Abstract
Primary T cell responses rely on the recruitment and proliferation of antigen-specific T cell precursors. The extent of expansion of each individual T cell clone may depend on (a) its frequency before immunization, (b) its proliferative capacity, and (c) the time at which it first encounters its cognate antigen. In this report, we have analyzed the relative contribution of each of these parameters to the shaping of immune repertoires in the T cell response specific for the epitope 170-179 derived from HLA-Cw3 and presented by Kd. By means of hemisplenectomy, we compared immune and naive repertoires in the same animal and found that the frequency of all expanded T cell clones was extremely low before immunization. In particular, the most expanded clones did not derive from high-frequency precursors. In addition, recruited T cells were found to proliferate at the same rate, irrespective of their T cell antigen receptor sequence. Finally, we showed that only T cells that encounter the antigen at early time points account for a significant part of the specific response. Therefore, the contribution of a T cell clone to the immune response is mostly determined by the time of its entry into the immune repertoire, i.e., the time of first cell division after antigen encounter.
Keywords: clonal expansion, CD8 T cells, primary response, antigen-specific repertoire, major histocompatibility complex–peptide tetramers
Adaptive responses to many pathogens rest upon the initial activation and expansion of antigen-specific CD8+ T lymphocytes which recognize foreign peptides presented in the context of self-MHC class I molecules (1). Elicited CD8+ T cells acquire effector functions and participate in the clearance of pathogen. It is of major interest to identify the factors that shape the composition of antigen-specific responses. In addition, understanding how a T cell response emerges is a prerequisite for rational approaches to vaccines. Until recently, the extent to which antigen-selected responses mobilize the peripheral T cell compartment has been difficult to address. The use of MHC–peptide tetrameric complexes has shown that the magnitude of several epitope-specific responses is much higher than previously thought (2–5). For instance, in BALB/c mice, >50% of the CD8+ cells are specific for the NP118-126 epitope at the peak of lymphocytic choriomeningitis virus infection (2). In humans, a high proportion of specific T cells has been observed during the primary response to EBV and in HIV-positive patients (6, 7). These massive expansions of CD8+ T cells arise from the proliferation of specific precursors which have undergone up to 15–25 cell divisions (8, 9).
In several systems, analysis of T cell immune responses has revealed that a few specific T cell clones dominate the repertoire. Using single-cell PCR, it has been demonstrated that the murine response against the transfected mastocytoma P815-CW3 is comprised of just a few clones (10). In the human T cell response against an influenza epitope, a large disparity in the relative contribution of individual T cell clones within the specific response has been observed (11). Our own work has shown that the relative contribution of each clone to the response against P815–HLA-A2 varies extensively: some clones are represented 20-fold more than others (12). The reason for such clonal dominance is unclear. Although several studies have compared TCR repertoires of primary and secondary responses (10, 13–17), little is known about evolution of the T cell repertoire during the massive expansion that follows the primary antigenic stimulation.
The abundance of a given clone in an immune individual may depend on (a) its frequency before immunization, (b) its proliferative capacity, and (c) the time at which it first encounters its cognate antigen. In this study, we have analyzed these three parameters and monitored T cell expansion during the response against the Kd-restricted epitope 170-179 derived from the HLA-Cw3 molecule. We show that the most abundant clones in the immune repertoire were not of particularly high frequency before immunization. Using several approaches, we have quantified the expansion of the antigen-specific repertoire and measured the proliferative capacity of various elicited T cell clones. We demonstrate that the early elicited T cells clones are homogeneously expanded during the primary response. Finally, we show that timing of antigen encounter has a dramatic impact on the extent of T cell expansion. Therefore, the major parameter that determines the abundance of a given clone in the immune response appears to be its time of initial antigen-driven activation.
Materials and Methods
Mice and Cell Line.
Male 8-wk-old DBA/2 (Ly 5.2) mice were purchased from IFFA-Credo. B6–Ly 5.1 mice were obtained from the Centre de Développement des Techniques Avancées pour L'Expérimentation Animale (Orleans, France). DBA/2–Ly 5.1 mice were generated by crossing B6–Ly 5.1 to DBA/2. After 10 backcrosses to DBA/2, an Ly 5.1 homozygote line was established. Immunizations were performed by intraperitoneal injection of 107 P815-CW3 transfectant tumor cells (18). Hemisplenectomy was performed 1 wk before immunization.
Antibodies and Flow Cytometry.
CyChrome-conjugated anti-CD8 (53-6.7) and FITC-conjugated anti-BV10 mAbs were purchased from PharMingen; FITC-conjugated anti-CD8, and biotinylated anti-B220 and anti-CD4 mAbs were purchased from Caltag. Biotinylated anti–Ly 5.1 (clone A20.1.7) was provided by Dr. A. Cumano (Pasteur Institute, Paris). Kd-CW3 tetrameric complexes were prepared as described previously (12). Cell samples were incubated for 1 h with PE-labeled Kd-CW3 tetramers, washed, and incubated with the indicated antibodies. Flow cytometry was performed on a FACScan™ and cell sorting on a FACStarPLUS™ (Becton Dickinson).
cDNA Synthesis.
RNA was extracted using Trizol reagent (GIBCO BRL) following the manufacturer's instruction, with the addition of 20 μg/ml of glycogen as carrier (Boehringer Mannheim). cDNA was synthesized using (dT)17 oligonucleotide and Moloney murine leukemia virus reverse transcriptase (GIBCO BRL).
Immunoscope Analysis.
Immunoscope analysis and BV10-, BC-, and BJ-specific primers have been described previously (19). In brief, PCR was performed on the indicated cDNA using a BV10- and a BC-specific primer. The PCR product was then subjected to run-off reactions using nested fluorescent primers specific either for BC or one BJ segment. Run-off products were resolved on an automated 373A sequencer (Perkin-Elmer). The size and the intensity of each band were recorded and then analyzed using Immunoscope software (19).
Cloning of TCR-β Rearrangements.
Analysis of the TCR-β rearrangements before immunization was performed as follows. Half of the spleen of a naive DBA/2 mouse was removed from the animal and prepared as a single-cell suspension. These splenocytes were depleted of CD4+ cells using biotinylated anti-CD4 antibody and streptavidin beads (Dynal). The cDNA was prepared and amplified using BV10- and BJ1.2-specific primers. TCR-β rearrangements displaying a 6 amino acid (aa)1–long CDR3β were separated on an 8% polyacrylamide 7 M urea gel as described (12) and cloned using the Topo TA cloning kit (Invitrogen). TCR-β rearrangements from immune animals were cloned after amplifying the indicated cDNA using a BV10-specific primer and either a BC- or a BJ1.2-specific primer.
Results
CW3-specific Repertoire in Immune Hemisplenectomized Animals.
DBA/2 mice, when injected intraperitoneally with P815-CW3 tumor cells, develop a massive CD8+ T cell response against the 170-179 epitope derived from HLA-Cw3 and presented by Kd (20). Peculiar features are observed in TCR usage among CW3-specific T cells, including exclusive usage of BV10, 6 aa–long CDR3β, preferential usage of BJ1.2, and a serine and a glycine at position 1 and 3 of the CDR3β (referred to as a SXG motif) (21). To determine whether the abundance of each immune T cell clone correlates with its frequency before immunization, we used hemisplenectomized mice and analyzed TCR-β chains in the immune population and in its naive counterpart.
Four naive DBA/2 mice, raised in specific pathogen– free conditions, were hemisplenectomized and immunized 1 wk later by intraperitoneal injection of 107 P815-CW3 tumor cells. After 13 d, PBLs were analyzed by flow cytometry using an anti-CD8 antibody and Kd-CW3 tetramers. As shown in Fig. 1 A, hemisplenectomized animals mount a strong CW3-specific T cell response: more than half of the CD8+ PBLs were stained with the tetramers, whereas no staining was detected on the PBLs from a naive animal. The magnitude of the response was comparable to that observed in normal DBA/2 mice. On day 19, mice were killed and the second half of the spleen was recovered. Splenocytes were triple stained with Kd-CW3 tetramers, anti-CD8 and anti-BV10 antibodies. Spleens were infiltrated with CW3-specific T cells since ∼30% of the CD8+ splenocytes were stained with the Kd-CW3 tetramers (Fig. 1 B). In addition, the response of hemisplenectomized animals was restricted to BV10 usage (Fig. 1 B) as usually observed for normal DBA/2 mice. Taken together, these observations demonstrate that hemisplenectomy does not alter the CW3-specific response and that the immune half of the spleen is a large source of specific T cells.
Figure 1.
Immune response of hemisplenectomized mice. (A) CW3-specific T cells are detected in the PBLs of immune hemisplenectomized mice. Hemisplenectomized DBA/2 mice were immunized intraperitoneally with 107 P815-CW3 tumor cells. At day 13, mice were bled from the tail vein, and PBLs were double stained with Kd-CW3 tetramers and anti-CD8 antibody. Histograms are gated on CD8+ cells and show a representative staining of an immune hemisplenectomized animal (solid line) and a naive mouse (dotted line). (B) CW3-specific response of hemisplenectomized animals is restricted to BV10 usage. At day 19, immune hemisplenectomized animals were killed. CD8+ enriched splenocytes were triple stained with Kd-CW3 tetramers and mAbs against CD8 and BV10. The profile is gated on CD8+ cells. (C) Purification of CW3-specific T cells. CW3-specific T cells were sorted on the basis of tetramer staining. Cell purity was checked immediately after sorting.
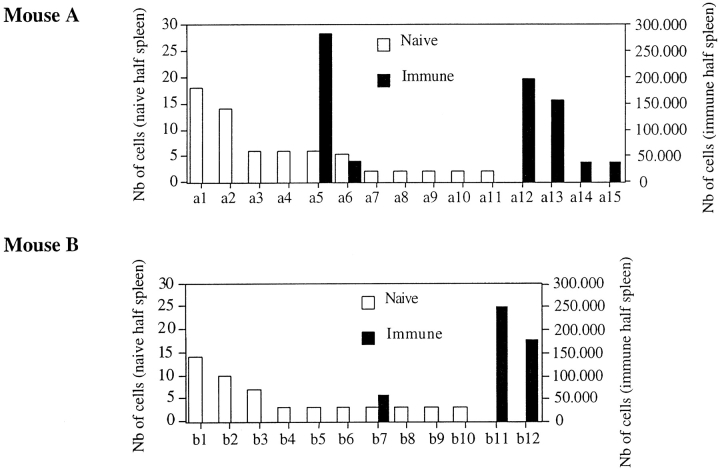
For two of these mice (mice A and B), we sorted the specific T cells on the basis of Kd-CW3 tetramer staining. Cell purity after sorting was ∼98% (Fig. 1 C). The cDNA was prepared and amplified using a BV10- and a BC-specific primer. The PCR product was cloned and sequenced. 10 and 6 distinct TCR-β rearrangements were identified in mice A and B, respectively (Table I). As expected from previous studies, all of these sequences displayed a 6 aa–long CDR3β and the SXG motif within their CDR3β. Moreover, the majority of the rearrangements (6/10 for mouse A, 3/6 for mouse B) used the BJ1.2 segment. Nucleotide and deduced aa sequences are listed in Table I. In addition, based on tetramer staining and sequence occurrence, we estimated the number of immune T cells bearing the indicated CDR3β sequences (Table I). This analysis revealed extensive differences in the abundance of the various specific T cell clones, varying from 4 × 104 to 3.7 × 105 cells.
Table I.
CDR3β Sequences of CW3-specific T Cells Derived from the Immune Half Spleen (Mice A and B)
| Sequence no. | CDR3β (amino acid) | BJ | CDR3β (nucleotide) | Occurrence | Estimated no. of cells* | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mouse A | ||||||||||
| hs.a5 | SRGSDY | TFG (1.2) | AGC CGG GGC TCC GAC TAC | 7/24 | 280,000 | |||||
| hs.a12 | SFGPDY | TFG (1.2) | AGC TTC GGA CCC GAC TAC | 5/24 | 200,000 | |||||
| hs.a13 | SQGSDY | TFG (1.2) | AGC CAG GGC TCC GAC TAC | 4/24 | 160,000 | |||||
| hs.a14 | SLGSDY | TFG (1.2) | AGC TTG GGC TCC GAC TAC | 1/24 | 40,000 | |||||
| hs.a6 | SQGSDY | TFG (1.2) | AGT CAG GGC TCC GAC TAC | 1/24 | 40,000 | |||||
| hs.a15 | SYGSDY | TFG (1.2) | AGC TAT GGC TCC GAC TAC | 1/24 | 40,000 | |||||
| hs.a16 | SLGNTL | YFG (1.3) | AGC CTT GGA AAT ACG CTC | 2/24 | 80,000 | |||||
| hs.a17 | STGERL | FFG (1.4) | AGC ACA GGC GAA AGA TTA | 1/24 | 40,000 | |||||
| hs.a18 | SLGQSL | YFA (1.6) | AGC TTG GGA CAA TCC CTC | 1/24 | 40,000 | |||||
| hs.a19 | SWGVEQ | YFG (2.7) | AGC TGG GGG GTT GAA CAG | 1/24 | 40,000 | |||||
| Mouse B | ||||||||||
| hs.b11 | SLGSDY | TFG (1.2) | AGC CTA GGC TCC GAC TAC | 4/16 | 250,000 | |||||
| hs.b12 | SRGSDY | TFG (1.2) | AGC CGA GGG TCC GAC TAC | 3/16 | 180,000 | |||||
| hs.b7 | SQGSDY | TFG (1.2) | AGT CAG GGC TCC GAC TAC | 1/16 | 60,000 | |||||
| hs.b13 | SLGEEV | FFG (1.1) | AGC TTG GGA GAG GAA GTC | 6/16 | 370,000 | |||||
| hs.b14 | SLGETL | YFG (2.3) | AGC TTG GGA GAA ACG CTG | 1/16 | 60,000 | |||||
| hs.b15 | SHGERL | FFG (1.4) | AGC CAC GGG GAA AGA TTA | 1/16 | 60,000 |
Absolute number of T cells bearing the indicated TCR-β sequence was calculated from tetramer staining and from sequence occurrence.
Tracking TCR-β Chain Frequencies before Immunization.
The majority of the CW3-specific T cells (79 and 50% in mice A and B, respectively) display BV10 and BJ1.2 usage together with a 6 aa–long CDR3β containing the SXG motif. We took advantage of these hallmarks to identify T cells which bear such TCR-β chains in the half of the spleen removed before immunization. We cloned and sequenced the TCR-β chains of CD8+ lymphocytes using the BV10–BJ1.2 combination and displaying a 6 aa–long CDR3β as described in Materials and Methods. On mouse A, 196 sequences were performed: 72 distinct nucleotide sequences were identified, among which only 11 contained the SXG motif. On mouse B, we found 10 distinct sequences containing the SXG motif (out of 86 sequences performed). These sequences are listed in Table II, along with the estimated number of T cells calculated from the sequence occurrence, and the percentage of BV10, BJ1.2, and 6 aa–long CDR3β usage in naive DBA/2 mice (12). Comparison of the abundance of the various TCR-β chains before and after immunization is shown in Fig. 2. There is no correlation between the abundance of a given TCR-β sequence in the immune animal and its frequency in the naive repertoire (Kendall rank correlation test, P < 0.05). These results demonstrate that even the most abundant clones in the immune response preexist at very low frequency in the naive repertoire.
Table II.
CDR3β Sequences Compatible with CW3 Specificity Derived from the Naive Half Spleen (Mice A and B)
| Sequence no.* | CDR3β (amino acid) | CDR3β (nucleotide) | Occurrence | Estimated no. of cells‡ | ||||
|---|---|---|---|---|---|---|---|---|
| Mouse A | ||||||||
| hs.a1 | SWGSDY | AGC TGG GGC TCC GAC TAC | 12/196 | 18 | ||||
| hs.a2 | SRGSDY | AGC AGG GGC TCC GAC TAC | 9/196 | 14 | ||||
| hs.a3 | SRGSDY | TCC CGG GGT TCC GAC TAC | 4/196 | 6 | ||||
| hs.a4 | SYGSDY | AGC TAC GGC TCC GAC TAC | 4/196 | 6 | ||||
| hs.a5 | SRGSDY | AGC CGG GGC TCC GAC TAC | 4/196 | 6 | ||||
| hs.a6 | SQGSDY | AGT CAG GGC TCC GAC TAC | 3/196 | 5 | ||||
| hs.a7 | SQGTDY | AGT CAG GGA ACC GAC TAC | 1/196 | 2 | ||||
| hs.a8 | SFGSDY | AGT TTT GGC TCC GAC TAC | 1/196 | 2 | ||||
| hs.a9 | SLGSDY | AGC TTA GGG TCC GAC TAC | 1/196 | 2 | ||||
| hs.a10 | SSGSDY | AGT TCA GGG TCC GAC TAC | 1/196 | 2 | ||||
| hs.a11 | SAGSDY | AGC GCC GGC TCC GAC TAC | 1/196 | 2 | ||||
| Mouse B | ||||||||
| hs.b1 | SQGSDY | AGC CAG GGC TCC GAC TAC | 4/86 | 14 | ||||
| hs.b2 | SYGSDY | AGC TAC GGC TCC GAC TAC | 3/86 | 10 | ||||
| hs.b3 | SSGSDY | AGT TCA GGG TCC GAC TAC | 2/86 | 7 | ||||
| hs.b4 | STGSDY | AGC ACA GGC TCC GAC TAC | 1/86 | 3 | ||||
| hs.b5 | SSGSDY | AGC TCA GGG TCC GAC TAC | 1/86 | 3 | ||||
| hs.b6 | SSGPDY | AGC TCA GGC CCC GAC TAC | 1/86 | 3 | ||||
| hs.b7 | SQGSDY | AGT CAG GGC TCC GAC TAC | 1/86 | 3 | ||||
| hs.b8 | SWGSDY | AGC TGG GGC TCC GAC TAC | 1/86 | 3 | ||||
| hs.b9 | SYGSDY | AGC TAC GGG TCC GAC TAC | 1/86 | 3 | ||||
| hs.b10 | SRGSDY | AGC CGC GGC TCC GAC TAC | 1/86 | 3 |
Only CDR3β sequences bearing the SXG motif are listed in this Table.
Absolute number of CD8+ T cells bearing the indicated CDR3β sequence was calculated from sequence occurrence and from the percentage of CD8+ T cells displaying BV10, BJ1.2, and 6 aa–long CDR3β in naive DBA/2 mice (reference 12).
Figure 2.
Comparison of TCR-β frequencies in the naive and the immune repertoire of the same mouse. For each immune and naive TCR-β sequence isolated from the hemisplenectomized mice, A and B (described in Tables I and II, respectively), we calculated the number (Nb) of T cells bearing this TCR-β chain in the naive or immune half spleen (see Tables I and II).
Monitoring the CW3-specific Repertoire.
As already mentioned, a large part of the CW3 response is comprised of T cells bearing the BV10, BJ1.2 gene segment and a 6 aa–long CDR3β. Other BJ segments may be used as well with a conserved 6 aa–long CDR3β (see Table II and reference 21). We performed CDR3 size distribution analyses (Immunoscope) on splenocytes from immunized DBA/2 animals using BV10- and BJ-specific primers. For several BV10–BJ combinations, we observed a clonal expansion corresponding to T cells bearing a 6 aa–long CDR3β (Fig. 3, top panels). We have previously shown that Kd-CW3 tetramers identify all CW3-specific T cells (12). Therefore, using Kd-CW3 tetramers, we sorted the CW3-specific T cells from the same animal and analyzed their CDR3β length. All the clonal expansions detected before sorting and corresponding to T cells bearing a 6 aa–long CDR3β were purified in the CW3-specific population, indicating that all of these expanded T cells were antigen specific (Fig. 3, bottom panels). The same experiment performed on other animals yielded the same conclusion although different BJs were involved, revealing individual variability in the response.
Figure 3.
Detection of CW3-specific T cells using CDR3 size distribution analysis. A DBA/2 mouse was immunized intraperitoneally with P815-CW3. At day 11, splenocytes were stained with Kd-CW3 tetramers and an anti-CD8 antibody, and CD8+ Kd-CW3 tetramer+ cells were sorted. Immunoscope analysis was performed on unsorted immune splenocytes (top panels) or on sorted CD8+ Kd-CW3 tetramer+ cells (bottom panels) using a BV10- and all BJ-specific primers. Before sorting, clonal expansions corresponding to T cells bearing a 6 aa–long CDR3β (arrows) were detected in various BV10–BJ profiles (BJ1.2, BJ1.4, BJ1.6, BJ2.1, BJ2.3, BJ2.5, and BJ2.7). The other profiles did not show clonal expansions (see, for example, the BV10– BJ2.2 profile). All of these clonal expansions were purified in the sorted population, indicating that they correspond to CW3-specific 15T cells.
Detection of the Early Expanded CW3 Repertoire.
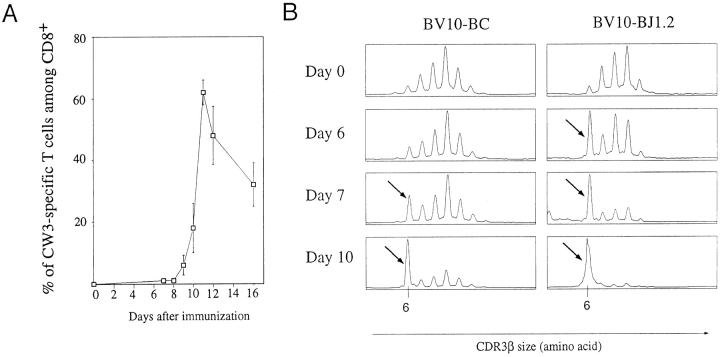
To follow antigen-specific T cell expansions occurring after P815-CW3 intraperitoneal injection, DBA/2 mice were immunized and bled at various time points. PBLs were double stained with Kd-CW3 tetramers and an anti-CD8 antibody. Fig. 4 A shows the kinetics of appearance of CW3-specific T cells from days 0 to 16. As described previously using BV10 staining, the peak of the response is reached at day 11, where >50% of the peripheral CD8+ are CW3 specific (20). Background level of staining was in the order of 0.5%, preventing detection of the specific subset before day 8. We performed an Immunoscope analysis on these blood samples using a BV10-specific primer and either a BC- or BJ1.2-specific primer. As shown in Fig. 4 B, the clonal expansion of T cells bearing 6 aa–long CDR3β could be detected on the BV10–BC profile from day 7. Moreover, CW3-specific T cells could be detected as soon as day 6 by using a BJ1.2-specific primer. Thus, Immunoscope analysis allowed us to detect CW3-specific T cells 5 d before the peak of the response.
Figure 4.
Monitoring the expansion of CW3-specific T cells after immunization. (A) Kinetics of CW3-specific response in the PBLs determined by tetramer staining. PBLs of immunized mice were double stained with Kd-CW3 tetramers and an anti-CD8 antibody. For each time point, results are means of two to six mice and correspond to the compilation of three independent experiments. (B) Emergence of CW3-specific response detected by Immunoscope. An immunized DBA/2 mouse was bled at various time points of the response. The CDR3β size distribution was analyzed by Immunoscope using a BV10-specific primer and either a BC- or BJ1.2-specific primer. The 6 aa–long CDR3β peaks correspond to CW3-specific T cells and are indicated by arrows.
Expansion of the Various Elicited T Cells during the Course of the Response.
As shown in Fig. 3, CDR3β size distribution analysis allowed us to monitor the CW3-specific repertoire. We took advantage of this observation to compare the outcome of various CW3-specific T cell populations bearing different BJ segments. Longitudinal analyses of the response were performed on three DBA/2 mice immunized intraperitoneally with P815-CW3 (referred to as mouse 1, 2, and 3) and bled at various time points. CDR3β size distribution was analyzed using all BV10–BJ combinations. Shown in Fig. 5 are the clonal expansions detected on day 6 for one mouse: they correspond to TCR-β chains bearing a 6 aa–long CDR3β (indicated by arrows in Fig. 5 and corresponding to BJ1.2, BJ1.4, BJ2.3, and BJ2.5 usage). All of these subsets went on proliferating, as revealed by the increase of the 6 aa peak in the BV10–BJ profiles on day 10. In mouse 1, five CW3-specific subpopulations were detected corresponding to BJ1.1, BJ1.2, BJ1.4, BJ2.3, and BJ2.4 usage. To compare the proliferative capacity of these subpopulations, we measured their expansion between days 8 and 10. We estimated that after day 7, the number of CW3-specific T cells in the blood samples was sufficient to allow accurate quantitation by PCR (on day 8, ∼200–300 CW3-specific T cells were collected in 100 μl of blood). Expansion index (EI) was calculated as the ratio between the surface area of the 6 aa peak to the surface area of the other peaks. Since only specific T cells modify the CDR3β size distribution, evolution of the EI directly reflects the expansion of the CW3-specific T cells. The EI of the total specific population can be calculated by analyzing BV10– BC profiles (Fig. 6 A). We also calculated the EI of the five CW3-specific subpopulations by analyzing BV10–BJ profiles. This approach allowed us to compare the proliferation of the CW3-specific T cells bearing different BJ segments. Remarkably, all five specific subsets are proliferating at the same rate (Fig. 6 B). Similar results were obtained analyzing CW3-specific subpopulations from mouse 2 (bled at days 7 and 10) and mouse 3 (bled at days 7, 9, and 11). In these two animals, four expanded subpopulations were identified corresponding to BJ1.2, BJ1.4, BJ2.3, and BJ2.5 (on day 11, BV10–BJ1.2 profile from mouse 3 displayed a single peak, preventing the calculation of the EI for this particular point). Taken together, these results demonstrate that the relative contribution of the various CW3-specific subpopulations to the response is conserved during the expansion.
Figure 5.
Conserved pattern of TCR usage during the course of T cell expansion. A DBA/2 mouse was bled at various time points after immunization. CDR3β size distribution analysis was performed using all 12 BV10–BJ combinations. Figure shows the profiles at days 0, 6, and 10. Note that CW3-specific expansions can already be detected on day 6 and correspond to four subpopulations using different BJ segments (indicated by arrows). These four subpopulations are further expanded on day 10, and no other expansion emerges.
Figure 6.
The various CW3-specific T cell subpopulations expand at the same rate. (A) Expansion of the CW3-specific T cells quantified by Immunoscope. An immunized DBA/2 mouse (mouse 1) was bled at days 8, 9, and 10. CDR3β size distributions were analyzed using a BV10-specific primer and either a BC- or BJ-specific primer. A peak corresponding to CW3-specific T cells is detected in the BV10–BC profile and in some BV10–BJ profiles (BJ1.1, BJ1.2, BJ1.4, BJ2.3, and BJ2.4). Figure shows the BV10–BC, BV10–BJ2.3, and BV10–BJ1.6 profiles. In the first two combinations, the darkened peaks correspond to CW3-specific T cells, whereas the BV10–BJ1.6 profiles do not show any clonal expansions. Expansion index (EI) was calculated as the ratio of the surface area of the 6 aa peak to the surface area of the rest of the profile and is indicated in the upper right corner of the profiles. (B) Kinetics of the expansion of the various CW3-specific subpopulations in mice 1–3. For each BV10–BJ profile that revealed CW3-specific T cells, we plotted the EI as a function of time. For mouse 1, the same analysis was performed using the BV10–BC profile, revealing the expansion of the total CW3-specific T lymphocytes. All subpopulations are expanding at the same rate.
Clonal Stability of the Antigen-specific Repertoire during T Cell Expansion.
To evaluate the expansion of various CW3-specific T cells at the clonal level, we analyzed the repertoire of TCR-β–specific sequences during the expansion. A P815-CW3–immunized DBA/2 mouse was bled at days 7 and 11. Tetramer staining indicated that between days 7 and 11, the absolute number of CW3-specific T cells had increased by ∼100-fold (Fig. 4 A). As shown in Fig. 4 B, Immunoscope analysis revealed that, on day 7, the vast majority of T cells bearing the BV10 and BJ1.2 segments were CW3 specific. Therefore, we amplified the cDNA prepared from blood samples using BV10- and BJ1.2-specific primers, then cloned and sequenced the PCR products. As expected, all sequences obtained on day 11 displayed the hallmarks of CW3 response, i.e., 6 aa–long CDR3β containing the SXG motif. This was also the case for almost all sequences (31/33) obtained on day 7; the two exceptions are likely due to the residual T cells displaying BV10–BJ1.2 usage without being CW3 specific (see Fig. 4 B). Of the 31 sequences from either day 7 or day 11, we identified 10 distinct nucleotide sequences in both samples (Table III). Therefore, the expanding repertoire on day 7 displays the same complexity as the repertoire at the peak (day 11) of the response (Table III). In addition, 25 out of 31 nucleotide sequences obtained from day 7 were identical to 20 out of 31 sequences from day 11. Taken together, these results demonstrate that the complexity as well as the clonal composition of the immune response are conserved during the expansion phase.
Table III.
Conserved Complexity and Clonal Composition of the CW3-specific Response during T Cell Expansion
| Percentage of specific T cells among CD8+ cells* | No. of specific T cells in total PBLs | No. of specific T cells in blood sample | No. of sequences performed | No. of distinct sequences | Common sequences‡ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 7 | 1 | 2,000 | 100 | 31 | 10 | 25/31 | ||||||
| Day 11 | 60 | 180,000 | 6,000 | 31 | 10 | 20/31 |
Percentage of CW3-specific T cells was determined by Kd-CW3 tetramer staining.
Abundance of nucleotide sequences common to both samples.
Time of Antigen Encounter Has a Major Impact on the Extent of T Cell Expansion.
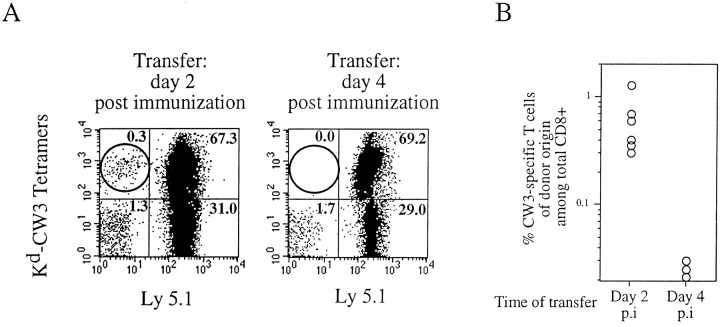
We next determined to what extent timing of initial activation affects the expansion of individual T cell clones. For that purpose, we monitored the expansion of naive T cells after transfer into immunized recipients. As a preliminary experiment, a naive Ly 5.1 DBA/2 mouse was injected intravenously with naive splenocytes (108) from Ly 5.2 DBA/2 mice and then immunized with P815-CW3. 2 wk later, splenocytes were analyzed by flow cytometry in order to determine the percentage of CW3-specific T cells among Ly 5.1+ and Ly 5.2+ populations. We found that 14 and 16% of Ly 5.1+ and Ly 5.2+ CD8+ cells, respectively, were stained with Kd-CW3 tetramers. This observation indicates that transferred T cells responded equivalently to endogenous T cells. We then compared the effect of transferring the naive T cells at various time points after immunization. DBA/2 (Ly 5.2) naive splenocytes (108) were injected intravenously into DBA/2 (Ly 5.1) mice immunized either 2 or 4 d before. 12 d after transfer, PBLs were triple stained with Kd-CW3 tetramers and anti-CD8 and anti–Ly 5.1 mAbs, and the contribution of T cells from donor origin to the response was determined. As shown in Fig. 7, when Ly 5.2+ splenocytes are transferred into a recipient immunized 2 d before, antigen-specific T cells from donor origin expand to 0.3–1.3% of the total T cells. In contrast, no specific expansion could be detected when donor cells were transferred into recipients immunized 4 d before. Therefore, a 48-h delay in antigen encounter dramatically affects the extent of clonal expansions.
Figure 7.
Time of antigen encounter has a major impact on T cell expansion. DBA/ 2–Ly 5.1 mice were immunized intraperitoneally with P815-CW3. Either 2 or 4 d after immunization (p.i), these animals were injected intravenously with splenocytes (108) from naive DBA/2–Ly 5.2 mice. 12 d after transfer, PBLs were triple stained with Kd-CW3 tetramers and anti-CD8 and anti–Ly 5.1 antibodies. (A) Detection of CW3- specific T lymphocytes in recipients (Ly 5.1) immunized either 2 or 4 d before splenocyte (Ly 5.2) transfer. Profiles are gated on CD8+ cells. (B) Expansions of antigen-specific T cells from donor origin are compiled from two independent experiments and plotted for each individual mouse as the percentage of the total CD8+ population.
Discussion
Numerous studies have focused on the composition of immune repertoires at or near the peak of T cell responses. A feature that has been extensively analyzed is the complexity of TCR usage (22). Some responses were found to be diverse, while others were strongly constrained (23). More scarce are the data on the complexity of the response in a given individual, i.e., the number of antigen-recruited T cell clones. Interestingly, when we and others analyzed the extent of antigen-specific CD8+ T cell response, large variations (20–50-fold differences) were observed in the relative abundance of the various recruited T cell clones (10–12). In fact, at least in these systems, a small number of clones are very efficiently expanded and contribute to the vast majority of the effectors. In contrast, many of the elicited T cells are poorly expanded and only account for a minor part of the response. The present work was undertaken in order to understand why so few clones are expanded and to identify the parameters that shape the clonal hierarchy of T cell response.
Why does a given clone become dominant in the immune response? One hypothesis might be that the composition of the immune repertoire is a magnified image of the naive repertoire. In other words, a dominant T cell clone in the response would be highly represented in the naive repertoire, and a large number of naive cells corresponding to the same T cell clone would be primed after immunization. In contrast, T cells derived from a single precursor could not contribute significantly to the immune response. We tested this hypothesis by using hemisplenectomized animals and compared TCR-β chains found in the CW3-specific immune repertoire with the TCR-β chains present before immunization. Clearly, our data do not support this hypothesis. In the hemisplenectomized animals, the immune T cell repertoire was isolated using Kd-CW3 tetrameric complexes and characterized by TCR sequencing. In the immune repertoire of mice A and B, we identified 10 and 6 CDR3β sequences, respectively. Their abundance was highly variable, indicating that some immune CTL clones are much more numerous than others (Table I). In the same mice, before immunization, we found that 11 and 10 TCR-β chain sequences (in mouse A and B, respectively) displayed the hallmark of CW3 specificity (BV10 and BJ1.2 usage, a 6 aa–long CDR3β, and the SXG motif). Most of these sequences (9/11 for mouse A, 9/10 for mouse B) are not found in the immune response. The fact that some CTL precursors bearing an appropriate β chain are not expanded during the response could be explained by a pairing with an α chain that does not provide CW3 specificity. It is also possible that some of the CW3-specific T cells are not expanded because they did not encounter the antigen. In the immune repertoire, several distinct nucleotide sequences displaying BV10, BJ1.2, and 6 aa–long CDR3β with the SXG motif were identified. Most of them (4/6 for mouse A, 2/3 for mouse B) were not observed in the naive spleen. This could be explained by the arrival of new T cell precursors from the thymus after the hemisplenectomy. However, immunization was performed only 1 wk after the hemisplenectomy, and the release of new T cells from the thymus has been shown to be low (24). Therefore, it is unlikely that most of the clones have emerged from the thymus in 1 wk. We favor the hypothesis that these cells were represented at extremely low frequencies (one to six cells in the entire animal) before immunization, preventing their detection by our approach. In contrast, some of these sequences are displayed by 2 × 105 cells in the immune animal (Fig. 2). The remaining immune sequences were also found in the half of the spleen removed before immunization. We estimated that about five to six cells bore the indicated TCR-β chains in the removed part of the spleen (Fig. 2). In the immune spleen, they have expanded to 4 × 104 and 2.8 × 105 cells (mouse A, sequences a5 and a6, respectively) and to 6 × 104 cells (mouse B, sequence b7). If the abundance of T cell clones in the immune repertoire was proportional to their frequencies before immunization, the TCR-β chains of the most abundant clones should be found at high frequency in the naive repertoire. This was not the case, and no correlation was observed between naive and immune TCR-β sequence frequencies. In conclusion, this analysis shows that abundant immune T cell clones are not of particularly high frequency before immunization.
Variations in the proliferative capacity of individual T cell clones could generate large differences in their contribution to the immune repertoire. If so, the fastest proliferating T cell clones would dominate the immune response. It is unknown whether a competition between the various T lymphocytes occurs during T cell expansion. In a recent study, it was proposed that during a secondary response, expansion of memory T cells is selective, leading to a narrowing of the T cell repertoire (15). Whether such a selection occurs during the primary expansion has not been addressed until now because of the lack of means to detect and characterize T cell repertoires at an early stage. In our experimental system, the sensitivity of Immunoscope analyses allows us to detect clonal expansions 5 d before the peak of the response. Conservation of the antigen-specific repertoire during the expansion was observed at two levels of resolution. First, by analyzing the CDR3β size profiles using various BJ primers, we showed that the pattern of TCR usage is conserved between days 6 and 10. Second, analysis of the clonal composition of the CW3 response revealed a remarkable conservation during the T cell expansion (Table III). If large differences exist in the proliferative capacity of the early detected clones, one would expect a narrowing of the repertoire as the T cells expand. In contrast, we showed that the complexity is conserved during the expansion (Table III). In good agreement with these results, we have quantified the expansion of various CW3-specific T cell subpopulations in the same animal and showed that all of them displayed a concurrent expansion, independent of their TCR sequences. We cannot formally exclude that all T cells specific for Kd-CW3 have the same avidity; however, this is unlikely, since CW3-specific clones use various TCR-α chains and to a lesser extent different TCR-β chains (21). In addition, differences in antigen recognition efficiency have been reported for various CW3-specific T cell clones (25). Therefore, in our system, TCR affinities do not play a major role once T cells have begun to proliferate.
It has been suggested that individual differences in the orientation of the CTL response against P815 could be explained by the stochastic timing of recruitment of the various epitope-specific T cells (26). We tested directly the impact of the timing of T cell recruitment on antigen-specific expansions. Using transfer experiments, we showed that a 48-h difference in timing of entry in the immune repertoire dramatically affects the extent of clonal expansion (Fig. 7). In addition, we found that each specific T cell clone preexists at very low frequency in the naive repertoire (generally less than five cells). Accordingly, it is unlikely that these rare T cells encounter the antigen simultaneously. Therefore, only the first elicited T cells could account for a significant part of the immune repertoire. Due to the short doubling time of proliferating T cells (estimated to be down to 6–8 h), a 1-d difference in T cell recruitment could lead to a 16-fold difference in the extent of expansion (2).
Several parameters could affect the clonal composition of the immune repertoires. One is the frequency of the various specific T cell precursors before immunization. Our analysis of naive and immune TCR-β chains in an hemisplenectomized mouse ruled out this possibility, since the most abundant clones in the immune animals were not the largest ones before immunization. An alternative hypothesis would be that various T cell precursors proliferate at different rates, depending for instance on their avidity for the MHC–peptide complex. Here, we showed a remarkable stability of the T cell repertoire during the expansion. Therefore, it appears that, once T cell clones have entered the immune repertoire, their respective contribution is conserved. Finally, we demonstrate that timing of T cell recruitment has a major impact on T cell expansion. Therefore, the major parameter that determines the hierarchy of the various elicited clones is their time of entry in the immune repertoire, i.e., the time of first cell division. Whether high-affinity T cells are first recruited because of a faster activation time or whether timing of recruitment is a purely stochastic event remains to be elucidated.
Acknowledgments
The authors wish to thank D. Laouini and A. Freitas for their skillful expert assistance in the animal experiments, and A. Cumano, J. Kanellopoulos, and I. Motta for critical reading of the manuscript.
This work was supported by grants from L'Association pour la Recherche sur le Cancer, La Ligue Nationale contre le Cancer (Subvention: Axe Immunologie des Tumeurs), and the European Community. P. Bousso was a recipient of a fellowship from the Délégation Générale de l'Armement.
Abbreviations used in this paper
- aa
amino acid(s)
- EI
expansion index
References
- 1.Townsend AR, Rothbard J, Gotch FM, Bahadur G, Wraith D, McMichael AJ. The epitopes of influenza nucleoprotein recognized by cytotoxic T lymphocytes can be defined with short synthetic peptides. Cell. 1986;44:959–968. doi: 10.1016/0092-8674(86)90019-x. [DOI] [PubMed] [Google Scholar]
- 2.Murali-Krishna K, Altman JD, Suresh M, Sourdive D, Zajac AJ, Miller JD, Slansky J, Ahmed R. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 3.Busch DH, Pilip IM, Vijh S, Pamer EG. Coordinate regulation of complex T cell populations responding to bacterial infection. Immunity. 1998;8:353–362. doi: 10.1016/s1074-7613(00)80540-3. [DOI] [PubMed] [Google Scholar]
- 4.Kuroda MJ, Schmitz JE, Barouch DH, Craiu A, Allen TM, Sette A, Watkins DI, Forman MA, Letvin NL. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus–infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I–peptide complex. J Exp Med. 1998;187:1373–1381. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallimore A, Glithero A, Godkin A, Tissot AC, Pluckthun A, Elliott T, Hengartner H, Zinkernagel R. Induction and exhaustion of lymphocytic choriomeningitis virus–specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I–peptide complexes. J Exp Med. 1998;187:1383–1393. doi: 10.1084/jem.187.9.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altman JD, Moss P, Goulder P, Barouch DH, McHeyzer WM, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 7.Callan MFC, Tan L, Annels N, Ogg GS, Wilson JDK, O'Callaghan CA, Steven N, McMichael AJ, Rickinson AB. Direct visualization of antigen-specific CD8+T cells during the primary immune response to Epstein-Barr virus in vivo. J Exp Med. 1998;187:1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butz EA, Bevan MJ. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity. 1998;8:167–175. doi: 10.1016/s1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMichael AJ, O'Callaghan CA. A new look at T cells. J Exp Med. 1998;187:1367–1371. doi: 10.1084/jem.187.9.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maryanski JL, Jongeneel CV, Bucher P, Casanova JL, Walker PR. Single-cell PCR analysis of TCR repertoires selected by antigen in vivo: a high magnitude CD8 response is comprised of very few clones. Immunity. 1996;4:47–55. doi: 10.1016/s1074-7613(00)80297-6. [DOI] [PubMed] [Google Scholar]
- 11.Naumov YN, Hogan KT, Naumova EN, Pagel JT, Gorski J. A class I MHC-restricted recall response to a viral peptide is highly polyclonal despite stringent CDR3 selection. Implications for establishing memory T cell repertoires in real-world conditions. J Immunol. 1998;160:2842–2852. [PubMed] [Google Scholar]
- 12.Bousso P, Casrouge A, Altman JD, Haury M, Kanellopoulos J, Abastado JP, Kourilsky P. Individual variations in the murine T cell response to a specific peptide reflect variability in naive repertoires. Immunity. 1998;9:169–178. doi: 10.1016/s1074-7613(00)80599-3. [DOI] [PubMed] [Google Scholar]
- 13.McHeyzer-Williams MG, Davis MM. Antigen-specific development of primary and memory T cells in vivo. Science. 1995;268:106–111. doi: 10.1126/science.7535476. [DOI] [PubMed] [Google Scholar]
- 14.Sourdive DJD, Muralikrishna K, Altman JD, Zajac AJ, Whitmire JK, Pannetier C, Kourilsky P, Evavold B, Sette A, Ahmed R. Conserved T cell receptor repertoire in primary and memory CD8 T cell responses to an acute viral infection. J Exp Med. 1998;188:71–82. doi: 10.1084/jem.188.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busch DH, Pilip I, Pamer EG. Evolution of a complex T cell receptor repertoire during primary and recall bacterial infection. J Exp Med. 1998;188:61–70. doi: 10.1084/jem.188.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker PR, Wilson A, Bucher P, Maryanski JL. Memory TCR repertoires analyzed long-term reflect those selected during the primary response. Int Immunol. 1996;8:1131–1138. doi: 10.1093/intimm/8.7.1131. [DOI] [PubMed] [Google Scholar]
- 17.Vijh S, Pamer EG. Immunodominant and subdominant CTL responses to Listeria monocytogenesinfection. J Immunol. 1997;158:3366–3371. [PubMed] [Google Scholar]
- 18.Maryanski J, Accolla R, Jordan B. H-2- restricted recognition of cloned HLA class I gene products expressed in mouse cells. J Immunol. 1986;136:4340–4347. [PubMed] [Google Scholar]
- 19.Pannetier C, Cochet M, Darche S, Casrouge A, Zöller M, Kourilsky P. The sizes of the CDR3 hypervariable regions of the murine T cell receptor β chains vary as a function of the recombined germ-line segments. Proc Natl Acad Sci USA. 1993;90:4319–4323. doi: 10.1073/pnas.90.9.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacDonald HR, Casanova JL, Maryanski JL, Cerottini JC. Oligoclonal expansion of major histocompatibility complex class I–restricted cytolytic T lymphocytes during a primary immune response in vivo: direct monitoring by flow cytometry and polymerase chain reaction. J Exp Med. 1993;177:1487–1492. doi: 10.1084/jem.177.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casanova J-L, Cerottini J-C, Matthes M, Necker A, Gournier H, Barra C, Widmann C, MacDonald HR, Lemonnier F, Malissen B, Maryanski JL. H-2 restricted cytolytic T lymphocytes specific for HLA display T cell receptors of limited diversity. J Exp Med. 1992;176:439–447. doi: 10.1084/jem.176.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pannetier C, Even J, Kourilsky P. T-cell repertoire diversity and clonal expansions in normal and clinical samples. Immunol Today. 1995;16:176–181. doi: 10.1016/0167-5699(95)80117-0. [DOI] [PubMed] [Google Scholar]
- 23.Casanova J-L, Maryanski JL. Antigen-selected T-cell receptor diversity and self-nonself homology. Immunol Today. 1993;14:391–394. doi: 10.1016/0167-5699(93)90140-G. [DOI] [PubMed] [Google Scholar]
- 24.Tough DF, Sprent J. Turnover of naive- and memory-phenotype T cells. J Exp Med. 1994;179:1127–1135. doi: 10.1084/jem.179.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casanova J-L, Martinon F, Gournier H, Barra C, Pannetier C, Regnault A, Kourilsky P, Cerottini J-C, Maryanski JL. T cell receptor selection by and recognition of two class I major histocompatibility complex–restricted antigenic peptides that differ at a single position. J Exp Med. 1993;177:811–820. doi: 10.1084/jem.177.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brichard VG, Warnier G, Van Pel A, Morlighem G, Lucas S, Boon T. Individual differences in the orientation of the cytolytic T cell response against mouse tumor P815. Eur J Immunol. 1995;25:664–671. doi: 10.1002/eji.1830250306. [DOI] [PubMed] [Google Scholar]