Abstract
Microsporidia constitute a group of extremely specialized intracellular parasites that infect virtually all animals. They are highly derived, reduced fungi that lack several features typical of other eukaryotes, including canonical mitochondria, flagella, and peroxisomes. Consistent with the absence of peroxisomes in microsporidia, the recently completed genome of the microsporidian Encephalitozoon cuniculi lacks a gene for catalase, the major enzymatic marker for the organelle. We show, however, that the genome of the microsporidian Nosema locustae, in contrast to that of E. cuniculi, encodes a group II large-subunit catalase. Surprisingly, phylogenetic analyses indicate that the N. locustae catalase is not specifically related to fungal homologs, as one would expect, but is instead closely related to proteobacterial sequences. This finding indicates that the N. locustae catalase is derived by lateral gene transfer from a bacterium. The catalase gene is adjacent to a large region of the genome that appears to be far less compact than is typical of microsporidian genomes, a characteristic which may make this region more amenable to the insertion of foreign genes. The N. locustae catalase gene is expressed in spores, and the protein is detectable by Western blotting. This type of catalase is a particularly robust enzyme that has been shown to function in dormant cells, indicating that the N. locustae catalase may play some functional role in the spore. There is no evidence that the N. locustae catalase functions in a cryptic peroxisome.
Microsporidia constitute a group of intracellular parasites that infect a wide variety of animals, as well as certain ciliates and gregarine apicomplexa. Outside their host cells, microsporidia exist as highly resistant spores that are protected by protein and chitin walls. In structure, microsporidian spores are dominated by organelles related to infection. The most prominent of these organelles is the polar filament, which is coiled tightly around the spore contents, or sporoplasm. When a spore is induced to germinate, the polar filament everts, becoming a hollow tube which is rapidly ejected from the spore. If this projectile tube pierces a host cell's membrane, the infective microsporidian sporoplasm can be injected directly into its cytosol (for a review, see reference 39).
Microsporidia possess a number of unusual characteristics that led to the idea that they were primitive, ancient eukaryotes (7). These traits include bacterium-sized ribosomes and fused 5.8S and large-subunit (LSU) rRNA genes (9, 20, 42), as well as the absence of several structures, such as mitochondria, flagella, and peroxisomes (39). The idea that these features indicate an ancient origin for microsporidia gained support from early phylogenetic trees, which placed microsporidia at the base of the eukaryotes, albeit with very long branches (4, 22, 23, 41). Given that long branches are the result of highly divergent sequences and can result in phylogenetic artifacts, this position raised some doubts about the ancient origin of microsporidia. These doubts were soon justified: as more sequence data accumulated from microsporidia and phylogenetic methods improved, less divergent microsporidian genes were shown to branch with fungi in the majority of analyses (3, 13, 14, 17, 18, 26, 27, 29, 35). Furthermore, when many of the original data sets that had indicated an early origin for microsporidia were reanalyzed with updated methods, an ancient origin was rejected (18). With the completion of the Encephalitozoon cuniculi genome, the phylogenetic relationship between microsporidia and fungi has been further solidified by the presence of numerous genes (24). The fungal origin of microsporidia has a significant impact on how we interpret their unusual characteristics. They no longer represent ancestral features but instead are indicative of the highly derived nature of these intracellular parasites. Indeed, the apparent absence of mitochondria in microsporidia has recently been discredited by the cytological demonstration that they possess cryptic mitochondria (43).
Nevertheless, microsporidia are still highly reduced, and there is still no cytological evidence for peroxisomes, or microbodies. In accordance with this observation, the complete genome of E. cuniculi lacks a gene for catalase (24), the characteristic enzyme often associated with this organelle. Catalase converts hydrogen peroxide into water and oxygen and is typically responsible for dealing with oxidative stress. It is predicted that E. cuniculi uses a unique manganese superoxide dismutase along with thioredoxin and glutathione peroxidases to deal with oxygen stress (24) and apparently does not need, or have, either catalase or a peroxisome.
Here we show that, in contrast to the genome of E. cuniculi, the genome of the microsporidian Nosema locustae contains a gene that codes for a group II large-subunit catalase. The gene is expressed, and the protein is present in spores. Unexpectedly, phylogenetic analyses indicate that the N. locustae catalase is not specifically related to peroxisomal or even fungal homologs but instead shares a close relationship with a specific group of proteobacterial sequences, indicating that it is derived by lateral gene transfer from a bacterium. The presence of this protein in a microsporidian raises a number of intriguing questions about the cellular location of catalase in N. locustae and the role of the enzyme in spores, highlighting the potential for metabolic adaptation via lateral gene transfer (2, 10, 33).
MATERIALS AND METHODS
DNA isolation, library construction, and sequencing.
N. locustae spores (ATCC 30860) were kindly provided by M&R Durango, Inc. (Bayfield, Colo.). Genomic DNA was isolated with the Plant DNeasy minikit (Qiagen), and multiple genomic libraries were constructed as part of an N. locustae random sequence survey (see reference 12 for a complete description of these methods). Randomly selected clones from all libraries were end sequenced with ABI BigDye chemistry. End sequences were compared with one another and against public databases by using the ESTid program (generously provided by M. Reith, Institute for Marine Biosciences, Halifax, Nova Scotia, Canada). The genomic fragment containing the catalase open reading frame (ORF) was identified based on end sequencing of a single clone that was subsequently found to be 5,378 bp long. This genomic fragment and all other fragments observed to overlap it were completely sequenced by primer walking, resulting in a genomic fragment of 13,209 bp and two smaller fragments of 3,530 and 3,466 bp that were clearly homologous but not identical. Multiple PCRs (using genomic DNA as a template) were carried out across the divergent regions to ensure that the variation truly occurred in the genome and was not a cloning artifact. All regions outside of the catalase ORF were assessed for coding potential with BLASTX and BLASTN searches.
RNA extraction and 3′ RACE.
N. locustae spores (350 mg) were ground under liquid nitrogen with a mortar and pestle, and the lysate was resuspended in 3 ml of TRIzol reagent (Invitrogen). RNA was purified from the resuspended lysate according to the manufacturer's protocol. Reverse transcription and 3′ rapid amplification of cDNA ends (RACE) were carried out with the First Choice RLM-RACE kit (Ambion). Total RNA from N. locustae spores was used as a template for first-strand synthesis with a poly(dT) 3′ adaptor oligonucleotide (Ambion) according to manufacturer's directions. Second-strand synthesis was carried out with the GeneAmp XL PCR kit (Perkin-Elmer) with a hot start with an N. locustae catalase-specific forward primer (GCTCACAACCTAGGCGTGAGGGAGC) and the 3′-RACE outer primer (Ambion). The 100-μl reaction mixture was initially incubated at 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 64°C for 30 s, and 72°C for 1 min. The single reaction product was gel isolated and cloned in TOPO pCR 2.1 (Invitrogen). Four positive clones were sequenced as described above. The ends of the 3′-RACE products were checked against the genomic sequence to ensure that the RACE products did not result from A tracts at the 3′ end of the catalase gene sequence. All products were derived from mRNA.
Antibody preparation and Western blotting.
Rabbit polyclonal antibodies were prepared commercially (AbCam) against a mixture of two synthetic peptides based on the N. locustae genomic sequence (CRLNGADPDFHRRDLW and GIQKDDGMVEIRDNC). Proteins were released from N. locustae spores by grinding them under liquid nitrogen as described above for RNA extraction. Approximately 2 ×109 crushed spores were resuspended in 200 μl of 0.15 M NaCl and stored at −20°C in an equal volume of protein sample buffer (62.5 mM Tris-HCl [pH 6.8], 0.01% bromophenol blue, 2% sodium dodecyl sulfate, 5% beta-mercaptoethanol, 10% glycerol). Proteins were separated on a sodium dodecyl sulfate-10% polyacrylamide resolving gel, with a 4% stacking gel. One lane of the gel was stained with Coomassie blue for size comparisons, and other samples were transferred to Hybond-P polyvinylidene difluoride membranes (Amersham) by electroblotting. Blots were blocked in a 5% skim milk-1× Tris-buffered saline [TBS]-0.1% Tween 20 solution and then incubated with either a 1:500 dilution of a rabbit anti-catalase antibody, or the preimmunization serum, in 1% skim milk-1× TBS-0.1% Tween 20 for 90 min. The final concentration of the anti-catalase antibody was 36 ng/ml. Blots were washed with 1% skim milk-1× TBS-0.1% Tween 20 and incubated with a secondary horseradish peroxidase-conjugated anti-rabbit immunoglobulin G antibody (Bio-Rad) for 90 min. Blots were washed in 1% skim milk-1× TBS-0.1% Tween and developed with Amersham ECL Western blotting reagents.
Phylogenetic methods.
The conceptual amino acid sequence of the N. locustae catalase was aligned with homologs representing all known classes of prokaryotic and eukaryotic catalases by using Clustal X (38). This 97-taxon alignment was manually edited, and all ambiguously aligned sites were removed for phylogenetic analyses, resulting in a data set of 316 characters. This data set corresponds closely to the core region used in other published phylogenetic analyses (30). Distances were calculated with TREE-PUZZLE 5.0 (36) by using the WAG substitution matrix and modeling site-to-site rate variation on a gamma distribution approximated with eight rate categories and an invariable-sites category. The shape parameter α and the proportion-of-invariable-sites parameter were estimated from the data. Trees were constructed by using weighted neighbor-joining and Fitch-Margoliash methods, with WEIGHBOR 1.0.1a and FITCH 3.6a, respectively (5; J. Felsenstein, PHYLIP [Phylogeny Inference Package], University of Washington, Seattle, 1993). Fitch-Margoliash trees were constructed with global rearrangements, and the input order was jumbled 10 times. One hundred resampled data sets were created with SEQBOOT (PHYLIP), and bootstrap distances were calculated with PUZZLEBOOT (shell script by A. Roger and M. Holder; www.tree-puzzle.de) by using the parameters estimated above. Bootstrap trees were calculated as described above, except for Fitch-Margoliash trees, where the global rearrangement option was not employed and the input order was not jumbled.
Analyses were also carried out with a smaller data set composed of only the group II catalases. By examining only this group of closely related sequences, the number of characters in the analysis was increased to 423, and a more comprehensive protein maximum likelihood analysis was undertaken by using ProML 3.6a (PHYLIP). Global rearrangements were carried out with the input order jumbled 10 times. The default substitution model (PAM250) was employed, as was the R option, implementing the rate categories and their associated probabilities calculated by TREE-PUZZLE (with eight rate categories and invariable sites). Protein maximum likelihood bootstrap trees were determined with a uniform rate (employing the R option proved too computationally intensive), global rearrangements, and the input order jumbled five times. This smaller data set was also analyzed by distance methods as described for the larger data set, except that global rearrangements and five input order jumbles were used to determine the Fitch-Margoliash bootstrap trees.
Nucleotide sequence accession numbers.
Nucleotide sequence data have been deposited in GenBank under accession number AY326437.
RESULTS AND DISCUSSION
Characterization of the N. locustae catalase gene and its genomic context.
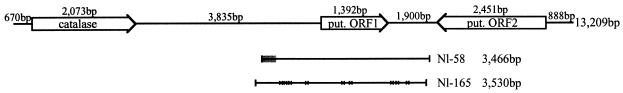
To examine the compaction of microsporidian genomes and to compare characteristics of the complete genome of E. cuniculi with a distantly related microsporidian, the locust parasite N. locustae has been the focus of an ongoing genomic sequencing survey. A 13,209-bp N. locustae genomic fragment comprised of three overlapping clones was found to contain a 2,073-bp ORF that, when compared to public databases, showed a high degree of sequence similarity to the catalase gene. The sequence lacks introns and, based on its inferred translation of 690 amino acids, appears to be a member of the oarge-subunit catalases, which include long amino- and carboxy-terminal extensions to the core conserved region recognized in all catalases (30).
In contrast to other regions of the N. locustae genome that have been characterized, where intergenic spaces range from ∼10 to ∼100 bp (12), the genomic context of the catalase gene is not at all compact (Fig. 1). For example, there is a 3,835-bp noncoding region directly downstream of the catalase gene. There are only two other putative genes in the 13.2-kb stretch, ORF1 and ORF2, which are also separated by 1,900 bp of apparently noncoding DNA. Based on BLASTX, these long putative intergenic regions do not contain evidence of pseudogenes, as no short regions exhibiting similarity to known genes could be identified. The ORF1 product is most similar to a hypothetical protein found in Dictyostelium discoideum, and the ORF2 product possesses weak similarity to inter-alpha-trypsin inhibitors from animals. Neither ORF has a clear homolog in the E. cuniculi genome. Although repetitive DNA is not common in microsporidian genomes, the distal ∼500 bp of the contig (upstream of putative ORF2) contains five 84-bp repeats.
FIG. 1.
Genomic context of the N. locustae catalase. The orientation and position of catalase, ORF1, and ORF2 are shown, as are the positions of the variant clones, Nl-58 and Nl-165. The region of decreased identity and individual base substitutions in comparison to the 13.2-kb contig are indicated by shading (Nl-58) and crosses (Nl-165). put., putative.
In addition to the scarcity of ORFs in this genomic region, another odd feature is the degree of divergence observed. Several clones were examined that were homologous but not identical to the region downstream of the catalase gene. Two such representative clones are indicated in Fig. 1. Designated Nl-165 (3,530 bp) and Nl-58 (3,466 bp), both clones are homologous to the same region. Nl-165 is identical to the large contig with the exception of nine substitutions, which occur in both noncoding regions and within ORF1, where several are nonsynonymous. Nl-58 is identical to the 13.2-kb contig over 88% of its length, but the identity drops to 73% in the terminal 425 bp of the clone. This region includes not only base substitutions but also insertions and deletions. Both clones are clearly homologous to, but substantially different from, the large contig, suggesting that variation might exist or that this region is present in multiple copies in the genome. It is possible that the clear demarcation between the regions with differing identities defines a recombination point and that differences arise via recombination within this region. As these two representative clones indicate, the region of the genome downstream of catalase is divergent, can withstand length variation, may be undergoing recombination, and apparently has a lower gene density than other regions of the N. locustae genome.
Catalase is expressed and present in N. locustae spores.
Microsporidian spores are largely dormant, but they must be highly resistant to environmental, mechanical, and chemical stress. Accordingly, catalase activity in the spore may potentially be very advantageous to the parasite. To determine whether the N. locustae catalase is expressed in spores, 3′ RACE was carried out on total RNA isolated from highly purified spore material. RACE products of the expected size were cloned, and the sequence of four clones revealed that all corresponded to catalase. Approximately one-quarter of the coding region of catalase mRNA was amplified, and three of the four clones examined were identical to the gene sequence. The fourth 3′-RACE product contained two single-base differences from the genomic sequence. All four products were identical in sequence to one another and to the genomic sequence in the 3′ untranslated regions, differing only slightly in length (46 to 49 nt), indicating that all transcripts have likely arisen from the same locus.
The presence of the catalase protein in N. locustae spores was also examined by Western blot analysis. Total protein preparations from purified spores were separated by electrophoresis and blotted, and the resulting blot was probed with antibodies raised against N. locustae catalase-specific peptides (Fig. 2). The estimated size of the reacting band was ∼81 kDa, which corresponds closely to the size for large-subunit catalases and to the predicted molecular mass (77.9 kDa) of the N. locustae catalase based on the inferred amino acid sequence. An identical blot probed with the preimmune serum did not produce a reaction (data not shown), confirming the specificity of the antibody. These results indicate that the catalase protein is also present in N. locustae spores. Consistent with the presence of the catalase protein in the spore, experiments conducted with active and inactivated spore extracts also indicate that the protein is enzymatically active (data not shown).
FIG. 2.

Western blot of N. locustae spore proteins. (a) Coomassie blue-stained total protein extracts from N. locustae spores. (b) Western blot of the protein extraction shown in lane a exposed to a rabbit polyclonal antibody raised against two N. locustae catalase peptides. The estimated size of the reacting band (81 kDa) correlates with the inferred molecular mass deduced from the gene sequence (77.9 kDa).
The N. locustae catalase is derived by lateral gene transfer from a bacterium.
Microsporidian gene sequences tend to be extremely divergent, a characteristic which originally led to their incorrect placement in phylogenetic trees and the assumption that microsporidia were among the most ancient eukaryotes (4, 22, 23, 41). As more molecular sequences were obtained from microsporidia and more accurate models of sequence evolution were developed for molecular analysis, phylogenetic trees clearly allied microsporidia with fungi (13, 18, 27). In fact, more recent comprehensive phylogenies indicate that microsporidia probably arose from within the fungal radiation (28, 29). Therefore, microsporidian genes inherited vertically (i.e., by descent) should be of fungal origin and branch with fungal homologs in phylogenetic trees. However, phylogenetic analyses suggest a very different origin for the N. locustae catalase.
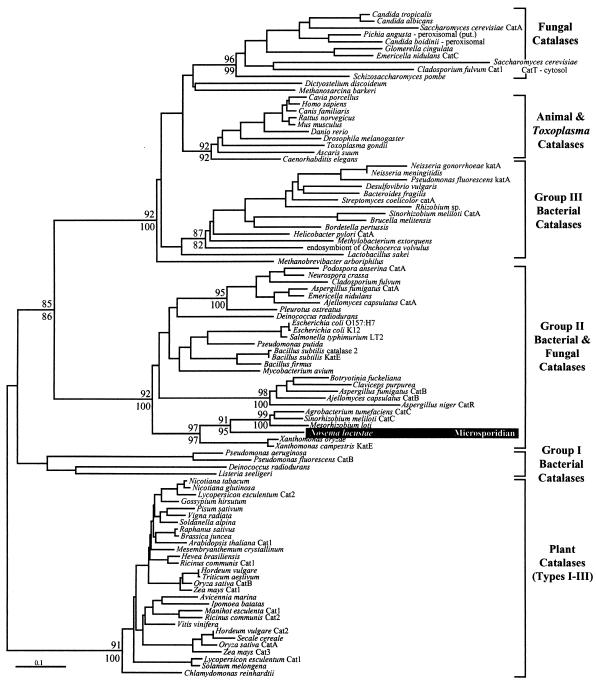
Previous catalase phylogenies identify five well-supported groups: a group of fungal and animal, mainly peroxisomal, catalases, two groups of bacterial catalases (designated group I and group III), a group composed of bacterial and fungal catalases (group II), and the distantly related plant catalases that are themselves subdivided into three groups (30, 40). All these related monofunctional catalases possess core regions that are highly conserved in sequence. However, the group II catalases from bacteria and fungi are unique in possessing lengthy extensions to the N and C termini of the conserved core and are often referred to as the large-subunit catalases (30).
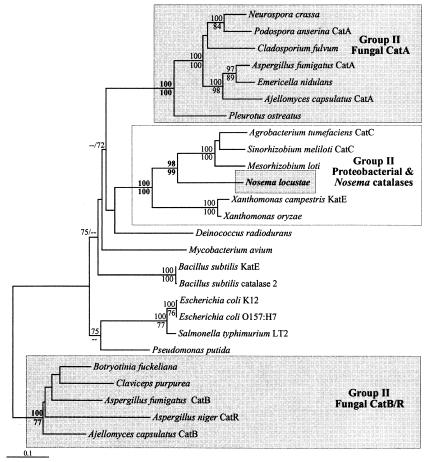
In all phylogenetic analyses, the N. locustae sequence shows no relationship to the conserved fungal homologs that are related to those of animals but instead diverges from within the group II large-subunit clade (Fig. 3), as anticipated based on the size of the protein. Although this clade includes fungal sequences, the N. locustae gene does not branch with these but rather branches specifically and strongly within a group of proteobacterial sequences (with which the N. locustae sequence shares 72 to 76% sequence identity). To more closely analyze the phylogenetic position of the N. locustae catalase, a comprehensive phylogenetic analysis, including additional characters, was undertaken with only the group II large-subunit catalase sequences (Fig. 4). Although the backbone of the tree describing the relationships among the subgroups of group II catalases is not well resolved, there are several highly supported internal subgroups. Reflecting the topology of the global tree in Fig. 3 and other previous catalase phylogenies are two highly supported subgroups of fungal group II catalases, CatA and CatB/R (6, 21, 30). However, the N. locustae sequence branches with neither of these groups. Instead, and in agreement with the global analysis, the N. locustae sequence branches within a group of proteobacterial sequences that is united to the exclusion of the other group II sequences with 100% bootstrap support. The closest relatives of the N. locustae sequence are catalases from three alpha-proteobacteria, an alliance with 98 to 99% bootstrap support. The phylogenetic position of the N. locustae catalase is thus strongly supported but is clearly at odds with the fungal ancestry of microsporidia. These results are best explained by a lateral gene transfer event between a member of the proteobacteria and an ancestor of N. locustae. The absence of a catalase homolog in E. cuniculi suggests that the lateral gene transfer event was fairly recent and likely occurred after the divergence of E. cuniculi and N. locustae. It is possible that lateral gene transfer has played a role in the relationships between the other fungal and bacterial group II catalases, and this subject has been addressed in a recent review (31).
FIG. 3.
Global phylogeny of catalase. The Fitch-Margoliash tree and bootstrap support for major nodes above 85% are indicated (Fitch-Margoliash at top, weighted neighbor-joining below). The major recognized divisions of catalase are indicated by brackets and names to the right, and the N. locustae sequence is shaded.
FIG. 4.
Phylogeny of the group II (LSU) catalase sequences. The Fitch-Margoliash tree is shown with bootstrap support above 70% indicated for both Fitch-Margoliash and protein maximum likelihood methods (above and below nodes, respectively). The fungal and N. locustae sequences are shaded.
In considering a potential case of lateral transfer where a gene of bacterial origin is ascribed to a eukaryotic source, it is critical to rule out the possibility that the sequence is derived from a contaminating bacterium. Such a situation has previously been demonstrated for catalase (16). In the present case, several lines of evidence dismiss the possibility of bacterial contamination and support the catalase gene's position within the genome of N. locustae. The spores have been thoroughly examined by light and transmission electron microscopies for the presence of bacteria, and none has been identified. Furthermore, in the more than 500 independent clones that have been sequenced from multiple genomic libraries constructed from spore material, no clones containing bacterial genomic sequences have been identified. In addition, the codon usage of the catalase gene has many of the biases typical of other N. locustae protein-coding genes (data not shown). The Western blots also demonstrate that a significant amount of protein is present in apparently axenic spores: the reaction to anti-catalase antibody has an intensity similar to that of controls based on other known microsporidian proteins (data not shown). The amplification of catalase by 3′ RACE is also strong evidence against contamination, since prokaryotic transcripts typically lack substantial poly(A) tails and are thus excluded from amplifications with poly(dT) primers. Direct confirmation of the authenticity of the N. locustae catalase comes from prepublication data of the Nosema locustae genome project at the Marine Biological Laboratory (released 12 December 2002; http://jbpc.mbl.edu/Nosema). According to these data, the catalase sequence is present in partially assembled contigs, and, furthermore, several ORFs upstream of the catalase gene possess significant similarity to E. cuniculi genes.
Microsporidian genomes, including the genome of N. locustae, are reduced and compact in nature, typically gene dense, and contain only short intergenic regions (12, 24). It is easy to imagine these characteristics hindering the introduction of foreign DNA, since they reduce the proportion of potential sites where a gene could be inserted without serious disruption to other genes. However, the region of the N. locustae genome surrounding catalase is significantly less compact than other regions of the genome (Fig. 1) and also appears to be poorly conserved. It is possible that these characteristics make this region of the genome better able to tolerate alterations and, by extension, make it a more likely region for the successful integration of genes by lateral transfer. If these factors had any impact on the transfer of the catalase gene, then the transfer must have occurred relatively recently.
Potential cellular location of the N. locustae catalase.
A defining feature of microsporidia, and one that led to their initial classification as primitive eukaryotes, is their apparent lack of typical eukaryotic cellular organelles such as mitochondria, flagella, and peroxisomes. However, as current data indicate, microsporidia are highly derived fungi, so such unique morphological characteristics are not ancestral but derived. Indeed, despite the long-held belief that microsporidia were amitochondriate, it is now known that they do in fact harbor a cryptic mitochondrion. The first evidence for this came from the characterization of nuclear-encoded proteins that were derived from the mitochondrion, HSP70, and pyruvate dehydrogenase (12, 15, 17, 35). Subsequently, the organelle was predicted based on the presence of numerous genes of putative mitochondrial origin in the complete genomic sequence of E. cuniculi (24), and ultimately the organelle was identified cytologically by the immunolocalization of mitochondrion-derived HSP70 in Trachipleistophora hominis (43). Peroxisomes, on the other hand, are still presumed to be absent, and the absence of catalase in the E. cuniculi genome was taken as direct evidence for the lack of this organelle (24). Therefore, the presence of catalase in N. locustae raises the possibility of a cryptic peroxisome.
Catalase in other eukaryotes is generally a peroxisomal matrix protein. Such proteins are posttranslationally imported into the organelle via one of two different cis-acting signals, peroxisomal targeting signal 1 (PTS1) and PTS2 (19). Examining the inferred amino acid sequence of the N. locustae catalase reveals no evidence for any sequence similar to the N-terminal PTS2 consensus. The extreme C terminus, on the other hand, carries the tripeptide RKI. Although this sequence does not strictly fall within the PTS1 consensus (S/A/C) (K/H/R)L, it does bear resemblance to other nonconsensus signals that are known to target peroxisomal proteins to the organelle, including those of fungi (1, 11, 15). Such variability in the targeting signal, taken together with the additional possibility of internal targeting signals, renders any judgment on the cellular location of this catalase difficult.
Perhaps more information about the localization of catalase in N. locustae can be gleaned from considering the phylogenetic results. The sequence falls with strong support into the group II large-subunit catalases (Fig. 3 and 4). None of the fungal group II sequences examined are peroxisome targeted. In some cases the protein has been localized to the cytosol or cell wall of spores, while in other cases the catalase protein has been found to be extracellular (21, 25, 44, 45). There is no evidence for a signal peptide at the N terminus of the N. locustae catalase, making it unlikely that the protein is secreted. Furthermore, catalase has also been shown to function in mitochondria in some organisms, yet there is no clear N-terminal mitochondrial targeting signal in the N. locustae sequence (although microsporidian mitochondrial targeting signals are not well understood). All in all, while the inferred protein sequence suggests that catalase is not peroxisomal, it does not conclusively reveal where catalase is found in N. locustae cells; determining the precise subcellular location of catalase awaits immunolocalization experiments.
Functional implications for N. locustae spores.
The microsporidian spore is the only stage of the parasite that is viable outside of a host cell. The spore is the infectious stage and is, by necessity, highly resistant to desiccation, mechanical and chemical damage, and a variety of other stresses. For these reasons, the presence and nature of an active catalase in N. locustae spores raise a number of interesting questions relating to its possible function and when it originated. The N. locustae catalase is a group II large-subunit enzyme. This type of catalase possesses a different heme group from other catalases and is more stable and resistant to inactivation than the small-subunit catalases (37). None of the fungal group II catalases have been localized to a peroxisome but instead, where they have been examined, are localized to the cytosol or cell wall or are secreted. Interestingly, this class of catalases has been shown to be expressed during stress responses in fungi, as well as during specific developmental stages such as sexual and asexual spore formation (6, 8, 25, 34). In those bacteria where large-subunit catalases are found, the gene is expressed during stationary phase, once again suggesting a possible role in protecting partially dormant cells (30, 32). We have demonstrated that the N. locustae catalase mRNA and protein are present in spores and that the enzyme is active. It is currently unclear whether catalase is expressed constitutively throughout the microsporidian life cycle or specifically during the spore stage. However, based on the characteristics and functions of other group II large-subunit catalases, it is tempting to speculate that the N. locustae enzyme allows the spore to deal with long-term oxidative stress in a manner unavailable to E. cuniculi—perhaps improving the spore's resistance to environmental stresses or prolonging spore viability. While there is no direct evidence for the cellular location of the N. locustae catalase, it is most likely not localized to a cryptic peroxisome. The absence of catalase in E. cuniculi and the lack of direct evidence for peroxisomes in any microsporidian (24), together with the phylogenetic evidence that the N. locustae catalase was secondarily derived from a proteobacterium, add up to suggest that microsporidia lost peroxisomes at some point in their evolution and that N. locustae has regained the activity of catalase though lateral gene transfer. Regardless of the cellular location or exact function of catalase in N. locustae, its presence highlights the important potential of lateral gene transfer on metabolic adaptation.
Acknowledgments
We acknowledge the Nosema locustae Genome Project, Marine Biological Laboratory at Woods Hole, funded by NSF award 0135272, for use of their unpublished sequences. We thank M&R Durango for the gift of spore material, M. Reith for the use and adaptations of ESTid, and J. M. Archibald and A. P. de Koning for discussion and comments on the manuscript.
This work was supported by a New Investigator Award in Molecular Pathogenic Mycology to P.J.K. from the Burroughs-Wellcome Fund (1001537). N.M.F. is supported by postdoctoral fellowships from the Canadian Institutes of Health Research (CIHR) and the Michael Smith Foundation for Health Research (MSFHR). P.J.K. is a scholar of the Canadian Institute of Advanced Research and New Investigator of the MSFHR and CIHR.
REFERENCES
- 1.Aitchinson, J. D., W. W. Murray, and R. A. Rachubinski. 1991. The carboxyl-terminal tripeptide Ala-Lys-Ile is essential for targeting Candida tropicalis trifunctional enzyme to yeast peroxisomes. J. Biol. Chem. 266:23197-23203. [PubMed] [Google Scholar]
- 2.Andersson, J. O., A. M. Sjögren, L. A. M. Davis, T. M. Embley, and A. J. Roger. 2003. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr. Biol. 13:94-104. [DOI] [PubMed] [Google Scholar]
- 3.Brown, J. R., and W. F. Doolittle. 1999. Gene descent, duplication, and horizontal transfer in the evolution of glutamyl- and glutaminyl-tRNA synthetases. J. Mol. Evol. 49:485-495. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. R., and W. F. Doolittle. 1995. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc. Natl. Acad. Sci. USA 92:2441-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruno, W. J., N. D. Socci, and A. L. Halpern. 2000. Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol. Biol. Evol. 17:189-197. [DOI] [PubMed] [Google Scholar]
- 6.Bussink, H. J., and R. Oliver. 2001. Identification of two highly divergent catalase genes in the fungal tomato pathogen, Cladosporium fulvum. Eur. J. Biochem. 268:15-24. [DOI] [PubMed] [Google Scholar]
- 7.Cavalier-Smith, T. 1983. A 6-kingdom classification and a unified phylogeny, p. 1027-1034. In H. E. A. Schenk and W. S. Schwemmler (ed.), Endocytobiology II: intracellular space as oligogenetic. Walter de Gruyter & Co., Berlin, Germany.
- 8.Chary, P., and D. O. Natvig. 1989. Evidence for three differentially regulated catalase genes in Neurospora crassa: effects of oxidative stress, heat shock, and development. J. Bacteriol. 171:2646-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curgy, J. J., J. Vávra, and C. P. Vivarès. 1980. Presence of ribosomal RNAs with prokaryotic properties in microsporidia, eukaryotic organisms. Biol. Cell. 38:49-51. [Google Scholar]
- 10.de Koning, A. P., F. S. Brinkman, S. J. Jones, and P. J. Keeling. 2000. Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol. Biol. Evol. 17:1769-1773. [DOI] [PubMed] [Google Scholar]
- 11.Didion, T., and R. Roggenkamp. 1992. Targeting signal of the peroxisomal catalase in the methylotrophic yeast Hansenula polymorpha. FEBS Lett. 303:113-116. [DOI] [PubMed] [Google Scholar]
- 12.Fast, N. M., and P. J. Keeling. 2001. Alpha and beta subunits of pyruvate dehydrogenase E1 from the microsporidian Nosema locustae: mitochondrion-derived carbon metabolism in microsporidia. Mol. Biochem. Parasitol. 117:201-209. [DOI] [PubMed] [Google Scholar]
- 13.Fast, N. M., J. M. Logsdon, Jr., and W. F. Doolittle. 1999. Phylogenetic analysis of the TATA box binding protein (TBP) gene from Nosema locustae: evidence for a microsporidia-fungi relationship and spliceosomal intron loss. Mol. Biol. Evol. 16:1415-1419. [DOI] [PubMed] [Google Scholar]
- 14.Germot, A., H. Philippe, and H. Le Guyader. 1997. Evidence for loss of mitochondria in microsporidia from a mitochondrial-type HSP70 in Nosema locustae. Mol. Biochem. Parasitol. 87:159-168. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, H., T. Didion, A. Thiemann, M. Veenhuis, and R. Roggenkamp. 1992. Targeting sequences of the two major peroxisomal proteins in the methylotrophic yeast Hansenula polymorpha. Mol. Gen. Genet. 235:269-278. [DOI] [PubMed] [Google Scholar]
- 16.Henkle-Duhrsen, K., V. H. O. Eckelt, G. Wildenburg, M. Blaxter, and R. D. Walter. 1998. Gene structure, activity and localization of a catalase from intracellular bacteria in Onchocerca volvulus. Mol. Biochem. Parasitol. 96:69-81. [DOI] [PubMed] [Google Scholar]
- 17.Hirt, R. P., B. Healy, C. R. Vossbrinck, E. U. Canning, and T. M. Embley. 1997. A mitochondrial Hsp70 orthologue in Vairimorpha necatrix: molecular evidence that microsporidia once contained mitochondria. Curr. Biol. 7:995-998. [DOI] [PubMed] [Google Scholar]
- 18.Hirt, R. P., J. M. Logsdon, Jr., B. Healy, M. W. Dorey, W. F. Doolittle, and T. M. Embley. 1999. Microsporidia are related to fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl. Acad. Sci. USA 96:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holroyd, C., and R. Erdmann. 2001. Protein translocation machineries of peroxisomes. FEBS Lett. 501:6-10. [DOI] [PubMed] [Google Scholar]
- 20.Ishihara, R., and Y. Hayashi. 1968. Some properties of ribosomes from the sporoplasm of Nosema bombycis. J. Invertebr. Pathol. 11:377-385. [Google Scholar]
- 21.Johnson, C. H., M. G. Klotz, J. L. York, V. Kruft, and J. E. McEwen. 2002. Redundancy, phylogeny and differential expression of Histoplasma capsulatum catalases. Microbiology 148:1129-1142. [DOI] [PubMed] [Google Scholar]
- 22.Kamaishi, T., T. Hashimoto, Y. Nakamura, Y. Masuda, F. Nakamura, K. Okamoto, M. Shimizu, and M. Hasegawa. 1996. Complete nucleotide sequences of the genes encoding translation elongation factors 1 alpha and 2 from a microsporidian parasite, Glugea plecoglossi: implications for the deepest branching of eukaryotes. J. Biochem. (Tokyo) 120:1095-1103. [DOI] [PubMed] [Google Scholar]
- 23.Kamaishi, T., T. Hashimoto, Y. Nakamura, F. Nakamura, S. Murata, N. Okada, K.-I. Okamoto, M. Shimzu, and M. Hasegawa. 1996. Protein phylogeny of translation elongation factor EF-1α suggests microsporidians are extremely ancient eukaryotes. J. Mol. Evol. 42:257-263. [DOI] [PubMed] [Google Scholar]
- 24.Katinka, M. D., S. Duprat, E. Cornillot, G. Méténier, F. Thomarat, G. Prensier, V. Barbe, E. Peyretaillade, P. Brottier, P. Wincker, F. Delbac, H. El Alaoui, P. Peyret, W. Saurin, M. Gouy, J. Weissenbach, and C. P. Vivarès. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414:450-453. [DOI] [PubMed] [Google Scholar]
- 25.Kawasaki, L., and J. Aguirre. 2001. Multiple catalase genes are differentially regulated in Aspergillus nidulans. J. Bacteriol. 183:1434-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keeling, P. J. 2003. Congruent evidence from alpha-tubulin and beta-tubulin phylogenies for a zygomycete origin of microsporidia. Fungal Genet. Biol. 38:298-309. [DOI] [PubMed] [Google Scholar]
- 27.Keeling, P. J., and W. F. Doolittle. 1996. Alpha-tubulin from early-diverging eukaryotic lineages and the evolution of the tubulin family. Mol. Biol. Evol. 13:1297-1305. [DOI] [PubMed] [Google Scholar]
- 28.Keeling, P. J., and N. M. Fast. 2002. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu. Rev. Microbiol. 56:93-116. [DOI] [PubMed] [Google Scholar]
- 29.Keeling, P. J., M. A. Luker, and J. D. Palmer. 2000. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol. Biol. Evol. 17:23-31. [DOI] [PubMed] [Google Scholar]
- 30.Klotz, M. G., G. R. Klassen, and P. C. Loewen. 1997. Phylogenetic relationships among prokaryotic and eukaryotic catalases. Mol. Biol. Evol. 14:951-958. [DOI] [PubMed] [Google Scholar]
- 31.Klotz, M. G., and P. C. Loewen. 2003. The molecular evolution of catalatic hydroperoxidases: evidence for multiple lateral transfer of genes between prokaryota and from bacteria into eukaryota. Mol. Biol. Evol. 20:1098-1112. [DOI] [PubMed] [Google Scholar]
- 32.Loewen, P. C., J. Switala, and B. L. Triggs-Raine. 1985. Catalases HPI and HPII in Escherichia coli are induced independently. Arch. Biochem. Biophys. 243:144-149. [DOI] [PubMed] [Google Scholar]
- 33.Müller, M. 1998. Enzymes and compartmentation of core energy metabolism of anaerobic eukaryotes—a special case in eukaryotic evolution?, p. 109-132. In G. H. Coombs, K. Vickerman, M. A. Sleigh, and A. Warren (ed.), Evolutionary relationships among protozoa. Chapman & Hall, New York, N.Y.
- 34.Navarro, R. E., M. A. Stringer, W. Hansberg, W. E. Timberlake, and J. Aguirre. 1996. catA, a new Aspergillus nidulans gene encoding a developmentally regulated catalase. Curr. Genet. 29:352-359. [PubMed] [Google Scholar]
- 35.Peyretaillade, E., V. Broussolle, P. Peyret, G. Méténier, M. Gouy, and C. P. Vivarès. 1998. Microsporidia, amitochondrial protists, possess a 70-kDa heat shock protein gene of mitochondrial evolutionary origin. Mol. Biol. Evol. 15:683-689. [DOI] [PubMed] [Google Scholar]
- 36.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 37.Switala, J., and P. C. Loewen. 2002. Diversity of properties among catalases. Arch. Biochem. Biophys. 401:145-154. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vávra, J., and J. I. R. Larsson. 1999. Structure of the microsporidia, p. 7-84. In M. Wittner and L. M. Weiss (ed.), The microsporidia and microsporidiosis. ASM Press, Washington, D.C.
- 40.von Ossowski, I., G. Hausner, and P. C. Loewen. 1993. Molecular evolutionary analysis based on the amino acid sequence of catalase. J. Mol. Evol. 37:71-76. [DOI] [PubMed] [Google Scholar]
- 41.Vossbrinck, C. R., J. V. Maddox, S. Friedman, B. A. Debrunner-Vossbrinck, and C. R. Woese. 1987. Ribosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature 326:411-414. [DOI] [PubMed] [Google Scholar]
- 42.Vossbrinck, C. R., and C. R. Woese. 1986. Eukaryotic ribosomes that lack a 5.8S RNA. Nature 320:287-288. [DOI] [PubMed] [Google Scholar]
- 43.Williams, B. A., R. P. Hirt, J. M. Lucocq, and T. M. Embley. 2002. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature 418:865-869. [DOI] [PubMed] [Google Scholar]
- 44.Witteveen, C. F. B., M. Veenhuis, and J. Visser. 1992. Localization of glucose oxidase and catalase activities in Aspergillus niger. Appl. Environ. Microbiol. 58:1190-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zancopé-Oliveira, R. M., E. Reiss, T. J. Lott, L. W. Mayer, and G. S. Deepe, Jr. 1999. Molecular cloning, characterization, and expression of the M antigen of Histoplasma capsulatum. Infect. Immun. 67:1947-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]