T cells (13). The other cell surface markers, however, were identical to those of mature CD8αα T cells (13). CD8-SP-IEL were all B220+ (unpublished data) and (Fig. 2, from left to right) c-kit−, CD25−, and IL-7R− (unpublished data). The DN population contained some c-kit+ and CD44+ cells (Fig. 2, left). Approximately 70% of these cells were B220+ (unpublished data) and all were CD25− (Fig. 2, right) and IL-7R− (unpublished data). Finally, DP cells were CD44+, B220+, c-kit−, and CD25− (unpublished data).
Thus, we were able to recover six different Lin− populations from gut isolates: Thy1+ and Thy1− from CP; and Thy1-SP, DP and DN cells, and CD8-SP from the epithelium. All of these populations had a lymphoblast morphology (unpublished data). The yields of Lin− CP were 69,000 ± 19,910, and of Lin− IEL 18,000 ± 5,613 (mean ± SE of five representative experiments).
The Kinetics of Generation of Lin− Subpopulations and TCR+ T Cells from BM Precursors.
Although CP harbor T-IEL precursors, it is not known if CP cells are the most immature precursors migrating from the BM to the gut. The sequence of development of CP cells into mature T-IEL is also unknown. An established method to identify precursor–product relationships during T cell differentiation is the study of the kinetics of generation of Lin− subpopulations after BM injection. We used this strategy to identify precursor–product relationships during gut differentiation. Ly5.1+ Rag2/γc− mice were thymectomized, irradiated, and injected with BM cells from Ly5.2+ nude mice. We followed Ly5.2+ donor cells in host mice at different times after transfer (Fig. 3).
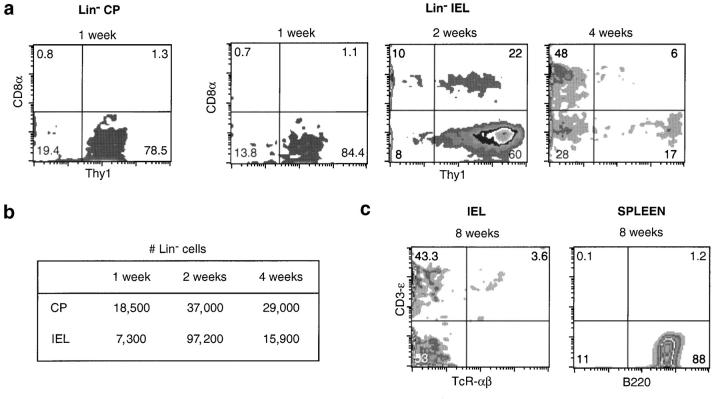
Figure 3.
The kinetics of generation of Lin− and TCR+ gut cells from BM precursors. Thymectomized, sublethally irradiated Ly5.1+ Rag2/γc−-deficient mice were injected with Ly5.2+ nude BM. The phenotype (a) and the absolute number (b) of Lin− Ly5.2+ donor cells in host mice at different times after transfer. (c) TCR+ cells in the IEL (left) and the spleen (right) of host mice, 2 mo after BM injection. Results are from one out of two experiments, giving the same results.
1 wk after BM injection we recovered Ly5.2+ donor cells in the gut of host mice (Fig. 3, A and B). All of these cells were Lin− (unpublished data) and 80–85% were Thy1+. Most were localized in CP, but rare cells were also present in the epithelium (Fig. 3 B). These results suggest that the most immature Lin− gut precursors to be generated from BM cells is Thy1+, and most likely initially homes to the CP.
The phenotype of Lin− CP cells did not change further (unpublished data) and the number increased to normal at 2 wk after BM injection (Fig. 3 B). In IEL, T cells were then absent (unpublished data) and the number of Lin− was higher than that of normal mice (Fig. 3 B). 60% Lin− IEL were yet Thy1-SP, but Thy1+ CD8+ were also present (20%) (Fig. 3 A). These populations correspond to 7–10% and 0.5–2% of Lin− IEL, respectively, in a normal mouse (Fig. 2). These results indicate that Thy1+ CD8+ DP-IEL are generated after Thy1-SP. In contrast, both CD8-SP cells (the most abundant Lin− population in normal mice) (Fig. 2) and DN cells were rare (Fig. 3 A).
By 1 mo after BM injection, Lin− IEL subpopulation distribution was the same as in normal mice. Most Lin− cells were CD8-SP and DN (Fig. 3 A). The number of Lin− IEL also decreased to normal (Fig. 3 B). Very rare T-IEL were then detected, their number increasing 2 mo after transfer (Fig. 3 C). All T-IEL were CD4− and CD8αβ− (unpublished data). In these hosts we could not detect mature T cells in the peripheral lymphoid organs (Fig. 3 C), confirming that the injected nude BM does not contain T cells (7).
The kinetics of generation of gut cells from BM precursors was therefore 1° Thy1-CP; 2° Thy1-SP-IEL; 3° DP-IEL; 4° CD8-SP-IEL; DN-IEL; and 5° T-IEL.
Comparison of CD3-ε, Rag-1, and Pre-Tα mRNA Expression between Lin− Cells from Thymus and Gut.
We next characterized the differentiation status and lineage commitment of different Lin− gut populations. We enumerated the cells expressing mRNAs coding for CD3-ε, expressed by pre-T cells, before TCR rearrangement (17), Rag-1, and the two isoforms of the pre-Tα (29). For the comparison with thymus differentiation, Lin− CD4− CD8− triple negative (TN) thymocytes from normal mice were also studied. Gated cells were enriched to 60–70% purity by a first sorting step and were then sorted to purity: contaminants, if present, represented less than 0.1%. Small numbers of cells were deposited into plates using an ADCU to study gene expression (16).
This study was performed in two steps. In the first step, cells were plated at the density of 500 cells/well and a large number of wells were screened for expression of these genes, together with the housekeeping gene HPRT. By using this approach, we could exclude the expression of certain genes. Negative results were obtained with many cells recovered in independent isolation runs and studied in multiple RT-PCRs. This initial analysis also allowed the identification of gene expressing cell types. In the second step, the latter populations were sorted in limiting dilution, or as single cells, and the frequency of gene expression determined by a single-cell RT-PCR method we previously described (16). Quantification of gene expression in limiting dilution requires the analysis of several hundred RT-PCRs in each population, and therefore cannot all be shown. A few examples of these RT-PCR reactions are in Fig. 4 (see figure legend). The results of quantification of gene expression (obtained from all RT-PCR data) are shown exclusively in Tables I and II.
Figure 4.
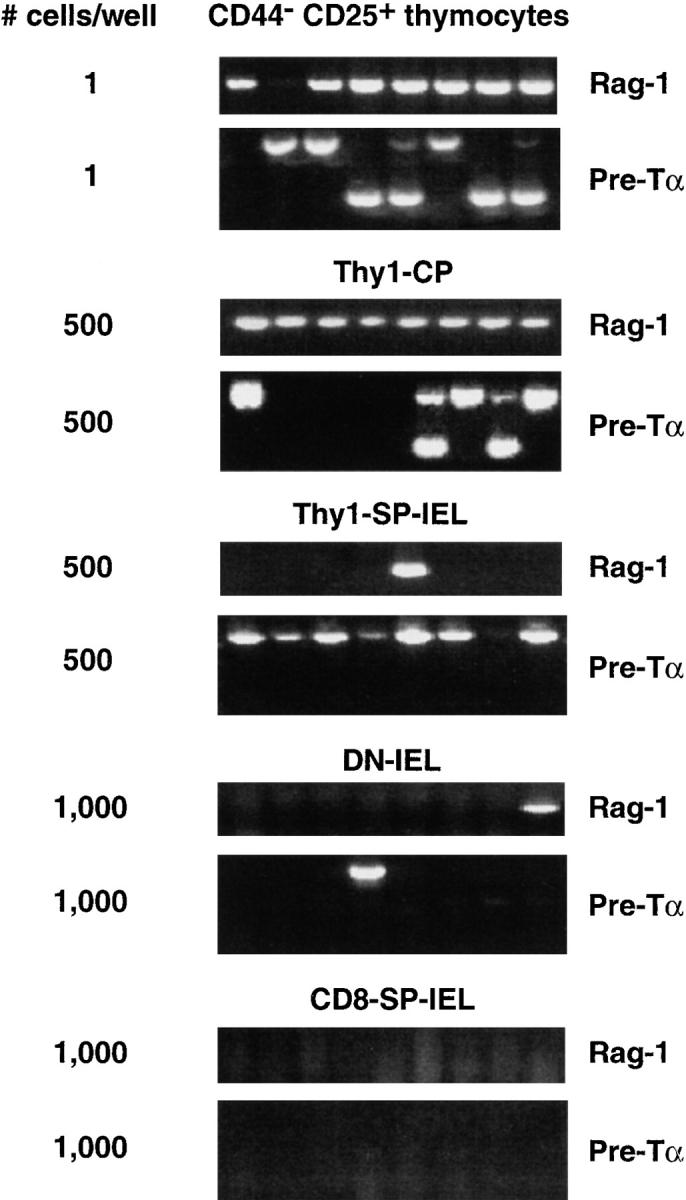
Results show a few examples of the RT-PCRs performed to quantify the expression of Rag-1 and pre-Tα mRNAs in thymus and gut precursors. All wells were positive for HPRT expression. These images illustrate a much higher expression of these genes in the thymus (where most individual cells express these genes) than in the gut (many wells containing 500 or 1,000 cells were negative). They also show that Lin− subpopulations express genes at different frequency: Rag-1 mRNA expression is much higher in CP (all wells containing 500 cells were positive) than in IEL. Maximal expression of pre-Tα is found in Thy1-SP IEL (all wells containing 500 cells were positive). We scored 320 Thy1-SP IEL cells expressing pre-Tα and only found the long form of pre-Tα, whereas both thymocytes and Thy1-CP cells express both forms. Please note that results of the quantification of gene expression (obtained from all RT-PCR data) are only shown in Tables I and II.
Table I.
Gene Expression in Lin−CD4−CD8− Thymocytes
| mRNA type | CD3-ε | Rag-1 | pre-Tα |
|---|---|---|---|
| % | % | % | |
| CD44+CD25− | 56 | 17 | ND |
| CD44+CD25+ | 96 | 72 | 59 |
| CD44−CD25+ | 96 | 83 | 75 |
| CD44−CD25− | 100 | 76 | 57 |
Lin−CD4−CD8− thymocytes from normal mice were sorted as individual cells and tested simultaneously for the expression of CD3-ε, Rag-1, pre-Tα, and HPRT (for plating efficiency). Results are from 48 individual cells in each supbpopulation. ND, not done.
Table II.
Gene Expression in Lin− Gut Cells
| Lin− CP
|
Lin− IEL
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Thy1−CD8− | Thy1-CP | Percent in CD3+ |
Thy1-SP | Percent in CD3+ |
DP | Percent in CD3+ |
DN | CD8-SP | |
| No. cells screeneda | 6,000 | 24,000 | 4,500 | 3,500 | 16,000 | 16,000 | |||
| CD3-εb | NEG | 6.3 (2.8) | 12.6 (5) | 15.5 (2) | 91 | 87 | |||
| Pre-Tαb | NEG | 0.14 (0.005) | 2.2 | 1.5 (1.6) | 12 | 0.14 (0.16) | 0.9 | NEGc | NEG |
| Rag-1b | NEG | 0.33 (0.1) | 5.2 | 0.02 (0.05) | 0.2 | 0.036 (0.007) | 0.2 | 0.006 (0.0013) | NEG |
Lin− gut cells were sorted at 500 cells/well and screened for CD3-ε, pre-Tα, and Rag-1 mRNA expression. NEG, not detected in all screened cells.
Cells were sorted in limiting dilution and the frequency of mRNA expressing cells were calculated according to the Poisson distribution (95% confidence limits are shown in brackets). Plating efficiency was evaluated by HPRT mRNA amplification.
Expression was too rare to allow frequency determination.
In the thymus, TN subpopulations expressed CD3-ε, Rag-1, and pre-Tα mRNAs (Table I). In 48 individual cells studied in each population, the lowest frequency of expression was found in CD44+ CD25− TN1 cells. CD3-ε mRNA was detected in 56% and Rag-1 in 17%. These results confirm that not all TN1 cells are committed to T cell differentiation, as this population can generate B, NK, and dendritic cell types (30). In more differentiated thymocytes, CD3-ε mRNA was detected in 96–100% of cells. Rag-1 (83%) and pre-Tα mRNA expression (75%) was maximal in CD44− CD25+ TN3 cells. We yet detected Rag-1 mRNA expression in 76% of TN4 cells, a population known to downregulate the amount of Rag-1 mRNA. Only 20% of these cells were reported to express Rag-1 mRNA by in situ hybridization (31). This difference is likely due to the far greater sensitivity of single cell RT-PCRs. We showed that 10 mRNA molecules/cell can be detected by this method (16).
Expression of these genes in CP cells showed a very different picture (Table II). Approximately 97% of these cells had no evidence of T cell commitment. Indeed, Thy1− cells, which constituted about half of the Lin− CPs (10) (Fig. 1 B), did not express mRNAs coding for CD3-ε, Rag-1, or pre-Tα. These genes were expressed by a minority of Thy1+ CP cells. CD3-ε mRNA was detected in 6% of cells, of which only 5% expressed Rag-1 mRNA. Pre-Tα mRNA expression was even less frequent (2%). These results indicate that only rare CP cells can undergo TCR rearrangements and that most cannot undergo β selection.
T cell precursors were much more abundant in Lin− IEL (Table II, Fig. 4). As compared with Thy1-CP cells, the frequency of cells expressing CD3-ε mRNA doubled in Thy1-SP and DP-IEL. Virtually all DN and CD8-SP-IEL expressed CD3-ε mRNA.
The highest frequency of pre-Tα mRNA expressers in the gut was found among CD3+ Thy1-SP-IEL (Table II, Fig. 4). It must be noted, however, that this maximal frequency (12%) was much lower than that found in the thymus (75%). Rare pre-Tα mRNA expressers were found in DP and DN cells, but not in CD8-SP cells.
We also found that only very rare Lin− IEL expressed Rag-1 mRNA (Table II, Fig. 4). These results were surprising relative to the situation described during thymus differentiation. In the thymus, recombinase expression is upregulated after β selection to allow TCR-α rearrangements to proceed (32). The highest levels of Rag-1 mRNA expression in the thymus are found among CD4+ CD8+ thymocytes (31) that lost pre-Tα expression. In contrast, in the gut the pre-Tα–negative populations did not upregulate Rag-1 expression; only very rare cells expressed Rag-1 mRNA.
TCR Rearrangements in Lin− Gut Cells.
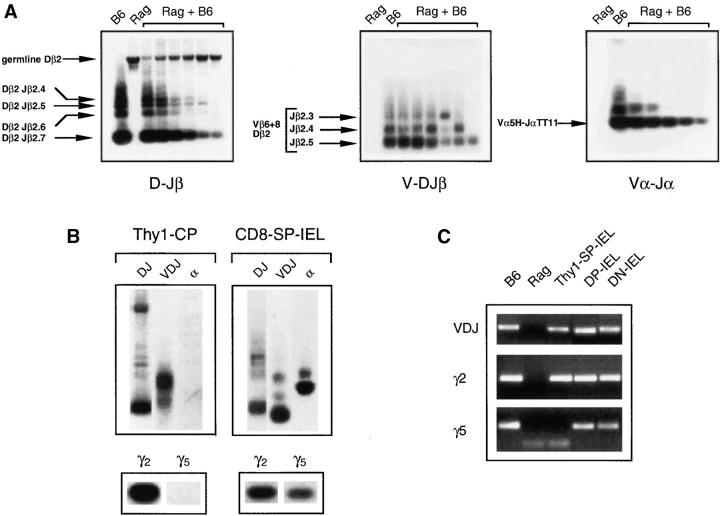
To further characterize Lin− cells we studied: TCR-β DJ rearrangements by a PCR method that permits the identification of the Dβ2-Jβ2.7 germline fragment, as well as all Dβ2-Jβ2 rearrangements (25); and TCR-β V(D)J and TCR-α VJ rearrangements by using a mixture of V-region primers together with a J-region primer (24, 26). These methods identify multiple rearrangements simultaneously, as each band corresponds to an individual type of rearrangement. They allow some quantification of the extent of rearrangements by counting the number of bands present, as well as by comparing the intensity of these bands with those found in mature T cells (see Materials and Methods). With respect to the TCR-γ locus, we studied two rearrangements: Vγ2-Jγ1 (γ2), which is abundant in thymus-derived TCR-γδ+ cells, and Vγ5-Jγ1 (γ5), which is prevalent in gut TCR-γδ+ IEL (33, 34). These methods previously used 200 ng/DNA per sample (26). We managed to reduce this amount to 60 ng (corresponding to ∼10,000/cells). Even then we could only characterize relatively abundant Lin− populations (Thy1-CP and CD8-SP-IEL), as minority populations cannot be recovered in such large numbers (see Materials and Methods).
To characterize the sensitivity of the method, we studied DNA from Rag-2−/− cells, mature CD8 TCR-αβ+ T cells, and serial twofold dilutions of mature CD8 TCR-αβ+ peripheral T cells in Rag-2−/− cells (Fig. 5 A). As expected we detected a TCR-β germline DJ band in Rag-2−/− cells, but no rearrangements. In mixtures containing Rag-2−/− and mature T cells DNA, we detected rearrangements when as little as 1% of the DNA originated from mature T cells (Fig. 5 A). To quantify rearrangements in Lin− gut T cells, they were compared with this titration and studied simultaneously.
Figure 5.
TCR rearrangements in Lin− gut populations. (A and B) Results are Southern blots of PCR products hybridized with 33P-labeled specific probes. All samples correspond to 104 cells, contained the same amount of DNA, and were studied simultaneously. (A) TCR-β DJ, V(D)J, and TCR-α VJ rearrangements were studied. DNA was extracted from: TCR-αβ CD8αβ T cells; Rag-2–deficient thymocytes; and serial twofold dilutions of mature T cells–DNA in Rag-2−/− cells DNA. (B) Thy1-CP and CD8-SP-IEL gut cells, studied simultaneously. We tested the same populations recovered from two or three independent sortings with similar results. (C) TCR rearrangements in minority IEL populations. Results show PCR products of 500 cells/sample amplified by two-nested PCR (27), in one out of four experiments, in which different samples of the same populations were tested. TCR-β V(D)J and Vγ2 rearrangements were found in all samples. In DP and DN cells we also detected Vγ5 rearrangements in all samples tested. These rearrangements were also detected when 200 cells/sample were amplified. In Thy1-SP-IEL we detected Vγ5 rearrangements in wells containing 500 cells in two out of four experiments, indicating that these rearrangements were probably quite rare. In one experiment we also detected TCR-α rearrangements in DN-IEL, whereas all other populations were negative (unpublished data). Please note that in these experimental conditions the most abundant rearrangements are preferentially amplified (27). PcR products usually show a single band, rather than multiple bands of rearrangements. PCR products were sequenced to ensure specificity.
In Thy1-CP cells, TCR-β locus rearrangements were incomplete (Fig. 5 B). The DJ germline band was present. The V(D)J rearrangements were rare when compared with the mature T cells rearrangements, shown in Fig. 5 A. TCR-α rearrangements were not detected. We were surprised to observe that γ2 (prevalent in the thymus and the periphery) was also the prevalent rearrangement in the gut Thy1-CP population.
In CD8-SP-IEL, rearrangements were more frequent and more mature (Fig. 5 B). The TCR-β locus–DJ germline band was absent, V(D)J rearrangements were frequent, and TCR-α chain rearrangements readily detected. Comparison between these cells (Fig. 5 B) and mature TCR-αβ cells (Fig. 5 A), showed similar levels of TCR-αβ rearrangements. The γ5 rearrangements were also readily detected. These results showed that CD8-SP-IEL were more mature than Thy1-CP.
We next examined rearrangements in minority populations. For that purpose we used a two-step nested PCR reaction that allows identification of rearrangements in very few cells (27). These populations were sorted at 200 and 500 cells/sample. Each sample was tested simultaneously for TCR-β locus DJ (unpublished data) and V(D)J rearrangements, TCR-γ2 and -γ5 rearrangements, and TCR-α rearrangements. These results are shown in Fig. 5 C.
Thy1-SP-IEL rather resembled Thy1-CP. We could always detect TCR-β V(D)J and γ2 rearrangements. In contrast, the γ5 rearrangements were rare in wells containing 500 cells, and we could detect these rearrangements in two of the four experiments performed. TCR-α rearrangements were not detected (unpublished data).
The DP- and DN-IEL were more similar to CD8-SP-IEL. The TCR-β V(D)J, and -γ2 and -γ5 rearrangements were detected in all tested samples. TCR-α rearrangements were absent. In DN-IEL, TCR-α rearrangements were detected (unpublished data).
To summarize, the extent of TCR rearrangements in Lin− populations showed that Thy1-CP cells were the most immature, and CD8-SP-IEL the most mature, population. The other Lin− IEL had intermediate characteristics.
The Effects of CD3-ε and Pre-Tα Deficiencies in Extrathymic Differentiation.
We identified T cell precursors in the gut in the same way as in the thymus, i.e., by their expression of CD3-ε mRNA. Previously published data suggests that this marker was adequate to identify gut precursors, since the CD3-εΔ5/Δ5 mutation blocked TCR rearrangements in Lin− IEL (13). It was conceivable, however, that whereas CD3 mutation blocked the transition between Thy1-CP and Lin− IEL, T cell maturation in CP could take place in the absence of CD3. To investigate this possibility, we characterized the Thy1-CP populations from CD3-εΔ5/Δ5–deficient mice (17) and found that TCR-β D and J fragments were in germline configuration, and V(D)J rearrangements were absent (Fig. 6 A). The CD3-εΔ5/Δ5 mutation also affected TCR-γ rearrangements. In some experiments these rearrangements were not detected at all. In others they were very rare (Fig. 6 A) when compared with Thy1-CP of normal mice (Fig. 5 B), and could only be revealed in blots after prolonged exposure. These results indicate that the CD3 complex was necessary for T cell differentiation in the gut, even in CP cells, and that CD3-ε mRNA was adequate to identify gut T cell precursors.
Figure 6.
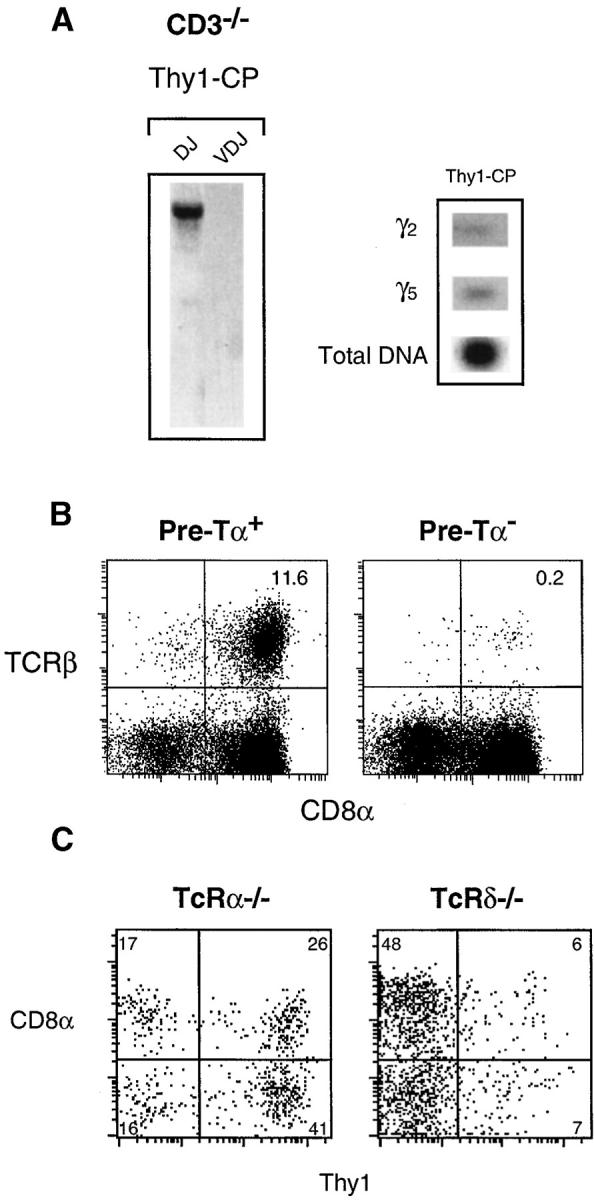
Analysis of gut populations in different mutant mice (A) TCR rearrangements in Lin− cells from CD3-εΔ5/Δ5 mice were studied as described in Fig. 5 B. The total amount of DNA (determined by amplification of the germline Cβ gene) is shown. Please note that samples from CD3-εΔ5/Δ5 mice contained the same amount of DNA and were amplified simultaneously with those from normal mice, as shown in Fig. 5 B. Results are from one of three independent sortings and show the experiment in which we detected the largest amounts of rearranged receptors. In one of these sortings, we could not detect TCR-γ rearrangements in CD3-εΔ5/Δ5–deficient cells. (B) TCR-β expression in CD8αα T-IEL from pre-Tα+/+ (left) and pre-Tα−/− mice (right), in one out of four experiments, with the same results. (C) Effects of TCR-α and -δ deficiencies in Lin− IEL. Results show the CD8/Thy1 phenotype of Lin− IEL in TCR-α– (left) and TCR-δ– (right) deficient mice in one out of eight (left), and two (right), experiments with similar results. In both mice, Thy1-SP-IEL were IL-7R− CD25− (unpublished data). These mutations did not change the phenotype of CP cells.
As pre-Tα expression was quite rare in Lin− gut cells, we also considered the hypothesis that local TCR-αβ generation could be independent of the pre-Tα expression. We examined the impact of the pre-Tα deficiency on extrathymic T cell differentiation by comparing the composition of the CD8αα IEL T cell pool in normal and pre-Tα–deficient mice (Fig. 6 B). In the thymus the pre-Tα deficiency reduces, but does not abrogate, TCR-αβ differentiation (14). In contrast, TCR-αβ+ CD8αα+ cells were virtually absent in pre-Tα–deficient mice (Fig. 6 B). These results demonstrate that TCR-αβ generation in the gut also requires pre-Tα expression. Moreover, they show that the production of TCR-αβ was even more dependent on pre-Tα expression in the gut than in the thymus.
Effects of TCR-α and TCR-δ Mutations on Lin− Gut Subpopulations.
Mutations of TCR chain genes were used to identify precursor/product relationships and checkpoints during thymus differentiation (19). We studied the impact of the TCR-α and TCR-δ mutations on Lin− gut cells. Neither modified the number nor the phenotype of CP cells (unpublished data). The TCR-α mutation induced major changes in Lin− IEL. Up to 70% of Lin− IEL from TCR-α−/− were Thy1+ (Fig. 6 C, left), whereas these cells are ∼10% of Lin− IEL in normal mice (Fig. 2). This major accumulation of Thy1-SP and DP-IEL in TCR-α–deficient mice suggests that TCR-α rearrangements favor the loss of Thy1 expression during gut differentiation. It also suggests that TCR-αβ precursors transit through Thy1-SP-IEL/DP-IEL–intermediate stages. In contrast, the TCR-δ mutation did not modify the distribution of Lin− IEL (Fig. 6 C, right).
Discussion
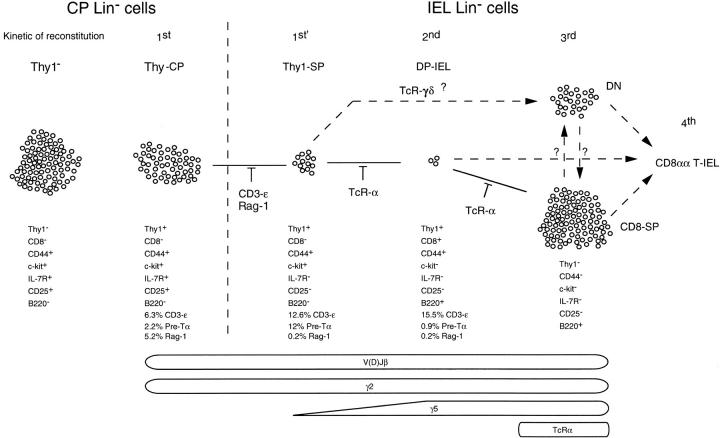
CD8αα+ T cells are a major component of the mature T cell pool that may have a main role in controlling antigenic challenge and/or ensuring the protection of the gut wall (35). These cells are very different from TCR-αβ+ thymus–derived cells (36). Their differentiation was thought to take place in CP, but recent data indicate a more complex process (10). Even though the generation of T cells in the thymus was studied in detail, the T cell differentiation pathways in the gut are unknown. This is due to the difficulty of isolating putative precursors, the very low yields, and the impossibility of inducing such cells to differentiate in vitro. In this study we used new isolation procedures to obtain CP cells, and methods that permit detailed, molecular characterization of very small numbers of cells. To establish putative precursor–product relationships, we also evaluated the kinetics of Lin− and gut T cell reconstitution after nude BM injection in thymectomized mice, and studied the impact of mutations affecting T cell development on Lin− gut cells. We identified six populations of Lin− gut cells with different phenotypes. A major c-kit+ CP population (lacking Thy1 and CD8 expression) was generated late after BM reconstitution and showed no molecular evidence of T cell differentiation. It is therefore likely that this population does not contain T cell precursors. The other five populations were implicated in the local differentiation process. They showed ample molecular evidence of ongoing T cell differentiation. They were modified by mutations affecting T cell differentiation only. They were generated from BM precursors before mature gut T cells in experimental conditions in which T cell generation can only be detected in the gut. Their phenotype, molecular characteristics, and the kinetics of their generation after BM injection, allowed a hierarchy of differentiation to establish, as summarized in Fig. 7.
Figure 7.
Characterization of the T cell differentiation process in the gut. This figure summarizes data on: relative size; localization; phenotype; gene expression; TCR rearrangements; the blockades induced by CD3-ε, Rag-2, and TCR-α mutations; and the kinetics of generation of Lin− gut populations. Based on these data, a hierarchy of differentiation is proposed.
Thy1-CP cells were the most immature. Their phenotype (Thy1+ CD8− CD44+ c-kit+ IL-7R+ CD25low) was as described in TN1/TN2 transition in the thymus (37). CD3-ε mRNA expression was rare. They expressed mRNAs coding for Rag-1 and pre-Tα, and included cells with incomplete TCR-β locus rearrangements and no TCR-γ5 or TCR-α rearrangements. They were detected in the gut 1 wk after BM injection.
The Thy1-SP-IEL rather resembled Thy1-CP cells. They shared several cell surface markers (Thy1+ c-kit+ CD44+) and similar TCR rearrangements (mostly TCR-β and -γ2). They differed from CP cells by the absence of CD25 and IL-7R, and by the increased frequency of pre-Tα and reduced frequency of Rag-1 expressions. In the thymus these modifications are associated with the TN2 to TN3 to TN4 transitions (14). By analogy with the thymus, these results suggest that Thy1-SP-IEL were more mature than Thy1-CP cells. This assumption was supported by the kinetics of generation of this population after BM transfer. Like Thy1-CP, they were detected 1 wk after BM injection, but their yield was lower. These reconstitution experiments also suggest that the Thy1-SP are the most immature precursor population in the epithelium. They were the first epithelial population to be detected after BM injection. At 1 and 2 wk after transfer, Thy1-SP cells were 90% and 60% of Lin− IEL, respectively (vs. ∼10% in normal mice). These results indicate that Thy1-SP were generated before other Lin− IEL.
The DP-IEL were closer to CD8-SP-IEL. They shared B220 expression and the presence of TCR-γ5 rearrangements. Pre-Tα chain and Rag-1 mRNA expression were rare. These characteristics suggest that the cell differentiation status was intermediate between Thy1-SP-IEL and CD8-SP-IEL. After BM injection they were detected later than Thy1-SP and were quite frequent 2 wk after BM injection (20% vs. 1–2% in normal mice).
CD8-SP cells recovered from the epithelium were the most mature population, and most resembled CD8αα T-IEL. Several phenotype characteristics were common (Thy1− c-kit− CD25− IL-7R− B220+). Virtually all were CD3-ε mRNA+, and did not express Rag-1 or pre-Tα mRNAs. TCR-β, -α, and -γ rearrangements were extensive. CD8-SP, however, differed from mature T cells, as many were Mac-1low (like TN thymocytes [28]) and CD44−. This latter characteristic indicated that CD8-SP cells were not postmature T cells that had lost all TCR surface expression as a result of intense stimulation (38). Moreover, CD8-SP were generated from BM precursors before mature T-IEL. Reconstitution of normal numbers of CD8-SP, however, was a late event, only detected 1 mo after BM injection. This data supports a relatively late ontogenic development of this population when compared with Thy1-SP and DP Lin− IEL.
Finally, DN-IEL were similar to CD8-SP. All were CD3-ε mRNA+, pre-Tα was absent, Rag-1 mRNA expression was very rare, and TCR-β, -γ2, and -γ5 rearrangements and TCR-α rearrangements were detected, as in CD8-SP. The kinetics of these cells' generation after BM reconstitution was also similar to that of CD8-SP. Normal distribution of DN-IEL was found 1 mo after BM transfer.
To identify checkpoints and differentiation pathways in the gut, we also tested the effect of several mutations on T cell differentiation (Fig. 7). None of the studied mutations affected Thy1-CP populations, further supporting their relative immaturity. IL-7R− CD25− Thy1-SP-IEL were absent in Rag2−/− and CD3-ε−/− mice (some Thy1-SP-IEL were present but they were IL-7R+ CD25+). These results indicate that the loss of IL-7R and CD25 expression in gut precursors required TCR rearrangements and/or CD3-ε expression, as in the thymus (37). They further support that Thy1-SP-IEL are more mature than Thy1-CP. The TCR-α mutation led to major accumulation of Thy1-SP and DP-IEL. These results suggest that αβ-lineage cells transit through these intermediate stages of differentiation, and that TCR-α rearrangements favor the transition from Thy1-SP-IEL/DP-IEL into DN/CD8-SP cells. They also support that Thy1-SP-IEL/DP-IEL are more immature than DN/CD8-SP cells. In contrast, TCR-δ deficiency did not modify the relative distribution of Lin− gut subpopulations. It is therefore likely that cells of the γδ lineage may transit directly from the Thy1-CP or Thy1-SP-IEL stage to the DN/CD8-SP stages in the gut. Similarly, most γδ-lineage cells in the thymus do not transit through the same intermediate stages of differentiation as αβ-lineage cells. They maturate before the TN3 stage.
Our results also provide a direct comparison of general characteristics of T cell differentiation in thymus and gut of normal mice. We found striking differences with regard to the frequency of pre-Tα and Rag-1 mRNA expression and the impact of pre-Tα and CD3-εΔ5/Δ5 deficiencies.
Only very rare Lin− cells in the gut expressed pre-Tα mRNA. The maximal frequency of pre-Tα mRNA expression (12% of CD3-ε mRNA+ Thy1-SP-IEL) was much lower than that found in the thymus (75% of CD3-ε mRNA+ CD25+ thymocytes). This low pre-Tα expression may reflect a predominance of precursors committed to TCR-γδ differentiation (39). In the thymus, the pre-TCR drives the expansion of TCR-β+ CD25+ thymocytes and their differentiation into CD4+ CD8+ cells of the TCR-αβ lineage, even in cells expressing in-frame TCR-γδ rearrangements (14, 15). Pre-TCR expression and β selection is responsible for the predominant thymus generation of TCR-αβ cells (14, 15). In the gut, only very rare Thy1+ cells would be able to undergo this expansion process that would hinder TCR-αβ generation. As we characterize all Lin− populations in the gut, we can conclude that β selection within the gut Lin− populations is a very rare event.
We also found that recombinase expression in CD3-ε mRNA–expressing gut cells was very low compared with the thymus. The distribution of Rag-expressing cells among gut precursors was also very different from that found in the thymus. The thymocyte precursors expressing the highest levels of recombinase at both mRNA and protein levels are CD4+ CD8+ DP cells that are TCRβ+, have undergone β selection, and no longer express pre-Tα mRNA (31). This upregulation of recombinases is required for TCR-α rearrangements to proceed (32). In the gut, the Lin− IEL populations that expressed TCR-β, but lacked detectable TCR-α rearrangements (TCR-β+α−) where pre-Tα was rare or absent, expressed Rag-1 mRNA at a very low level. In these circumstances, the number of precursors present in the gut able to undergo TCR-α rearrangements, and differentiate into TCR-αβ cells, is very small.
The patterns of pre-Tα and Rag-1 mRNA expression in the gut Lin− cells thus suggest a very low frequency of TCR-αβ generation during local differentiation.
We cannot exclude that some gut CD8αα+ T cells also originate from precursors that differentiate outside the gut and express pre-Tα or Rag elsewhere. A low frequency of β selection and TCR-α rearrangements, however, justifies for the first time the very unusual features of CD8αα TCR+ repertoires (40): the prevalence of TCR-γδ cells; the late ontogenic development of TCR-αβ cells, which are very rare in young mice and only accumulate with age; and the very restricted clonal diversity of TCR-αβ cells in the gut. Indeed, only ∼100 different clones are present in the gut whereas peripheral TCR-αβ cell repertoires are highly diverse (41). It is unlikely that this oligoclonality is antigen driven. In the peripheral lymphoid organs a background of diverse rearrangements is always seen during an immune response, and this background is absent in the gut (40). Moreover, these clones are totally different between individual animals, even in the same antigenic environment, belonging to the same litter, and bred in the same cage (40). A random and quite rare TCR-αβ generation would generate this type of individual variation.
Thymus and gut differentiation also differed in their requirements for CD3 expression. CD3 deficiencies have no effect on TCR-β, -γ, and -δ rearrangements in early thymocyte precursors (17, 42). Even CD3-ζ CD3-εΔ5/Δ5 multideficient CD25+ thymocyte precursors (that do not express CD3-ζ-ε-δ chains and have a partial γ deficiency) undergo TCR-β, -γ, and -δ rearrangements at a normal frequency, ruling out redundant effects of the various CD3 components on TCR rearrangements in the thymus (42). In contrast, the CD3-εΔ5/Δ5 mutation was associated with a complete blockade of TCR-β locus rearrangements (13) and a marked reduction in TCR-γ rearrangements in the gut that we found to affect even the most immature precursors. The reason for this difference is unknown. It is conceivable that CD3 components, by participating in TCR-γδ or pre-TCR assembly (15), may be required to ensure the survival of rearranged cells in the gut. Alternatively, the CD3 components may play a role in initiating TCR rearrangements, independently of or before their function as a component of the pre-TCR or of the mature γδ-TCR. A similar role was described for Igβ during VH-DJ recombination in the course of B cell development (43).
Finally, our data raise questions concerning the role of CP. As these structures have been assimilated to a “local thymus,” it was surprising that the first molecular characterization of Lin− CP cells showed such little evidence of T cell differentiation (10). Only germline TCR-β transcripts and rare CD3-ε transcripts were detected. These cells were reported to lack TCR-β and γ rearrangements and pre-Tα transcripts (10). In contrast, we detected TCR-β, γ rearrangements, and pre-Tα transcripts in Thy1-CP cells. The differences between this data and our results are likely methodological. Our molecular characterization was exhaustive and efficient enough to detect the rare CP cells that are undergoing T cell differentiation. Contrary to previous studies, we looked at DJ rearrangements that precede V-DJ rearrangements (14, 37), we used mixtures of V primers (rather than a single Vβ), and we looked at γ2 rearrangements (prevalent in CP cells). We used single cell RT-PCR and specific reverse transcription to detect pre-Tα and Rag-1 mRNA expression, which is an approach that can identify 10 mRNA molecules (16). We also subdivided CP cells, allowing selection of the Thy1-CP cells that include T cell precursors and exclude Thy1− CP cells that showed no signs of T cell commitment. This population was included in other studies (10). Our quantitative analysis, however, showed that only 3% of CP cells were clearly T cell precursors. The function of the remaining 97% of CP cells is yet to be established. We previously showed that CP cells express GATA-2 (44), a transcription factor expressed by multipotent hematopoietic progenitors (45). The complex CP structure may thus contain precursor cell types with other differentiation potentials, and may have a major role in the local generation of hematopoietic cells other than T lymphocytes. A likely candidate is the Lin− mast cell precursor, whose phenotype is similar to that of Lin− Thy1-CP (46). In vitro, Lin− gut cells can generate mast cells (47).
To summarize, the molecular characterization of Lin− populations recovered from the gut wall shows that the differentiation processes in thymus and gut did not overlap. The peculiar features of each differentiation process explains the characteristics of the respective mature T cell progenies. In the thymus, the high pre-Tα expression (favoring β selection) and the high recombinase expression (allowing frequent rearrangements) lead to the generation of a very diverse TCR-αβ repertoire (41). The rarity of pre-Tα expression and the very low recombinase activity in precursors isolated from the gut, may lead to rare β selection and hinder TCR-α rearrangements. These local characteristics explain why TCR-αβ CD8αα T-IEL have a very restricted diversity (40) and only accumulate with age (48, 49). The mechanisms underlying the unique role of CD3 components in gut differentiation are unknown, but may be related to differences in signaling cascades in gut precursors (50, 51).
Acknowledgments
We are grateful to D. Guy-Grand for help, F. Vasseur, A.-C. Waché, and V. Pasqualetto for help with tissue sections, and A. Freitas, A. Sarukhan, and C. Tanchot for reading the manuscript.
F. Lambolez was supported by a grant from the Association Claude Bernard (Ligue pour la Recherche contre le Cancer). This work was supported by the Agence Nationale de Recherches sur le SIDA.
Footnotes
Abbreviations used in this paper: BM, bone marrow; CP, cryptopatches; DN, double negative; DP, double positive; IEL, intraepithelial lymphocytes; Lin−, lineage markers negative; Rag, recombination activating gene; SP, single positive; T-IEL, gut intraepithelial T lymphocytes; WL, wall lymphocytes.
References
- 1.Lefrancois, L., and T. Goodman. 1989. In vivo modulation of cytolytic activity and Thy-1 expression in TCR-gamma delta+ intraepithelial lymphocytes. Science. 243:1716–1718. [DOI] [PubMed] [Google Scholar]
- 2.Guy-Grand, D., J.P. DiSanto, P. Henchoz, M. Malassis-Seris, and P. Vassalli. 1998. Small bowel enteropathy: role of intraepithelial lymphocytes and of cytokines (IL-12, IFN-gamma, TNF) in the induction of epithelial cell death and renewal. Eur. J. Immunol. 28:730–744. [DOI] [PubMed] [Google Scholar]
- 3.Rocha, B., P. Vassalli, and D. Guy-Grand. 1991. The V beta repertoire of mouse gut homodimeric α CD8+ intraepithelial T cell receptor α/β + lymphocytes reveals a major extrathymic pathway of T cell differentiation. J. Exp. Med. 173:483–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guy-Grand, D., N. Cerf-Bensussan, B. Malissen, M. Malassis-Seris, C. Briottet, and P. Vassalli. 1991. Two gut intraepithelial CD8+ lymphocyte populations with different T cell receptors: a role for the gut epithelium in T cell differentiation. J. Exp. Med. 173:471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arstila, T., T.P. Arstila, S. Calbo, F. Selz, M. Malassis-Seris, P. Vassalli, P. Kourilsky, and D. Guy-Grand. 2000. Identical T cell clones are located within the mouse gut epithelium and lamina propia and circulate in the thoracic duct lymph. J. Exp. Med. 191:823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guy-Grand, D., B. Rocha, P. Mintz, M. Malassis-Seris, F. Selz, B. Malissen, and P. Vassalli. 1994. Different use of T cell receptor transducing modules in two populations of gut intraepithelial lymphocytes are related to distinct pathways of T cell differentiation. J. Exp. Med. 180:673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rocha, B., P. Vassalli, and D. Guy-Grand. 1994. Thymic and extrathymic origins of gut intraepithelial lymphocyte populations in mice. J. Exp. Med. 180:681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanamori, Y., K. Ishimaru, M. Nanno, K. Maki, K. Ikuta, H. Nariuchi, and H. Ishikawa. 1996. Identification of novel lymphoid tissues in murine intestinal mucosa where clusters of c-kit+ IL-7R+ Thy1+ lympho-hemopoietic progenitors develop. J. Exp. Med. 184:1449–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito, H., Y. Kanamori, T. Takemori, H. Nariuchi, E. Kubota, H. Takahashi-Iwanaga, T. Iwanaga, and H. Ishikawa. 1998. Generation of intestinal T cells from progenitors residing in gut cryptopatches. Science. 280:275–278. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki, K., T. Oida, H. Hamada, O. Hitotsumatsu, M. Watanabe, T. Hibi, H. Yamamoto, E. Kubota, S. Kaminogawa, and H. Ishikawa. 2000. Gut cryptopatches: direct evidence of extrathymic anatomical sites for intestinal T lymphopoiesis. Immunity. 13:691–702. [DOI] [PubMed] [Google Scholar]
- 11.Lin, T., G. Matsuzaki, H. Yoshida, N. Kobayashi, H. Kenai, K. Omoto, and K. Nomoto. 1994. CD3−CD8+ intestinal intraepithelial lymphocytes (IEL) and the extrathymic development of IEL. Eur. J. Immunol. 24:1080–1087. [DOI] [PubMed] [Google Scholar]
- 12.Bruno, L., B. Rocha, A. Rolink, H. von Boehmer, and H.R. Rodewald. 1995. Intra- and extra-thymic expression of the pre-T cell receptor alpha gene. Eur. J. Immunol. 25:1877–1882. [DOI] [PubMed] [Google Scholar]
- 13.Page, S.T., L.Y. Bogatzki, J.A. Hamerman, C.H. Sweenie, P.J. Hogarth, M. Malissen, R.M. Perlmutter, and A.M. Pullen. 1998. Intestinal intraepithelial lymphocytes include precursors committed to the T cell receptor alpha beta lineage. Proc. Natl. Acad. Sci. USA. 95:9459–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Boehmer, H., I. Aifantis, O. Azogui, J. Feinberg, C. Saint-Ruf, C. Zober, C. Garcia, and J. Buer. 1998. Crucial function of the pre-T-cell receptor (TCR) in TCR beta selection, TCR beta allelic exclusion and alpha beta versus gamma delta lineage commitment. Immunol. Rev. 165:111–119. [DOI] [PubMed] [Google Scholar]
- 15.Aifantis, I., O. Azogui, J. Feinberg, C. Saint-Ruf, J. Buer, and H. von Boehmer. 1998. On the role of the pre-T cell receptor in alphabeta versus gammadelta T lineage commitment. Immunity. 9:649–655. [DOI] [PubMed] [Google Scholar]
- 16.Veiga-Fernandes, H., U. Walter, C. Bourgeois, A. McLean, and B. Rocha. 2000. Response of naive and memory CD8+ T cells to antigen stimulation in vivo. Nat. Immunol. 1:47–53. [DOI] [PubMed] [Google Scholar]
- 17.Malissen, M., A. Gillet, L. Ardouin, G. Bouvier, J. Trucy, P. Ferrier, E. Vivier, and B. Malissen. 1995. Altered T cell development in mice with a targeted mutation of the CD3-epsilon gene. EMBO J. 14:4641–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinkai, Y., G. Rathbun, K.P. Lam, E.M. Oltz, V. Stewart, M. Mendelsohn, J. Charron, M. Datta, F. Young, and A.M. Stall. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 68:855–867. [DOI] [PubMed] [Google Scholar]
- 19.Mombaerts, P., A.R. Clarke, M.A. Rudnicki, J. Iacomini, S. Itohara, J.J. Lafaille, L. Wang, Y. Ichikawa, R. Jaenisch, and M.L. Hooper. 1992. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 360:225–231 (erratum published 360:491). [DOI] [PubMed] [Google Scholar]
- 20.Itohara, S., P. Mombaerts, J. Lafaille, J. Iacomini, A. Nelson, A.R. Clarke, M.L. Hooper, A. Farr, and S. Tonegawa. 1993. T cell receptor delta gene mutant mice: independent generation of alpha beta T cells and programmed rearrangements of gamma delta TCR genes. Cell. 72:337–348. [DOI] [PubMed] [Google Scholar]
- 21.Soudais, C., T. Shiho, L.I. Sharara, D. Guy-Grand, T. Taniguchi, A. Fischer, and J.P. Di Santo. 2000. Stable and functional lymphoid reconstitution of common cytokine receptor gamma chain deficient mice by retroviral-mediated gene transfer. Blood. 95:3071–3077. [PubMed] [Google Scholar]
- 22.Guy-Grand, D., B. Cuenod-Jabri, M. Malassis-Seris, F. Selz, and P. Vassalli. 1996. Complexity of the mouse gut T cell immune system: identification of two distinct natural killer T cell intraepithelial lineages. Eur. J. Immunol. 26:2248–2256. [DOI] [PubMed] [Google Scholar]
- 23.Delassus, S., I. Titley, and T. Enver. 1999. Functional and molecular analysis of hematopoietic progenitors derived from the aorta-gonad-mesonephros region of the mouse embryo. Blood. 94:1495–1503. [PubMed] [Google Scholar]
- 24.Rodewald, H.R., K. Awad, P. Moingeon, L. D'Adamio, D. Rabinowitz, Y. Shinkai, F.W. Alt, and E.L. Reinherz. 1993. Fc gamma RII/III and CD2 expression mark distinct subpopulations of immature CD4−CD8− murine thymocytes: in vivo developmental kinetics and T cell receptor β chain rearrangement status. J. Exp. Med. 177:1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodewald, H.R., K. Kretzschmar, S. Takeda, C. Hohl, and M. Dessing. 1994. Identification of pro-thymocytes in murine fetal blood: T lineage commitment can precede thymus colonization. EMBO J. 13:4229–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin, S.D., S.J. Anderson, K.A. Forbush, and R.M. Perlmutter. 1993. A dominant-negative transgene defines a role for p56lck in thymopoiesis. EMBO J. 12:1671–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aifantis, I., J. Buer, H. von Boehmer, and O. Azogui. 1997. Essential role of the pre-T cell receptor in allelic exclusion of the T cell receptor beta locus. Immunity. 7:601–607 (erratum published 7:895). [DOI] [PubMed] [Google Scholar]
- 28.Godfrey, D.I., A. Zlotnik, and T. Suda. 1992. Phenotypic and functional characterization of c-kit expression during intrathymic T cell development. J. Immunol. 149:2281–2285. [PubMed] [Google Scholar]
- 29.Barber, D.F., L. Passoni, L. Wen, L. Geng, and A.C. Hayday. 1998. The expression in vivo of a second isoform of pT alpha: implications for the mechanism of pT alpha action. J. Immunol. 161:11–16. [PubMed] [Google Scholar]
- 30.Wu, L., M. Antica, G.R. Johnson, R. Scollay, and K. Shortman. 1991. Developmental potential of the earliest precursor cells from the adult mouse thymus. J. Exp. Med. 174:1617–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson, A., W. Held, and H.R. MacDonald. 1994. Two waves of recombinase gene expression in developing thymocytes. J. Exp. Med. 179:1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagaoka H., W. Yu, and M.C. Nussenzweig. 2000. Regulation of RAG expression in developing lymphocytes. Curr. Opin. Immunol. 12:187–190. [DOI] [PubMed] [Google Scholar]
- 33.Takagaki, Y., A. DeCloux, M. Bonneville, and S. Tonegawa. 1989. Diversity of gamma delta T-cell receptors on murine intestinal intra-epithelial lymphocytes. Nature. 339:712–714. [DOI] [PubMed] [Google Scholar]
- 34.Livak, F., M. Tourigny, D.G. Schatz, and H.T. Petrie. 1999. Characterization of TCR gene rearrangements during adult murine T cell development. J. Immunol. 162:2575–2580. [PubMed] [Google Scholar]
- 35.MacDonald, T.T., M. Bajaj-Elliott, and S.L. Pender. 1999. T cells orchestrate intestinal mucosal shape and integrity. Immunol. Today. 20:505–510. [DOI] [PubMed] [Google Scholar]
- 36.Rocha, B., D. Guy-Grand, and P. Vassalli. 1995. Extrathymic T cell differentiation. Curr. Opin. Immunol. 7:235–242. [DOI] [PubMed] [Google Scholar]
- 37.Shortman, K., and L. Wu. 1996. Early T lymphocyte progenitors. Annu. Rev. Immunol. 14:29–47. [DOI] [PubMed] [Google Scholar]
- 38.Lantelme, E., B. Palermo, L. Granziero, S. Mantovani, R. Campanelli, V. Monafo, A. Lanzavecchia, and C. Giachino. 2000. Cutting edge: recombinase-activating gene expression and V(D)J recombination in CD4+CD3low mature T lymphocytes. J. Immunol. 164:3455–3459. [DOI] [PubMed] [Google Scholar]
- 39.Tanamachi, D.M., T. Hanke, H. Takizawa, A.M. Jamieson, and D.R. Raulet. 2001. Expression of natural killer receptor alleles at different Ly49 loci occurs independently and is regulated by major histocompatibility complex class I molecules. J. Exp. Med. 193:307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regnault, A., A. Cumano, P. Vassalli, D. Guy-Grand, and P. Kourilsky. 1994. Oligoclonal repertoire of the CD8 αα and the CD8 αβ TCR-α/β murine intestinal intraepithelial T lymphocytes: evidence for the random emergence of T cells. J. Exp. Med. 180:1345–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arstila, T.P., A. Casrouge, V. Baron, J. Even, J. Kanellopoulos, and P. Kourilsky. 1999. A direct estimate of the human alphabeta T cell receptor diversity. Science. 286:958–961. [DOI] [PubMed] [Google Scholar]
- 42.Ardouin, L., J. Ismaili, B. Malissen, and M. Malissen. 1998. The CD3-γδε and CD3-ϑ/η modules are each essential for allelic exclusion at the T cell receptor beta locus but are both dispensable for the initiation of V to (D)J recombination at the T cell receptor-β, -γ, and -δ loci. J. Exp. Med. 187:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong, S., and M.C. Nussenzweig. 1996. Regulation of an early developmental checkpoint in the B cell pathway by Ig beta. Science. 272:411–414. [DOI] [PubMed] [Google Scholar]
- 44.Lambolez, F., and B. Rocha. 2001. Molecular characterization of gut T cell precursors in euthymic and athymic mice. Adv. Exp. Med. Biol. 495:15–24. [DOI] [PubMed] [Google Scholar]
- 45.Tsai, F.Y., G. Keller, F.C. Kuo, M. Weiss, J. Chen, M. Rosenblatt, F.W. Alt, and S.H. Orkin. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 371:221–226. [DOI] [PubMed] [Google Scholar]
- 46.Rodewald, H.R., M. Dessing, A.M. Dvorak, and S.J. Galli. 1996. Identification of a committed precursor for the mast cell lineage. Science. 271:818–822. [DOI] [PubMed] [Google Scholar]
- 47.Guy-Grand, D., M. Dy, G. Luffau, and P. Vassalli. 1984. Gut mucosal mast cells. Origin, traffic, and differentiation. J. Exp. Med. 160:12–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poussier, P., and M. Julius. 1994. Thymus independent T cell development and selection in the intestinal epithelium. Annu. Rev. Immunol. 12:521–553. [DOI] [PubMed] [Google Scholar]
- 49.Steege, J.C., W.A. Buurman, and P.P. Forget. 1997. The neonatal development of intraepithelial and lamina propria lymphocytes in the murine small intestine. Dev. Immunol. 5:121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mallick-Wood, C.A., W. Pao, A.M. Cheng, J.M. Lewis, S. Kulkarni, J.B. Bolen, B. Rowley, R.E. Tigelaar, T. Pawson, and A.C. Hayday. 1996. Disruption of epithelial gamma delta T cell repertoires by mutation of the Syk tyrosine kinase. Proc. Natl. Acad. Sci. USA. 93:9704–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Page, S.T., N.S. van Oers, R.M. Perlmutter, A. Weiss, and A.M. Pullen. 1997. Differential contribution of Lck and Fyn protein tyrosine kinases to intraepithelial lymphocyte development. Eur. J. Immunol. 27:554–562. [DOI] [PubMed] [Google Scholar]