Abstract
Determinants of membrane targeting of Rho proteins were investigated in live cells with green fluorescent fusion proteins expressed with or without Rho-guanine nucleotide dissociation inhibitor (GDI)α. The hypervariable region determined to which membrane compartment each protein was targeted. Targeting was regulated by binding to RhoGDIα in the case of RhoA, Rac1, Rac2, and Cdc42hs but not RhoB or TC10. Although RhoB localized to the plasma membrane (PM), Golgi, and motile peri-Golgi vesicles, TC10 localized to PMs and endosomes. Inhibition of palmitoylation mislocalized H-Ras, RhoB, and TC10 to the endoplasmic reticulum. Although overexpressed Cdc42hs and Rac2 were observed predominantly on endomembrane, Rac1 was predominantly at the PM. RhoA was cytosolic even when expressed at levels in vast excess of RhoGDIα. Oncogenic Dbl stimulated translocation of green fluorescent protein (GFP)-Rac1, GFP-Cdc42hs, and GFP-RhoA to lamellipodia. RhoGDI binding to GFP-Cdc42hs was not affected by substituting farnesylation for geranylgeranylation. A palmitoylation site inserted into RhoA blocked RhoGDIα binding. Mutations that render RhoA, Cdc42hs, or Rac1, either constitutively active or dominant negative abrogated binding to RhoGDIα and redirected expression to both PMs and internal membranes. Thus, despite the common essential feature of the CAAX (prenylation, AAX tripeptide proteolysis, and carboxyl methylation) motif, the subcellular localizations of Rho GTPases, like their functions, are diverse and dynamic.
Keywords: Rho, Rac, Cdc42hs, RhoGDI, green fluorescent protein
Introduction
Rho proteins are Ras-related GTPases that regulate a variety of cellular processes. More than fifteen mammalian Rho proteins have been described including RhoA–E and G, Rac1–3, two isoforms of Cdc42hs, and TC10. Originally identified as genes homologous to Ras, a great amount of interest in Rho proteins was awakened when Ridley and Hall 1992 discovered that RhoA, Rac1, and Cdc42hs differentially regulate the actin cytoskeleton (Ridley et al. 1992; Nobes and Hall 1995). Subsequently, it was recognized that signaling pathways for transcriptional activation, including the JNK/stress-activated protein kinase (Coso et al. 1995; Minden et al. 1995) and p38 (Minden et al. 1995) mitogen-activated protein kinase cascades and the serum response factor (SRF) pathway (Hill et al. 1995), are regulated by Rho family GTP-ases. Moreover, Ras-mediated cellular transformation is dependent on Rho GTPases (Qiu et al. 1995). Other cellular processes shown to be regulated by Rho GTPases include the phagocyte NADPH oxidase (Abo et al. 1991; Knaus et al. 1991), macrophage phagocytosis (Caron and Hall 1998), endocytosis (Lamaze et al. 1996; Leung et al. 1999; Jou et al. 2000), epithelial cell polarization (Kroschewski et al. 1999), and morphogenesis (Barrett et al. 1997).
Like all regulatory GTPases, Rho proteins are activated by guanine nucleotide exchange factors (GEFs). A large number of GEFs for Rho proteins have been identified, most of which contain a domain homologous to one found to have GEF activity in the Dbl oncogene (Cerione and Zheng 1996). The list of effectors of Rho proteins has expanded even more rapidly than the list of GEFs and includes numerous kinases and actin regulatory proteins (Aspenström 1999). Two classes of negative regulators of Rho proteins have been described. Like Ras proteins, Rho proteins have relatively slow intrinsic rates of GTP hydro-lysis that can be accelerated by a family of GTPase activating proteins (Lamarche and Hall 1994). Finally, a class of proteins shown to inhibit the release of GDP from Rho proteins, designated guanine nucleotide dissociation inhibitors (GDIs), have been described (Fukumoto et al. 1990). Perhaps more important than their effects on the GDP/GTP cycle is the ability of GDIs to solubilize membrane-associated Rho proteins (Isomura et al. 1991).
Like Ras proteins and G protein γ subunits, Rho GTPases are synthesized as cytosolic proteins but have the capacity to associate with membranes by virtue of a series of posttranslational modifications of a COOH-terminal CAAX (prenylation, AAX tripeptide proteolysis, and carboxyl methylation) motif (Clarke 1992). Unlike Ras proteins, prenylated Rho proteins can be sequestered in the cytosol by their interaction with RhoGDI, and the capacity to cycle on and off membranes conferred by this interaction is thought to be integral to their biological activity. Consistent with this extra variable, the subcellular localization of Rho family proteins has proven to be more complex than that of Ras proteins (Boivin and Beliveau 1995), perhaps reflecting the more varied functions of Rho proteins. Rho proteins have been localized in cytosol (Abo et al. 1991; Knaus et al. 1991; Adamson et al. 1992), plasma membranes (PMs) (Boivin and Beliveau 1995), including cholesterol-rich microdomains (Michaely et al. 1999), subplasmalemmal actin mesh (Robertson et al. 1995), Golgi (Erickson et al. 1996), endosomes (Adamson et al. 1992), multivesicular bodies (Robertson et al. 1995), and nuclei (Baldassare et al. 1997). Moreover, upon activation, Rho proteins have been shown to translocate from cytosol to membranes (Philips et al. 1993; Boivin and Beliveau 1995; Fleming et al. 1996; Kranenburg et al. 1997).
Prenylation of the CAAX motif targets proteins specifically to the endomembrane where they are proteolyzed and methylated (Choy et al. 1999). Ras proteins require a second signal for transport from the endomembrane to the PM. For N-Ras and H-Ras, this signal consists of one or two cysteines upstream of the CAAX motif in the hypervariable region that are modified by palmitic acid. In the case of K-Ras4B, the second signal is a polybasic region adjacent to the CAAX motif (Hancock et al. 1990). These two types of second signals engage distinct pathways to the PM, one that involves vesicular transport (N- and H-Ras) and one that does not (K-Ras4B) (Choy et al. 1999; Apolloni et al. 2000). Although RhoB is believed to be palmitoylated (Adamson et al. 1992) and other members of the Rho family have polybasic regions upstream of their CAAX motifs, the function of the secondary membrane targeting motif has remained largely unexplored. Indeed, the requirement for engaging transport pathways to the PM other than that afforded by binding cytosolic RhoGDI is not established.
Examination of the subcellular localization of green fluorescent protein (GFP)-tagged Ras proteins in live cells recently revealed unexpected results that forced a reevaluation of Ras trafficking (Choy et al. 1999). We have applied this technology to seven Rho family proteins and fragments, mutants, and chimeras thereof to analyze determinants of their subcellular distribution. We found that although the hypervariable region determines to which membrane compartment each protein is targeted, this targeting is regulated by binding to RhoGDI in the case of RhoA, Rac1, Rac2, and both isoforms of Cdc42hs but not RhoB or TC10. Although RhoB localized like H-Ras to the PM and Golgi, TC10 localized to PMs and endosomes. Also, we show that RhoA is sequestered in the cytosol by a RhoGDI-dependent and -independent mechanisms, that RhoGDI binding is not affected by the length of the prenyl chain (farnesylation versus geranylgeranylation), and that palmitoylation blocks RhoGDI binding. Finally, we were surprised to find that point mutations that render RhoA, Cdc42hs, or Rac1 either constitutively active or dominant negative, abrogate binding to RhoGDI and redirect expression to both PM and internal membranes.
Materials and Methods
Cell Culture and Transfection
COS-1, CHO, NIH3T3, MDCK, and ECV304 cells were obtained from American Type Culture Collection. Porcine aortic endothelial (PAE) cells were obtained from Lena Claesson-Welsh (Chidren's Hospital, Boston, MA). All cells were grown in DME containing 10% FBS (Cellgro) at 5% CO2 and 37°C. Metabolic labeling with [3H]methyl-l-methionine (Amersham Pharmacia Biotech) for methylation assays was performed in methionine-free media, and metabolic labeling with 16-[125I]iodohexadecanoic acid (Marilyn Resh, Memorial Sloane-Kettering Cancer Institute, New York, NY) for palmitoylation was performed with dialyzed FCS. For microscopy, cells were plated, transfected, and imaged in the same 35-mm culture dish that incorporated a no. 1.5 glass coverslip–sealed 15-mm cut out on the bottom (MatTek). All transfections were performed 1 d after plating at 50% confluence using SuperFect™ according to the manufacturer's instructions (QIAGEN). Unless otherwise noted, 0.5 μg of DNA was used for each 35-mm dish or 2 μg for each 10-cm dish. In some experiments, brefeldin A (BFA) or 2-bromopalmitate (2BP) (Sigma-Aldrich) was added at the time of transfection. Stably expressing cell lines were established by selecting transfected cells with 0.8 mg/ml G418 (Life Technologies) for ≥2 wk followed by fluorescence-activated cell sorting for GFP expression. Cells were then G418 selected for another 2 wk. Unless otherwise noted, for coexpression of RhoGDIα, a 1:1 plasmid/DNA ratio was used for coimmunoprecipitation and a 2:1 ratio (RhoGDIα/GTPase) was used for imaging. Control transfections omitting RhoGDIα contained an equivalent amount of vector DNA. Transiently transfected cells were analyzed 1 d after transfection.
Plasmids
cDNAs for the placental isoform of Cdc42hs (pCdc42hs), Rac1, Rac2, RhoA, RhoB, and RhoGDIα were obtained from Richard Cerione (Cornell University, Ithaca, NY), Gary Bokoch (Scripps Research Institute, La Jolla, CA), and Alan Hall (University College London, London, UK). TC10 was cloned from human teratocarcinoma cDNA as described previously (Drivas et al. 1990). The brain isoform of Cdc42hs (bCdc42hs) was cloned from a human fetal brain cDNA library (CLONTECH Laboratories, Inc.) by PCR using 5′-GCGCGAATTCTGATGCAGACAATTAAGTGT forward and 5′-GCGGGGCCCTTAGAATATACAGCACTTCCT reverse primers. The cDNA for each GTPase was subcloned into pEGFP-C3 (CLONTECH Laboratories, Inc.) either by restriction digest or by PCR amplification of the ORF using primers that incorporated desired restriction sites (e.g., 5′ Hind III and 3′ EcoRI). Truncations and point mutations were generated by PCR and cloned into pEGFP-C3. cDNAs encoding chimeric proteins were generated by two step PCR in which 5′ and 3′ fragments with the desired complementary overhangs were generated in the first round of amplification and mixed and joined with a second round of amplification. All constructs were verified by sequencing.
Fluorescence Microscopy
Cells for indirect immunofluorescence were plated into 6-well trays (6 × 35 mm) at a density of 2 × 105 cells per well, containing four coverslips per well. For colocalization studies, the cells were transfected with GFP-tagged Rho proteins the next day using SuperFect™. Cells on coverslips were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 or 0.5% saponin, or fixed and permeabilized with ice-cold methanol/acetone (1:1, vol/vol) and blocked with 1% milk/0.5% Tween in PBS. For localization of endogenous proteins, the cells were stained with polyclonal antisera to Cdc42hs, RhoA, RhoB, or RhoGDIα (Santa Cruz Biotechnology, Inc.) or an anti-Rac1 mAb (Transduction Laboratories). For colocalization, transfected cells were stained with antisera to mannosidase II (Keley Moremen, University of Georgia, Atlanta, GA), or lysosome-associated membrane protein (LAMP)1 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) followed by Texas red–conjugated secondary antisera (Jackson ImmunoResearch Laboratories), and mounted on glass slides with Mowiol. For colocalization in live cells, BODIPY TR-ceramide (10 μg/ml) or Texas red–conjugated transferrin (100 μg/ml) (Molecular Probes) was incubated with the cell for 30 min before imaging. Live cells were examined 12–24 h after transfection with an Axioscope epifluorescence microscope (63× PlanApo 1.4 NA objective; ZEISS) equipped with a Princeton Instruments cooled charge-coupled device (CCD) camera and MetaMorph™ digital imaging software (Universal Imaging Corp.) or a 510 laser scanning confocal microscope (100× PlanApo 1.4 NA objective; ZEISS). Digital images were processed with Adobe Photoshop® v5.0.
Subcellular Fractionation
COS-1 cells were grown to confluence, scraped into 2 ml of hypotonic buffer (10 mM Hepes, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.25 M sucrose, 2 mM PMSF, 10 μg/ml each chymotrypsin, pepstatin, and antipain, 27 μg/ml aprotinin) and disrupted with a ball-bearing homogenizer using 30 passes. Homogenates were cleared of nuclei and unbroken cells (2,500 g, 5 min) and total membrane (P100) and cytosol (S100) were separated by centrifugation (100,000 g, 90 min). Cytosolic and membrane fractions (1 vs. 4 cell equivalents) were analyzed by SDS-PAGE and immunoblotting using polyclonal antisera to RhoGDIα (1:500), RhoA (1:200), RhoB (1:200), or Cdc42 (1:200), or an mAb to Rac1 (1:200) followed by rabbit anti–mouse Ig (1:1,000) and then 125I–protein A (2.5 × 10−4 mCi/ml). Immunolocalized proteins were visualized and quantitated by a PhosphorImager (Molecular Dynamics).
Coimmunoprecipitation
MDCK cells plated in a 10-cm plate were transiently transfected with pEGFP-C3 containing the desired Rho GTPase insert and either pcDNA3.1-RhoGDIα or pcDNA3.1 lacking an insert (1:1 ratio of plasmid DNA). 1 d after transfection, the plates were lysed in 500 μl 1× RIPA (1% Triton X-100, 50 mM Tris, pH 7.4, 150 mM NaCl, 2 mM PMSF, 10 μg/ml each chymotrypsin, pepstatin, and antipain, 27 μg/ml aprotinin). Debris was removed by centrifugation. The supernatant was mixed with an anti-GFP mAb (Boehringer) at 1:500 for 1 h at 4°C. Protein G–agarose beads (20 μl of 1:1 suspension in 1× RIPA; CLONTECH Laboratories, Inc.) was then added for 1 h. Agarose beads were washed two times with 1× RIPA and two times with PBS. Proteins were eluted with 30 μl SDS sample buffer containing 0.5% β-mercaptoethanol (CLONTECH Laboratories, Inc.) and analyzed by immunoblot for RhoGDIα (21–30-kD region; 1:200 anti-RhoGDIα antiserum) and GFP-GTPase (42–66-kD region; 1:200 anti-GFP antiserum) using 125I–protein A (2.5 × 10−4 mCi/ml in phosphotungstic acid) as a secondary reagent. Immunolocalized proteins were visualized and quantitated by a PhosphorImager.
Quantitation of Endogenous Rho GTPases
Recombinant Rac1, Cdc42hs, RhoA, and RhoGDIα were produced as glutathione S-transferase (GST) fusion proteins (pGEX2T) in Escherichia coli and cleaved from GST with thrombin. The concentrations of each recombinant protein were determined by densitometric analysis of Coomassie blue–stained gels using BSA as a standard. MDCK, COS, and ECV cells were plated on 10-cm plates and grown to confluence. Cells were lysed in 250 μl of SDS sample buffer containing 0.5% β-mercaptoethanol (CLONTECH Laboratories, Inc.). Various amounts of lysate (2–10 μl) were analyzed alongside titrated recombinant protein standards by 14% Tris-glycine SDS-PAGE (Novex) and immunoblotted using antisera to Cdc42hs, Rac1, RhoA, or RhoGDIα followed by 125I–protein A. Immunodetected proteins were quantitated by a PhosphorImager, and cellular concentrations of the various proteins were calculated.
SRF Assay
COS-1 cells were plated in 6-well plates. The next day the cells were transfected using SuperFect™ with 0.2 μg/well SRE-Luciferase plasmid DNA (Promega), 0.4 μg/well of pEGFP DNA containing inserts for wild-type pCdc42hs, pCdc42hs61L, or pCdc42hs12V, and 0.8 μg/well of DNA from either pcDNA3.1 or pcDNA3.1-RhoGDIα. After 3–4 h of recovery, cells were subjected to serum starvation overnight. The next day each well was lysed in 100 μl CCLR detergent. Debris was removed by centrifugation, and 5 μl of each supernatant was mixed with 100 μl Luciferase Assay Reagent (Promega). Luminescence was measured in a luminometer. 20 μl of each supernatant was analyzed by 14% Tris-glycine SDS-PAGE and immunoblotted using antisera to GFP or RhoGDIα and 125I–protein A, and immunolocalized proteins were quantitated by a PhosphorImager. Luminometer determinations were normalized to GTPase expression.
Online Supplemental Material
Four QuickTime® supplemental videos are available at http://www. jcb.org/cgi/content/full/152/1/111/DC1 showing time-lapse (150× speed) images of GFP-H-Ras, GFP-RhoB, GFP-TC10, and GFP-Cdc42hs expressed in COS-1 cells. Time-lapse digital epifluorescent images were captured from live cells with MetaMorph™ imaging software and converted to avi video files that were edited with Adobe Premiere™ v4.0 and compiled as QuickTime® movies.
Results
Differential Localization of GFP-tagged Rho GTPases
To determine the subcellular location of Rho GTPases in live cells, we tagged the NH2 terminus of seven full-length Rho proteins with GFP and expressed them by transient transfection in a variety of cell lines. MDCK cells (Fig. 1) proved especially informative because, when grown to confluence, they assume a semicolumnar, nonoverlapping morphology such that the PM at the edge of the cell is viewed tangentially, and localization of GFP-tagged proteins in this compartment is easily scored. As we reported previously using this system, GFP-K-Ras4B (Fig. 1 a) was expressed in a pattern easily distinguished from GFP-H-Ras (Fig. 1 b) in that, although the former localized only to the PM, the latter localized to the PM and a discrete perinuclear structure that we have identified as Golgi (Choy et al. 1999). GFP-tagged Rho GTPases were observed to localize in at least five distinct patterns.
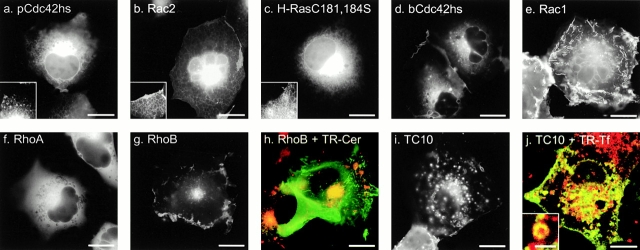
Figure 1.
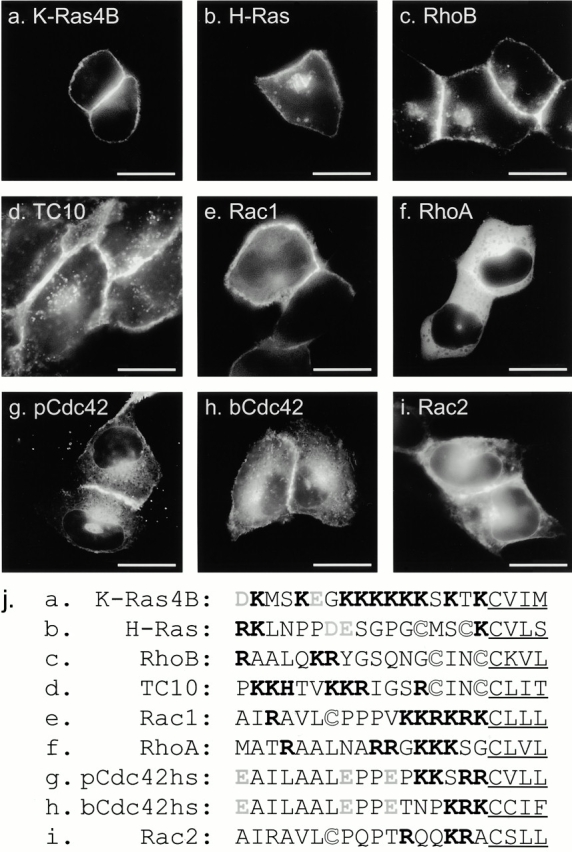
Localization of GFP-tagged Ras and Rho proteins in MDCK cells. (a–i) MDCK cells were transiently transfected with GFP-tagged constructs of the indicated GTPase and imaged alive 24 h later by digital epifluorescence microscopy using a cooled CCD camera. (j) Listed are the hypervariable regions of each GTPase with the CAAX motif underlined, potential palmitoylation sites shown in outline, acidic residues (gray), and basic residues (bold). Bars, 10 μm.
GFP-RhoB (Fig. 1 c) localized in a pattern similar to that of GFP-H-Ras with prominent fluorescence in the PM and in a discrete perinuclear structure. Sensitivity to BFA and colocalization with mannosidase II, βCOP (data not shown), and BODIPY ceremide C6 (Fig. 2 h) confirmed that this structure was Golgi. GFP-TC10 (Fig. 1 d) also localized prominently on the PM and on internal membranes, but in contrast to GFP-H-Ras and GFP-RhoB, the intracellular localization of GFP-TC10 was on cytoplasmic vesicles that colocalized with the endosomal markers LAMP (not shown) and internalized transferrin (see Fig. 2 i). These motile vesicles present throughout the cell accumulated in the perinuclear region overlapping the Golgi but could be distinguished from Golgi by morphology, markers, and lack of sensitivity to BFA. Inspection of the hypervariable regions (Fig. 1 j) of H-Ras, RhoB, and TC10 revealed a CXXC PM targeting motif immediately upstream of the CAAX motif. Unlike H-Ras and RhoB, TC10 has a polybasic region adjacent to the CXXC motif and thus has two potential membrane targeting motifs. The CXXC motif has been shown to be a site for dual palmitoylation in H-Ras (Hancock et al. 1989). We have observed incorporation of label into GFP-RhoB and GFP-TC10 in cells metabolically labeled with 16-[125I]iodohexanoic acid (Webb et al. 2000; data not shown), indicating that, like H-Ras, RhoB and TC10 are palmitoylated. Thus, these data suggest that although palmitoylation of CAAX proteins alone regulates steady-state accumulation in the Golgi, palmitoylation in conjunction with a polybasic region mediates association with endosomes.
Figure 2.
(a–i) Localization of GFP-tagged Ras and Rho proteins in COS-1 cells. COS-1 cells were transfected with the indicated GFP fusion protein and imaged as in Fig. 1. The insets (a–c) show an enlargement of the ER. (h) Cells transfected with GFP-RhoB were treated with BODIPY TR-ceremide for 30 min before imaging, such that the Golgi appears red in nontransfected cells, and colocalization with GFP-RhoB could be scored as yellow. (j) Cells transfected with GFP-TC10 were treated with Texas red–conjugated transferrin for 30 min before imaging, revealing colocalization in endosomes. Motile peri-Golgi vesicles carrying GFP-RhoB (g) and endosomes carrying GFP-TC10 (i) were best compared by time-lapse imaging as QuickTime® movies available at http://www.jcb.org/cgi/content/full/152/1/111/DC1). Compare also the QuickTime® movies of GFP-H-Ras and GFP-pCdc42hs. Bars, 10 μm.
In contrast to GFP-RhoB and GFP-TC10, GFP-Rac1 was observed predominantly in the PM (Fig. 1 e) in a pattern similar to that of GFP-K-Ras4B. Like K-Ras4B, Rac1 has a polybasic region immediately upstream of its CAAX motif, suggesting that this type of second signal engages a distinct trafficking pathway that does not permit steady-state accumulation on internal membranes.
Although RhoA has a polybasic region that is only one amino acid shorter than that of Rac1 (Fig. 1 j), GFP-RhoA was observed almost exclusively in the cytosol, as revealed by numerous negatively imaged organelles (Fig. 1 f) similar to those observed in the cytoplasm of cells expressing GFP alone. Although there was some amorphous perinuclear accumulation of GFP-RhoA, no distinct membrane structures, like those seen with GFP-RhoB, were observed. Most striking was the absence of localization of GFP-RhoA at the PM, despite levels of expression that equaled or exceeded that of the other GFP-tagged GTPases. The cytosolic localization of GFP-RhoA suggests that it is actively retained in the cytosol. Although binding to RhoGDI could account for this retention, the membrane localization of GFP-Rac1 and GFP-pCdc42hs, also known to bind RhoGDI, suggests that other factors may be involved.
To function as a PM targeting motif, the polybasic region of K-Ras4B has been shown to require a net positive charge of four or more (Hancock et al. 1990). Therefore, neither the placental nor the brain isoform of Cdc42hs (pCdc42hs and bCdc42hs), nor Rac2, have obvious PM targeting signals upstream of their CAAX motifs (Fig. 1 j). Nevertheless, each of these constructs, when tagged with GFP and overexpressed, localized, to some extent, at the PM, suggesting a weak second signal. However, more striking than the PM expression was expression of each of these constructs on internal membranes, similar to that observed with GFP-tagged mutants of H-Ras and K-Ras4B that lack the secondary targeting signal (Choy et al. 1999). GFP-pCdc42hs (Fig. 1 g) and GFP-bCdc42hs (Fig. 1 h) localized on the nuclear envelope, on ER emanating from the nuclear envelope, and in the Golgi region. Expression of these constructs on the Golgi was confirmed by colocalization with mannosidase II and BODIPY ceremide C6 (not shown), consistent with prior localization of pCdc42hs in a BFA-sensitive compartment (Erickson et al. 1996). However, localization in the Golgi region was more diffuse than that observed with GFP-H-Ras and GFP-RhoB, apparently due to expression in the surrounding ER. GFP-Rac2 had a similar pattern of localization; however, less protein was observed at the PM, and the ER pattern was more pronounced (Fig. 1 i). These subtle differences suggest that determinants other than a strong polybasic region or a palmitoylation site can influence membrane localization.
To determine if these differential localizations were cell type specific, an identical analysis was performed in COS-1 (Fig. 2), CHO, and ECV cells (not shown) with similar results. Although MDCK cells allowed efficient detection of PM-associated proteins, subconfluent COS-1 cells are more spread and proved superior for analyzing localization on intracellular membranes. Using COS-1 cells, the endomembrane localization of GFP-pCdc42hs (Fig. 2 a) and GFP-Rac2 (Fig. 2 b) was evident, with localization on the nuclear envelope, ER, and Golgi in a pattern indistinguishable from that of GFP-H-RasC181,184S (Fig. 2 c), a mutant that lacks palmitoylation sites. GFP-bCdc42hs also localized predominantly on the Golgi, nuclear envelope, and ER but revealed slightly more expression in PMs and peri-Golgi vesicles (Fig. 2 d). GFP-Rac1 was predominantly expressed on the PM of COS-1 cells with both peripheral and dorsal lamellipodia easily visualized (Fig. 2 e) in a pattern similar to that observed for GFP-K-Ras4B (Choy et al. 1999). However, unlike GFP-K-Ras4B, GFP-Rac1 was also observed in the endomembrane of COS-1 cells, especially the nuclear membrane (Fig. 2 e) and, in some cells, the nucleoplasm (not shown). The distinct expression patterns of GFP-RhoA (Fig. 2 f), GFP-RhoB (Fig. 2g and Fig. h), and GFP-TC10 (Fig. 2i and Fig. j) in COS-1 cells matched that in MDCK cells: cytosolic versus PMs and Golgi versus PMs and endosomes.
Each GFP fusion protein, when expressed in COS-1 cells, was a substrate in vivo for prenylcysteine-directed COOH methyltransferase (pcCMT; not shown). Thus, despite the various steady-state localizations, each construct had access to the endomembrane since this is the only compartment in which pcCMT is expressed (Dai et al. 1998).
COS-1 cells also afforded superior views of the peri-Golgi vesicles previously reported for GFP-H-Ras (Choy et al. 1999). Similar vesicles were observed with GFP-RhoB. These were best viewed by time-lapse microscopy (QuickTime® movies are available at http://www.jcb.org/cgi/content/full/152/1/111/DC1). The peri-Golgi vesicles illuminated by GFP-H-Ras and GFP-RhoB were smaller (<0.1 μm) than the endosomes illuminated by GFP-TC10, moved faster, and did not colocalize with LAMP or internalized transferrin. The tubulovesicular structures illuminated with GFP-H-Ras and GFP-RhoB were similar in morphology and kinetics to the peri-Golgi vesicles observed with the secretory protein VSVG-GFP (Presley et al. 1997). Both the smaller vesicles observed with GFP-H-Ras and GFP-RhoB and the endosomes illuminated by GFP-TC10 were observed to move to and from the Golgi region in a saltatory fashion along linear tracks consistent with microtubules.
Rho Protein Hypervariable Regions Determine Membrane Localization
To determine if, like Ras proteins, the hypervariable region of Rho GTPases contains all of the targeting information necessary to regulate localization on membranes, we tagged the isolated hypervariable regions of pCdc42hs, bCdc42hs, Rac1, and RhoA with GFP (designated GFP-GTPase-tail). GFP-pCdc42hs-tail, GFP-bCdc42hs-tail, and GFP-Rac1-tail were expressed in patterns identical to those of the full-length GFP fusion proteins (Fig. 3, a–c). In contrast, GFP-RhoA-tail (Fig. 3 d) localized in a pattern strikingly distinct from that of the full-length protein, one that was indistinguishable from that of GFP-pCdc42hs and GFP-bCdc42hs. Inspection of the hypervariable region of RhoA (Fig. 1 j) reveals a polybasic region one residue weaker than that of Rac1 and two weaker than that of K-Ras4B, such that it may not constitute a strong PM targeting motif. Thus, the hypervariable region of RhoA appears to be functionally equivalent to that of the Cdc42hs isoforms. GFP-RhoAaa73–193 (Fig. 3 e), a construct with a 73–amino acid NH2-terminal truncation, was expressed in a pattern identical to GFP-RhoA-tail, indicating that the NH2-terminal third of the protein is required for sequestration in the cytosol either by direct involvement in protein–protein interaction or by allowing proper folding. To further confirm that the hypervariable domains were sufficient to determine membrane localization of Rho GTPases, we constructed a GFP-tagged chimera consisting of the NH2-terminal 188 amino acids of TC10 and the COOH-terminal 20 amino acids of pCDC42hs. The localization of this construct (Fig. 3 f) was distinct from that of GFP-TC10 and identical to that of GFP-pCDC42hs, confirming that the hypervariable domain is necessary and sufficient to determine membrane localization. From these data we conclude that the hypervariable domains contain all of the information required to differentially direct Rho GTPases to membrane compartments and that RhoA is unique among the Ras and Rho proteins surveyed in that it contains information in its sequence upstream of the hypervariable domain that causes it to be efficiently sequestered in the cytosol, even when overexpressed.
Figure 3.
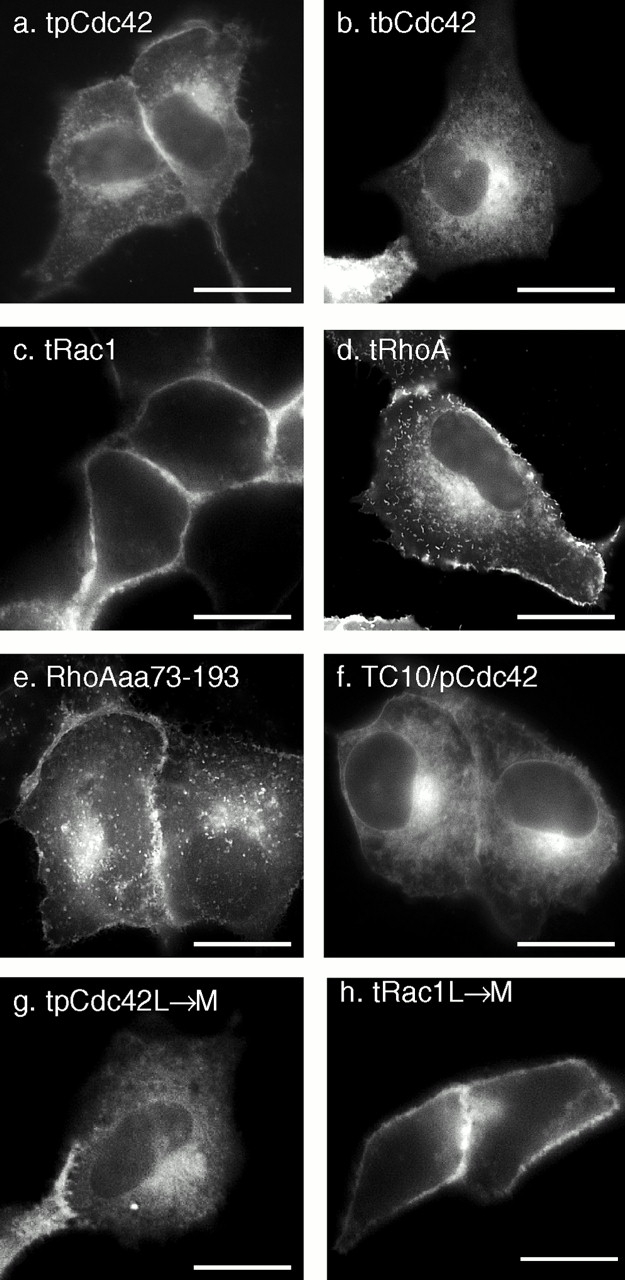
The hypervariable regions of Rho GTPases are sufficient for differential membrane targeting which is independent of the length of the prenyl chain. MDCK cells were transiently transfected with the indicated constructs that included GFP extended at the COOH terminus with the hypervariable regions (last 20 amino acids) of pCdc42hs (a), bCdc42hs (b), Rac1 (c), and RhoA (d) or amino acids 73–193 of RhoA (e). (f) A doubly chimeric fusion protein consisting of GFP followed by amino acids 1–188 of TC10 and then amino acids 172–191 of pCdc42hs. (g and h) The last amino acid of the hypervariable regions of pCdc42hs (g) and Rac1 (h) were changed from L to M to switch modification from geranylgeranylation to farnesylation. Bars, 10 μm.
To determine if the membrane localizations of the GFP fusion proteins were dependent on the geranylgeranyl modification, we constructed mutants in which the COOH-terminal L of GFP-pCdc42hs-tail and GFP-Rac1-tail were changed to M, such that the proteins would instead be modified by a farnesyl group. The localizations of the farnesylated constructs were identical to those of the geranylgeranylated versions (Fig. 3g and Fig. h), indicating that the length of the prenyl chain does not affect membrane targeting.
Expression Level Affects Localization of GFP-tagged Rho GTPases
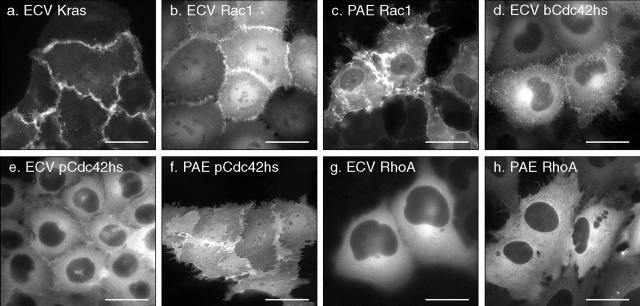
The subcellular localizations described above were determined by transient transfection using the pEGFP vector that results in overexpression of the GFP-tagged protein. Although this system is informative with regard to differential membrane targeting, the results may not accurately reveal the localization of endogenous Rho GTPases because overexpression may overwhelm the capacity of endogenous RhoGDI to regulate localization. To address this problem, we established, using neomycin and cytofluorometric selection, cell lines that stably express GFP-tagged Rho proteins at four- to eightfold lower levels (determined by cytofluorimeter and immunoblot) than those observed in transient transfections (Fig. 4). Like cells transiently transfected with GFP-Rac1, ECV uroepithelial cells stably expressing GFP-Rac1 (Fig. 4 b) showed predominant PM localization, particularly in peripheral lamellipodia. However, unlike the transiently transfected cells, the cell lines revealed a greater proportion GFP-Rac1 in the cytosol and nucleoplasm, consistent with sequestration from membranes by binding to RhoGDI. ECV cells stably expressing GFP-K-Ras4B at similar levels revealed only PM localization (Fig. 4 a), consistent with the lack of a GDI-like chaperon for K-Ras4B. PAE cells stably expressing GFP-Rac1 revealed localization in PM lamellipodia, cytosol, and nuclear envelope but exclusion from the nucleoplasm (Fig. 4 c), suggesting that the nuclear localization is cell type specific. Similarly, the localization of GFP-bCdc42hs in stably transfected ECV cells paralleled that of the transiently transfected fusion protein except for considerably more cytosolic fluorescence (Fig. 4 d). Both ECV (Fig. 4 e) and PAE (Fig. 4 f) cells stably expressing GFP-pCdc42hs revealed significantly more cytoplasmic localization than that of the transiently overexpressed GFP fusion protein. Interestingly, although GFP-pCdc42hs did not enter the nucleoplasm of ECV cells, it labeled this compartment in PAE cells (Fig. 4 f), confirming the cell type dependence of nuclear localization. Finally, both ECV (Fig. 4 g) and PAE (Fig. 4 h) cells stably expressing GFP-RhoA reveal a pattern of localization identical to that observed by transient transfection: cytoplasmic expression with enhancement in the Golgi region without distinct membrane localization. Thus, transient overexpression of GFP-tagged Rac1 and Cdc42hs underestimates the cytosolic pool but reveals expression in the same membrane compartments observed in cells expressing levels of protein closer to endogenous.
Figure 4.
Localization of K-Ras4B and Rho GTPases in stably transfected cells. Stable transformants of ECV304 or PAE cells expressing the indicated GFP-tagged fusion proteins were selected with G418 and cytofluorometry and imaged alive by digital epifluorescence microscopy using a cooled CCD camera. Bars, 10 μm.
To validate the localization of GFP-tagged Rho GTPases, we performed indirect immunofluorescence analysis on MDCK cells (Fig. 5 a). Although the morphology and resolution of membrane compartments in the fixed permeabilized cells were markedly inferior to those observed in live cells and, more important, the localization patterns were significantly affected by the method of fixation and permeabilization, this analysis had the advantage of permitting localization of endogenous proteins. RhoA was localized diffusely throughout the cytoplasm with some enhancement around the nucleus and no PM staining (Fig. 5 a, i). In cells fixed with paraformaldehyde and permeabilized with Triton X-100, RhoB was observed in vesicular structures (Fig. 5 a, ii) similar to those reported previously, using similar experimental conditions, as early endosomes (Adamson et al. 1992). However, when these cells were permeabilized with saponin, RhoB was localized in a discrete perinuclear compartment (Fig. 5 a, iii) that colocalized with Golgi markers (not shown) and corresponded to the structure seen in live cells transfected with GFP-RhoB (Fig. 1 c). Cdc42hs was localized in the cytoplasm and was markedly enhanced in the Golgi region (Fig. 5 a, iv), consistent with both the previous studies of endogenous Cdc42hs (Erickson et al. 1996) and with the localization of GFP-Cdc42hs (Fig. 1g and Fig. h). Rac1 was localized in the PM as well as the cytoplasm (Fig. 5 a, v). RhoGDI was observed exclusively in the cytosol revealing negatively imaged organelles (Fig. 5 a, vi).
Figure 5.
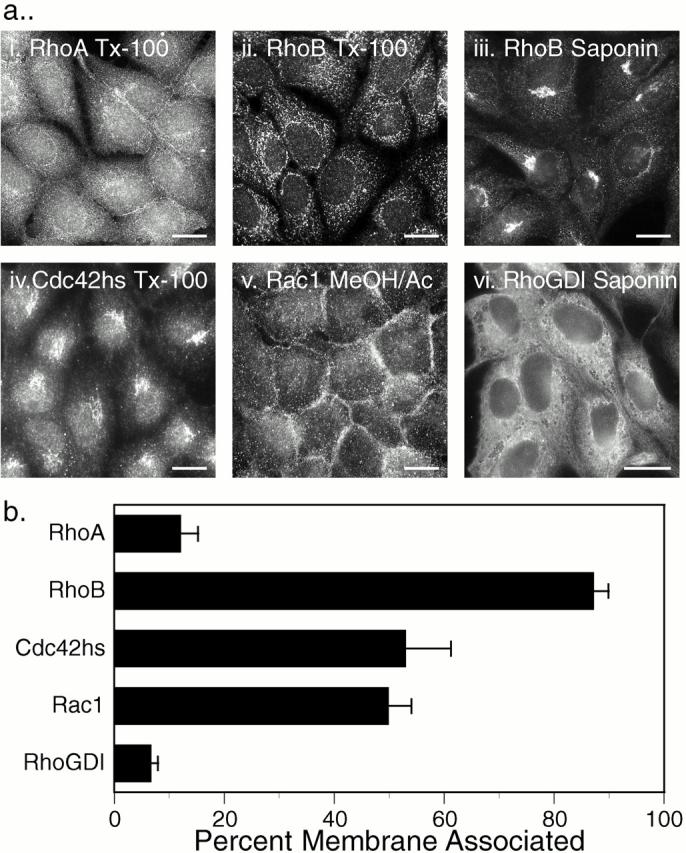
Localization of endogenous Rho GTPases and RhoGDI. (a) Indirect immunofluorescence: untransfected MDCK cells were grown on coverslips, fixed with 3% paraformaldehyde (i–iv, and vi) or ice-cold methanol/acetone (1:1; v), permeabilized with Triton X-100 (i, ii, and iv) or saponin (iii and vi), and stained for the indicated protein as described in Materials and Methods. (b) Subcellular fractionation: COS-1 cell cytosolic and membrane fractions were analyzed by immunoblot for the indicated proteins, and relative amounts were quantitated by a PhosphorImager. Results are expressed as the percentage of the endogenous protein detected in the membrane fraction and are given as mean ± SEM (n = 8). Bars, 10 μm.
To further validate these results, we determined localization of endogenous Rho proteins and RhoGDI in COS-1 cells by subcellular fractionation (Fig. 5 b). Although RhoGDI and RhoA were almost entirely soluble, 87 ± 3% of RhoB was detected in the membrane fraction, confirming both the localization of the GFP-tagged proteins and the indirect immunofluorescent localization of the endogenous counterparts. In contrast to these extremes, endogenous Rac1 and Cdc42hs were equally partitioned between soluble and membrane fractions, consistent with the capacity for both membrane and RhoGDI binding. Thus, the localization of endogenous Rho GTPases paralleled that of the corresponding GFP-tagged proteins, validating the strikingly distinct subcellular localizations of members of this family of GTPases.
Translocation of GFP-tagged Rho GTPases to the PM
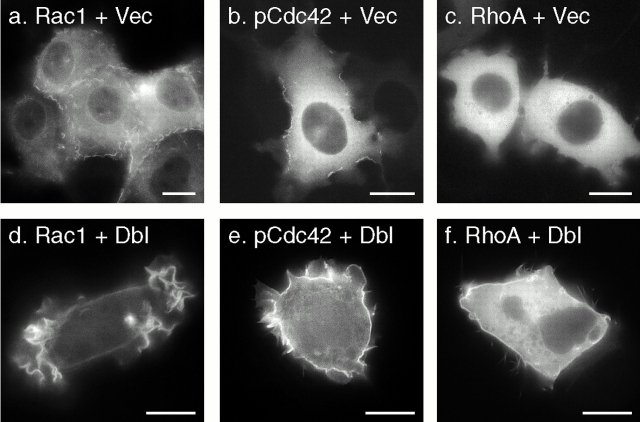
Rho GTPases have been shown to translocate to membranes upon activation (Philips et al. 1993; Boivin and Beliveau 1995; Fleming et al. 1996; Kranenburg et al. 1997). However, most published reports of such translocation relied on cell fractionation into S100 versus P100 pools and thereby could not distinguish which membranes were involved. To determine if translocation could be observed in live cells, we stimulated ECV cells stably transfected with GFP-Rac1, GFP-pCdc42hs, or GFP-RhoA by transiently expressing oncogenic Dbl, a GEF for Cdc42hs and RhoA. As expected, transient expression of Dbl, but not a vector control, induced marked lamellipodia and filopodia formation (Fig. 6), the former presumably mediated by indirect stimulation of Rac1 through activated Cdc42hs. In cells transfected with Dbl, a portion of GFP-Rac1 (Fig. 6 d) and GFP-pCdc42hs (Fig. 6 e) redistributed from the cytosol to the lamellipodia membrane. Although fluorescence in the Dbl-stimulated lamellipodia could be observed with any GFP-tagged protein that labeled PMs (e.g., GFP-Rac1-tail), accumulation of GFP-Rac1 and GFP-pCdc42hs on lamellipodia was accompanied by clearing of cytosolic fluorescence. A similar, but less pronounced, PM translocation with less cytosolic clearing was observed with GFP-RhoA (Fig. 6 f). Similar results were obtained in transient transfections when Dbl was cotransfected with the GFP-tagged Rho protein (not shown). Thus, the GFP-tag did not interfere with recruitment to the PM of Rho GTPases. More important, these data indicate that despite intrinsic affinity of GFP-Cdc42hs and GFP-RhoA-tail for endomembrane, Dbl-stimulated translocation is predominantly directed to the PM, the compartment most likely involved in remodeling of the actin cytoskeleton.
Figure 6.
Translocation of Rho GTPases to lamellipodia. ECV304 cells stably expressing GFP-tagged Rac1 (a and d), pCdc42hs (b and e), or RhoA (c and f) were transiently transfected with oncogenic Dbl (d–f) or vector alone (a–c) and imaged alive after 24 h as in the legend to Fig. 4. Bars, 10 μm.
Binding of GFP-tagged Rho GTPases to RhoGDI In Vivo
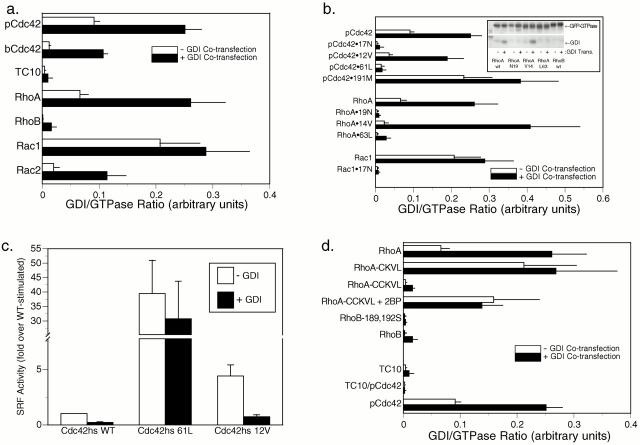
The increase in the cytosolic pool of GFP-Rac1 and GFP-Cdc42hs but not GFP-RhoA in stably transfected cell lines relative to that of transiently transfected cells suggested that the localization of these proteins was regulated by RhoGDI binding in vivo and that the buffering capacity of RhoGDI could be overcome for Rac1 and Cdc42hs but not RhoA. To test this hypothesis directly, we expressed GFP-tagged Rho GTPases at different levels with and without cooverexpression of RhoGDIα (Fig. 7). Although at high levels of expression, GFP-pCdc42hs, GFP-bCdc42hs, GFP-Rac1, and GFP-Rac2 were predominantly observed in membranes (Fig. 7, a ii, c ii, d ii, and e ii), as described above, at lower levels of expression these proteins were largely cytosolic (Fig. 7, a i, c i, d i, and e i). When these proteins were cooverexpressed at high levels with RhoGDIα, the localization was shifted to a cytosolic pattern (Fig. 7, a iii, c iii, d iii, and e iii). From these data we conclude that, as expected, RhoGDIα is a regulator of Cdc42hs and Rac localization. Also, we conclude that the GFP tag at the NH2 terminus of Rho GTPases does not interfere with binding to RhoGDIα. Thus, coexpression of GFP-tagged Rho GTPases and RhoGDIα serves as a powerful assay of in vivo binding.
Figure 7.
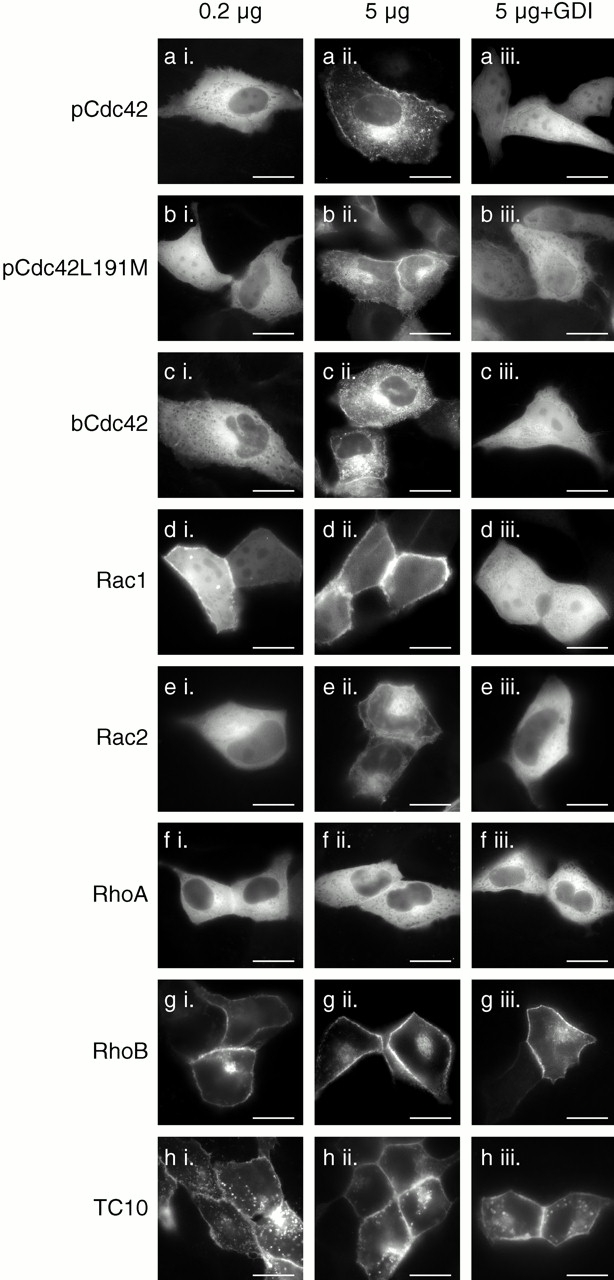
Relative expression levels of Rho GTPases and RhoGDIα affect the localization of some, but not all, Rho proteins. MDCK cells were transiently transfected with 0.2 μg (a i–h i) or 5 μg (a ii–h iii) of plasmid DNA encoding the indicated GFP-tagged GTPase along with 10 μg of vector (pcDNA3.1) DNA (a ii–h ii) or pcDNA3.1-RhoGDIα DNA (a iii–h iii). 24 h after transfection, the cells were imaged alive as in the legend to Fig. 1. Bars, 10 μm.
We used this assay to determine if the geranylgeranyl modification rather than a farnesyl modification is required for binding to RhoGDI in vivo. We constructed a GFP-tagged pCdc42hs mutant in which the COOH-terminal L was switched to M to promote farnesylation. We confirmed that the mutant was farnesylated in our system by demonstrating that, in contrast to GFP-pCdc42hs, membrane localization of GFP-pCdc42hsL191M was blocked by a farnesyltransferase inhibitor (not shown). The expression pattern of GFP-pCdc42hsL191M was identical to that of GFP-pCdc42hs. Both low level expression and cooverexpression with RhoGDIα resulted in cytosolic expression (Fig. 7 b). Thus, despite the ability of the prenyl binding grove of RhoGDIα to accommodate the 20-carbon prenyl chain (Hoffman et al. 2000), the shorter 15-carbon prenyl chain is sufficient for binding.
As expected, the cytosolic pattern of GFP-RhoA was altered neither by expression at low levels nor by cooverexpression of RhoGDIα (Fig. 7 f). The basis for the increased buffering capacity of the cytosol for GFP-RhoA relative to GFP-Rac and GFP-Cdc42hs was explored by determining the relative stoichiometry of RhoA, Rac1, Cdc42hs, and RhoGDI in three cell lines (Table ). Although the endogenous levels of RhoGDI exceeded those of each of the three GTPases individually, the molar sum of RhoA, Rac1, and Cdc42hs was approximately equal to the molar amount of RhoGDI in each of the three cell types, suggesting coordinated regulation between the production of these GTPases and their cytosolic chaperon. Although the near unity of the RhoA plus Rac1 plus Cdc42hs/RhoGDIα molar ratio explains why overexpressing GFP-Rac and GFP-Cdc42hs leads to membrane localization, it does not explain why overexpressing GFP-RhoA does not and suggests that factors other than RhoGDIα contribute to maintaining GFP-RhoA in the cytosol.
Table 1.
Relative Content of Rho GTPases and RhoGDIα in Three Cell Lines
| GDI | RhoA | Rac | Cdc42 | RhoA + Rac + Cdc42 | ||||
|---|---|---|---|---|---|---|---|---|
| Cell line | ng/106 cells | ng/106 cells | GDI/GTPase | ng/106 cells | GDI/GTPase | ng/106 cells | GDI/GTPase | GDI/GTPase |
| MDCK | 283 ± 27 | 56 ± 14 | 5.6 ± 1.1 | 124 ± 27 | 2.3 ± 0.3 | 53 ± 18 | 7.4 ± 2.6 | 1.1 ± 0.1 |
| COS1 | 179 ± 55 | 34 ± 16 | 7.4 ± 1.9 | 82 ± 10 | 2.0 ± 0.6 | 26 ± 11 | 11.6 ± 5.3 | 1.1 ± 0.2 |
| ECV | 154 ± 30 | 50 ± 15 | 3.4 ± 0.7 | 82 ± 14 | 1.8 ± 0.3 | 49 ± 21 | 5.3 ± 2.0 | 0.8 ± 0.1 |
The fact that the molar amount of RhoGDIα was roughly equivalent to that of the sum of RhoA, Rac, and Cdc42hs presented a conundrum in that it suggested that no free RhoGDIα would be available to bind other Rho family GTPases. Therefore, we employed our in vivo RhoGDIα binding assay to determine the binding capacity of two other Rho GTPases, GFP-RhoB (Fig. 7 g) and GFP-TC10 (Fig. 7 h). The localization of these constructs was changed neither by expression at low levels nor cooverexpression of RhoGDIα, suggesting that these proteins do not bind RhoGDIα in vivo. Thus, binding to RhoGDIα is not a universal characteristic of Rho family GTPases.
Palmitoylation Regulates Membrane Targeting and RhoGDI Binding
Recently, 2BP was reported to be an effective inhibitor of protein palmitoylation in vivo (Webb et al. 2000). Therefore, we used 2BP to determine the role of palmitoylation in the membrane targeting of RhoB and TC10. We have shown that a palmitoylation-deficient mutant of GFP-H-Ras, GFP-HrasC181,184S, is targeted to the ER and Golgi but not the PM (Choy et al. 1999) (Fig. 2 c). COS-1 cells expressing GFP-H-Ras in the presence of 2BP revealed a pattern of expression identical to that of GFP-HrasC181,184S (Fig. 8, a and b), thus validating the utility of 2BP in localization assays. The specificity of 2BP for palmitoylated proteins was confirmed by showing that the localization of GFP-Rac1 is unaffected by treatment with 2BP (data not shown). The effect of 2BP on GFP-RhoB was similar to its effect on GFP-H-Ras, causing mislocalization of the fusion protein to the nuclear envelope and ER and blocking PM expression (Fig. 8c and Fig. d). In some cells treated with 2BP, GFP-RhoB accumulated in the cytosol obscuring localization in the ER (not shown). These patterns of expression were identical to those revealed by expression of GFP-RhoB-C189,192S (Fig. 8 e), confirming that the targeting of RhoB to the Golgi and PM, like that of H-Ras, is dependent on palmitoylation and that in the absence of this modification the protein accumulates on ER.
Figure 8.
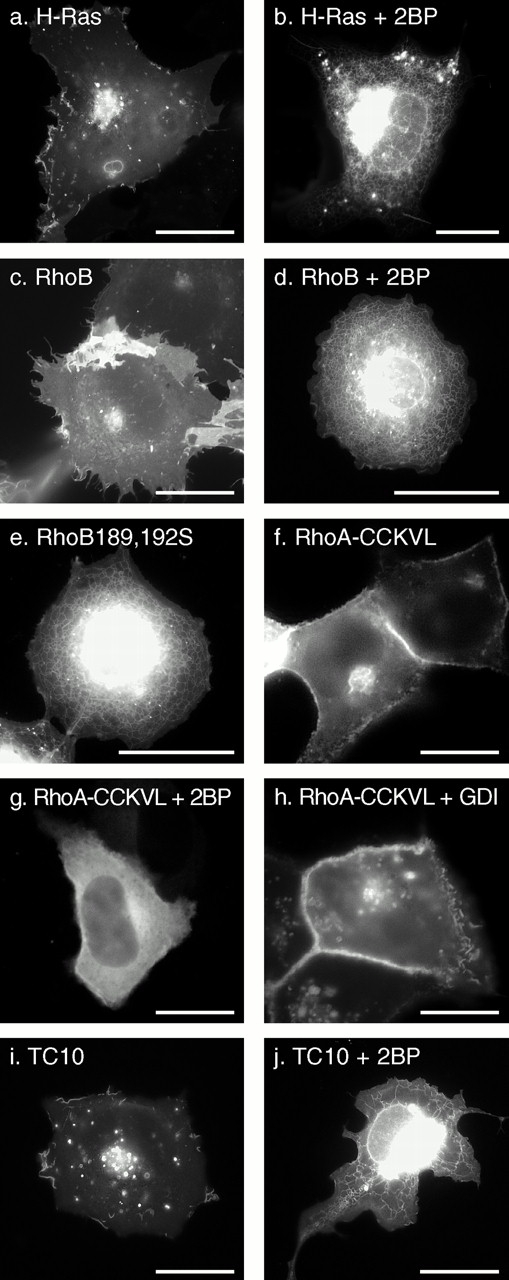
The role of palmitoylation on localization and RhoGDIα binding of Rho GTPases. COS-1 (a–e, i, and j) or MDCK (f–h) cells were transiently transfected with the indicated GFP-tagged GTPase in the absence (a, c, e, f, h, and i) or presence (b, d, g, and j) of 25 μM 2BP and imaged alive after 24 h as described in the legend to Fig. 1. (f–h) GFP-tagged RhoA in which the last four amino acids (CLVL) were switched to the last five amino acids of RhoB (CCKVL) was expressed without (f) or with (g) 2BP or coexpressed with excess (1:2 DNA ratio) RhoGDIα (h). Bars, 10 μm.
To confirm that the striking difference in the localization of GFP-RhoA versus GFP-RhoB was a consequence of palmitoylation in the secondary targeting motif of RhoB, we constructed GFP-tagged RhoA/RhoB chimeras. We switched the CAAX motif alone of GFP-RhoA (CLVL) with that of RhoB (CKVL) and, as expected, found no effect on localization (not shown). In contrast, when we switched the RhoA CAAX motif with that of RhoB but included the cysteine that occurs NH2-terminal to this motif in RhoB (CCKVL), the localization of the chimera was dramatically changed to that of GFP-RhoB (Fig. 8 f). Moreover, expression of this construct in the presence of 2BP converted the expression pattern to that of RhoA (Fig. 8 g). Thus, a single palmitoylatable cysteine in the hypervariable domain is sufficient to overcome sequestration in the cytosol and redirect RhoA to an expression pattern similar to that of GFP-H-Ras and GFP-RhoB.
Because sequestration of RhoA in the cytosol is likely to be due, in part, to binding to RhoGDIα, and because a palmitoyl group cannot be accommodated in the crystal structure of geranylgeranylated Cdc42hs bound to RhoGDIα (Hoffman et al. 2000), we hypothesized that palmitoylation would block RhoGDIα binding. Cooverexpression of RhoGDIα had no effect on the RhoB-like localization of GFP-RhoA-CCKVL (Fig. 8 h), and RhoGDIα could be coimmunoprecipitated with GFP-RhoA but not GFP-RhoA-CCKVL (Fig. 10 d). Thus, palmitoylation blocks binding to RhoGDIα.
Figure 10.
Differential binding of Rho GTPases to RhoGDIα. (a, b, and d) COS-1 cells were transiently transfected with the indicated GFP-tagged GTPase without (white bars) or with (black bars) cotransfection with equal amounts of DNA encoding RhoGDIα. After 24 h, each GTPase was immunoprecipitated from cell lysates with an anti-GFP antibody, and immunoprecipitates were immunoblotted (b, inset) for both GFP and RhoGDIα, using 125I–protein A and PhosphorImager analysis to quantitate immunoprecipitated proteins. Binding indices are shown in arbitrary units as the ratio of RhoGDIα/GFP-GTPase PhosphorImager volumes and are plotted as the mean ± SEM of n ≥ 4. (a) Wild-type (WT) Rho proteins, (b) pCdc42hs, RhoA, and Rac1 mutation series, (d) chimeric GTPases, and the effect of palmitoylation (introduction of a palmitoylation site into RhoA, RhoA-CCKVL, with or without 2BP, and the removal of palmitoylation sites from RhoB, RhoB189,192S). (c) COS-1 cells were transiently transfected with GFP-tagged Cdc42hs wild-type, Cdc42hs61L, or Cdc42hs12V, and an SRF luciferase reporter construct without (white bars) or with (black bars) coexpression of RhoGDIα. Serum-starved cells were lysed after 16 h, and luciferase activity was measured and normalized to that stimulated by Cdc42hs wild-type in the absence of RhoGDIα. Equivalent expression of Cdc42hs mutants was confirmed by anti-GFP immunoblot. Results shown represent the mean ± SEM, n = 4. The effect of RhoGDIα is significant to P < 0.0001 for Cdc42hs wild-type), P < 0.01 for Cdc42hs12V, and insignificant for Cdc42hs61L.
To determine the role of palmitoylation in targeting TC10 to PMs and endosomes, we expressed GFP-TC10 in the absence or presence of 2BP (Fig. 8i and Fig. j). In the presence of the inhibitor, GFP-TC10 was mislocalized, like GFP-H-Ras and GFP-RhoB, to the nuclear envelope, ER, and Golgi. These data confirm that TC10 is palmitoylated and that palmitoylation is required for targeting to PMs and endosomes.
Differential RhoGDI Binding In Vivo of Activated and Dominant Negative Alleles of Rho GTPases
GTP binding stimulates release in vitro of Rho GTPases from RhoGDI and promotes association with membranes (Isomura et al. 1991; Philips et al. 1993; Bokoch et al. 1994). We used our fluorescence-based assay in live cells to determine if this occurs in vivo. We tagged with GFP a set of well characterized mutants of Rho GTPases in which the guanine nucleotide binding state is altered and expressed them in MDCK cells with or without cooverexpression of RhoGDIα (Fig. 9 a). As expected, GFP-pCdc42hs-61L, an activated allele of pCdc42hs defective in GTPase activity, was localized only in membrane compartments, and this localization was unaffected by cooverexpression of RhoGDIα. In contrast, although GFP-pCdc42hs-12V, another activated allele, when expressed alone was predominantly membrane associated, cooverexpression of RhoGDIα resulted in sequestration in the cytosol, indicating binding. Thus, although both the 61L and 12V mutants of Cdc42hs are dominant active alleles in functional assays, they differ in their capacity to bind RhoGDIα.
Figure 9.
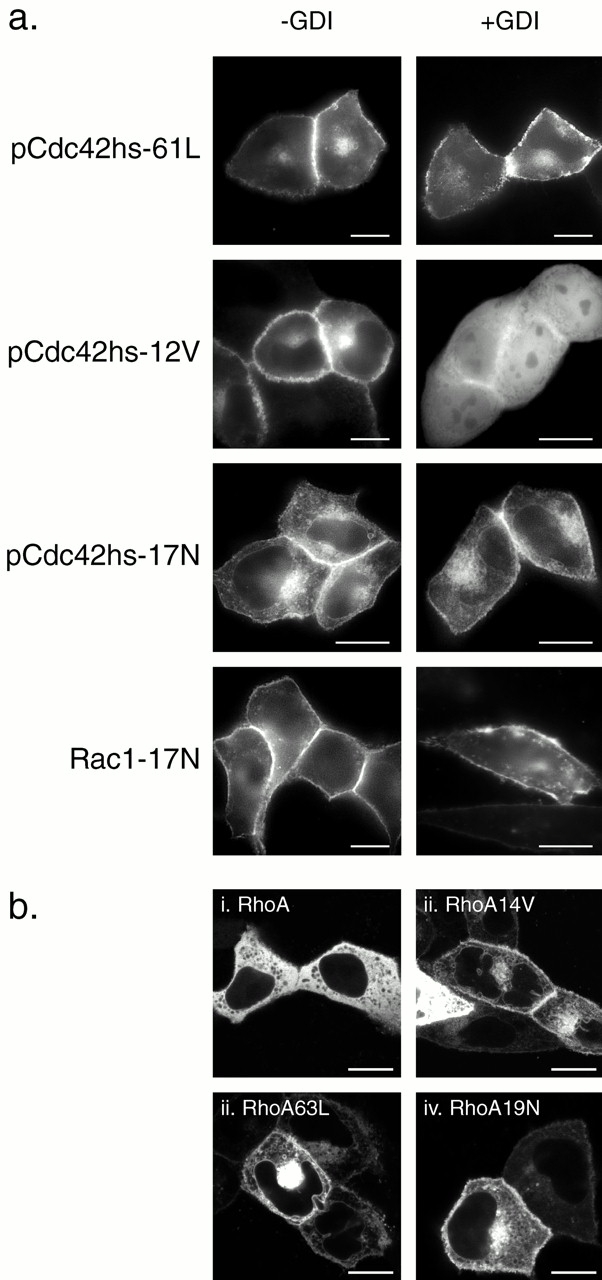
Dominant negative and some dominant active mutants of Rho proteins do not bind RhoGDIα in vivo. (a) MDCK cells were transiently transfected with the indicated GFP-tagged Rho GTPase mutant without (−GDI) or with (+GDI) cotransfection with RhoGDIα as described in the legend to Fig. 7 and imaged alive after 24 h as described in the legend to Fig. 1. (b) MDCK cells were transiently transfected with the indicated GFP-tagged RhoA allele and imaged alive after 24 h with a Zeiss 510 LSC inverted microscope. Note negatively imaged organelles (i) versus positively imaged organelles (ii–iv). Bars, 10 μm.
Dominant negative alleles of Ras-related GTPases have been thought to be locked in a GDP-bound state, despite recent evidence suggesting that they may be nucleotide-free (Strassheim et al. 2000). One might predict that a GDP-locked Rho GTPase would have a higher affinity for RhoGDI, a molecule that was first identified functionally to inhibit the release of GDP (Fukumoto et al. 1990). Therefore, we were surprised to find that GFP-pCdc42hs17N was expressed only in membrane compartments even when cooverexpressed with RhoGDIα (Fig. 9 a). To confirm the effect of the 17N mutation on RhoGDIα binding, we examined GFP-Rac1-17N and found that it too failed to bind RhoGDIα (Fig. 9 a).
To determine if the analogous mutations of RhoA would similarly affect localization, we prepared equivalent GFP-tagged constructs and localized them in live cells using confocal microscopy (Fig. 9 b). As observed with epifluorescence, confocal analysis confirmed that GFP-RhoA was predominantly cytosolic as scored by diffuse fluorescence with distinct, negatively outlined organelles. In striking contrast, GFP-RhoA-14V, GFP-RhoA-63L, and GFP-RhoA-19N were all localized on both PMs and internal membranes, including the nuclear membrane and numerous vesicles. Little cytosolic fluorescence was observed. Cooverexpression of RhoGDIα had no effect on the localization of GFP-RhoA-63L or GFP-RhoA-19N but induced partial relocalization of GFP-RhoA-14V to the cytosol (data not shown), concordant with the results observed for GFP-tagged pCdc42hs mutants. Thus, any of these three single amino acid changes was sufficient to release RhoA from sequestration in the cytosol, a process mediated in part by RhoGDI binding.
To verify the differential binding in vivo of Rho GTPases and their various mutants to RhoGDIα as determined by fluorescence, we performed coimmunoprecipitation (Fig. 10). Using this method, we observed that, although GFP-RhoA, GFP-Rac1, GFP-Rac2, GFP-pCdc42hs, and GFP-bCdc42hs were capable of binding RhoGDIα, GFP-RhoB and GFP-TC10 were not (Fig. 10 a). Coimmunoprecipitation analysis of the nucleotide binding and prenylation mutants recapitulated the fluorescence assay: 12/14V mutants bound RhoGDIα but neither 61/63L nor 17/19N mutants bound to the chaperon, and the farnesylated GFP-pCdc42hsL191M bound RhoGDIα at least as well as the wild-type geranylgeranylated form (Fig. 10 b).
To validate these results with a functional assay, we analyzed the effect of overexpressing RhoGDIα on SRF-dependent transcriptional activation by dominant active alleles of pCdc42hs (Fig. 10 c). SRF was activated more efficiently by pCdc42hs-61L than by pCdc42hs-12V, suggesting that, indeed, the former is more deficient in GTPase activity. Moreover, although pCdc42hs-12V-mediated activation was markedly inhibited by overexpression of RhoGDIα, pCdc42hs-61L-mediated activation was unaffected by RhoGDIα, confirming a lack of binding.
Coimmunoprecipitation also confirmed that the introduction of a palmitoylatable cysteine adjacent to the CAAX motif of RhoA completely abrogated RhoGDIα binding and that inhibition of palmitoylation with 2BP restored binding (Fig. 10 d). Finally, like TC10, the chimera consisting of 188 amino acids of TC10 fused to the 20–amino acid hypervariable domain of pCdc42hs but did not bind RhoGDIα (Fig. 10 d), suggesting that palmitoylation is not the only factor that prevents TC10 from binding to the chaperon. These data thereby confirm the observations that, despite a high degree of sequence homology to RhoA and Cdc42hs respectively, neither RhoB nor TC10 bind RhoGDIα and that palmitoylation inhibits binding to RhoGDIα.
Discussion
The signals that target Ras proteins to membranes have been extensively characterized (Hancock et al. 1990; Choy et al. 1999), but those that target Rho family GTPases to membranes are incompletely understood. The CAAX motif, shared by Ras and Rho family proteins, signals for prenylation which in turn targets the protein to the ER where it encounters the RCE1 protease (Schmidt et al. 1998) and pcCMT (Dai et al. 1998). Although N-Ras and H-Ras then traffic by vesicular transport to the PM via the Golgi, K-Ras4B takes an alternative, yet uncharacterized path (Choy et al. 1999; Apolloni et al. 2000). The signal for engagement of each of these pathways is contained in the so-called “second signal” that lies adjacent to the CAAX motif and consists of either cysteines that are sites of palmitoylation (N-Ras and H-Ras) or a polybasic region (K-Ras4B). The trafficking of Rho family GTPases is more complex because several members of this family can bind to RhoGDIα, a ubiquitously expressed chaperon that has the capacity to retain COOH-terminally processed Rho proteins in the cytosol. Although prenylation has been shown to be required for binding to RhoGDI (Hori et al. 1991), it is unclear how proteolysis, methylation, and palmitoylation affect this interaction. Moreover, the nature of the second signals for most Rho family GTPases has not been established.
In this study, we used GFP fusion proteins to analyze the membrane targeting and RhoGDI binding in live cells of seven Rho family GTPases. We found that although both isoforms of Cdc42hs, Rac1, Rac2, and RhoA bind, RhoGDIα, RhoB, and TC10 do not. Moreover, we found that palmitoylation of RhoA blocked RhoGDI binding and that the GTP/GDP state of those proteins that can bind RhoGDI profoundly influenced binding in vivo. When each Rho protein, except RhoA, was expressed at levels that overcame the binding capacity of RhoGDI, they localized to a variety of membranes as determined by their hypervariable domains. RhoB, like H-Ras, is palmitoylated and targeted to PMs, Golgi, and peri-Golgi vesicles. TC10, which has both a palmitoylation site and a polybasic region, is targeted to PMs and endosomes. Rac1, which has a strong polybasic region, is targeted like K-Ras4B primarily to the PM. Both isoforms of Cdc42hs and Rac2, which have weak polybasic regions, remain predominantly in the endomembrane, although some protein is expressed at the PM. Neither localization nor RhoGDI binding was affected by substituting farnesylation for geranylgeranylation of Cdc42hs. When RhoA was truncated by 73 amino acids from the NH2 terminus, it behaved like Cdc42hs, consistent with its relatively weak polybasic region, suggesting that this GTPase has an NH2-terminal domain that promotes retention in the cytosol.
Because it permits localization in live cells, the use of GFP fusion proteins to localize Rho GTPases offers several distinct advantages over previously employed methods. These include markedly superior resolution of endomembrane structures, avoidance of fixation artifacts, and the ability to directly observe dynamic localizations. The localizations of Rac1, pCdc42hs, and RhoA determined by GFP fusions were generally consistent with previous studies. Endogenous Rac1 has been localized by cell fractionation to both cytosol and crude membranes (Boivin and Beliveau 1995; Michaely et al. 1999) and shown to translocate from cytosol to membranes in response to PDGF (Fleming et al. 1996). Indirect immunofluorescence of myc-tagged Rac1V12 showed expression on PMs and internal membranes (Jou et al. 2000). Cdc42hs has also been reported to localize in the cytosol and membrane pellet (Bokoch et al. 1994; Boivin and Beliveau 1995; Bilodeau et al. 1999) and, by indirect immunofluorescence, to Golgi (Erickson et al. 1996). Our localization of GFP-pCdc42hs expressed in excess of RhoGDI to the ER and nuclear envelope in addition to Golgi suggests that fluorescence in live cells is more sensitive. Our localization is consistent with the recent observation that Cdc42hs binds the γ-subunit of the coatomer complex (γCOP) (Wu et al. 2000) since γCOP cycles between ERs and Golgi. Although RhoA has been reported in both cytosol and membrane pellets (Boivin and Beliveau 1995) and has been colocalized with caveolin-1 in PM microdomains by immunogold electron microscopy (Michaely et al. 1999), indirect immunofluorescence analysis has shown RhoA to be cytosolic (Adamson et al. 1992; Kranenburg et al. 1997). Our observation that activation of Rac1, pCdc42hs, and RhoA in live cells by coexpressing Dbl results in translocation to membrane ruffles is consistent with previous reports of translocation to membranes (Fleming et al. 1996; Kranenburg et al. 1997).
In neutrophils, Rac2 regulates assembly of the NADPH oxidase essential for host defense and, in resting cells, is in the cytosol in a 1:1 complex with RhoGDI (Knaus et al. 1991). Upon activation of Rac2 in vitro (Philips et al. 1993) and upon activation of neutroplasts (Philips et al. 1995), Rac2 translocates to a membrane fraction that has been interpreted as a PM. However, a method for separating PMs from other light membranes by fractionation of neutrophils has not been developed. Given the localization of GFP-Rac2 expressed in excess of RhoGDI predominantly in endomembranes, this interpretation will have to be reevaluated. The NADPH oxidase is thought to be assembled in vivo on phagosome membranes, derived from PMs; however, given the diffusing capacity of reactive oxygen species, assembly on endomembrane may be effective in microbial killing.
Our localization of GFP-RhoB to PMs, Golgi, and peri-Golgi vesicles distinct from endosomes contrasts with the prior localization of RhoB to endosomes. Microinjected overexpressed myc-tagged RhoB was observed in paraformaldehyde-fixed cells permeabilized with Triton X-100 to be located in cytoplasmic vesicles that partially colocalized with transferrin-loaded vesicles and partially colocalized with distinct mannose 6-phosphate receptor–positive vesicles but not with Lucifer yellow–labeled vesicles (Adamson et al. 1992). We localized endogenous RhoB by indirect immunofluorescence and found two distinct staining patterns (see Fig. 4). When paraformaldehyde-fixed cells were permeabilized by Triton X-100, we observed a vesicular pattern similar to the one reported earlier (Adamson et al. 1992). In contrast, when we permeabilized with saponin, we observed localization in a juxtanuclear structure that colocalized with Golgi markers and corresponded to the structure we observed in live cells expressing GFP-RhoB (Fig. 1 and Fig. 2). These observations suggest that the earlier assignment of RhoB to endosomes may have resulted from a fixation artifact. Interestingly, in saponin-permeabilized cells, RhoD colocalized precisely with internalized transferrin (Murphy et al. 1996), suggesting that this Rho family GTPase is endosomal, but in Triton X-100, permeabilized cells RhoD and RhoB colocalized only partially in the perinuclear area (Adamson et al. 1992). Our interpretation of the GFP-RhoB data as arguing against endosomal localization was strengthened by our observation that GFP-TC10 colocalized with internalized transferrin in live cells in relatively large motile vesicles (Fig. 2) and in fixed cells with LAMP. Thus, in live cells the endosomal compartment was marked by GFP-TC10 and did not overlap with the membrane compartments illuminated with GFP-RhoB. The effects of activated or dominant negative alleles of TC10 on endosomal function have yet to be tested. However, localization on endosomes is not required for the regulation of endosome function since, in addition to RhoD (Murphy et al. 1996), Rac1 (Lamaze et al. 1996; Jou et al. 2000), RhoA (Lamaze et al. 1996; Leung et al. 1999), RhoB (Gampel et al. 1999), and Cdc42hs (Kroschewski et al. 1999) have all been implicated in regulating endosomal trafficking.
As expected, both isoforms of Cdc42hs, Rac1, Rac2, and RhoA bound RhoGDIα as determined by both the in vivo fluorescence assay and by coimmunoprecipitation. However, although Cdc42hs, Rac1, and Rac2 could be readily expressed at levels that overcame the capacity of RhoGDI to retain these molecules in the cytosol such that their various membrane localizations became evident, RhoA remained cytosolic even when expressed at very high levels. This difference is not likely due to a higher affinity of RhoA for the available endogenous pool of RhoGDIα since our measurements indicated (see Table ) that RhoGDIα is expressed at a level equivalent to that of the sum of Cdc42hs, Rac1, and RhoA and that even if RhoA had a high enough affinity for RhoGDIα to displace bound Cdc42hs and Rac1, the total pool of RhoGDIα has only a three- to eightfold molar excess over endogenous RhoA, a deficit clearly overcome by our transient transfections. Other known isoforms of RhoGDI are not likely to account for the retention of overexpressed GFP-RhoA in the cytosol since RhoGDIβ is expressed only in hematopoietic cells and has a 10-fold lower affinity for RhoA, Rac1, and Cdc42hs (Gorvel et al. 1998), and RhoGDIγ is expressed mainly in brain and pancreas, binds RhoA with low affinity, and is itself targeted to membranes by virtue of a hydrophobic NH2-terminal domain (Zalcman et al. 1996; Adra et al. 1997). The possibility that overexpressed RhoA remains cytosolic because it overwhelms the capacity for posttranslational processing is unlikely since GFP extended with the COOH-terminal 20 amino acids of RhoA was observed in membranes as was a GFP-tagged NH2-terminal truncation mutant (amino acids 73–193) of RhoA. The latter observation suggests that an NH2-terminal domain is responsible for RhoGDI-independent sequestration in the cytosol, either directly through protein–protein interaction or indirectly by affecting the folding of the protein.
The failure of RhoB and TC10 to bind RhoGDIα was unexpected since these GTPases are highly homologous to RhoA and Cdc42hs, respectively. Nevertheless, our data are consistent with the observation that RhoB could not be extracted from membranes by RhoGDIα (Bilodeau et al. 1999). The recently solved crystal structure of RhoGDIα in complex with geranylgeranylated Cdc42hs reveals a hydrophobic binding pocket for the geranylgeranyl isoprenoid (Hoffman et al. 2000). Because the structure cannot accommodate an adjacent palmitoyl group, we tested the hypothesis that palmitoylation blocked binding. Indeed, incorporation of a palmitoylation site into RhoA abrogated binding to RhoGDIα. However, this could not explain entirely the lack of binding of RhoB and TC10, since mutants that lacked palmitoylation sites did not bind RhoGDIα. The protein–protein interface determined by the cocrystalization of Cdc42hs and RhoGDIα revealed that amino acids 103, 104, 184, and 186 of Cdc42hs (HHKR) form hydrogen bonds with specific residues of RhoGDIα (Hoffman et al. 2000). Although the corresponding surface of Rac1 (HHKK) would be predicted to support these polar interactions, neither that of TC10 (EYVK) nor RhoB (HFQG) is a good substitute, suggesting a mechanism of diminished affinity. The lack of interaction with RhoGDIα suggests that the trafficking of RhoB and TC10 is likely distinct from that of the other members of the Rho family and perhaps similar to that of Ras proteins.
The reversal of relative affinities for RhoGDIα versus membranes that we observed in dominant active mutations of RhoA and Cdc42hs is consistent with previous studies of the capacity of RhoGDIα to extract the GDP-bound but not the GTP-bound form of Rho proteins from membranes (Isomura et al. 1991) and supports the model of cycling on and off membranes as part of the activation cycle. Our data add an important new aspect to this model in that they show that the membranes to which activated Rho proteins are targeted include endomembrane. However, although Dbl stimulated association of Rho GTPases with membranes predominantly involved with the PM, the intrinsically activated Rho mutants localized on the PM and endomembrane, raising the possibility that dominant active alleles of Rho proteins may not accurately substitute for stimulation through endogenous pathways. Our observation that, although RhoA63L and pCdc42hs61L do not interact at all with RhoGDIα, RhoA14V and pCdc42hs12V retain some RhoGDI binding capacity, is consistent with the hypothesis that the former, like the Rap1 Q61 mutant, is GTPase-dead whereas the latter, like Rap1 G12 mutants, is sensitive to GAP and intrinsic GTPase activity; therefore RhoGDIα can bind the GDP-bound pool and sequester molecules in the inactive state. Alternatively, since in vitro GTP-bound Cdc42hs binds to RhoGDIα as well as the GDP-bound form (Nomanbhoy and Cerione 1996), the difference may result from the different amino acid substitutions rather than the GTP/GDP state. Many functional studies have used 61L and 12V mutants of Rho proteins interchangeably. Our data suggest that these mutations are not identical in localization or in activity.
The membrane localization of 17/19N dominant negative mutants of RhoA, pCdc42hs, and Rac1 and their failure to bind RhoGDIα was unexpected since these molecules have been thought to be locked in a GDP-bound state that would be expected to have high affinity for RhoGDIα. However, recent evidence suggests that rather than existing in a GDP-bound state in complex with RhoGDIα, RhoA-19N is nucleotide-free and does not coprecipitate with RhoGDIα (Strassheim et al. 2000). The nucleotide-free state is predicted to have the highest affinity for its cognate GEF and such a conformation is thereby consistent with the model of dominant negative activity mediated by sequestration of GEF molecules. Since it is unlikely that a Rho GTPase can interact with a Dbl family GEF while bound to RhoGDIα, the lack of binding to RhoGDIα of 17/19N mutants and the resulting membrane localization may play a significant role in their mode of action, e.g., bringing them into proximity with membrane-associated GEFs. Alternatively, since most Dbl family GEFs are believed to function on membranes, it is possible that the membrane localization of 17/19N mutants and their inability to bind RhoGDIα in vivo are both mediated by high affinity binding to membrane-associated Dbl family proteins. In either case, the previously unappreciated endomembrane localization of 17/19N mutants that we observed is likely to play a significant role in their mechanism of action.
The most striking result of our analysis of GFP-tagged Rho proteins in live cells is the diversity of localizations despite a common pathway of posttranslational modification. Our data indicate that these localizations are regulated primarily by determinants in the hypervariable region upstream of the CAAX motif and by the capacity to bind RhoGDI. However, the relatively subtle differences in localization observed between proteins with very similar hypervariable regions and RhoGDI binding capacities, e.g., pCdc42hs versus Rac2, suggest that other factors can influence localization. The diversity of localization matches the diversity of function reported for the Rho family of proteins. In some cases localization suggests function. For example, the ER and Golgi localization of pCdc42hs is consistent with its ability to bind γCOP and regulate ER to Golgi transport (Wu et al. 2000). In other cases, e.g., the ability of Cdc42hs to regulate filopodia formation at the PM, the relationship between localization and function remains obscure. Nevertheless, elucidation of the biology of Rho GTPases will require an understanding of their intracellular targeting.
Acknowledgments
We thank Arie Abo, Gary Bokoch, Richard Cerione, Adrienne Cox, Alan Hall, and Danny Manor for providing cDNAs. We thank Marianne Feoktistov and Pheobe Recht for expert technical assistance.
This work was supported by National Institutes of Health research grants GM55279, AI36224, General Clinical Research Center grant M01 RR00096 and training grant T32 GM07308; The Burroughs Wellcome Fund; and the National Science Foundation Major Instrumentation Award 9977430.
Footnotes
The online version of this article contains supplemental material.
Abbreviations used in this paper: 2BP, 2-bromopalmitate; BFA, brefeldin A; CCD, charge-coupled device; GDI, guanine nucleotide dissociation inhibitor; GEF, guanine nucleotide exchange factor; GFP, green fluorescent protein; GST, glutathione S-transferase; LAMP, lysosome-associated membrane protein; PAE, porcine aortic endothelial; pcCMT, prenylcysteine-directed COOH methyltransferase; PM, plasma membrane; SRF, serum response factor.
References
- Abo A., Pick E., Hall A., Totty N., Teahan C.G., Segal A.W. Activation of the NADPH oxidase involves the small GTP-binding protein p21rac1 . Nature. 1991;353:668–670. doi: 10.1038/353668a0. [DOI] [PubMed] [Google Scholar]
- Adamson P., Paterson H.F., Hall A. Intracellular localization of the P21rho proteins. J. Cell Biol. 1992;119:617–627. doi: 10.1083/jcb.119.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adra C.N., Manor D., Ko J.L., Zhu S., Horiuchi T., Van Aelst L., Cerione R.A., Lim B. RhoGDIgammaa GDP-dissociation inhibitor for Rho proteins with preferential expression in brain and pancreas. Proc. Natl. Acad. Sci. USA. 1997;94:4279–4284. doi: 10.1073/pnas.94.9.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apolloni A., Prior I.A., Lindsay M., Parton R.G., Hancock J.F. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 2000;20:2475–2487. doi: 10.1128/mcb.20.7.2475-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspenström P. Effectors for the Rho GTPases. Curr. Opin. Cell Biol. 1999;11:95–102. doi: 10.1016/s0955-0674(99)80011-8. [DOI] [PubMed] [Google Scholar]
- Baldassare J.J., Jarpe M.B., Alferes L., Raben D.M. Nuclear translocation of RhoA mediates the nitrogen-induced activation of phospholipase D involved in nuclear envelope signal transduction. J. Biol. Chem. 1997;272:4911–4914. doi: 10.1074/jbc.272.8.4911. [DOI] [PubMed] [Google Scholar]
- Barrett K., Leptin M., Settleman J. The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell. 1997;91:905–915. doi: 10.1016/s0092-8674(00)80482-1. [DOI] [PubMed] [Google Scholar]
- Bilodeau D., Lamy S., Desrosiers R.R., Gingras D., Beliveau R. Regulation of Rho protein binding to membranes by rhoGDIinhibition of releasing activity by physiological ionic conditions. Biochem. Cell Biol. 1999;77:59–69. [PubMed] [Google Scholar]
- Boivin D., Beliveau R. Subcellular distribution and membrane association of Rho-related small GFP-binding proteins in kidney cortex. Am. J. Physiol. 1995;269:F180–F189. doi: 10.1152/ajprenal.1995.269.2.F180. [DOI] [PubMed] [Google Scholar]
- Bokoch G.M., Bohl B.P., Chuang T. Guanine nucleotide exchange regulates membrane translocation of rac/rho GTP-binding proteins. J. Biol. Chem. 1994;269:31674–31679. [PubMed] [Google Scholar]
- Caron E., Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;282:1717–1721. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
- Cerione R.A., Zheng Y. The Dbl family of oncogenes. Cur. Opin. Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- Choy E., Chiu V.K., Silletti J., Feoktistov M., Morimoto T., Michaelson D., Ivanov I.E., Philips M.R. Endomembrane trafficking of rasthe CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- Clarke S. Protein isoprenylation and methylation at carboxyl terminal cysteine residues. Annu. Rev. Biochem. 1992;61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- Coso O.A., Chiariello M., Yu J.C., Teramoto H., Crespo P., Xu N., Miki T., Gutkind J.S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- Dai Q., Choy E., Chiu V., Romano J., Slivka S., Steitz S., Michaelis S., Philips M.R. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J. Biol. Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- Drivas G.T., Shih A., Coutavas E., Rush M.G., D'Eustachio P. Characterization of four novel ras-like genes expressed in a human teratocarcinoma cell line. Mol. Cell. Biol. 1990;10:1793–1798. doi: 10.1128/mcb.10.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J.W., Zhang C., Kahn R.A., Evans T., Cerione R.A. Mammalian Cdc42 is a Brefeldin A-sensitive component of the Golgi apparatus. J. Biol. Chem. 1996;271:26850–26854. doi: 10.1074/jbc.271.43.26850. [DOI] [PubMed] [Google Scholar]
- Fleming I.N., Elliott C.M., Exton J.H. Differential translocation of Rho family GTPases by lysophosphatidic acid, endothelin-1, and platelet-derived growth factor. J. Biol. Chem. 1996;271:33067–33073. doi: 10.1074/jbc.271.51.33067. [DOI] [PubMed] [Google Scholar]
- Fukumoto Y., Kaibuchi K., Hori Y., Fujioka H., Araki S., Ueda T., Kikuchi A., Takai Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene. 1990;5:1321–1328. [PubMed] [Google Scholar]
- Gampel A., Parker P.J., Mellor H. Regulation of epidermal growth factor receptor traffic by the small GTPase rhoB. Curr. Biol. 1999;9:955–958. doi: 10.1016/s0960-9822(99)80422-9. [DOI] [PubMed] [Google Scholar]
- Gorvel J.P., Chang T.C., Boretto J., Azuma T., Chavrier P. Differential properties of D4/LyGDI versus RhoGDIphosphorylation and rho GTPase selectivity. FEBS Lett. 1998;422:269–273. doi: 10.1016/s0014-5793(98)00020-9. [DOI] [PubMed] [Google Scholar]
- Hancock J.F., Magee A.I., Childs J.E., Marshall C.J. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- Hancock J.F., Paterson H., Marshall C.J. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- Hill C.S., Wynne J., Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- Hoffman G.R., Nassar N., Cerione R.A. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- Hori Y., Kikuchi A., Isomura M., Katayama M., Miura Y., Fujioka H., Kaibuchi K., Takai Y. Post-translational modifications of the COOH-terminal region of the rho protein are important for its interaction with membranes and the stimulatory and inhibitory GDP/GTP exchange proteins. Oncogene. 1991;6:515–522. [PubMed] [Google Scholar]
- Isomura M., Kikuchi A., Ohga N., Takai Y. Regulation of binding of rhoB p20 to membranes by its specific regulatory protein, GDP dissociation inhibitor. Oncogene. 1991;6:119–124. [PubMed] [Google Scholar]
- Jou T.S., Leung S.M., Fung L.M., Ruiz W.G., Nelson W.J., Apodaca G. Selective alterations in biosynthetic and endocytic protein traffic in Madin-Darby canine kidney epithelial cells expressing mutants of the small GTPase Rac1. Mol. Biol. Cell. 2000;11:287–304. doi: 10.1091/mbc.11.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus U.G., Heyworth P.G., Evans T., Curnutte J.T., Bokoch G.M. Regulation of phagocyte oxygen radical production by the GTP-binding protein Rac 2. Science. 1991;254:1512–1515. doi: 10.1126/science.1660188. [DOI] [PubMed] [Google Scholar]
- Kranenburg O., Poland M., Gebbink M., Oomen L., Moolenaar W.H. Dissociation of LPA-induced cytoskeletal contraction from stress fiber formation by differential localization of RhoA. J. Cell Sci. 1997;110:2417–2427. doi: 10.1242/jcs.110.19.2417. [DOI] [PubMed] [Google Scholar]
- Kroschewski R., Hall A., Mellman I. Cdc42 controls secretory and endocytic transport to the basolateral plasma membrane of MDCK cells. Nat. Cell Biol. 1999;1:8–13. doi: 10.1038/8977. [DOI] [PubMed] [Google Scholar]
- Lamarche N., Hall A. GAPs for rho-related GTPases. Trends Genet. 1994;10:436–440. doi: 10.1016/0168-9525(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Lamaze C., Chuang T.H., Terlecky L.J., Bokoch G.M., Schmid S.L. Regulation of receptor-mediated endocytosis by Rho and Rac. Nature. 1996;382:177–179. doi: 10.1038/382177a0. [DOI] [PubMed] [Google Scholar]
- Leung S.M., Rojas R., Maples C., Flynn C., Ruiz W.G., Jou T.S., Apodaca G. Modulation of endocytic traffic in polarized Madin-Darby canine kidney cells by the small GTPase RhoA. Mol. Biol. Cell. 1999;10:4369–4384. doi: 10.1091/mbc.10.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaely P.A., Mineo C., Ying Y., Anderson R.G.W. Polarized distribution of endogenous Rac1 and RhoA at the cell surface. J. Biol. Chem. 1999;274:21430–21436. doi: 10.1074/jbc.274.30.21430. [DOI] [PubMed] [Google Scholar]
- Minden A., Lin A., Claret F.X., Abo A., Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- Murphy C., Saffrich R., Grummt M., Gournier H., Rybin V., Rubino M., Auvinen P., Lutcke A., Parton R.G., Zerial M. Endosome dynamics regulated by a Rho protein. Nature. 1996;384:427–432. doi: 10.1038/384427a0. [DOI] [PubMed] [Google Scholar]
- Nobes C., Hall A. Rho, Rac and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Nomanbhoy T.K., Cerione R.A. Characterization of the interaction between RhoGDI and Cdc42Hs using fluorescence spectroscopy. J. Biol. Chem. 1996;271:10004–10009. doi: 10.1074/jbc.271.17.10004. [DOI] [PubMed] [Google Scholar]
- Philips M.R., Pillinger M.H., Staud R., Volker C., Rosenfeld M.G., Weissmann G., Stock J.B. Carboxyl methylation of ras-related proteins during signal transduction in neutrophils. Science. 1993;259:977–980. doi: 10.1126/science.8438158. [DOI] [PubMed] [Google Scholar]
- Philips M.R., Feoktistov A., Pillinger M.H., Abramson S.B. Translocation of p21rac2 from cytosol to plasma membrane is neither necessary nor sufficient for neutrophil nadph oxidase activity. J. Biol. Chem. 1995;270:11514–11521. doi: 10.1074/jbc.270.19.11514. [DOI] [PubMed] [Google Scholar]
- Presley J.F., Cole N.B., Schroer T.A., Hirschberg K., Zaal K.J.M., Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Qiu R.G., Chen J., Kirn D., McCormick F., Symons M. An essential role for Rac in Ras transformation. Nature. 1995;374:457–459. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Paterson H.F., Johnston C.L., Diekmann D., Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Robertson D., Paterson H.F., Adamson P., Hall A., Monaghan P. Ultrastructural localization of ras-related proteins using epitope-tagged plasmids. J. Histochem. Cytochem. 1995;43:471–480. doi: 10.1177/43.5.7537292. [DOI] [PubMed] [Google Scholar]
- Schmidt W.K., Tam A., Fujimura-Kamada K., Michaelis S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl. Acad. Sci. USA. 1998;95:11175–11180. doi: 10.1073/pnas.95.19.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassheim D., Porter R.A., Phelps S.H., Williams C.L. Unique in vivo associations with SmgGDS and RhoGDI and different guanine nucleotide exchange activities exhibited by RhoA, dominant negative RhoA (Asn-19), and activated RhoA (Val-14) J. Biol. Chem. 2000;275:6699–6702. doi: 10.1074/jbc.275.10.6699. [DOI] [PubMed] [Google Scholar]
- Webb Y., Hermida-Matsumoto L., Resh M.D. Inhibition of protein palmitoylation, raft localization, and T-cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J. Biol. Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- Wu W.J., Erickson J.W., Lin R., Cerione R.A. The γ-subunit of the coatomer complex binds Cdc42 to mediate transformation. Nature. 2000;405:800–804. doi: 10.1038/35015585. [DOI] [PubMed] [Google Scholar]
- Zalcman G., Closson V., Camonis J., Honore N., Rousseau-Merck M.F., Tavitian A., Olofsson B. RhoGDI-3 is a new GDP dissociation inhibitor (GDI). Identification of a non-cytosolic GDI protein interacting with the small GTP-binding proteins RhoB and RhoG. J. Biol. Chem. 1996;271:30366–30374. doi: 10.1074/jbc.271.48.30366. [DOI] [PubMed] [Google Scholar]