Abstract
Quantitative analysis of the relationship between virus expression and disease outcome has been critical for understanding HIV-1 pathogenesis. Yet the amount of viral RNA contained within an HIV-expressing cell and the relationship between the number of virus-producing cells and plasma virus load has not been established or reflected in models of viral dynamics. We report here a novel strategy for the coordinated analysis of virus expression in lymph node specimens. The results obtained for patients with a broad range of plasma viral loads before and after antiretroviral therapy reveal a constant mean viral (v)RNA copy number (3.6 log10 copies) per infected cell, regardless of plasma virus load or treatment status. In addition, there was a significant but nonlinear direct correlation between the frequency of vRNA+ lymph node cells and plasma vRNA. As predicted from this relationship, residual cells expressing this same mean copy number are detectable (frequency <2/106 cells) in tissues of treated patients who have plasma vRNA levels below the current detectable threshold (<50 copies/ml). These data suggest that fully replication-active cells are responsible for sustaining viremia after initiation of potent antiretroviral therapy and that plasma virus titers correlate, albeit in a nonlinear fashion, with the number of virus-expressing cells in lymphoid tissue.
Keywords: HIV-1 infection, quantitative RT-PCR, lymph node biopsy, in situ hybridization, HIV pathogenesis
The amount of HIV-1 in blood correlates with the risk of clinical progression to AIDS (1, 2) and provides a useful assay to evaluate responses to antiretroviral therapy (3, 4). However, a large proportion of the virus in the body (as measured by detection of viral [v]RNA)1 is present in lymphoid tissues (5). There is limited and conflicting quantitative information available regarding the correlation between tissue and blood HIV-1 pools, the extent and distribution of residual HIV-infected cells in tissue compartments after the extensive reduction in plasma HIV RNA associated with effective therapy, and the quantitative relationship between the number of virus-expressing cells and plasma virus load (5–9). A critical component in formulating an accurate and comprehensive model of HIV pathogenesis based on these parameters is the analytical distinction between vRNA inside infected cells and the extracellular virions trapped in the follicular dendritic cell (FDC) network of lymph node germinal centers (6, 7). Although lymphoid tissue contains much more vRNA than blood, the technical challenges of quantitative analysis are compounded by the relative difficulty of obtaining adequate lymphoid tissue and the inherent complexity of tissue compared with blood plasma. To address these issues, and because the quantitative assessment of different viral compartments in vivo is crucial to understanding the complex dynamic relationships between cellular populations and different forms of viral genetic material, we have developed an alternative strategy to measure the amount of vRNA within tissue compartments. Three sets of analyses were performed on the same fresh-frozen tissue specimen: a sensitive in situ hybridization (ISH) procedure (10, 11); quantitative, competitive RT-PCR (QC-RT-PCR) analysis of the bulk tissue; and QC-RT-PCR analysis of individual cells isolated from tissue at limiting dilution.
Materials and Methods
Lymph Node Tissue Processing.
After appropriate informed consent, subjects underwent an excisional lymph node biopsy under local anesthesia in an outpatient operating room. The tissue was obtained from levels 3 and 4 of the posterior cervical chain, with the second biopsy obtained from the contralateral site. In each case, the largest palpable lymph node was selected for excision. The freshly excised lymph node tissue was immediately cut into pieces, and a measured portion was snap frozen under OCT compound as a block of tissue; the remaining portion was separated into a single-cell suspension by mechanical disaggregation. The single-cell suspension was counted and used for several different analyses, including the preparation of microtiter wells used for a limiting dilution analysis (LDA) of vRNA-expressing cells.
Production of Probes for ISH.
Probes for ISH of HIV-1 RNA were produced from three separate plasmids (covering sequences 225–3378, 5546–7308, and 7308–9107; sequence data available from EMBL/GenBank/DDBJ under accession no. L02317) that were linearized, and single-stranded RNA molecules were produced by in vitro transcription. During the transcription, digoxigenin-UTP ([uridine 5′-triphosphate]; Boehringer Mannheim) or 35S-UTP was directly incorporated into the sequence as described (12). For the digoxigenin-labeled probes, three separate antisense probes were made, quantified by trace labeling with 3H during transcription, and hydrolyzed for 10 min in 20 mM NaHCO3, pH 10.2, to yield labeled fragments of ∼300 bp. The three probe preparations were then mixed at equal molar amounts and used at 0.1 fm/μl in hybridization buffer for ISH. The procedure was performed as previously reported (11–13). The color reaction was stopped by rinsing the slides in Tris-EDTA buffer, pH 8.0, and the sections mounted in aquamount.
Quantitative Morphometric Analysis.
The frequency of HIV RNA+ cells was counted by direct microscopic observation, using a calibrated ocular grid, by an observer blinded to the other analyses performed on the same specimen. The frequency of individual vRNA+ cells was counted in multiple adjacent sections, and the total area occupied by lymphoid tissue and density of cells was determined in each section. In each case, the number of HIV RNA+ cells and the total lymphoid tissue area was determined for each section. The number of adjacent sections analyzed depended on the frequency of positive cells and ranged from 5 to 100 sections per lymph node. The density of cells was determined by counting nuclei in an adjacent hematoxylin and eosin– stained section for each case. Among these specimens, the mean number of cells (nuclei)/mm2 was 12,500. By comparing the number of positive cells with the total amount of lymphoid tissue examined, the frequency of positive cells per 106 total cells was calculated. ISH analysis of lymph node tissue from HIV− individuals, using riboprobes with the same HIV-1 sequences in sense orientation or probes specific for murine sequences, failed to identify any positively stained cells (data not shown).
QC-RT-PCR.
HIV RNA was quantitated by a modification of the procedure described previously (14). The synthetic competitor for HIV analysis was constructed by ligation of the 5′ and 3′ respective oligonucleotide templates into the general cloning vector pQPCR1 as described (14). This pQPCR.HRV2 competitor contained several primer sets, one of which was a site in the HIV-1 pol gene designed to amplify a 415-bp fragment extending from position 2713 to 3127 in the HIV genome (based on the position number available from EMBL/GenBank/DDBJ under accession no. K03455). All competitors were used to generate single-stranded competitor RNA molecules, which were purified based on a specific hybridization tag 3′ to the reverse primer cassette and quantitated by trace labeling with a known specific activity of 3H-UTP.
The single tube quantitative PCR technique, a modification of the multi-tube quantitative PCR technique (14), was performed by adding GITC extract containing an unknown amount of HIV-1 RNA to a competitor cocktail containing five different synthetic HIV competitors at a series of concentrations differing by 0.4 log10 multiples. For example, competitor cocktail 3 consists of competitor A at log10 3 (1,000 copies), competitor B at log10 3.4 (2,510 copies), etc. Each competitor differed from the others by an internal 25-bp segment of DNA that is the basis for differential hybridization and detection. Extraction, cDNA generation, and quantitative PCR were performed as described (14). The enzyme immunoassay detection procedure was also carried out essentially as described (14), except that each PCR product was plated into 12 microtiter wells instead of 4. Two wells each were probed with detection oligonucleotides for the HIV-1 gene and stuffer detection oligos specific for competitors A, B, C, D, and E. Calculation of endpoints was performed as described (14). The pol gene primers used were: upstream oligo, 5′-CATACAATACTCCAGTATTTGCCA; and pol downstream oligo, 5′-AAGTCAGATCCTACATACAAATCA. PCR conditions for pol amplification were: 0.8 μM primers, 2.6 mM Mg2+, and 50°C annealing for 35 cycles. The detection oligonucleotide for hybridization of the internal HIV sequence was 5′-TGGATGTGGGTGATGCATATTTTTCAGTTC; each different competitor species was detected with a distinct 25-mer sequence using standard hybridization conditions (14).
For determination of the bulk amount of vRNA in tissue, 20–40 4-μm sections were cut from the frozen tissue block adjacent to other sections used to perform ISH and immunohistochemical analysis. This material was immediately solubilized by adding 10 μl GITC/section to a tube containing the frozen sections and was used for the QC-RT-PCR analysis as described above (termed tissue homogenate).
LDA.
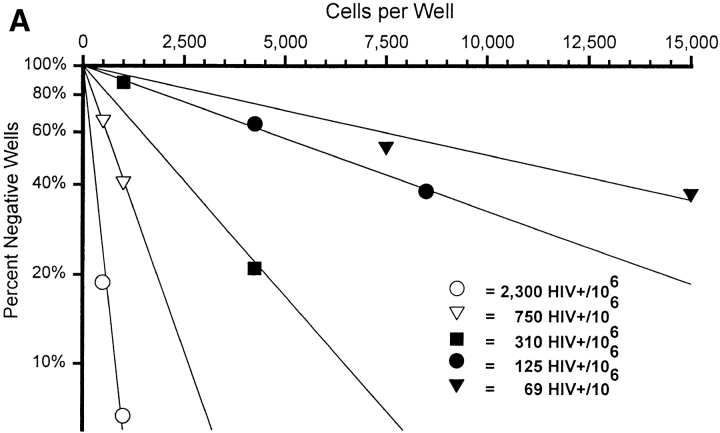
A measured portion of each lymph node biopsy specimen was separated in the operating suite and transported immediately to the containment lab in ice cold balanced salt solution. The tissue was separated into a single-cell suspension by standard mechanical disaggregation techniques, washed, and counted. Limiting diluted wells were prepared by dispensing 500, 1,000, 5,000, or 10,000 cells into 36 replicate microtiter wells in 10 μl HBSS with 10% FCS, and then 100 μl GITC was added to each well. Individual wells were analyzed by QC-RT-PCR as described using a competitor cocktail with the lowest individual competitor concentration at 1,000 copies/reaction. The logarithm of the fraction of negative wells (<1,000 vRNA) was plotted against the number of cells per well. Regression lines passing through the origin were calculated and used to estimate the precursor frequency of positive cells. When the frequency of negative wells is >40%, most of the positive wells contain only a single positive cell and, therefore, the copy number directly determined by the QC-RT-PCR procedure in those positive wells is also a direct measure of the copy number per replication-active cell in vivo.
Results
Patient Characteristics.
The analysis contained herein was applied to cervical lymph node biopsy specimens obtained from nine patients with a wide spectrum of plasma HIV RNA levels, six of whom had serial biopsies performed before and after the introduction of highly active antiretroviral therapy (combinations of HIV-1 protease inhibitors and two reverse transcriptase inhibitors). All of the subjects were in relatively advanced stages of HIV disease, with <350 CD4 T cells/μl. The plasma vRNA levels and the absolute blood CD4 cell counts concurrent with the biopsies, the timing of the biopsies relative to therapy, the specific antiretroviral regimens involved, and the quantitative results obtained in this analysis are shown in Table I.
Table I.
Comparison of Three Independent Methods of Analysis of HIV RNA in Lymph Node Tissue
Summary of quantitative analysis of HIV vRNA in a series of patients undergoing excisional lymph node biopsies. Subjects are identified by a four-letter code, and information concerning drug treatment at the time of the biopsy, the time after the change in therapy before the second (or third) biopsy, the plasma viral load, and CD4 T cell count determined on a blood specimen obtained the same day the biopsy is given. Results obtained from the ISH analysis of tissue sections are shown as a histologic score (Neg to 4+) for germinal center–associated vRNA signal, and the frequency of individual vRNA positive cells expressed per million lymph node cells examined. Results from LDA (see Fig. 3) are given as frequency of cells scored positive by QC-RT-PCR (Fig. 3 A) and mean (±SEM) of absolute copies measured in each positive well at limiting dilution (Fig. 3 B). The right-most column shows the results of analysis of a series of adjacent frozen sections obtained from the face of the tissue block used for ISH analysis that were analyzed for the bulk amount of vRNA by QC-RT-PCR. The measured amount of vRNA in this extract is expressed per million cells homogenized, which was calculated using the measured area (mm2) of each section, the density of cells/mm2, the thickness of the section (4 μm), and an estimate of 12 μm for the average diameter of a lymphocyte. Thus, the denominators for both the bulk tissue amount of vRNA by QC-RT-PCR and the frequency of vRNA+ cells by ISH are the same, such that the simple ratio of these measurements yields an estimate of the vRNA copies per positive cell that is completely independent of the result obtained from the LDA. For the average copy number per cell by bulk analysis, only those samples that were scored as negative for FDC-associated virus were included. The determination of copy number per cell by the two methods was not significantly different using the Mann–Whitney Rank sum test or Student's t test, comparing either the cases with or without significant FDC signal, respectively. In those cases in which there was significant FDC-associated vRNA detected by ISH, this bulk tissue estimate of vRNA copies per cell is higher than either the LDA determined estimate or the bulk tissue estimate on those specimens without FDC vRNA signal.
not done.
Validation of the QC-RT-PCR Technique.
Fundamental to these studies was the development of a procedure to accurately quantify the HIV vRNA from tissue specimens. Using primers specific to various regions of the HIV genome detecting only full length, unspliced message (see Materials and Methods), a modification of the previously described QC-RT-PCR assay was employed (14). To verify the accuracy of our QC-RT-PCR assay, we compared this assay to two other independent methods for HIV RNA quantitation, using a series of plasma specimens obtained from patients in various clinical stages of HIV infection as well as a standard HIV RNA preparation obtained from the National Institutes of Health AIDS reagent repository. The results of this analysis (Fig. 1) demonstrated excellent concordance between the UAB QC-RT-PCR and the Roche Amplicor HIV Monitor® assay (15), whereas comparison with the branched chain DNA (bDNA; Chiron) assay (16) using the same specimens revealed the previously described slight systematic bias toward lower viral load results in the bDNA assay (data not shown). Thus, the independently constructed QC-RT-PCR assay provides results equivalent to the Roche assay over a wide range of RNA concentrations. By using dilutions of specimens containing varying concentrations of HIV vRNA, this QC-RT-PCR (14) assay has a sensitivity of ∼100 copies/reaction (data not shown).
Figure 1.
Plasma samples from HIV-infected patients were obtained and immediately separated into 1-ml aliquots, which were analyzed by each of two methods with blinded results. In addition, a standard preparation of HIV RNA was obtained from the National Institute of Allergy and Infectious Diseases AIDS Research and Reference Reagent Program and analyzed by the UAB QC-RT-PCR procedure. Roche Monitor® assay was performed as per manufacturer's instructions. Specimens with viral loads >5 × 105/ml were analyzed after appropriate dilution. The UAB QC-RT-PCR procedure was performed as described in Materials and Methods.
Limiting Dilution QC-RT-PCR Analysis of Lymph Node Biopsy Specimens Reveals a Constant Mean Viral RNA Copy Number per Cell.
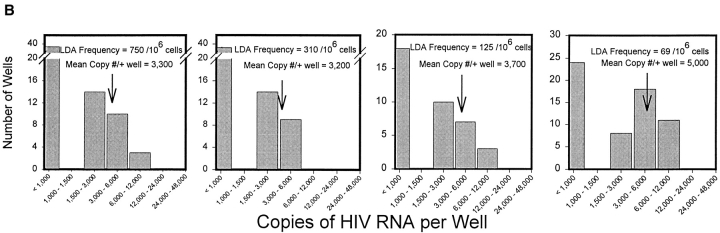
The goal of these studies is to accurately quantitate the amount of virus contained in HIV-infected cells and to cross-validate the results using independent procedures. The sensitivity of RT-PCR allows single-cell analysis; therefore, the current study began by assessing the vRNA content of individual HIV-infected cells. To directly determine the frequency and vRNA content of lymph node cells actively producing virus, five individual biopsy specimens obtained before combination antiretroviral therapy were subjected to LDA. Both the frequency of individual cells expressing detectable vRNA (Fig. 2 A) and the copy number per cell (copies/well at limiting dilution; Fig. 2 B) were directly determined. The mean copy number of vRNA per cell for each patient is remarkably consistent (3,900 or 3.6 log10 copies/cell [range: 3.5–3.7 log10 copies/cell]; Table I).
Figure 2.
Analysis of HIV RNA+ cells by LDA and single-cell UAB QC-RT-PCR. (A) LDA for the frequency of HIV vRNA–expressing cells on five untreated patients described in Table I. After dilution of lymph node cells in 96-well microtiter plates, each well was harvested and QC-RT-PCR performed. For the graph in A, wells were scored positive or negative for HIV vRNA only. The number of lymph node cells diluted per well is plotted along the x-axis, and the percent negative wells by QC-RT-PCR is plotted along the y-axis. The frequency of HIV vRNA–expressing cells per 106 lymph node cells was calculated by Poisson statistics. (B) Frequency histograms for four of the LDAs graphed in A for quantification of copies per well. The range of copies per well for each bar on the x-axis represents 0.3 log10 multiples of copies of HIV vRNA. Only positive wells (>1,000 copies) were used for the calculation of mean HIV vRNA copies per well. See Table I for summary of other patient data and calculations of HIV vRNA quantity in different pools.
The Frequency of HIV vRNA–expressing Cells Is Comparable by LDA and ISH.
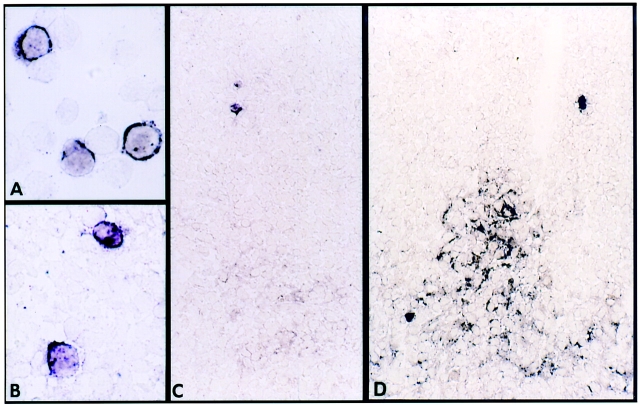
To corroborate the QC-RT-PCR data, the frequency of vRNA+ cells was also determined by ISH analysis using digoxigenin-labeled riboprobes as previously described (10, 11). As observed by others using 35S-labeled riboprobes (5, 6, 17–19), HIV RNA is present in lymphoid tissue in two distinct patterns: within individual cells (Fig. 3, A and B) and between cells in the FDC network of germinal centers (Fig. 3, C and D). The frequency of vRNA+ cells we observed by ISH is comparable to the frequency of vRNA+ cells determined by the independently performed LDA method on adjacent portions of the same tissue specimen (Table I). In this series of patients, most with relatively advanced HIV disease, many of the lymph nodes demonstrated negligible amounts of ISH signal associated with germinal centers. Nevertheless, in each specimen, morphological germinal centers were identified by immunohistochemistry (data not shown). In several cases, the ISH procedure was repeated with 35S-labeled HIV riboprobes and with proteinase K–digested tissue as described (6, 17), with results equivalent to those of digoxigenin-labeled probes for both frequency of individual vRNA+ cells and detection of FDC virions (data not shown). The relative intensity and extent of germinal center–associated virus was assessed on a semiquantitative visual scale (Table I), but no attempt to precisely calculate numbers of vRNA molecules was made based on this image analysis. Although several of these tissue specimens from advanced HIV disease patients (CD4 <200 cells/μl) had negligible FDC-associated vRNA signal, in this analysis the quantification of vRNA was carried out exclusively using the standardized soluble vRNA assay by QC-RT-PCR.
Figure 3.
Representative photomicrographs of ISH for HIV RNA. (A) Suspension of normal PBMC infected with HIV-1 in vitro. (B) HIV RNA+ cell identified in lymph node frozen tissue section. (C) Lymph node section of patient HADE showing HIV RNA in germinal center of lymph node scored as 1+ and two individual positive single cells. (D) Lymph node section of patient TRRA showing HIV RNA in germinal center of lymph node scored as 3+ and two individual positive single cells.
The HIV RNA Mean Copy Number per Cell Is Similar between LDA and Bulk Tissue Analysis When FDC-associated Virus Is Minimal.
To cross-validate the HIV RNA mean copy number per cell by LDA, a further independent analysis of the vRNA content of homogenized tissue was performed on each biopsy specimen. Tissue homogenates from serial sections from the same tissue block used for the ISH analysis were made as described. The amount of vRNA present in tissue homogenates, representing both individual vRNA-expressing infected cells and free virions trapped in FDC structures, was then directly determined using the QC-RT-PCR procedure and is expressed in Table I as HIV RNA copies/million lymph node cells (see Bulk Tissue Analysis). The total amount of tissue vRNA was then divided by the number of HIV-expressing cells detected by ISH and the results listed as HIV RNA Copies/Cell bulk/ ISH in Table I. Consistent with the LDA data, when FDC-associated virus was not detected (Neg), the HIV RNA Copies/ISH+ cell hovered around 5,000 copies. When significant FDC-associated virus was detected by ISH, then the HIV RNA copies/ISH+ cell was much higher (Table I; see patients BABI, EVJE, and TRRA). Thus, the detection of FDC-associated virions is not simply dependent on the sensitivity of the ISH analysis. Direct measurement of total tissue vRNA detects significantly more vRNA than can be accounted for by the frequency of individual vRNA+ cells only in those cases where FDC signal is detected by qualitative ISH methodology.
The Frequency of HIV vRNA–expressing Cells Is Related to the Plasma Viral Load.
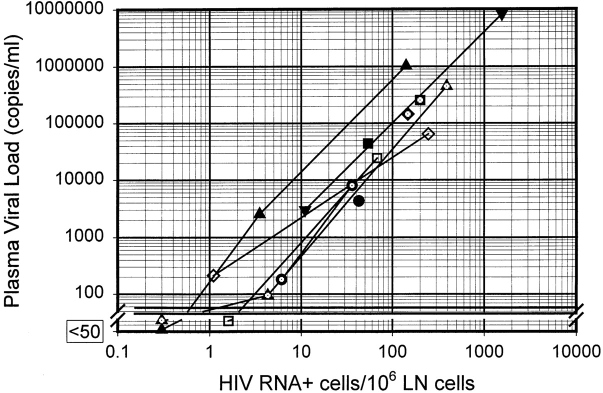
The relatively constant mean vRNA content per cell implies a constant instantaneous production of HIV virions (instantaneous burst size). Although the actual burst size represents the total virus production over the lifetime of the cell and depends upon several factors (see below), one prediction of a constant burst size would be a direct correlation between the number of infected cells and the plasma viral load over a broad range of values. To investigate this possibility, we compared the frequency of vRNA+ cells per million lymph node cells determined by ISH with the plasma viral load of the patient at the time of the biopsy (Fig. 4). As predicted, there was a direct and highly significant correlation between the logarithm of the vRNA+ cell frequency in lymph node tissue and the logarithm of the plasma vRNA. The slope of this relationship is relatively constant, whether the data derives from different patients (slope = 1.6; Pearson product moment correlation coefficient (r) = 0.89; P < 0.0005) with widely varying viral loads (a 6 log10 range) before protease inhibitor therapy or from patients undergoing serial biopsies before and after potent therapy (Fig. 4, mean of individual slopes = 1.6; points connected by lines). The overall relationship among the logarithms of these values is highly significant (slope = 1.6; r = 0.95; P < 10−8). Although the relationship is consistent over a wide range of viral load values, the direct correlation is not 1:1 (that is, the slope of the log–log plot ≠ 1). This result differs from predictions based on standard mathematical models of viral dynamics (20, 21). In other words, a 1 log10 drop in the number of vRNA+ cells in lymph node tissue is associated with a 1.6 log10 drop in vRNA in plasma. The interpretation of this observation remains open for speculation, but the consistency of this direct relationship over such a broad range of viral load values is a novel observation that must be accounted for in quantitative models of viral dynamics.
Figure 4.
Relationship of plasma viral load to HIV vRNA–expressing cells in lymph node tissue. Each point represents a separate biopsy specimen paired with the plasma viral load at time of biopsy. HIV vRNA+ cells were measured by ISH, and plasma viral loads were determined by the Roche Monitor® ultra-sensitive assay. The lines connecting points with the same symbols represent the same patient before and after HAART, as shown in Table I. Overall regression line (using the log-transformed data) of these data: slope = 1.6; r = 0.95 (P < 10−8).
Persistence of vRNA-expressing Cells in Lymph Node Tissue Despite Undetectable Plasma Viral Load.
In each of the three biopsy specimens obtained from subjects on stable highly active antiretroviral therapy (HAART) with a plasma viral value <50 copies/ml, we continued to detect individual vRNA+ cells by ISH, albeit at extremely low frequency. This analysis required examination of 50–100 sections spaced at least 12 μm apart to detect the few positive cells. The quantification of the total vRNA from the interspersed sections used for ISH analysis divided by the frequency of vRNA+ cells detected by ISH indicated a copy number per cell similar to that obtained during episodes of high viral replication (Table I). Although this estimate is not highly precise due to the low number of total cells detected, these data indicate that the majority of detectable vRNA molecules in lymph node tissue of subjects on HAART is contained within a few individual cells. Because the copy number estimate for these residual cells is similar to that measured by both LDA and bulk tissue analysis in subjects not treated with antiretroviral drugs, it is likely that these very few cells represent fully replication-active cells, rather than some form of abortive transcriptional activity.
Discussion
This coordinated analysis of HIV vRNA by three interlocking but independent methods (ISH, QC-RT-PCR of bulk tissue, and LDA) provides cross-validation of the results. First, we found that the frequency of positive cells determined by ISH is equivalent to the frequency determined independently by LDA (Table I). Thus, the sensitivity of ISH for detecting individual vRNA+ cells was directly validated by the completely independent method of LDA, which has not been performed when using other ISH methods. Second, when there was negligible FDC-associated virus detected by ISH, the copy number of HIV RNA per cell determined by LDA is equivalent to the vRNA copies present in bulk tissue divided by the frequency of cells. In contrast, when significant FDC-associated vRNA was detected by ISH, this ratio was higher than the independent estimate of copy number determined by LDA (Table I). These data imply that the ISH procedure using digoxigenin-labeled riboprobes is not missing significant amounts of FDC-associated virus when it is present. If ISH was underestimating the extent of FDC-associated virus, then the copy number calculated based on the bulk tissue method would not match the result obtained by LDA. As the quantification of vRNA copies is not performed by image analysis of the ISH signal but rather by the standardized soluble QC-RT-PCR assay, this quantification does not directly depend on the sensitivity of detection by ISH. Even if the intensity of the FDC-associated ISH signal is weaker using this ISH procedure, this potential problem does not affect the analysis based on cross-validation of independent methods. For example, the first biopsy of a subject with advanced disease (Modified Card Sorting Test [MCST], CD4 count = 27; viral load = 1.1 × 106; Table I) showed negligible FDC signal. The frequency of individual positive cells was essentially equivalent by two independent methods (140/106 cells by ISH and 125/106 cells by LDA). The copy number per positive cell was also equivalent by two independent methods (3,700 copies/cell by LDA and 4,100 copies/cell by the ratio of total tissue RNA divided by the frequency of identified cells). If there was significant FDC virus in this tissue specimen, then this cross-validation would not have been observed. Indeed, when significant qualitative FDC signal was observed, there was significantly more total vRNA in tissue than could be accounted for by the frequency of individual positive cells (Table I; see subjects BABI, EVJE, and TRRA before HAART). Thus, this analysis is internally validated by independent methods, with the quantitation depending on the QC-RT-PCR analysis of soluble RNA rather than image analysis. The measurement of soluble RNA has been standardized using both internal and external standards and is comparable to the Roche Amplicor HIV Monitor® assay (15) (Fig. 1). This analytical approach demonstrates that each vRNA- expressing cell in patient lymph node tissue contains, on average, ∼3.6 log10 (or ∼4000) copies of HIV RNA. This result is independent of the frequency of infected cells, the plasma viral load or CD4 count, or the form or potency of the antiretroviral being administered at the time of the biopsy.
Our determination of the HIV RNA copies per cell is significantly higher than that of previous results based on image analysis using 35S-labeled probes (17, 18, 22, 23). Furthermore, in a recent report, image analysis of lymph node specimens from patients with acute and early HIV infection did not demonstrate a relationship between infected cell frequency in tissue and plasma viral load (24). Although previous investigators have emphasized the major predominance of FDC-associated virions over intracellular virus, in the current analysis the degree of trapping of virions in lymph node germinal centers is less than previously reported (17, 22). It is likely that this discrepancy in absolute quantification of copies per detectable vRNA+ cell is due to the standardization of the method used to determine copy number. Molar quantification using image analysis of 35S-based ISH relies on complete efficiency of RNA preservation and probe access in fixed, paraffin-embedded tissue and 100% efficiency of grain production in the emulsion. These assumptions have not be independently validated. In separate experiments not reported here, both of these potential problems appear to be significant. To examine the efficiency of grain production in the emulsion for 35S-labeled probe ISH, we measured the frequency of infected cells and counted grains over each positive cell in populations of HIV-1 infected PBMC. Calculation of copy number by grain count, using the specific activity of the probe and time of emulsion exposure as described (17), generated a significantly lower estimate of copy number per cell than direct QC-RT-PCR analysis of 10,000 such cells (corrected for the fraction that was vRNA+). By this analytical approach, detection of silver grains is only about 5% efficient; in other words, determination of the probe specific activity in a well scintillation counter is more efficient than silver grain production in a very thin emulsion.
To examine the potential loss of signal by tissue block fixation and paraffin embedding, the expression of murine Ig μ heavy chain mRNA was compared in fresh-frozen tissue versus formalin-fixed, paraffin-embedded tissue. Both small B cells in primary follicles and sinusoidal plasma cells were detected by digoxigenin-labeled riboprobes in frozen tissue, whereas only the plasma cells were clearly detected in formalin-fixed, paraffin-embedded tissue. Thus, previous reports that used the image analysis method of RNA quantitation may have two systematic errors that lead to an underestimation of the amount of cell-associated virus.
Another aspect of the precision and sensitivity of ISH analysis is the discrimination of cells with positive ISH signal from cellular debris that nonspecifically binds the labeled riboprobe. Although debris of irregular shape and size larger than a cell can be discriminated with both techniques, the immunoenzymatic signal is detected directly in the same focal plane as the individual cell in thin section, unlike detection of grains deposited in a photographic emulsion above the tissue section. This additional criterion of morphological analysis allows discounting of rare bits of debris that are similar in size and shape to individual cells but localized out of the plane of section using the digoxigenin system. Perhaps previous reports of significant decreases in the grain count per cell with therapy (17) may be due to inclusion of a higher fraction of artifactual localized accumulations of grains with lower grain density in the setting of a very low absolute frequency of HIV positive cells.
Although interpretation of the relative sensitivity of various histochemical techniques is controversial and has not been cross-validated by multiple independent laboratories, the analysis presented in this report is not fundamentally dependent upon the quantitative sensitivity of the ISH procedure. Rather than simply assume that the frequency of cells detected by ISH was correct, we directly confirmed this frequency by an independent LDA method. Furthermore, the paucity of FDC signal intensity in some of these specimens was directly confirmed by analysis of a bulk quantity of RNA in these tissues. Thus, unlike other analyses of HIV RNA in tissue, this analysis depends on quantitative agreement between several independent analytical methods rather than sole reliance on ISH methodology.
In addition to the novel methodological approach, the biological implications of the findings presented here are significant. The invariant mean HIV RNA copy number per cell regardless of treatment history or stage of disease implies that infected cells in tissue maintain a constant instantaneous burst size. The actual burst size is dependant on the life span of the replication-active cell and rate of virion budding in addition to the cell-associated vRNA pool size. We hypothesize that viral production at the single-cell level may be an all-or-none phenomenon, analogous to the situation described for T cell cytokine gene expression (10, 13, 25). The consistency of vRNA amount in productively infected cells during treatment with antiretroviral drugs is consistent with the mechanism of action of these agents, which inhibit either reverse transcription or maturation of viral particles rather than transcriptional activity. Although the precision of the estimate of copy number per identified vRNA+ cell in the few cases with very low cellular frequency is not sufficient to rule out a modest change after therapy, a substantial change of >10-fold as reported previously (17) is not supported by this analysis.
Independent of the measurement of absolute copies of vRNA present in each cell, the frequency of such cells is highly correlated with plasma viral load. The surprising observation that this relationship is nonlinear (slope of log–log plot ≠ 1) requires some modification of the standard models of viral dynamics (20, 21). One potential explanation for this observation is that the production rate of plasma virus is significantly modified by the presence of antiretroviral drugs. Several lines of evidence argue against this possibility. First, the mechanism of action of these agents is a blockade of de novo infection, either by inhibition of reverse transcription or extracellular maturation of virion particles rather than transcriptional inhibition or virion budding. Second, our demonstration of a roughly constant amount of vRNA in individual cells argues that the amount of intracellular vRNA awaiting assembly and budding is not altered by these agents. Finally, the same nonlinear relationship is found when comparing different individual subjects before therapy but with differing viral load set points. Thus, the nonlinear character of this relationship cannot arise from alteration of viral production rate per cell induced by antiretroviral therapy.
Another potential explanation is that clearance of viral particles is a direct function of viral load. To account for the extent of nonlinearity over the range of viral load values examined, the clearance rate must vary by ∼100-fold in a consistent relationship to viral load. Although antibody-mediated clearance rate may be more efficient for low virus load and may be saturated at high virus load, it seems unlikely that such a tremendous variation in clearance rate could account for these observations.
Alternatively, we favor a hypothesis that focuses on the different in vivo tissue compartments containing replication-active cells and their relative contribution to the systemic plasma viral load. It is possible that most of the infected cells are present in spleen and gut rather than lymph nodes due to the sheer volume of these compartments. As the T cell zones in lymph nodes are single, relatively contiguous areas unlike the larger spleen and gut tissue environments, the density of infection in these areas may be consistently higher at any given steady state level. The effect of decreased efficiency of cell-to-cell transfer of infectious virus induced by antiretroviral drugs may thus have a lesser effect on the lymph node microenvironment than on the bulk of the lymphoid tissue present in spleen and gut. A comprehensive, quantitative analysis of various tissue compartments containing infected cells in rhesus macaques infected with SIV variants may provide a direct experimental test of this model.
A clinically relevant prediction based on the observed relationship between vRNA+ cell frequency and plasma viral load is that cells actively replicating viral proteins persist after the substantial decline in plasma viremia observed during combination antiretroviral therapy. The correlation of plasma virus load with the frequency of vRNA+ cells over the six log range of quantifiable viral load suggests that this relationship is quite robust. The intercept of this relationship at “undetectable” plasma viral load is ∼1 vRNA+ cell/million lymph node tissue cells. Given the large lymphoid tissue mass present in the entire body (estimated at 1011 cells), a reasonable interpretation of these data is that ∼100,000 cells with viral replication potential could persist in an infected subject, but the virions produced by these few cells would be cleared too quickly to achieve a detectable steady state concentration in plasma. The extrapolation of this relationship to plasma viral load values of <50 copies/ml is validated by our direct observation of cells containing full length viral transcripts within tissue specimens from three patients with concomitant “undetectable” plasma viremia.
The origin and fate of the few residual vRNA+ cells in patients with substantial drug-induced suppression of viral replication is of fundamental importance for understanding HIV disease pathogenesis and developing further therapeutic alternatives for HIV-infected individuals. If these cells represent new rounds of de novo cellular infection, the implications concerning the eventual emergence of drug- resistant mutants are significant. Alternatively, these few cells could represent in vivo activation of latently infected cells, a process not blocked by HAART. If the virions produced by such activated, latently infected cells do not complete the viral life cycle due to effective antiretroviral therapy, new mutations will not be fixed in the viral population. The apparent extremely low clearance rate of such latently infected cells in subjects on prolonged HAART (26–28) may be attributable to a decreased antigen-driven immune clearance mechanism of these cells (29) rather than a complete failure of these cells to ever become transcriptionally active. Whatever the biological origin of these few residual vRNA+ cells, their persistence may result in rapid reignition of high level viral replication after discontinuation of antiretroviral drugs. Consistent with such a conceptual model are anecdotal reports that patients who discontinue combination antiretroviral therapy, even after many months of apparently complete viral suppression in the blood, experience a rapid return of viremia. Attempts to accurately determine the status of HIV infection in aggressively treated hosts must take into account both the residual pool of latently infected cells and the presence of persistent active viral replication in lymphoid tissue cells. Although combination antiretroviral regimens can reduce plasma HIV RNA levels below current detection limits, our data demonstrate that this parameter is inadequate to measure the extent to which potentially infectious virus remains in other sites (such as lymphoid tissue). Therapeutic strategies that attempt to completely eradicate HIV must be based on firm, quantitative information concerning both blood and tissue sources and the biological nature of residual HIV-1 virus.
Acknowledgments
The authors thank Dr. Ronald Mitsuyasu and Richard Pollard and the AIDS Clinical Trials Group 328 team for the leadership of the clinical trial protocol; the study subjects; and Debra Horton, Julie Decker, Ya Fen Ji, Jimin Li, Jesus F. Salazar-Gonzalez, Li Fang Li, Cindy James, Rebecca Gilleland, Connie Miller, and Mark MacEwen for technical assistance.
Abbreviations used in this paper
- FDC
follicular dendritic cell
- HAART
highly active antiretroviral therapy
- ISH
in situ hybridization
- LDA
limiting dilution analysis
- QC-RT-PCR
quantitative, competitive RT-PCR
- UTP
uridine 5′-triphosphate
- vRNA
viral RNA
Footnotes
This work was partially supported by Chiron Corp. and the Adult AIDS Clinical Trials Group.
References
- 1.Mellors J, Rinaldo C, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 2.O'Brien W, Hartigan P, Martin D. Changes in plasma HIV-1 RNA and CD4+lymphocyte counts and the risk of progression to AIDS. Rapid and simple PCR assay for quantitation of HIV-1 RNA in plasma: application to acute retroviral infection. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter CC, Fischl MA, Hammer SM, Hirsch MS, Jacobsen DM, Katzenstein D, Montaner JS, Richman DD, Saag MS, Schooley RT, et al. Antiretroviral therapy for HIV infection in 1996. Recommendations of an international panel. International AIDS Society-USA. JAMA. 1996;276:146–154. [PubMed] [Google Scholar]
- 4.Saag MS, Holodniy M, Kuritzkes DR, O'Brien WA, Coombs R, Poscher ME, Jacobsen DM, Shaw GM, Richman DD, Volberding PA. HIV viral load markers in clinical practice. Nat Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 5.Pantaleo G, Graziosi C, Demarest JF, Butini L, Montroni M, Fox CH, Orenstein JM, Kotler DP, Fauci AS. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature. 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 6.Fox CH, Tenner-Racz K, Racz P, Firpo A, Pizzo PA, Fauci AS. Lymphoid germinal centers are reservoirs of human immunodeficiency virus type 1 RNA. J Infect Dis. 1991;164:1051–1057. doi: 10.1093/infdis/164.6.1051. [DOI] [PubMed] [Google Scholar]
- 7.Pantaleo G, Graziosi C, Butini L, Pizzo PA, Schnittman SM, Kotler DP, Fauci AS. Lymphoid organs function as major reservoirs for human immunodeficiency virus. Proc Natl Acad Sci USA. 1991;88:9838–9842. doi: 10.1073/pnas.88.21.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pantaleo G, Graziosi C, Fauci AS. The immunopathogenesis of human immunodeficiency virus infection. N Engl J Med. 1993;328:327–335. doi: 10.1056/NEJM199302043280508. [DOI] [PubMed] [Google Scholar]
- 9.Embretson J, Zupancic M, Ribas JL, Burke A, Racz P, Tenner-Racz K, Haase AT. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature. 1993;362:359–364. doi: 10.1038/362359a0. [DOI] [PubMed] [Google Scholar]
- 10.Bucy RP, Karr L, Huang GQ, Li J, Devore-Carter D, Honjo K, Lemons A, Murphy KM, Weaver CT. Single-cell analysis of cytokine gene co-expression during naive CD4+T cell phenotype development. Proc Natl Acad Sci USA. 1995;92:7565–7569. doi: 10.1073/pnas.92.16.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karr L, Panoskaltsis-Mortari A, Li J, Devore-Carter D, Weaver CT, Bucy RP. In situhybridization for cytokine mRNA with digoxigenin labeled riboprobes: sensitivity of detection and double label applications. J Immunol Methods. 1995;182:93–106. doi: 10.1016/0022-1759(95)00027-8. [DOI] [PubMed] [Google Scholar]
- 12.Panoskaltsis-Mortari A, Bucy RP. In situ hybridization with digoxigenin-labeled RNA probes: facts and artifacts. Biotechniques. 1995;18:300–307. [PubMed] [Google Scholar]
- 13.Bucy RP, Panoskaltsis-Mortari A, Huang GQ, Li J, Karr L, Ross M, Russel JH, Murphy KM, Weaver CT. Heterogeneity of single cell cytokine gene expression in clonal T cell populations. J Exp Med. 1994;180:1251–1262. doi: 10.1084/jem.180.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hockett RD, Janowski KM, Bucy RP. Simultaneous quantitation of multiple cytokine mRNAs by RT-PCR utilizing plate based EIA methodology. J Immunol Methods. 1995;187:273–285. doi: 10.1016/0022-1759(95)00195-5. [DOI] [PubMed] [Google Scholar]
- 15.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of HIV-1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Y, Ho DD, Todd J, Kokka R, Urdea M, Lifson JD, Piatak M, Jr, Chen S, Hahn BH, Saag MS. Clinical evaluation of branched DNA signal amplification for quantifying HIV type 1 in human plasma. AIDS Res Hum Retroviruses. 1995;11:353–361. doi: 10.1089/aid.1995.11.353. [DOI] [PubMed] [Google Scholar]
- 17.Haase AT, Henry K, Zupancic M, Sedgewick G, Faust RA, Melroe H, Cavert W, Gebhard K, Staskus K, Zhang Z, et al. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science. 1996;274:985–989. doi: 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 18.Cavert W, Notermans DW, Staskus K, Wietgrefe SW, Zupancic M, Gebhard K, Henry K, Zhang ZQ, Mills R, McDade H, et al. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science. 1997;276:960–964. doi: 10.1126/science.276.5314.960. [DOI] [PubMed] [Google Scholar]
- 19.Orenstein JM, Fox C, Wahl SM. Macrophages as a source of HIV during opportunistic infections. Science. 1997;276:1857–1861. doi: 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- 20.Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 21.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 22.Peng H, Reinhart TA, Retzel EF, Staskus KA, Zupancic M, Haase AT. Single cell transcript analysis of human immunodeficiency virus gene expression in the transition from latent to production infection. Virology. 1995;206:16–27. doi: 10.1016/s0042-6822(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 23.Wong JK, Gunthard HF, Havlir DV, Zhang ZQ, Haase AT, Ignacio CC, Kwok S, Emini E, Richman DD. Reduction of HIV-1 in blood and lymph nodes following potent antiretroviral therapy and the virologic correlates of treatment failure. Proc Natl Acad Sci USA. 1997;94:12574–12579. doi: 10.1073/pnas.94.23.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pantaleo G, Cohen OJ, Schacker T, Vaccarezza M, Graziosi C, Rizzardi GP, Kahn J, Fox CH, Schnittman SM, Schwartz DH, et al. Evolutionary pattern of human immunodeficiency virus (HIV) replication and distribution in lymph nodes following primary infection: implications for antiviral therapy. Nat Med. 1998;4:341–345. doi: 10.1038/nm0398-341. [DOI] [PubMed] [Google Scholar]
- 25.Negulescu PA, Shastri N, Cahalan MD. Intracellular calcium dependence of gene expression in single T lymphocytes. Proc Natl Acad Sci USA. 1994;91:2873–2877. doi: 10.1073/pnas.91.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 27.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 28.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bucy RP. Immune clearance of HIV-1 replication active cells: a model of two patterns of steady state HIV infection. AIDS Res Hum Retroviruses. 1999;15:223–227. doi: 10.1089/088922299311394. [DOI] [PubMed] [Google Scholar]