Abstract
Chagas' disease is a major health and economic problem caused by the protozoan Trypanosoma cruzi. Multiple independently evolving clones define a complex parasite population that can be arranged into two broad genetic lineages termed T. cruzi I and II. These lineages have different evolutionary origin and display distinct ecological and biological traits. Here we describe a novel molecule termed TSSA for trypomastigote small surface antigen that provides the first immunological marker allowing discrimination between lineages. TSSA is a surface, glycosylphosphatidyl inositol (GPI)-anchored mucin-like protein, highly antigenic during the infection. TSSA sequences from different parasite isolates reveal a population dimorphism that perfectly matches with the two T. cruzi lineages. Interestingly, this dimorphism is restricted to the central region of the molecule, which comprises the immunodominant B cell epitopes. This sequence variability has a major impact on TSSA antigenicity, leading to no immunological cross-reactivity between both isoforms for antibodies present either in immunization or infection sera. Furthermore, the absolute seroprevalence for TSSA in confirmed Chagasic patients is restricted to T. cruzi II isoform, strongly suggesting that human infections are due to this particular subgroup. Even though association of T. cruzi II with Chagas' disease has been proposed based on molecular markers, this is the first immunological evidence supporting this hypothesis. The implications of these results for the future research on Chagas' disease could be envisaged.
Keywords: Trypanosoma cruzi, mucin-like, lineage dimorphism, Chagas' disease, serodiagnosis
Introduction
Trypanosoma cruzi is the etiological agent of Chagas' disease, an endemic illness that affects 18 million people in Latin America (1). The parasite alternates its life cycle between vertebrates and insect vectors, with different developmental stages involved in each host (2). Within the reduviid vector two forms of the parasite can be observed: replicative epimastigotes and metacyclic trypomastigotes. The latter form brings the infection into humans when released on the skin or mucosa with the depositions of the bug. After cell invasion, metacyclic trypomastigotes differentiate into the replicative amastigote form that, after several divisions, differentiates into bloodstream trypomastigotes. The latter stage is able to invade a wide variety of nucleated cells, thus propagating the infection. The cycle closes when the hematophagous vector ingests circulating trypomastigotes with its blood meal.
After an acute phase characterized by a high parasitemia, the infection progress to a chronic stage, where parasites are barely detected. In 35% of the human cases pathological signs such as myocarditis and neurodegenerative effects on the digestive system would appear, thus leading to Chagas' disease (1). The incidence of acute cases in humans is steadily declining in a number of countries mainly due to the successful control of vectorial transmission (1). However, as T. cruzi develops a life-long infection in humans, these people can serve as parasite reservoirs throughout their lifetime. Thus, the risk of congenital and/or horizontal transmission by infected blood transfusion may become a major problem in nonendemic regions, increased by the migration of people from endemic areas in South and Central America to the developed countries (1). In this context, the identification of new immunodominant parasite molecules reliable for serodiagnosis is desirable (3).
T. cruzi is grossly divided into two divergent genetic groups or lineages, called T. cruzi I and II that include all typed strains and cloned stocks thus far isolated (4–6), although further subdivisions are possible (7, 8). This population structure is a consequence of both its divergent evolutionary history (9) and its clonal rather than sexual propagation (10). Diversity among parasite isolates has been early noted, mainly based on biochemical, ecological, and epidemiological data (11). More recently, several molecular markers have been identified that allow the conformation of two broad lineages and a more accurate discrimination between isolates. These markers include the 24SαrRNA gene and its promoter sequence (5, 12), the intergenic region of the tandemly repeated mini-exon gene (5) and microsatellite DNA (13). Current biological and epidemiological data provide evidence for a strong association of T. cruzi II with human disease whereas T. cruzi I is preferentially detected in the sylvatic cycle (affecting mainly American marsupials and edentates; references 6 and 14). However, a state-of-the-art demonstration of this assumption is still lacking.
TcMUC is a complex T. cruzi family of genes that resemble vertebrate mucin genes (15, 16), comprising 500 to 700 members per haploid genome (17, 18). Accordingly to the structure of the deduced proteins three groups of genes were conformed (17). All of them code for short Thr, Ser, and Pro-rich proteins that share highly homologous N- and COOH-terminal regions, encoding for an endoplasmic reticulum targeting signal and a glycosylphosphatidyl inositol (GPI)* anchor attachment signal, respectively (17). Divergences that account for their classification on different groups arise mainly in their central domains. Mucin molecules belonging to group I were identified on the surface of the vertebrate stages of T. cruzi and have Thr(8)-Lys-Pro(2) tandem repeats in the central region that become highly O-glycosylated in vivo (19). The products of group II display nonrepeated unique central sequences and their expression at the protein level have never been confirmed in the parasite. Nevertheless the deduced products are rich enough in Thr, Ser, and Pro residues as to support its mucin-like denomination (17, 20). Finally, group III yielded a very small-deduced product that displays no significant homologies with anything in the sequences databases (17).
In the present work, we show that TcMUC group III is actually conformed by a dimorphic single-copy gene, with two alleles distributed among the parasite isolates coinciding with the two broad lineages postulated for T. cruzi (5, 6). The expression of its encoded protein was verified in the surface of the cell-derived trypomastigotes. Therefore, we named it trypomastigote small surface antigen (TSSA) given that the product is highly immunogenic during T. cruzi infections in humans and laboratory animals. By using peptide scanning we demonstrated that the Ab recognition was mainly restricted to epitope(s) contained within a single 10-amino acid long peptide. Interestingly, sequence differences observed for the two alleles found in the species were mainly clustered within this antigenic region and lead to no immunological cross-reactivity between both isoforms. More important, the high prevalence (100% among confirmed Chagasic individuals and >87% among potentially Chagasic individuals) of anti-TSSA Abs is exclusively attributed to T. cruzi II isoform. Thus, we propose TSSA as the first immunological marker of T. cruzi genetic subgroups that correlates human infection with T. cruzi II.
Materials and Methods
Parasites and Experimental T. cruzi Infection Sera.
The CL Brener genome project reference clone of T. cruzi was used for the antigen characterization (21). The following strains or cloned stocks were used for DNA preparation and analysis: CL, RA, Y, Dm28c, Tulahuen2, SilvioX10 (5), AWP, Mg, ITN, UP (22), CA-I/69, CA-I/72, Miranda (23), Peru (12), Tulahuen0 (24), GER, #2538, and #2378 (cloned stocks isolated from chronic Chagasic patients from Brazil (GER) and Argentina (#2538 and #2378), our unpublished results). Sera from BALB/c mice infected by the intraperitoneal route with a nonlethal dose of 103 CL Brener bloodstream trypomastigotes and bled at different times postinfection were provided by Dr. S. Leguizamón (Depto. de Microbiología, Facultad de Medina, Universidad de Buenos Aires, Buenos Aires, Argentina). Additional sera from BALB/c infected mice were provided by Dr. M.M. Teixeira (Departamento de Parasitologia, Instituto de Ciências Biomédicas, Universidade de São Paulo, São Paulo, Brazil). Sera from Wistar rats infected with 106 SilvioX10 or Miranda bloodstream trypomastigotes by the intravenous route were provided by Dr. J. Bua (Instituto Nacional de Parasitología “Dr. M. Fatala Chaben,” Buenos Aires, Argentina). Further sera from rabbits infected with different T. cruzi isolates were a generous gift from Dr. D. Sánchez (IIB-INTECH, UNSAM, Buenos Aires, Argentina).
Human Sera Survey.
Sera were collected from 460 people living in different endemic and nonendemic geographic areas in Argentina (66 potentially Chagasic individuals and 10 non-Chagasic individuals), Brazil (112 potentially Chagasic individuals, 123 confirmed Chagasic individuals, and 126 non-Chagasic individuals) and Chile (23 potentially Chagasic individuals). Some of the Brazilian confirmed Chagasic patients were coinfected with HIV. Brazilian human serum samples were provided by Dr. D.T. Covas (Fundação Hemocentro de Ribeirão Preto, Ribeirão Preto, Brazil), Dr. A. Fragata Filho (Instituto Dante Pazzanese de Cardiologia, São Paulo, Brazil), and Dr. M.A. Shikanai-Yasuda (Instituto de Medicina Tropical, Universidade de São Paulo, São Paulo, Brazil). Argentinean serum samples were purchased from the Instituto Nacional de Parasitología “Dr. M. Fatala Chaben.” Chilean serum samples were a gift from Dr. J. Gonzales (Unidad de Parasitologia, Universidad de Antofagasta, Antofagasta, Chile). All sera were diagnosed by three independent assays (ELISA using total parasite homogenate, indirect immunohemaglutination [IHA], and indirect immunofluorescence [IFI]). In addition, some patients were also tested by chemiluminescent-ELISA (CL-ELISA, see below) using an epimastigote preparation as antigen (25) and/or for the presence of T. cruzi in the total blood by hemoculture (26). Individuals were grouped as potentially Chagasic (those rendering positive results only for conventional serology) or confirmed Chagasic (those rendering positive results both for conventional serology and hemoculture).
Expression of TSSA as a GST-fusion Protein and Antisera Development.
A fragment of both tssa-I (from Dm28c) and tssa-II (from CL Brener) genes was amplified by PCR using the oligonucleotides PsMUC/Bam (5′-CGTGGATCCTTGGTGCTCGCCCTGTGCT-3′) and DGS/Eco (5′-CGGGAATTCAGCTGCCGAGGCTGCCGTC-3′; Fig. 1). The products were BamHI/EcoRI digested and cloned into pGEX-2T (Amersham Pharmacia Biotech). Expression and purification of the ensuing GST-TSSA-I and GST-TSSA-II chimeric proteins were done as described elsewhere (19). For the production of specific antisera, 100 μg of either GST-TSSA were emulsified in CFA (Sigma-Aldrich) and injected into rabbits by the subcutaneous route. Two boosters of 50 μg each in IFA were given at days 30 and 45 postinjection. A similar immunization schedule was followed with mice (C3H/HeN) but using 25, 5, and 5 μg doses, respectively, by the intraperitoneal route.
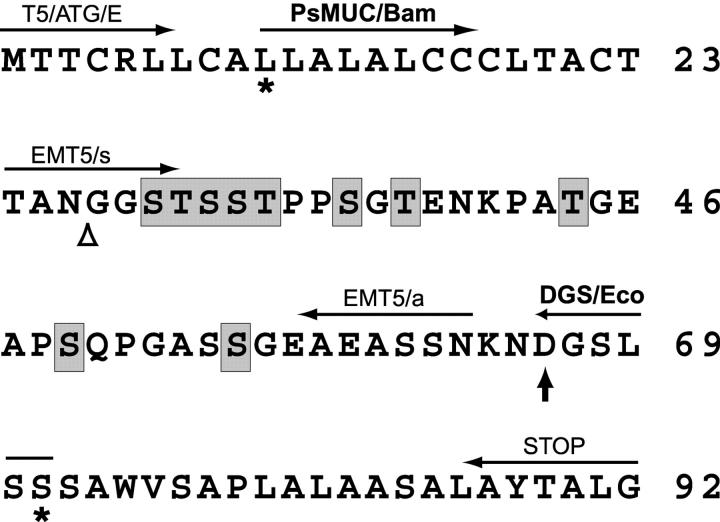
Figure 1.
Primary structure and features of the TSSA. The deduced amino acid sequence of TSSA obtained from the CL Brener gene is indicated. Ser and Thr residues predicted to be O-glycosylated are shaded. The predicted signal peptide cleavage site (open triangle) and the putative GPI anchor addition (vertical arrow) are indicated. Sequence between asterisks indicates the fragment expressed as GST-fusion. The position and names of the oligonucleotides used in this work are indicated above the corresponding spanned protein sequence.
Purification of TSSA-II–specific Abs from Chagasic Sera.
Purified GST-TSSA-II (1 mg) was immobilized on a NHS-activated column (HiTrap®; Amersham Pharmacia Biotech) following manufacturer's guidelines. Samples (100 μl each) of 10 Chagasic sera selected on the basis of their high reactivity against GST-TSSA-II were pooled, diluted to 10 ml in Tris-buffered saline 0.15 M pH 7.6 containing 0.1% Tween 20 (TBS-T), and applied onto the GST-TSSA-II column. After extensive washing with TBS, affinity-retained Abs were eluted with 5 ml of glycine 0.1 M pH 3 and immediately neutralized with a few drops of Tris-HCl, 1 M, pH 8.
ELISA.
Polystyrene ELISA microplates (Maxisorp; Nunc) were coated with 100 ng of the indicated antigen in PBS. Blocking of the plates were made in TBS-T supplemented with 5% nonfat dried milk. Appropriate dilutions of serum samples in the same solution were incubated for 1 h at room temperature and captured Abs were evaluated by the addition of secondary Abs coupled to HRPO (1:10,000 dilution; Dako) followed by 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (Roche Diagnostics). Plates were read at 405 nm in a microplate reader (Bio-Rad Laboratories). Samples obtained from healthy patients or animals were used as negative controls. Serum samples giving OD values higher than the mean obtained for the negative population + 3 SD were recorded as positive. Alternatively, anti-TSSA Abs were evaluated by the more sensitive CL-ELISA as described previously (25, 27). For this assay, plates were coated either with 10 or 20 ng of the recombinant molecules and sera from Chagasic and non-Chagasic individuals or T. cruzi–infected animals tested as indicated.
SPOT Assays.
Immobilized peptide SPOTs (28) were processed as described by the manufacturers (Sigma-Genosys) with subtle modifications. Briefly, membranes were blocked overnight in casein based blocking reagent (Sigma-Genosys) supplemented with 1% nonfat dried milk. Sera were diluted 1:100 in the same solution and incubated for 3 h. After extensive washings, secondary Abs coupled to HRPO were added and reaction developed by chemiluminescence with WestPico SuperSignal substrate (Pierce Chemical Co.).
DNA and RNA Purification and Hybridization.
DNA was prepared from T. cruzi epimastigotes by proteinase K-phenol/chloroform extraction (15). Parasite RNA was prepared from the different developmental stages (obtained as described in reference 15) using TRIzol® (Life Technologies) according to manufacturer's guidelines. After nucleic acid immobilization onto Z-probe membranes (Bio-Rad Laboratories) Southern and Northern blot assays were performed as described elsewhere (15). The tssa-II–specific probe was synthesized by PCR using the oligonucleotides EMT5/s (5′-ACAGCGAATGGTGGGTCT-3′) and EMT5/a (5′-TTTGAGGAGGCTTCTGCTTC-3′) (Fig. 1). Hybridization was performed in SSC/SDS/Denhardt solution and washings performed under high stringency conditions (65°C, 0.1× SSC, 0.1% SDS) as described (15).
Cloning of tssa from Different T. cruzi Isolates.
The tssa alleles were cloned by PCR using 100 ng of genomic DNA of the Tulahuen2, Dm28c, Y, RA, CA-I/72, and Silvio×10 isolates as template. Two reactions were necessary to isolate the complete genes given that the TcMUC family has highly conserved ends (17). In a first set of reactions the oligonucleotide T5/ATG/E (5′-CGGAATTCATGACTACGTGCCGTCT-3′) was used in combination with the oligonucleotide EMT5/a and, in a second reaction, the oligonucleotide STOP (5′-TCAGCCCAGAGTGGTGTACGC-3′) was used in combination with the oligonucleotide EMT5/s (Fig. 1). The products were cloned in pGEM® T-Easy (Promega) and fully sequenced using the Sequenase 2.0 kit (United States Biochemical Corp.). The complete sequences of the deduced proteins shown for each strain in Fig. 4 B are a mosaic of these two sequences except for CL Brener.
Figure 4.
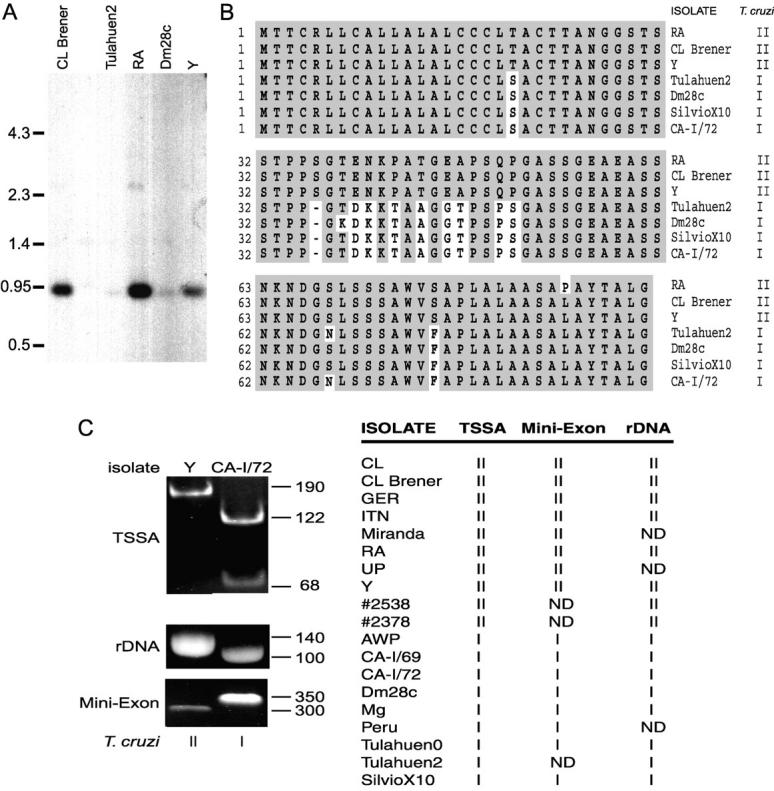
Inter-strain variability of TSSA. (A) Genomic DNA from the indicated T. cruzi strains or cloned stocks was digested by ApaLI and hybridized under stringent conditions with the tssa probe. Size markers (in kpb) are indicated on the left. (B) Comparison of the deduced TSSA amino acid sequences from the indicated parasite isolates. Isolates were classified as T. cruzi I or II according to reference 5. Amino acid identities with the CL Brener sequence are shadowed. Hyphens were introduced for better alignment. (C) tssa and lineage typing of T. cruzi strains and cloned stocks. A fragment of the tssa gene was amplified from DNA samples of the indicated isolates, digested with PvuII and resolved by PAGE. From the same samples the rDNA and mini-exon intron were amplified and resolved in agarose gels. An example of the pattern observed for each lineage as determined in this work for tssa and already described for the rDNA and mini-exon (reference 5) is shown at left. The results for all the isolates are summarized in the table at right. ND, not determined.
tssa, Mini-Exon, and rDNA Gene-typing of T. cruzi Isolates.
A fragment of the tssa alleles from several isolates was amplified from genomic samples using the oligonucleotides T5/ATG/E and EMT5/a as described above. The amplification product was treated with PvuII for 3 h and subsequently resolved in a 10% polyacrylamide/TBE gel followed by ethidium bromide staining. In addition, aliquots of the same genomic samples were used as templates for amplification of the intergenic region of the mini-exon genes and the divergent domain of the 24SαrRNA gene as described (5).
PI-PLC Treatment.
Cell-derived trypomastigotes (2 × 108) were washed twice in PBS and resuspended in 500 μl of D-MEM (Life Technologies). Parasites were incubated for 3 h at 37°C with or without the addition of 4 U of phosphatidylinositol-specific phospholipase C (PI-PLC) from Bacillus cereus (Sigma-Aldrich), washed in PBS, and pellets and concentrated conditioned media aliquots prepared for Western blotting analysis.
Parasite Lysates and Western Blotting.
PBS-washed parasites were resuspended to 106 parasites per μl in TBS 1% Triton X-100 with 1 mM PMSF, 0.5 mM N−α−p-tosyl-l-lysine chloromethyl ketone, and 100 μg/ml DNase (all from Sigma-Aldrich). SDS was added to 1% and lysates boiled for 5 min. Total lysates corresponding to 2 × 107 parasites of each stage were separated by SDS-PAGE and transferred to nitrocellulose membranes (Amersham Pharmacia Biotech; reference 29). Filters were probed with an anti GST-TSSA-II serum and revealed using HRPO-conjugated secondary Abs followed by chemiluminescence detection.
Indirect Immunofluorescence.
A drop of parasites resuspended at 5 × 106 per ml in PBS was layered onto a 3-aminopropyltriethoxysilane (Sigma-Aldrich) coated round coverslips and let stand for 30 min at room temperature. Parasites were fixed with the nonpermeabilizing reagent 4% paraformaldehyde (29) for 30 min. Blocking (1 h incubation) and Ab dilutions were in PBS containing 2% BSA and 5% normal rabbit serum. Blocking buffer was supplemented with 25 mM NH4Cl. Anti GST-TSSA-II mouse serum diluted 1:100 was allowed to react for 1 h followed by washing in PBS 2% BSA. FITC-conjugated anti-mouse IgG (Fab′)2 raised in rabbit (Dako) were added for additional 60 min and washed as above. Coverslips were mounted in 50% glycerol in PBS and observed and photographed using an Olympus IX50 inverted fluorescence microscope.
Results
Description of a TSSA Belonging to the TcMUC Family.
As a first step toward the characterization of the group III of TcMUC gene-family, three cDNA clones obtained from the CL Brener clone (17) were completely sequenced. Two further sequences from CL Brener were downloaded from GenBank: EST TENU3264 and GSSTc10735 (GenBank/EMBL/DDBJ accession nos. AI077116 and AZ049892, respectively). They all seemed to be derived from the same gene given that no nucleotide differences were detected among them. The open reading frame spanned 276 bp rendering a 92-amino acid long deduced product (Fig. 1). Comparison of this product with those from TcMUC groups I and II showed a predicted 26-amino acid long signal peptide at the NH2 terminus (30), 73% identical to the consensus of the family (17). Likewise, the last 27 amino acid residues were compatible with a GPI-anchoring signal with 85% identity to the one described for groups I and II (17). As these two regions were shown to be functional for TcMUC genes in T. cruzi (19) the mature product could be predicted with some confidence. Therefore, this protein would be 40-amino acid long after these two regions were intracellularly processed, with a M r of 3,747 D and a pI of 4.13. The mature protein has a high content of hydroxyamino acids (32.5% Ser + Thr), Pro (12.5%), Gly (15%), and Ala (12.5%) residues. This biased amino acid composition, along with the existence of 10 putative O-glycosylation sites (31; Fig. 1), suggests that this gene encode for a T. cruzi mucin-like molecule. As will be shown below, this is a surface antigen expressed in the trypomastigote stage and was so named TSSA after trypomastigote small surface antigen.
Genomic Organization and Gene Expression of tssa.
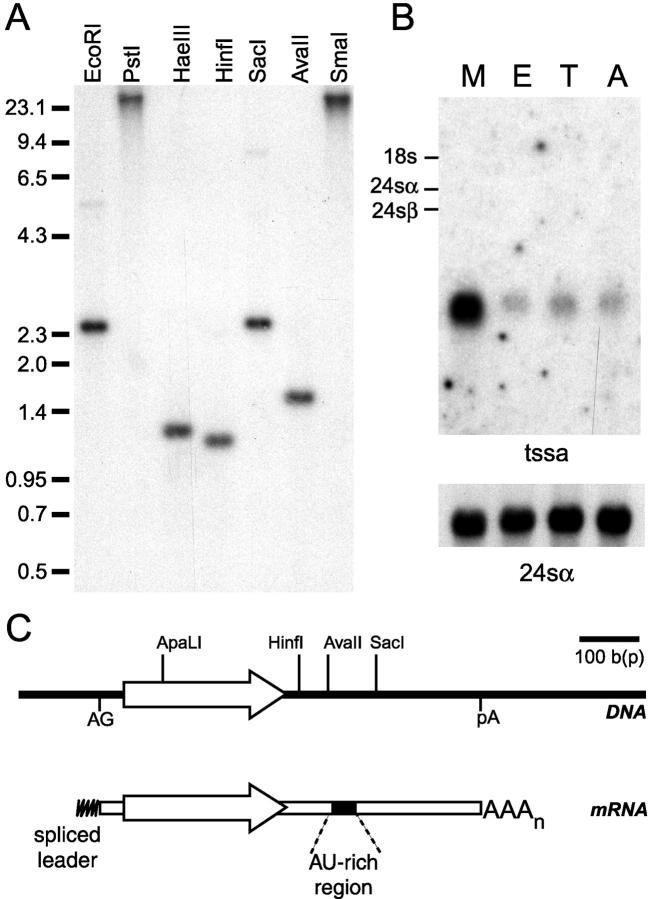
The genomic organization of tssa was analyzed by Southern blot using a probe that encompassed the central region of the CL Brener tssa gene. This probe did not cross-hybridize with genes from groups I and II of the TcMUC family as it excludes any conserved region (17). The hybridization pattern was compatible with the existence of a single gene per haploid genome (Fig. 2 A). In addition to the Southern blot, two further evidences reinforced the previous statement. First, a search in a T. cruzi genomic sequence survey database (18) using the tssa central region as query rendered only one hit (GSSTc10735). Second, hybridization of a T. cruzi CL Brener cosmid library ordered in a high-density array (32) with the same probe produced 0.8 positive signals when normalized per haploid genome (17). Altogether, these results strongly suggest that tssa is a single-copy gene at least in the CL Brener clone.
Figure 2.
Genomic organization and expression of tssa. (A) Southern blot of genomic DNA from T. cruzi CL Brener (1 μg/lane) digested using the indicated enzymes, transferred to membranes, and hybridized under stringent conditions with the tssa probe. Size markers (in kpb) are indicated on the left. (B) Northern blot of total T. cruzi CL Brener RNA purified from metacyclic trypomastigotes (M), epimastigotes (E), cell-derived trypomastigotes (T), and amastigotes (A) using the tssa probe under stringent conditions. The position of the rRNA components are shown on the left as size indication (reference 46). Control for assessing similar lane loading was performed by using the 24SαrRNA probe. (C) In scale scheme of the genomic CL Brener tssa locus (above) and its derived mature mRNA as deduced from DNA and cDNA sequencing and restriction mapping. The recognition sites for restriction enzymes used in the Southern blotting are indicated. AG, splice leader acceptor site; pA, polyadenylation acceptor site.
The same probe was used in a Northern blot assay revealing that the tssa mRNA is present in all the developmental stages of the parasite (Fig. 2 B). The steady-state levels of tssa mRNA varied among the stages being more abundant in metacyclic trypomastigotes. This expression pattern contrasted with that observed for the TSSA protein, which is mostly expressed in cell-derived trypomastigotes (Fig. 3, A and B). It should be noted, however, that posttranscriptional regulation is the rule in T. cruzi gene expression (33, 34) and some examples have been described in which protein abundance did not correlate with steady-state mRNA levels (35). As this seems to be the case for tssa, the 3′ untranslated region (UTR) of its transcript was analyzed for possible posttranscriptional regulatory elements placed in cis. The scheme presented in Fig. 2 C shows the presence of AU-rich elements found in the tssa 3′ UTR that were already shown to regulate the stability of mucin-gene transcripts in T. cruzi (36, 37). Although only structural, the antecedents in the field allow the presumption that posttranscriptional regulation plays an important role in the expression of the tssa gene.
Figure 3.
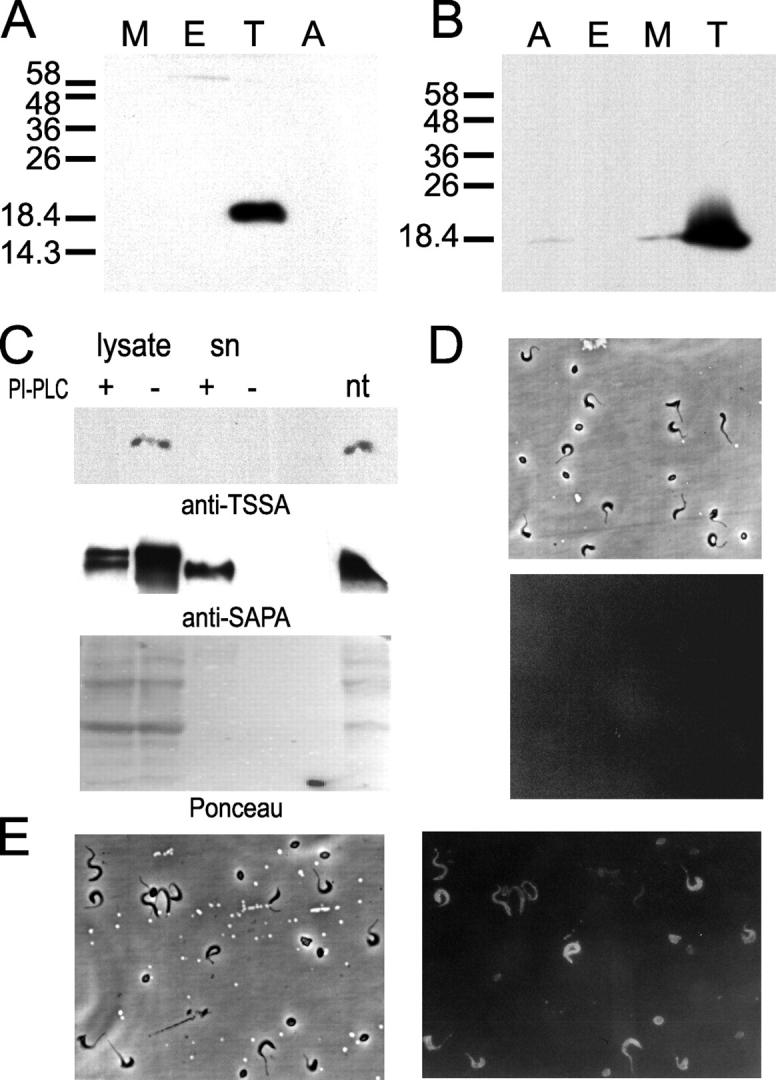
Stage-specific expression of TSSA. (A and B) Western blot of total lysates from T. cruzi (CL Brener clone) metacyclic trypomastigotes (M), epimastigotes (E), cell-derived trypomastigotes (T), and amastigotes (A). Approximately 2 × 107 parasites were loaded in each lane of a SDS-PAGE gel and assayed by Western blotting with an anti-GST-TSSA serum raised in mouse (A) or with a pool of anti-TSSA affinity-purified Abs from Chagasic sera (B). In both panels M r markers (in kD) are indicated on the left. (C) Intact live cell–derived trypomastigotes were treated (+) or not (−) with B. cereus PI-PLC. Normal morphology and motility was controlled by microscopic observation before and after the incubation time. Parasites were lysed and aliquots from total lysates and supernatants (sn) were sequentially probed with an anti–GST-TSSA serum raised in mouse and an anti-GST-SAPA serum raised in rabbit by Western blotting. Ponceau staining of the membrane is shown to assess similar lane loading. nt, nontreated parasites. (D and E) A mixture of cell-derived trypomastigotes, amastigotes, and intermediate forms were analyzed by indirect immunofluorescence. Parasites were probed using normal mouse serum (D), or an anti-GST-TSSA serum raised in mouse (E) and photographed (180× magnification) under phase contrast and fluorescence.
Detection and Characterization of the TSSA Product.
A fragment of the coding region of the tssa allele obtained from the CL Brener clone (Fig. 1) was expressed as a GST fusion and used to raise specific antisera in rabbits and mice. Both kinds of antisera were tested against total parasite lysates by Western blotting and reacted against a single 20-kD band present in cell-derived trypomastigote lysates (Fig. 3 A, and data not shown). Due to the discrepancy between the predicted and observed M r, anti-GST-TSSA Abs were affinity-purified from a pool of TSSA-reactive Chagasic sera and used to probe T. cruzi lysates to ascertain the specificity of the signal. These Abs recognized the same 20-kD protein in cell-derived trypomastigote lysates although a faint signal was observed also in amastigotes and metacyclic trypomastigotes (Fig. 3 B).
As the deduced protein had a putative GPI anchor signal (Fig. 1), the presence of such modification in TSSA was evaluated by PI-PLC treatment on intact cell-derived trypomastigotes. The addition of PI-PLC caused the disappearance of the signal in lysates from treated parasites when probed with an anti-GST-TSSA serum in Western blotting (Fig. 3 C). Nevertheless, the band was not recovered in the culture supernatant. As a control, the same filter was probed with an antiserum raised against shed acute-phase antigen (SAPA), a cell-derived trypomastigote surface antigen known to be GPI-anchored and readily shed into the medium by the same PI-PLC treatment (38). SAPA was solubilized and detected in its intact form in the supernatant after the treatment (Fig. 3 C). So, it seems that the failure to detect soluble TSSA in the filter was due to the inability of the released protein to bind to nitrocellulose membranes.
The surface localization of TSSA was further assessed by indirect immunofluorescence on nonpermeabilized parasites. The surface of cell-derived trypomastigotes was clearly labeled by anti-TSSA Abs but not by a mouse normal serum (Fig. 3, D and E). Broad trypomastigote forms were more strongly labeled than slender trypomastigotes. On the other hand, amastigotes were only faintly labeled (Fig. 3 E). Axenic culture-derived insect parasite stages (epimastigotes and metacyclic trypomastigotes) gave no reaction by this technique (data not shown). Along with results obtained from the Western blotting it is concluded that TSSA is a surface antigen mainly expressed in the cell-derived trypomastigote stage.
Interstrain Variability of TSSA.
The analysis of several T. cruzi isolates by Southern blotting with the tssa probe indicated that the genomic organization and gene copy number was conserved among isolates. However, differences in the signal intensity were noted when comparing the isolates, which might denote certain degree of heterogeneity within this locus (Fig. 4 A). To clarify this issue, tssa alleles were cloned and sequenced from Y and RA strains (both rendering strong signals on the Southern blot) and Dm28c and Tulahuen2 isolates (that rendered weak signals on the Southern blot; Fig. 4 A). The tssa gene from SilvioX10 and CA-I/72, not assayed in the Southern blot, were also cloned and sequenced. The resulting deduced proteins showed that two forms of tssa existed within the species yielding two different kinds of TSSA proteins (Fig. 4 B). Even though minor changes were observed in the NH2-terminal peptide and the putative GPI anchor, the inter-strain variability was mainly focused to the central region. Interestingly, this dimorphism seemed to correlate with the two major evolutive lineages proposed for T. cruzi (5, 6; Fig. 4 B). To further evaluate this possibility tssa alleles from several T. cruzi strains or cloned stocks were typed by endonuclease restriction after PCR amplification of their central regions. A unique PvuII site exclusively present in the variable region of the tssa allele from T. cruzi I isolates was used as diagnostic (Fig. 4 C). In addition, aliquots of the same DNA samples from the different isolates were typed as T. cruzi I or II by analyzing the PCR product length from the intergenic region of the tandemly repeated mini-exon gene and the variable region of the 24SαrRNA gene as originally described (5). As shown in Fig. 4 C, the tssa dimorphism perfectly correlated with the existence of these two lineages. Hence, the tssa forms were now called tssa-I and -II and their encoded proteins TSSA-I and TSSA-II as being present in T. cruzi I and II, respectively.
Ab Recognition of TSSA by Chagasic Patients and Experimentally Infected Animals.
The antigenicity of TSSA during the infection was first assessed using Chagasic patients' serum samples obtained from different countries. In a first series of assays only GST-TSSA-II was used to search for specific Abs. Each group of samples was heterogeneous, and included people living in endemic and nonendemic areas, most probably parasitized by different T. cruzi strains and showing diverse Chagas' disease-associated pathology. The prevalence for TSSA-II specific Abs among Argentinean potentially Chagasic sera was 95.4% (63 out of 66 sera tested) as judged by ELISA (Table I). The overall TSSA-II reactivity among Brazilian Chagasic sera was ∼92% (215 out of 235 sera) and 100% when only hemoculturepositive samples (n = 123) were considered (Table I), thus in the range of other immunodominant molecules currently tested for Chagas' disease serodiagnosis (3, 39). Values from Chilean serum samples (87% positive) were in the same line. None of the 126 normal serum samples assayed were reactive for TSSA-II (Table I).
Table I.
Seroprevalence of Anti–TSSA-I and Anti–TSSA-II Abs in Populations from Distinct Geographical Origins
| Percentage of reactive sera
|
|||||||
|---|---|---|---|---|---|---|---|
| Origin | Serum panel | n | Conventional serology (IHA, IFI, ELISA) reactivity (%) |
EpEx CL-ELISAareactivity (%) | Hemoculture | Anti–TSSA-Ib | Anti–TSSA-II |
| Argentina | Chagasic | 66 | 100 | ND | ND | 3.0 | 95.4 |
| Non-Chagasic | 10 | 0 | ND | ND | 0 | 0 | |
| Brazil | Chagasicc | 112 | 100 | 98.2 | ND | 5.4 | 82.1 |
| Chagasicd | 48 | 100 | 100 | + | 0 | 100 | |
| Chagasice | 75 | 100 | 100 | + | 2.7 | 100 | |
| Non-Chagasic | 126 | 0 | 0 | ND | 0 | 0 | |
| Chile | Chagasicc | 23 | NDf | 100 | ND | 8.7 | 87.0 |
Sera were grouped according to the country of origin and diagnosis criteria and tested at 1:200 dilution for TSSA-I and TSSA-II reactivity by ELISA (Argentinean samples) and/or CL-ELISA (Brazilian and Chilean samples).
EpEx CL-ELISA, chemiluminescent ELISA test using an epimastigote (Tulahuén strain) preparation as antigen (reference 25).
Positive sera showed very low reactivity against TSSA-I, with reading (absorbance or relative luminescent unit) values close to the cutoff value for the specific test. Sera reactive for TSSA-I were also positive for TSSA-II, with reading values for the latter higher than those observed for the former (see also Fig. 6 C).
Potentially Chagasic patients.
Confirmed Chagasic patients.
Confirmed Chagasic patients coinfected with HIV.
ND, not determined.
The antigenicity of TSSA was also evaluated by searching for specific Abs in sera from laboratory animals experimentally infected with different T. cruzi isolates. Most of the sera tested from T. cruzi II-infected animals (7 out of 7 rabbits, 22 out of 24 mice, and 2 out of 2 rats) were highly reactive to GST-TSSA-II indicating a marked antigenicity. However, a startling observation was that none of the animals infected with T. cruzi I isolates reacted against GST-TSSA-II (addressed more in detail below). To assess the persistence of anti–TSSA-II Abs during the course of the infection, a follow up study was performed in BALB/c mice experimentally infected with the CL Brener clone. All of them (n = 12) showed a peak of TSSA-II–specific Abs that coincided with the moment of parasitemia declining and, thereafter, circulating Abs could be detected up to 4 mo postinfection at a fairly constant level (about half of the peak value, data not shown). The lineage dependence recognition observed for animal experimental infections along with the striking high prevalence of anti–TSSA-II Abs in human infections (Table I) prompted us to study in more detail the antigenicity of and the immune response against both TSSA isoforms.
Mapping of Linear B Cell Restricted Epitopes in TSSA-II.
Linear B cell epitopes present in TSSA-II were analyzed by the SPOT method (28). Overlapping sequences with two- or five-amino acid residue offset were synthesized onto membranes in a defined array with 15 peptides covering almost the whole TSSA-II deduced protein. These filters were sequentially tested with two independent Chagasic sera and a pool of 10 Chagasic sera, rendering almost identical results (Fig. 5). It was observed that the central region of TSSA-II contained all the linear antigenic determinants detected by Chagasic sera, whereas the proposed signal peptides and both N- and COOH-terminal portions of the mature protein gave no reaction. Normal human sera tested by the same technique rendered negative results (Fig. 5).
Figure 5.
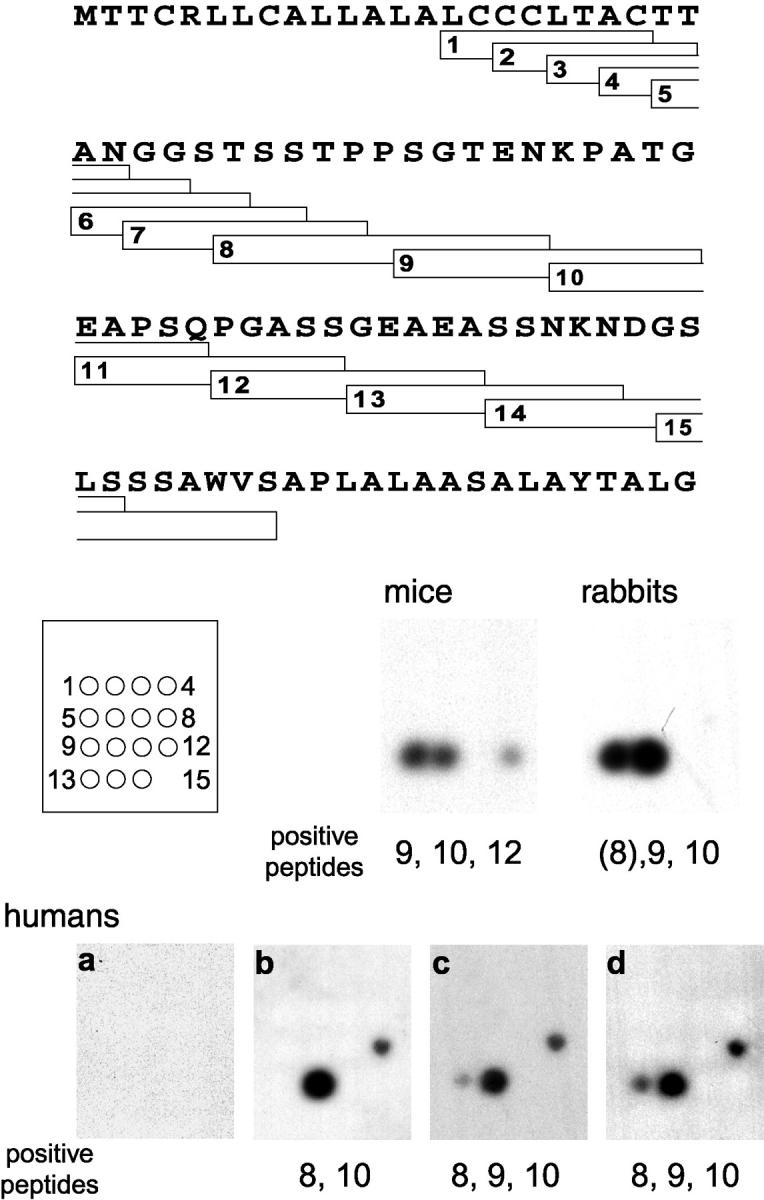
Mapping of linear epitopes from TSSA. Peptides were custom-synthesized by the SPOT technique in a defined array. The region spanned by each peptide, along with its identification number and position in the microarray is indicated. Filters were probed with normal human (a) or Chagasic sera (b and c). A pool of 10 Chagasic sera (d) was also used. The same filter was probed with serum pools of eight CL Brener-infected mice and six RA-infected rabbits.
Within the central region, every Chagasic sera highlighted a major epitope spanning the sequence of peptide 10 (40-KPATGEAPSQ-49; Fig. 5). Interestingly, the anti–TSSA-II Ab response displayed by T. cruzi II–infected mice and rabbits was also mainly driven to the same sequence (Fig. 5), suggesting that the immunodominance of the epitope(s) included within this peptide is not a species-specific phenomenon. Another minor epitope, restricted to humans, might be ascribed to peptide 8 (29-TSSTPPSGTEN-39). However, peptide 9 (35-SGTENKPATG-44) encompassing the proximal and distal half of these two sequences, respectively, displayed only a weak signal in two out of three assays (Fig. 5).
Epidemiological Analysis of TSSA.
One striking feature concerning TSSA is the strain dimorphism depicted in Fig. 4 B. Variability was restricted to the central region of the molecule, and involved the two major antigenic peptides (#10 and #8) defined above (Fig. 5). Amino acid sequence variation included one single residue deletion and several nonconservative replacements (in particular the exchange of 42Pro for Thr) that might have a major impact on the antigenic structure of TSSA. To evaluate this possibility, we first analyzed the antigenic cross-reactivity between recombinant TSSA isoforms. For this purpose, we expressed the central (antigenic) portion of TSSA-I from Dm28c cloned stock (typed as T. cruzi I, Fig. 4 C; and reference 5) as GST fusion as already done for TSSA-II (Fig. 1). Both recombinant molecules were probed by ELISA with different anti-TSSA sera obtained from hyperimmune rabbits and mice. As summarized in Fig. 6 A, anti–TSSA-I sera did not recognize the TSSA-II molecule even at 1:100 dilution. The reciprocal experiment confirmed this, as anti–TSSA-II sera did not recognize TSSA-I either, suggesting that both molecules display no major immunological cross-reactivity.
Figure 6.
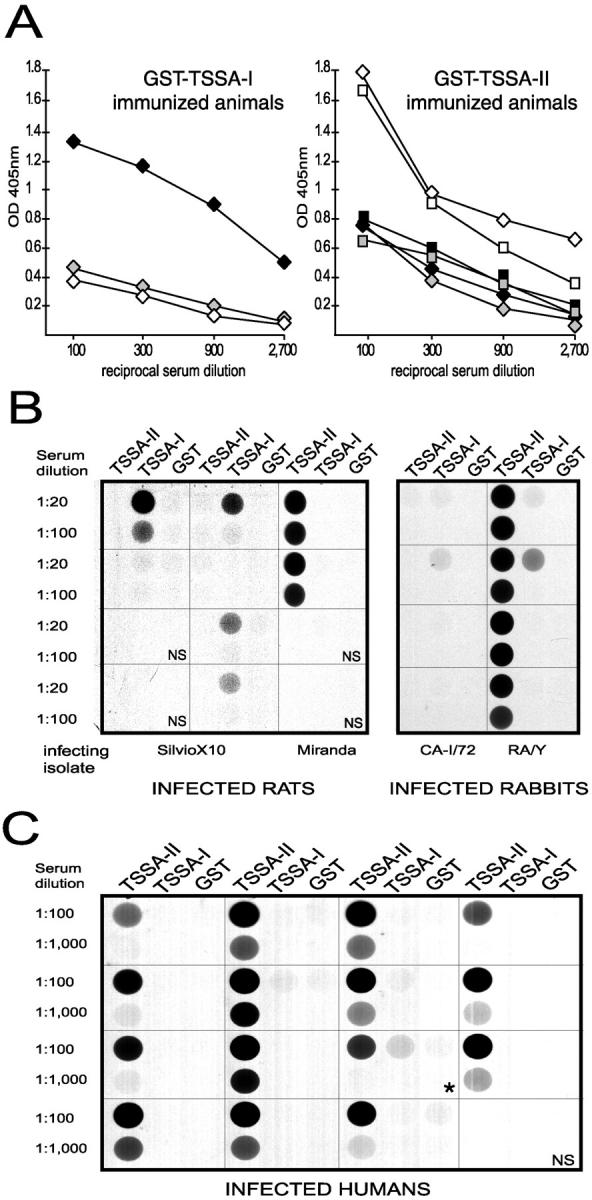
Analysis of antigenic cross-reactivity between TSSA isoforms. (A) Reactivity of mouse (diamonds) and rabbit (squares) sera immunized with the indicated protein against GST (gray symbols), GST-TSSA-I (black symbols), and GST-TSSA-II (white symbols) recombinant proteins. Each serum was tested in triplicate and mean OD405nm values are plotted (SD values did not exceed 10% of mean values in any case; not shown) against reciprocal serum dilution. One out of two experiments with similar results is shown. The accurate loading of ELISA plates was verified with an anti-GST polyclonal Ab (data not shown). (B and C) CL-ELISA assays showing representative examples of the reactivity displayed by sera from experimentally-infected animals (B) and Chagasic individuals (C) against both GST-TSSA-I and GST-TSSA-II recombinant proteins. NS, normal serum. Parasite isolate used to infect each animal is indicated below. In the RA/Y lane, the two upper samples were obtained from RA-infected animals while the two lower samples from Y-infected animals. Asterisk (*) exemplifies one Chagasic serum showing mixed anti–TSSA-I/anti–TSSA-II antibody recognition.
We extended this analysis to the actual infection by probing sera from animals experimentally infected with parasite isolates belonging to T. cruzi I or II. As noticed before, a vast majority of the sera (31 out of 33) from animals infected with T. cruzi II isolates clearly recognized TSSA-II, irrespectively of the species analyzed (Table II and Fig. 6 B). On the other hand, recognition of the TSSA-I molecule showed some species dependence for animals infected with T. cruzi I isolates (Table II and Fig. 6 B). While just 3 out of 12 rabbits detected TSSA-II, 3 out of 5 mice and 5 out of 8 rats were clearly reactive at 1:20 dilution. Notwithstanding this, lack of immunological cross-reaction was still the case as the majority of the animals reacted only to the specific TSSA form present in the lineage they were infected with (Table II). Only one of the RA (T. cruzi II)-infected rabbits cross-recognized the TSSA-I protein, though this reaction was considerably weaker than that displayed against the TSSA-II molecule (Fig. 6 B). Thus, as previously demonstrated for the animals immunized with the recombinant molecules (Fig. 6 A), the in vivo Ab response against the TSSA protein core also displayed a marked specificity.
Table II.
Seroprevalence of Anti–TSSA-I and Anti–TSSA-II Abs in Infected Animals
| Species | Infecting T. cruzi isolate | T. cruzi group | n | Anti–TSSA-I positive | Anti–TSSA-II positive |
|---|---|---|---|---|---|
| Rabbit | CA-I, CA-I/72, Tulahuen, AWP | I | 12 | 3 | 0 |
| RA, UP | II | 7 | 1b | 7 | |
| Mouse | Laura, Geora, FMSa, Joséa, SEa | I | 5 | 3 | 0 |
| CL Brener, RA, Piam 16a, BSIMTa | II | 24 | 0 | 22 | |
| Rat | SilvioX10 | I | 8 | 5 | 0 |
| Miranda | II | 2 | 0 | 2 |
Sera were grouped according to the species and infecting T. cruzi strain (T. cruzi I or II) and tested at serial dilution for TSSA-I and TSSA-II reactivity by CL-ELISA using 20 ng/well of GST-TSSA-I or GST-TSSA-II as coating antigen.
Serum samples kindly provided by Dr. M.M Teixeira whose infecting strains were gene-typed as T. cruzi I and II using the 24SarRNA and the mini-exon gene (reference 5).
Reactivity displayed by this serum against TSSA-I was significantly lower than that observed against TSSA-II (see also Fig. 6 B).
Interestingly, Chagasic sera showed a similar Ab specificity when tested against both TSSA isoforms at variable serial dilutions as indicated (Table I and Fig. 6 C). The seroprevalence of anti-TSSA-II Abs among potentially Chagasic patients was estimated in 87% (175 out 201), whereas only 5% (10 out of 201) of these sera displayed a mixed recognition. Among confirmed Chagasic patients (n = 123), seroprevalence of anti–TSSA-II was 100%, whereas only 2% (2 out of 123) of these sera displayed a mixed recognition (Table I). All of the latter samples rendered a much stronger reaction with TSSA-II than with TSSA-I (Fig. 6 C). On the other hand, the few potentially Chagasic sera that did not recognize TSSA-II neither reacted against TSSA-I (data not shown). Altogether, these results indicate that: (a) every human infection with T. cruzi involves at least one strain belonging to T. cruzi II; and (b) none of the Chagasic patients analyzed throughout this work showed an exclusive T. cruzi I infection, though a minor proportion of T. cruzi I/T. cruzi II coinfections could exist.
Discussion
TcMUC is a complex T. cruzi gene-family whose diversity is still not well understood (16, 17). In the present work we have identified and characterized the product of TcMUC group III as a small membrane antigen expressed by the infective cell-derived trypomastigote (Fig. 3, A and B). This protein, termed TSSA, is encoded by a single-copy gene (Fig. 2 A) that is dimorphic among T. cruzi isolates correlating with the two broad lineages previously described for the species (5, 6; Fig. 4, B and C). A preliminary biochemical characterization of TSSA was done on the CL Brener protein since it has been chosen as the genome project reference clone and the description of its gene products would be of great interest in the postgenomic phase (18, 21).
On its COOH terminus, TSSA displayed a hydrophobic region compatible with a GPI-anchoring signal (17). Furthermore, this sequence is 85% identical to the previously described in MUC-R (a tagged version of a TcMUC gene), which rendered a GPI-anchored mucin when transfected into T. cruzi (19). Amino acid conservation included the critical tripeptide Asp-Gly-Ser, postulated to function as the ω, ω +1, and ω +2 positions for lipid attachment in GPI-anchored proteins from T. cruzi and Trypanosoma brucei (40, 41). Two further evidences suggest that TSSA is GPI-anchored to the parasite surface. First, immunofluorescence of nonpermeabilized parasites clearly labeled the surface of cell-derived trypomastigotes, specially the broad forms (Fig. 3 E). Slender trypomastigotes and amastigotes were faintly labeled (Fig. 3 E) while the insect-derived stages were negative (data not shown). Broad trypomastigotes might correspond to intermediate forms (2), suggesting that the expression of TSSA is narrowed to defined periods of the trypomastigote development. Second, treatment of intact cell-derived trypomastigotes with PI-PLC resulted in the lost of the signal on parasite lysates when probed with an anti-TSSA serum in Western blot (Fig. 3 C). Unexpectedly, this signal was not recovered in the culture media. The possibility of generalized proteolytic degradation during PI-PLC treatment is unlikely since another GPI-anchored molecule (SAPA) was recovered in its intact form in the supernatant (Fig. 3 C). It has been recently demonstrated that certain GPI-anchored proteins present in protozoan and mammalian cells suffer a drastic effect on their antigenicity after the removal of the GPI lipid moieties, suggesting that they might undergo conformational changes (42). Whether this phenomenon might be operating in our system is uncertain but it is noteworthy the presence of strong linear epitopes on TSSA (Fig. 5) whose recognition should not, in principle, be affected by protein conformation. One more likely explanation is that TSSA might be posttranslationally modified in vivo, in particular by the addition of O-linked oligosaccharides as suggested by its primary sequence (31; Fig. 1). Therefore, the cleavage of its lipid moieties would yield a highly hydrophilic, soluble TSSA that no longer binds to nitrocellulose filters, as previously observed for the T. cruzi trypomastigote total mucins (43, 44). The same explanation could also account for the observed discrepancies between the predicted and the observed SDS-PAGE mobility (Fig. 3, A and B). Altogether, these results are compatible with TSSA being GPI-anchored to the surface of the cell-derived trypomastigote stage.
Genomic analyses of the tssa locus revealed the existence of two alleles that code for two distinct forms of the protein (Fig. 4 B). Interestingly, the distribution of these alleles correlated with the two major lineages proposed for the species (5, 6). As shown in Fig. 4 C, lineage classification of 19 T. cruzi isolates by our tssa gene-typing assay showed a perfect correlation with two of the most commonly used methods: mini-exon and rDNA PCR polymorphism (5). Accordingly, it can be proposed that the tssa-I allele is restricted to T. cruzi I isolates (and encodes the TSSA-I isoform) and tssa-II is restricted to T. cruzi II isolates (and encodes the TSSA-II protein).
In addition to its potential use as a reliable serodiagnostic molecule (Table I) and a genetic tool for T. cruzi typification, TSSA provides with the first immunological marker allowing the lineage identification of infecting isolates on serum samples. Amino acid variations between TSSA-I and TSSA-II are mainly restricted to their central regions (Fig. 4 B), which turned out to comprise the major B cell antigenic determinants (Fig. 5). These sequence changes have a major impact on TSSA antigenicity, leading to negligible cross-reactivity between both isoforms. As such, anti–TSSA-I hyperimmune sera did not recognize recombinant TSSA-II and viceversa (Fig. 6 A). Counterpart in vivo evidence was provided by sera from experimentally infected animals (Table II and Fig. 6 B) and from Chagasic patients (Fig. 6 C). Sera from animals infected with T. cruzi II isolates showed an exclusive recognition toward the TSSA-II molecule. In the same line, although weak anti–TSSA-I specific responses are elicited during the infection with T. cruzi I parasites, a proportion of these animals (3 out of 12 rabbits, 3 out of 5 mice, and 5 out of 8 rats) did elicit significant anti–TSSA-I Abs that were unable to cross-recognize TSSA-II (Table II and Fig. 6 B). The decreased amounts of Abs detectable with the TSSA-I recombinant molecule can be attributed to differences in the course of infection caused by both kinds of parasite lineages. It has been established that T. cruzi I isolates develop a mild infection in laboratory animals, characterized by low parasitemia and low morbidity rate, thus leading to decreased overall reactivity against parasite antigens (6). Alternatively, it may be proposed that TSSA-I is less immunogenic than TSSA-II or undergoes major posttranslational modifications in vivo leading to a decreased Ab response against the TSSA-I protein backbone. We do not favor the latter hypothesis because the similarities between the primary sequences of both proteins (Fig. 4 B) are strong enough to assume that posttranslational modifications would be similar in kind and extension. Further biochemical characterization of both TSSA isoforms should help to clarify this issue.
The major finding of the present work was that, even though anti–TSSA-I Abs can be readily detected in T. cruzi I experimental infections (Table II), the seroreactivity displayed by Chagasic patients was clearly shifted toward the recognition of TSSA-II (Table I and Fig. 6 C). As shown, 175 out of 201 (87%) of potentially Chagasic sera and 100% of confirmed Chagasic sera (n = 123) showed specific reactivity against TSSA-II, whereas only 12 out of 324 (3.7%) total Chagasic serum samples displayed a mixed recognition (Table I). All of the latter sera, that might have been underestimated due to the low titers of Ab directed toward TSSA-I peptide epitopes elicited during T. cruzi I infections (Table II), exhibited a much stronger anti–TSSA-II Ab response (Fig. 6 C). Therefore, they can represent detection of minor cross-reactivities due to Ab populations directed against common epitopes in both TSSA isoforms, mostly in high responsive patients. More likely, they can be interpreted as genuine T. cruzi I/T. cruzi II coinfection cases (45). It is worth noting that none of the Chagasic sera analyzed so far showed a restricted recognition toward TSSA-I. These simple results have a great epidemiological value as it indicates that strains belonging to T. cruzi II (or even a subgroup of this lineage) are by far the more common in producing human infections and, hence, Chagas' disease. On the other hand, they strongly argue against human infection cases exclusively due to T. cruzi I strains. Although molecular markers and biological evidences existed associating T. cruzi II with the domestic cycle of the infection and T. cruzi I with the sylvatic cycle (6, 14), TSSA provides with the first antigenic marker supporting this hypothesis. This fact should be kept in mind for reevaluating previous data obtained in animals experimentally infected with T. cruzi I isolates and, more important, for choosing the appropriate strains to develop animal models to study human pathogenesis and treatments.
Acknowledgments
The authors would like to thank Profs. M. Shikanai-Yasuda, V. Amato-Neto (Instituto de Medicina Tropical da Universidade de São Paulo), M.M. Teixeira (Universidade de São Paulo), A. Fragata Filho (Instituto Dante Pazzanese de Cardiologia, São Paulo), J. Gonzales (Universidad de Antofagasta, Chile), J. Bua, and D. Sánchez for kindly providing some of the serum samples used in this study. We are also indebted to D. Sánchez for the T. cruzi DNA samples, F. Fraga for his technical assistance, and I. Cuevas for the 24SαrRNA probe. Experiments with infected mice were carried out by Dr. M.S. Leguizamón, from the Departmento de Microbiología, Facultad de Medicina, Universidad de Buenos Aires, Argentina. Helpful suggestions on the manuscript made by Drs. D. Sánchez, O. Campetella, and J.J. Cazzulo are also appreciated.
This work has been supported by grants from the Ministerio de Salud, Argentina; the International Atomic Energy Agency and the UNDP/World Bank/WHO Special Program for Research and Training in Tropical Diseases (TDR), World Health Organization. The research of A.C.C. Frasch was supported in part by an International Research Scholar grant from the Howard Hughes Medical Institute and a fellowship from the John S. Guggenheim Memorial Foundation. J.M. Di Noia and C.A. Buscaglia are post-doctoral fellows and A.C.C. Frasch is a researcher from the CONICET, Argentina. I.C. Almeida is a research fellow of the Conselho Nacional de Desenvolvimento científico e Tecnológico (CNPq) and is supported by a grant (no. 98/10495-5) from the Fundação de Amparo à Pesquisa do estado de São Paulo (FAPESP).
J.M. Di Noia's present address is MRC Laboratory of Molecular Biology, PNAC Unit, Hills Road, Cambridge CB2 2QH, UK.
Footnotes
Abbreviations used in this paper: GPI, glycosylphosphatidyl inositol; TSSA, trypomastigote small surface antigen.
References
- 1.WHO. 1997. World Health Organization. Tropical Disease Research, progress 1995–96, ed. WHO Publications, Geneva, Switzerland.
- 2.Tyler, K.M., and D.M. Engman. 2001. The life cycle of Trypanosoma cruzi revisited. Int. J. Parasitol. 31:472–481. [DOI] [PubMed] [Google Scholar]
- 3.da Silveira, J.F., E.S. Umezawa, and A.O. Luquetti. 2001. Chagas disease: recombinant Trypanosoma cruzi antigens for serological diagnosis. Trends Parasitol. 17:286–291. [DOI] [PubMed] [Google Scholar]
- 4.Macedo, A.M., and S.D.J. Pena. 1998. Genetic variability of Trypanosoma cruzi: implications for the pathogenesis of Chagas' disease. Parasitol. Today. 14:119–124. [DOI] [PubMed] [Google Scholar]
- 5.Souto, R.P., O. Fernandes, A.M. Macedo, D.A. Campbell, and B. Zingales. 1996. DNA markers define two phylogenetic lineages of Trypanosoma cruzi. Mol. Biochem. Parasitol. 83:141–152. [DOI] [PubMed] [Google Scholar]
- 6.Zingales, B., B.S. Stolf, R.P. Souto, O. Fernandes, and M.R. Briones. 1999. Epidemiology, biochemistry and evolution of Trypanosoma cruzi lineages based on ribosomal RNA sequences. Mem. Inst. Oswaldo Cruz. 94(Suppl. 1):159–164. [DOI] [PubMed] [Google Scholar]
- 7.Machado, C.A., and F.J. Ayala. 2001. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. USA. 98:7396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawashita, S.Y., G.F. Sanson, O. Fernandes, B. Zingales, and M.R. Briones. 2001. Maximum-likelihood divergence date estimates based on rRNA gene sequences suggest two scenarios of Trypanosoma cruzi intraspecific evolution. Mol. Biol. Evol. 18:2250–2259. [DOI] [PubMed] [Google Scholar]
- 9.Briones, M.R., R.P. Souto, B.S. Stolf, and B. Zingales. 1999. The evolution of two Trypanosoma cruzi subgroups inferred from rRNA genes can be correlated with the interchange of American mammalian faunas in the Cenozoic and has implications to pathogenicity and host specificity. Mol. Biochem. Parasitol. 104:219–232. [DOI] [PubMed] [Google Scholar]
- 10.Tibayrenc, M., P. Ward, A. Moya, and F.J. Ayala. 1986. Natural populations of Trypanosoma cruzi, the agent of Chagas disease, have a complex multiclonal structure. Proc. Natl. Acad. Sci. USA. 83:115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miles, M.A., A. Souza, M. Povoa, J.J. Shaw, R. Lainson, and P.J. Toye. 1978. Isozymic heterogeneity of Trypanosoma cruzi in the first autochthonous patients with Chagas' disease in Amazonian Brazil. Nature. 272:819–821. [DOI] [PubMed] [Google Scholar]
- 12.Nunes, L.R., M.R. de Carvalho, and G.A. Buck. 1997. Trypanosoma cruzi strains partition into two groups based on the structure and function of the spliced leader RNA and rRNA gene promoters. Mol. Biochem. Parasitol. 86:211–224. [DOI] [PubMed] [Google Scholar]
- 13.Oliveira, R.P., N.E. Broude, A.M. Macedo, C.R. Cantor, C.L. Smith, and S.D. Pena. 1998. Probing the genetic population structure of Trypanosoma cruzi with polymorphic microsatellites. Proc. Natl. Acad. Sci. USA. 95:3776–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark, C.G., and O.J. Pung. 1994. Host specificity of ribosomal DNA variation in sylvatic Trypanosoma cruzi from North America. Mol. Biochem. Parasitol. 66:175–179. [DOI] [PubMed] [Google Scholar]
- 15.Di Noia, J.M., D.O. Sánchez, and A.C. Frasch. 1995. The protozoan Trypanosoma cruzi has a family of genes resembling the mucin genes of mammalian cells. J. Biol. Chem. 270:24146–24149. [DOI] [PubMed] [Google Scholar]
- 16.Freitas-Junior, L.H., M.R. Briones, and S. Schenkman. 1998. Two distinct groups of mucin-like genes are differentially expressed in the developmental stages of Trypanosoma cruzi. Mol. Biochem. Parasitol. 93:101–114. [DOI] [PubMed] [Google Scholar]
- 17.Di Noia, J.M., I. D'Orso, L. Aslund, D.O. Sánchez, and A.C. Frasch. 1998. The Trypanosoma cruzi mucin family is transcribed from hundreds of genes having hypervariable regions. J. Biol. Chem. 273:10843–10850. [DOI] [PubMed] [Google Scholar]
- 18.Agüero, F., R.E. Verdun, A.C. Frasch, and D.O. Sánchez. 2000. A random sequencing approach for the analysis of the Trypanosoma cruzi genome: general structure, large gene and repetitive DNA families, and gene discovery. Genome Res. 10:1996–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollevick, G.D., J.M. Di Noia, M.L. Salto, C. Lima, M.S. Leguizamón, R.M. de Lederkremer, and A.C. Frasch. 2000. Trypanosoma cruzi surface mucins with exposed variant epitopes. J. Biol. Chem. 275:27671–27680. [DOI] [PubMed] [Google Scholar]
- 20.Van Klinken, B.J., J. Dekker, H.A. Buller, and A.W. Einerhand. 1995. Mucin gene structure and expression: protection vs. adhesion. Am. J. Physiol. 269:G613–G627. [DOI] [PubMed] [Google Scholar]
- 21.Zingales, B., M.E. Pereira, K.A. Almeida, E.S. Umezawa, N.S. Nehme, R.P. Oliveira, A. Macedo, and R.P. Souto. 1997. Biological parameters and molecular markers of clone CL Brener: the reference organism of the Trypanosoma cruzi genome project. Mem. Inst. Oswaldo Cruz. 92:811–814. [DOI] [PubMed] [Google Scholar]
- 22.Sánchez, D.O., A.C. Frasch, A.E. Carrasco, S.M. González-Cappa, E.D. de Isola, and A.O. Stoppani. 1984. Rapid evolution of kinetoplast DNA mini-circle subpopulations in Trypanosoma cruzi. Mol. Biochem. Parasitol. 11:169–178. [DOI] [PubMed] [Google Scholar]
- 23.Engel, J.C., J.A. Dvorak, E.L. Segura, and M.S. Crane. 1982. Trypanosoma cruzi: biological characterization of 19 clones derived from two chronic chagasic patients. I. Growth kinetics in liquid medium. J. Protozool. 29:555–560. [DOI] [PubMed] [Google Scholar]
- 24.Di Noia, J.M., G.D. Pollevick, M.T. Xavier, J.O. Previato, L. Mendoça-Previato, D.O. Sánchez, and A.C. Frasch. 1996. High diversity in mucin genes and mucin molecules in Trypanosoma cruzi. J. Biol. Chem. 271:32078–32083. [DOI] [PubMed] [Google Scholar]
- 25.Almeida, I.C., D.T. Covas, L.M. Soussumi, and L.R. Travassos. 1997. A highly sensitive and specific chemiluminescent enzyme-linked immunosorbent assay for diagnosis of active Trypanosoma cruzi infection. Transfusion. 37:850–857. [DOI] [PubMed] [Google Scholar]
- 26.Luz, Z.M., M.G. Coutinho, J.R. Cançado, and A.U. Krettli. 1994. Hemoculture: sensitive technique in the detection of Trypanosoma cruzi in chagasic patients in the chronic phase of Chagas disease. Rev. Soc. Bras. Med. Trop. 27:143–148. [DOI] [PubMed] [Google Scholar]
- 27.Almeida, I.C., E.G. Rodrigues, and L.R. Travassos. 1994. Chemiluminescent immunoassays: discrimination between the reactivities of natural and human patient antibodies with antigens from eukaryotic pathogens, Trypanosoma cruzi and Paracoccidioides brasiliensis. J. Clin. Lab. Anal. 8:424–431. [DOI] [PubMed] [Google Scholar]
- 28.Frank, R., and R. Doring. 1988. Simultaneous multiple peptide synthesis under continuous flow conditions on cellulose paper discs as segmental solid synthesis. Tetrahedron. 44:6031–6040. [Google Scholar]
- 29.Harlow, E., and D. Lane. 1988. Antibodies. A Laboratory Manual. Cold Spring Harbour Laboratory, New York.
- 30.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- 31.Gupta, R., H. Birch, K. Rapacki, S. Brunak, and J.E. Hansen. 1999. O-GLYCBASE version 4.0: a revised database of O-glycosylated proteins. Nucleic Acids Res. 27:370–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanke, J., D.O. Sánchez, J. Henriksson, L. Aslund, U. Pettersson, A.C. Frasch, and J.D. Hoheisel. 1996. Mapping the Trypanosoma cruzi genome: analyses of representative cosmid libraries. Biotechniques. 21:686–693. [DOI] [PubMed] [Google Scholar]
- 33.Graham, S.V. 1995. Mechanisms of stage-regulated gene expression in Kinetoplastida. Parasitol. Today. 11:217–223. [DOI] [PubMed] [Google Scholar]
- 34.Vanhamme, L., and E. Pays. 1995. Control of gene expression in trypanosomes. Microbiol. Rev. 59:223–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomas, A.M., and J.M. Kelly. 1996. Stage-regulated expression of cruzipain, the major cysteine protease of Trypanosoma cruzi is independent of the level of RNA1. Mol. Biochem. Parasitol. 76:91–103. [DOI] [PubMed] [Google Scholar]
- 36.Di Noia, J.M., I. D'Orso, D.O. Sánchez, and A.C. Frasch. 2000. AU-rich elements in the 3′-untranslated region of a new mucin-type gene family of Trypanosoma cruzi confers mRNA instability and modulates translation efficiency. J. Biol. Chem. 275:10218–10227. [DOI] [PubMed] [Google Scholar]
- 37.D'Orso, I., and A.C. Frasch. 2001. Functionally different AU- and G-rich cis-elements confer developmentally regulated mRNA stability in Trypanosoma cruzi by interaction with specific RNA-binding proteins. J. Biol. Chem. 276:15783–15793. [DOI] [PubMed] [Google Scholar]
- 38.Pollevick, G.D., J.L. Affranchino, A.C.C. Frasch, and D.O. Sánchez. 1991. The complete sequence of a shed acute-phase antigen of Trypanosoma cruzi. Mol. Biochem. Parasitol. 47:247–250. [DOI] [PubMed] [Google Scholar]
- 39.Umezawa, E.S., S.F. Bastos, M.E. Camargo, L.M. Yamauchi, M.R. Santos, A. Gonzalez, B. Zingales, M.J. Levin, O. Sousa, R. Rangel-Aldao, and J.F. da Silveira. 1999. Evaluation of recombinant antigens for serodiagnosis of Chagas' disease in South and Central America. J. Clin. Microbiol. 37:1554–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez, M.I., S.B. Boscardin, R.C. Ruiz, S.W. Han, G.S. Paranhos-Baccala, N. Yoshida, R.A. Mortara, and J.F. da Silveira. 1999. Heterologous expression of a Trypanosoma cruzi surface glycoprotein (gp82) indicates that requirements for glycosylphosphatidylinositol anchoring are different in mammalian cells and this trypanosome. Mem. Inst. Oswaldo Cruz. 94:527–530. [DOI] [PubMed] [Google Scholar]
- 41.Moran, P., and I.W. Caras. 1994. Requirements for glycosylphosphatidylinositol attachment are similar but not identical in mammalian cells and parasitic protozoa. J. Cell Biol. 125:333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butikofer, P., T. Malherbe, M. Boschung, and I. Roditi. 2001. GPI-anchored proteins: now you see ‘em, now you don’t. FASEB J. 15:545–548. [DOI] [PubMed] [Google Scholar]
- 43.Schenkman, S., M.A. Ferguson, N. Heise, M.L. de Almeida, R.A. Mortara, and N. Yoshida. 1993. Mucin-like glycoproteins linked to the membrane by glycosylphosphatidylinositol anchor are the major acceptors of sialic acid in a reaction catalyzed by trans-sialidase in metacyclic forms of Trypanosoma cruzi. Mol. Biochem. Parasitol. 59:293–303. [DOI] [PubMed] [Google Scholar]
- 44.Almeida, I.C., M.A. Ferguson, S. Schenkman, and L.R. Travassos. 1994. Lytic anti-alpha-galactosyl antibodies from patients with chronic Chagas' disease recognize novel O-linked oligosaccharides on mucin-like glycosyl-phosphatidylinositol-anchored glycoproteins of Trypanosoma cruzi. Biochem. J. 304:793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vago, A.R., L.O. Andrade, A.A. Leite, D. d'Avila Reis, A.M. Macedo, S.J. Adad, S. Tostes, Jr., M.C. Moreira, G.B. Filho, and S.D. Pena. 2000. Genetic characterization of Trypanosoma cruzi directly from tissues of patients with chronic Chagas disease: differential distribution of genetic types into diverse organs. Am. J. Pathol. 156:1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Arruda, M.V., F.C. Reinach, W. Colli, and B. Zingales. 1990. Sequence of the 24S alpha ribosomal RNA gene and characterization of a corresponding pseudogene from Trypanosoma cruzi. Mol. Biochem. Parasitol. 40:35–41. [DOI] [PubMed] [Google Scholar]