Abstract
Srb11p-Srb10p is the budding yeast C-type cyclin-cyclin-dependent kinase that is required for the repression of several stress response genes. To relieve this repression, Srb11p is destroyed in cells exposed to stressors, including heat shock and oxidative stress. In the present study, we identified Ask10p (for activator of Skn7) by two-hybrid analysis as an interactor with Srb11p. Coimmunoprecipitation studies confirmed this association, and we found that, similar to Srb11p-Srb10p, Ask10p is a component of the RNA polymerase II holoenzyme. Ask10p is required for Srb11p destruction in response to oxidative stress but not heat shock. Moreover, this destruction is important since the hypersensitivity of an ask10 mutant strain to oxidative stress is rescued by deleting SRB11. We further show that Ask10p is phosphorylated in response to oxidative stress but not heat shock. This modification requires the redundant mitogen-activated protein (MAP) kinase kinase Mkk1/2 but not their normal MAP kinase target Slt2p. Moreover, the other vegetative MAP kinases—Hog1p, Fus3p, or Kss1p—are not required for Ask10p phosphorylation, suggesting the existence of an alternative pathway for transducing the Pkc1p→Bck1→Mkk1/2 oxidative stress signal. In conclusion, Ask10p is a new component of the RNA polymerase II holoenzyme and an important regulator of the oxidative stress response. In addition, these results define a new role for the Pkc1p MAP kinase cascade (except the MAP kinase itself) in transducing the oxidative damage signal directly to the RNA polymerase II holoenzyme, thereby bypassing the stress-activated transcription factors.
In response to oxidative damage, the cell induces enzymes able to inactivate reactive oxygen species (ROS; e.g., superoxide dismutases and catalases) or that can reduce protein disulfides (e.g., thioredoxin [for a review, see reference 39]). In addition, a more general class of stress response genes, the heat shock genes, are also induced in response to oxidative stress to aid in maintaining cell viability through their chaperone activity (10). Therefore, two critical regulatory systems must merge for the cell to mount a successful response to oxidative stress. First, the damage must be recognized and the signal transduced to the nucleus. Second, transcription factor activity must then be influenced by this signal to elicit the proper gene expression program.
Several signal transduction pathways that initiate the proper gene expression program in response to environmental challenges have been identified (for a review, see reference 18). For example, activation of the mitogen-activated protein (MAP) kinase Hog1p allows the cell to adjust to a hyperosmotic environment (44). Similarly, Slt2p, the MAP kinase activated by the protein kinase C Pkc1p→Bck1p→Mkk1p/Mkk2p pathway, is necessary to maintain cell wall integrity during growth in hypotonic medium (31). In addition to MAP kinase cascades, a “two-component” regulatory system responds to heat shock and oxidative stress (40). The two-component system utilizes a histidine kinase sensor and its effector protein, which is known as a response regulator (26, 43). In budding yeast, Ssk1p and Skn7p are response proteins of the Sln1p histidine kinase (3, 4, 33). Skn7p is a transcription factor that regulates cell homeostasis in response to hypertonic medium and oxidative damage (27, 29, 34).
In general, signal transduction pathways alter gene expression by affecting the activity of transcription factors. For example, Slt2p controls transcription factors that mediate cell cycle progression (Swi4-Swi6 [37]) or the stress response (Rlm1p [12, 24]). Similarly, stress response factors Msn2 and Msn4p require Hog1p for activation (16, 38). However, the role of signal transduction pathways in regulating components of the RNA polymerase II (RNA Pol II) holoenzyme itself is much less well understood. The Snf1p AMP kinase, which signals switches in carbon source availability, has been shown to associate with the Mediator complex of RNA Pol II (28), although no substrate has been identified to date. In addition, the yeast C-type cyclin (Ume3p/Srb11p) and its cyclin-dependent kinase (Cdk) Ume5p/Srb10p (6, 35, 53) are components of the Mediator (35). This cyclin-Cdk represses the transcription of several stress response genes, including SSA1 (6) and CTT1 (20). To relieve this repression, Srb11p is destroyed in response to environmental stress, including heat shock and oxidative stress (6, 7), suggesting that it is a target of a stress-activated signaling pathway.
Previous studies have found that Plc1p, a homologue of the mammalian phospholipase Cγ (14), is required for the oxidative-stress-induced destruction of Srb11p (7). However, the steps downstream of Plc1p controlling Srb11p destruction remain unknown. The present study identified the putative transcription factor Ask10p as an interactor of Srb11p that is required for the rapid destruction of the cyclin in response to oxidative stress. This destruction is important since deletion of SRB11 suppresses the oxidative-damage hypersensitivity phenotype exhibited by ask10 mutants. Finally, Ask10p is phosphorylated in response to oxidative stress, and this modification requires the redundant MAP kinase kinases Mkk1p/Mkk2p. These findings indicate that the Pkc1p MAP kinase pathway transmits an oxidative stress signal to the RNA Pol II holoenzyme through phosphorylation of Ask10p.
MATERIALS AND METHODS
Media and strains.
The yeast strains used in these studies are listed in Table 1. Strains SKY191 and SKY473 used in the two-hybrid studies are described elsewhere (48). The hog1, slt2/mpk1, fus3, and kss1 mutants were obtained from the Research Genetics deletion collection. Cultures were grown in synthetic dextrose (SD) medium (20 g of dextrose, 1.7 g of yeast nitrogen base, 5 g of ammonium acetate, and 15 g of agarose [if necessary] per liter, supplemented with an amino acid mix plus 1% tryptophan, 1% uracil, and 1% histidine as needed). Two-hybrid plates were prepared by omitting dextrose from the SD medium and adding 2% galactose, 1% raffinose, 0.02% X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid; Diagnostic Chemicals, Ltd.), and 100 μg of G418/ml (final concentration) as indicated. All time course cultures were grown in SD medium lacking leucine to select for the maintenance of LEU2-marked plasmids.
TABLE 1.
Yeast strains used in this study
| Straina | Genotype |
|---|---|
| RSY471 | MATα ade2-1 his3-1 leu2-3,112 2lexAop::LEU2 trp1-1 ura3-1 |
| RSY867 | MATα ade2-1 his3-1 leu2-3,112 2lexAop::LEU2 trp1-1 ura3-1 ask10::TRP1 |
| RSY868 | MATα ade2-1 his3-1 leu2-3,112 2lexAop::LEU2 trp1-1 ura3-1 ask10::TRP1 srb11::URA3 |
| RSY869 | MATα ade2-1 his3-1 leu2-3,112 2lexAop::LEU2 2 trp1-1 ura3-1 ASK10::3HA::TRP1 |
| RSY797 | MATα ade2-1 his3-1 leu2-3,112 2lexAop::LEU2 trp1-1 ura3-1 srb11::TRP1 |
| RSY940 | MATα ade2-1 his3-1 leu2-3,112 2lexAop::LEU2 trp1-1 ura3-1 yap1::his5+ |
| RSY941 | MATα ade2-1 his3-1 leu2-3,112 2lexAop::LEU2 trp1-1 ura3-1 yap1::his5+srb11::TRP1 |
| RSY870 | MATα ade2-1 his3-1 leu2-3,112 2lexAop::LEU2 trp1-1 ura3-1 srb11::URA3 ASK10::3HA::TRP1 |
| RSY903 | MATα ade2-1 his3-1 leu2-3,112 2lexAop::LEU2 trp1-1 ura3-1 KAN::GAL1::ASK10::3HA::TRP1 |
All strains were from the present study.
Plasmids.
pLR101 and pLR102 contain the myc epitope-tagged wild-type SRB11 allele or the A110V mutant under the control of the ADH1 promoter (6) inserted into pRS315 (50), respectively. The SRB11 two-hybrid DNA-binding domain fusion gene was constructed by treating the 1.5-kbp EcoRI fragment from pKC239 (6) containing the myc-tagged SRB11 allele with Klenow polymerase to produce blunt ends and inserting this fragment into the PvuII site of pGBS9A (48). Srb11p can activate transcription when fused to a DNA-binding domain due to its ability to associate with the RNA polymerase II (RNA Pol II) holoenzyme (6). Therefore, the holoenzyme interaction domain was mutated (pKC227) (8) prior to fusion to the cI DNA-binding domain under the control of the ADH1 promoter (pTC300). The myc epitope-tagged SRB11 allele was placed under the control of the GAL1 promoter in pYES2 (Invitrogen, Inc.), yielding pKC333. pJG-45 (19) contains the GAL1 promoter driving the expression of a yeast genomic library fused to a cassette consisting of a nuclear localization signal, a transcription activation domain, and a hemagglutinin (HA) epitope tag. The cIop-GusA and cIop-LYS2 reporter constructs are described elsewhere (48). An HA epitope-tagged derivative of ASK10 was constructed by first integrating the 3HA-his5 cassette (36) at the 3′ end of a genomic ASK10. This genomic allele was then amplified by PCR and inserted into pRS316 (50), yielding pAK3.
Chromosomal deletion/epitope tagging.
The generation of a deletion, chromosomally HA epitope-tagged, or GAL1-inducible ASK10 allele was accomplished by using PCR-mediated strategies with plasmids pFA6a-TRP1, pFA6a-HA-TRP1, or pFA6a-TRP1-PGAL-3HA, respectively, as a template (36). The YAP1 deletion strains were constructed by an identical method utilizing pFA6a-His3MX6. These modified alleles were verified by PCR analysis of genomic DNA (data not shown). The HA epitope-tagged Ask10p derivative was functional, as determined by complementation assays using oxidative stress hypersensitivity as the assay.
Stress sensitivity assays.
The strains with the indicated genotypes were grown to mid-log phase (5 × 106 cells/ml) and serially diluted 1:10. The dilutions were spotted onto rich plates or a onto rich plate freshly prepared containing 0.4 mM H2O2. The H2O2 and heat shock plates were incubated for 2 days at 30 and 37°C, respectively, and then photographed.
Western blot analyses.
Heat shock and oxidative-stress time course experiments were conducted, and protein extracts were prepared as previously described (6, 7). Srb11p-myc or Ask10p-HA levels were monitored by Western blot analysis of immunoprecipitates from 250 μg of soluble protein. Coimmunoprecipitation studies were conducted with 1 mg of soluble protein with the antibodies, as indicated. Holoenzyme immunoprecipitations with Rpb1p and TFIIS were performed as described previously (8). Antibodies directed against Rpb1p and TFIIS were generous gifts of J. Jaehning (University of Colorado Health Sciences Center, Denver). Ask10p-HA or Srb11p-myc signals were visualized with an anti-mouse secondary antibody conjugated to alkaline phosphatase and chemiluminescence by using the CDP-star reagent (Tropix). Signal quantitation was obtained by using the LAS-1000 chemiluminescence system (Fuji, Inc.) and Image Pro software. Lines were generated by linear regression analysis with r > 0.9.
Phosphatase assays.
The ASK10-HA strain was grown to mid-log phase (5 × 106 cells/ml) and treated with H2O2 (0.4 mM), and samples were harvested at the times indicated in the text. Extracts were prepared, and 1 mg of soluble protein was immunoprecipitated overnight at 4°C with anti-HA antibodies. The immunoprecipitates were collected on protein A-Sepharose beads and washed four times with buffer 3 (50 mM Tris-HCl [pH 7.4], 250 mM NaCl, 5 mM EDTA, 0.1% NP-40), followed by a final wash with phosphatase buffer (50 mM Tris-HCl [pH 7.5], 0.1 mM Na2EDTA, 5 mM dithiothreitol, 2 mM MnCl2). After the last wash, the pellet was divided in two; one half was treated with 2 U of lambda phosphatase (New England Biolabs), and the other half was treated with phosphatase plus inhibitors (5 mM EDTA, 5 mM Na2EGTA, 10 mM sodium pyrophosphate, and 0.1 mM sodium orthovanadate). Samples were incubated at 37° for 30 min, and protein A-Sepharose beads were collected by centrifugation. Ask10p phosphorylation was assayed by mobility shift using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western analysis as described above.
RESULTS
Srb11p associates with Ask10p in vivo.
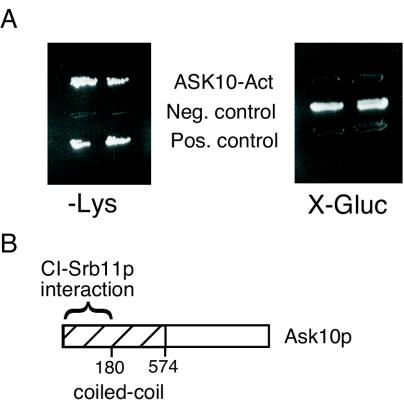
To identify proteins that regulate Srb11p destruction, two-hybrid analysis was performed (19). A cI-SRB11 bait plasmid (pTC300) was introduced into the haploid strain SKY191 and mated to SKY473, which harbored a galactose-inducible genomic library fused to a synthetic activation domain (a gift from E. Golemis, Fox Chase Cancer Center). The diploids were tested for activation of two reporter genes (cIop-LYS2 and cIop-GusA) on medium supplemented with galactose to induce the library plasmid (48). In two successive searches, Ask10p, was isolated from the screen as a protein that activated both reporter genes (Fig. 1A). Ask10p is a 127-kDa protein that was originally identified as a high-copy enhancer of Skn7-dependent transcription (42). Sequence analysis revealed that Ask10p is a member of a gene family with predicted family members found in budding (Ynl1215p and Ypr115p) and fission (NP_5946315P) yeast. Comparing Ask10p to the database did not reveal any specific functional motifs. The amino half of the protein contains a predicted coiled-coil domain; a motif known to mediate protein-protein interactions (Fig. 1B). Sequence analysis of the three interacting clones revealed that only the amino-terminal portion of the protein (amino acids 1 to 276) was always present, suggesting that Ask10p binds Srb11p through its coiled-coil domain.
FIG. 1.
The coiled-coil domain of Ask10p associates with Srb11p in a two-hybrid analysis. (A) The activation domain fusion library plasmid containing Ask10p (ASK10-Act), negative control (activation domain alone), or positive control (synthetic activation domain fused to cI DNA-binding domain) was introduced into a strain expressing cI-Srb11p, along with the cIop-LYS2 and cIop-GusA reporter genes. Activation of the reporter genes allows growth on medium lacking lysine (−Lys) and cleaves the chromergic agent X-Gluc. (B) Ask10p interaction region. Schematic of Ask10p illustrating a predicted coiled-coil domain (stripped region). The region required for Srb11p binding is indicated. Amino acid boundries of these domains are indicated.
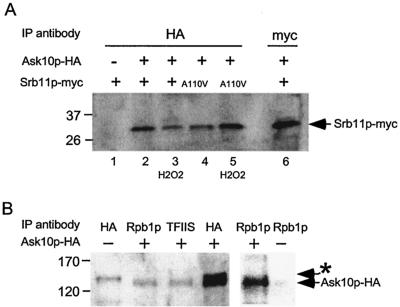
To verify the Srb11p-Ask10p interaction in vivo, coimmunoprecipitation studies were performed. A strain containing a chromosomally HA epitope-tagged ASK10 allele (RSY869) and the parental control strain (RSY471) were transformed with pLR101 (myc epitope-tagged ADH1-SRB11 expression plasmid). The transformants were grown to mid-log phase and harvested, and protein extracts were prepared (see Materials and Methods for details). The extracts were immunoprecipitated with HA monoclonal antibodies, and the immunoprecipitates were analyzed by Western blot analysis probing for Srb11p with the myc antibody. A band specific for myc-tagged Srb11p was observed in the strain containing pLR101 (Fig. 2A, lane 2) versus the control lacking the epitope-tagged ASK10 (lane 1). These findings, in combination with the two-hybrid results, indicate that Ask10p and Srb11p associate in vivo.
FIG. 2.
Ask10p binds Srb11p and the RNA Pol II holoenzyme in vivo. (A) Extracts were prepared from cultures under nonstress conditions (lanes 2, 4, and 6) or 30 min after oxidative stress (lanes 3 and 5) and contained Srb11p-myc, Ask10-HA (+), the Srb11pA110V mutant (A110V), or vector control (−) as indicated. Srb11p-myc immunoprecipitation (lane 6) serves as a size standard, and lane 1 controlled for nonspecific association of Srb11p-myc to protein A-Sepharose beads. (B) Extracts prepared from the ASK10-HA strain RSY869 (+) or parental control (−) were immunoprecipitated with the antibodies specific for RNA Pol II holoenzyme components Rpb1p and TFIIS as indicated. The Ask10p-specific signal and a nonspecific band (asterisk) are indicated. The negative control for nonspecific interaction of Rpb1p antibodies in a strain lacking Ask10p-HA (right panel) was overexposed to detect even minor cross-reactivity. Molecular weight markers are indicated (in kilodaltons) on the left sides of both panels.
Ask10p associates with Rpb1p and TFIIS in vivo.
Previous studies in our laboratory and others have found that Srb11p associates with the RNA Pol II holoenzyme (8, 35). The association of Ask10p with Srb11p prompted the question of whether Ask10p is also a holoenzyme component. To test this possibility, extracts prepared from RSY869 containing the HA-tagged ASK10 allele were immunoprecipitated with antibodies directed against two components of the RNA polymerase holoenzyme: Rpb1p and TFIIS (17). The immunoprecipitates were collected with protein A-Sepharose beads and subjected to Western blot analysis probing for Ask10p. These experiments revealed the presence of Ask10p in both Rpb1p and TFIIS immunoprecipitates (Fig. 2B) but not in control extracts, indicating that Ask10p is complexed with these proteins in vivo. These results indicate that Ask10p is a previously unknown component of the RNA Pol II holoenzyme and are consistent with the previous report that Ask10p regulates transcription (42).
Ask10p is required for Srb11p destruction in response to oxidative stress but not heat shock.
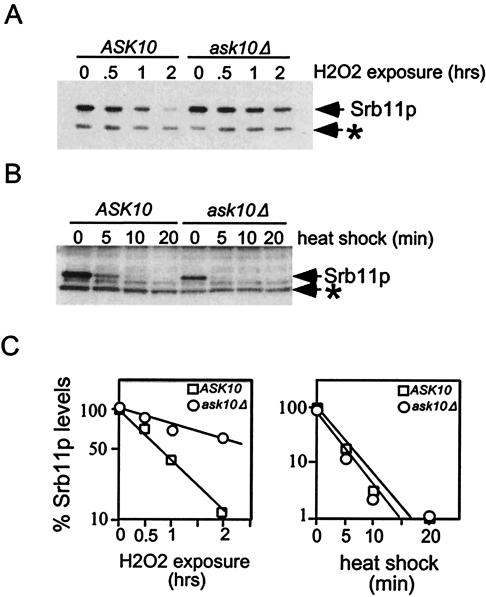
Ask10p was originally identified by its ability to stimulate Skn7p-dependent transcription (42). Skn7p activates genes that respond to environmental changes, including oxidative stress and heat shock (40). We have previously demonstrated that Srb11p is destroyed in response to both types of stress (6, 7). Therefore, we tested whether Ask10p is required for Srb11p destruction in cells subjected to either oxidative damage or heat shock. Wild-type (RSY471) and ask10Δ deletion strains (RSY867) containing the myc-tagged SRB11 expression plasmid (pKC333) were grown to 5 × 106 cells/ml and then exposed to 0.4 mM hydrogen peroxide. Previous studies in our laboratory have demonstrated that overexpression of Srb11p has no impact on its regulation compared to the normally expressed protein (6). Samples were taken prior to, and after, H2O2 treatment, and Srb11p levels were monitored by Western blot analysis. Similar to previous results (7), Srb11p levels were significantly reduced in the wild-type control after a 2-h exposure to H2O2 (Fig. 3A, quantitated in 3C). However, in the ask10 mutant strain, the decay rate of Srb11p levels was approximately threefold slower than in the wild type (3 h versus 0.9 h, respectively). The half-life for Srb11p under “normal” growth conditions was calculated to be 1.5 h (6). These results indicate that Ask10p is required for normal Srb11p destruction in response to oxidative stress.
FIG. 3.
Ask10p is required for Srb11p destruction in response to oxidative stress. (A) Oxidative stress. Wild-type (RSY471) and ask10Δ (RSY867) strains harboring the SRB11-myc expression construct pKC333 were grown to mid-log phase (0 h), H2O2 was added to 0.4 mM, and time points taken as indicated (in hours). Srb11p levels were monitored (arrow) in extracts prepared from these samples (see Materials and Methods for details). (B) Heat shock. This experiment was performed as described above except the cultures were shifted to 37°C and time points were taken (minutes). (C) Quantitation of Srb11p decay kinetics. The chemiluminescent signals derived in Panels A and B were quantitated and plotted on log scale versus time. Decay curves were generated by linear regression analysis (r ≥ 0.9). The asterisk indicates a nonspecific cross-reacting protein.
We next determined whether Ask10p is also required for the heat-induced destruction of Srb11p. The wild type and ask10Δ mutant harboring pLR101 were grown to mid-log phase at 30°C and then transferred to 37°C, and time points were taken. As previously observed (6), Srb11p levels declined quickly after heat shock in the wild-type culture, going below the limits of detection by 10 min (Fig. 3B, quantitated in 3C). Similar to the wild type, the ask10Δ mutant efficiently destroyed Srb11p, indicating that Ask10p is not required for the rapid destruction of this cyclin in response to heat shock. These results indicate that Ask10p is required for Srb11p degradation in response to oxidative stress but not in response to heat shock. These findings suggest that the oxidative stress and heat shock response pathways are separate but converge at Srb11p.
Ask10p-dependent destruction of Srb11p is important for cell survival in response to oxidative stress.
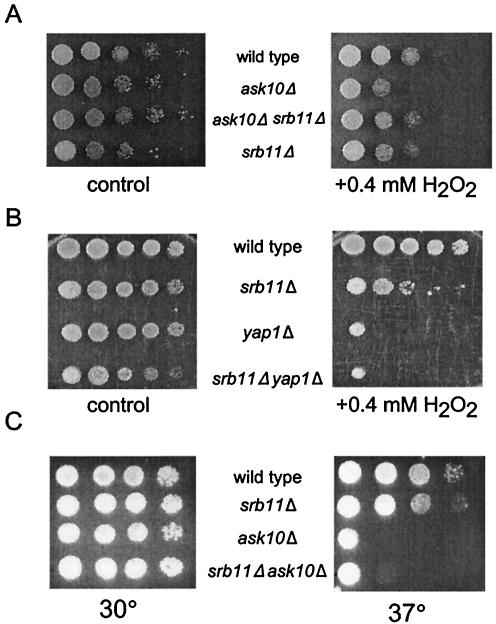
Previous studies indicated an important role for Srb11p destruction for cell viability when cells are exposed to oxidative stress (7). Although Ask10p has been implicated in the Sln1-Ssk1 two-component system (42), which is also required for normal viability in response to H2O2 (51), its requirement for maintaining cell viability after exposure to oxidative damage has not been determined. A plate assay was used to test the sensitivity of each strain to oxidative damage (see Materials and Methods for details). Wild-type and ask10Δ strains were spotted onto rich medium containing 0.4 mM H2O2, and cell viability was assessed after a 2-day incubation at 30°C. These experiments indicate that cell survival was reduced at least 10-fold in ask10Δ mutants compared to the control (Fig. 4A). If the failure to destroy Srb11p is responsible (at least partially) for the hypersensitivity of ask10Δ mutants to H2O2, then deleting the cyclin should suppress this growth defect. Indeed, the ask10Δ srb11Δ double mutant exhibited H2O2 sensitivity to a level observed for the wild-type control. One interpretation of these results is that the Ask10p-dependent destruction of Srb11p is important to maintain cell viability after ROS-induced stress. Alternatively, deleting SRB11 could nonspecifically enhance viability in response to oxidative damage. To address this question, Yap1p, a transcription factor required for ROS-responsive gene induction, was deleted in an srb11 mutant. As previously reported (52), yap1 mutants are hypersensitive to oxidative stress (Fig. 4B). However, the yap1Δ srb11Δ double mutant did not exhibit enhanced viability on H2O2-containing plates, suggesting that srb11 mutations do not provide general protection from oxidative stress. These results indicate a new role for Ask10p in the oxidative stress response and suggest that the Ask10p-dependent destruction of Srb11p is important for protecting the cell from reactive oxygen damage.
FIG. 4.
Ask10p is required for the cellular response to oxidative stress and heat shock. (A) Ask10p regulates the oxidative stress response through Srb11p. Wild-type (RSY471), ask10Δ (RSY867), srb11Δ (RSY870), ask10Δ srb11Δ (RSY868), yap1Δ (RSY940), and yap1Δ srb11Δ (RSY941) strains were grown to mid-log phase, diluted, and spotted onto rich medium containing 0.4 mM H2O2 as indicated. The plates were incubated for 2 days at 30°C and then photographed. (B) Ask10p and Yap1p function independently. The strains (listed in panel A) with the indicated genotypes were treated as described above (C) Ask10p is required for growth at elevated temperature. The strains described in panel A were diluted (1:10) and spotted onto rich medium, followed by incubation at 30 or 37°C for 2 days prior to being photographed.
Ask10p is required for cell survival in response to heat shock independent of Srb11p.
Ask10p has been implicated in enhancing Skn7p-dependent activation and, as shown here, is required for the oxidative stress response. In addition to ROS, Skn7p is involved in responding to several types of stress, including heat shock. To test whether Ask10p plays a more general role in cellular response to stress, the viability of an ask10Δ mutant under heat shock conditions was determined. Wild-type and ask10Δ mutant strains were grown at 30°C in rich medium to mid-log phase, serially diluted (1:10), and spotted onto duplicate rich solid medium. These plates were incubated at 30 and 37°C overnight and then inspected for growth. Interestingly, the viability of the ask10Δ mutant at 37°C was reduced by 4 orders of magnitude compared to the wild-type control (Fig. 4C). No difference in plating efficiency was observed on the 30°C plate, indicating that the growth defect was due to the increased temperature. Consistent with our earlier finding that Ask10p is not required for heat-induced destruction of Srb11p, deleting SRB11 was not able to rescue the temperature sensitivity of the ask10 mutant. Taken together, these experiments indicate that Ask10p is also required for growth at a high temperature. However, this requirement is independent of the Srb11p status, suggesting that Ask10p does not solely function to mediate cyclin destruction.
Ask10p association to Srb11p is independent of the stress-activated degron.
Ask10p is required for the destruction of Srb11p in response to oxidative stress but not heat shock. Our previous studies identified an element defined by two separate single amino acid substitutions (A110V and E170K) that mediates oxidative stress-induced destruction of Srb11p (7). To determine whether the increased stability observed with Srb11pA110V was due to a defect in Ask10p binding, coimmunoprecipitation experiments were performed. The Srb11pA110V-myc expression plasmid was introduced into the strain harboring the HA-tagged ASK10 allele, and extracts prepared from this culture were immunoprecipitated with HA antibody. The immunoprecipitates were blotted and probed with the myc antibody recognizing Srb11pA110V-myc. These experiments showed that Srb11pA110V, like the wild-type cyclin, associated with Ask10p under normal growing conditions (Fig. 2A, lane 4). We conclude that the A110V mutation does not significantly alter the association of Ask10p under nonstress conditions. We next determined whether the A110V mutation affected the association of the cyclin to Ask10p after exposure to oxidative damage. Extracts were prepared from cultures after a 30-min oxidative stress treatment and subjected to coimmunoprecipitation studies as just described. Immunoprecipitating Ask10p-HA retained similar amounts of Srb11pA110V-myc, as observed prior to stress treatment (Fig. 2A, lane 5). These results suggest that Ask10p functions through another element on Srb11p or that the coimmunoprecipitation experiments are unable to detect more subtle changes in Ask10p-Srb11p interactions.
Ask10p is hyperphosphorylated in response to H2O2 stress.
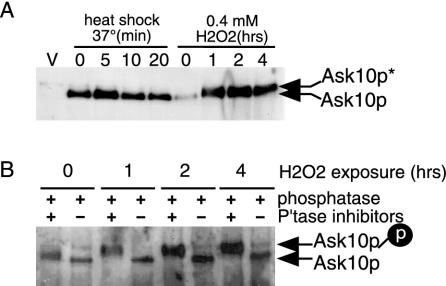
In most organisms, exposure to stress activates protein kinase cascades that transduces the signal to the nucleus to effect changes in the gene expression program (47). Therefore, we sought to determine whether Ask10p itself is phosphorylated in cells exposed to ROS. To test this hypothesis, Ask10p-HA levels were monitored by Western blotting of immunoprecipitates after H2O2 treatment as described above. The results showed that Ask10p levels do not change in response to H2O2 treatment (Fig. 5A). However, Ask10p mobility appears to be upshifted after H2O2 addition to the medium (asterisk). Interestingly, this upshift was not observed when cells were exposed to heat shock, suggesting that this effect is not a general property of stressed cells. To determine whether this alteration in mobility is due to phosphorylation, the protein A immunoprecipitation pellets from each time point were split and treated with either λ-phosphatase or λ-phosphatase plus phosphatase inhibitors (see Materials and Methods for details). Ask10p-HA was recovered and analyzed by Western blot by using polyacrylamide gel electrophoresis conditions that enhanced the migration difference between the two species of Ask10p. These studies revealed that, prior to the addition of H2O2 (T = 0), Ask10p is upshifted since treatment with phosphatase collapses this band into a faster-migrating form (Fig. 5B). These results indicate that Ask10p is phosphorylated even under “nonstress” conditions. Exposure to ROS resulted in more-extensive upshifting of Ask10p, indicating the presence of a hyperphosphorylated species, which was confirmed by phosphatase treatment. These results provide evidence that Ask10p is hyperphosphorylated specifically in response to H2O2 stress but not in response to heat shock. Furthermore, when combined with the results of the coimmunoprecipitation studies described above, these results implicate Ask10p as a nuclear target of an oxidative-stress signaling pathway.
FIG. 5.
Ask10p is phosphorylated in response to oxidative stress. (A) Ask10p mobility is altered by oxidative stress. Western blot analysis of HA immunoprecipitates derived from cultures of RSY869 harboring a chromosomally epitope-tagged allele of ASK10 subjected to either heat shock (37°C) or oxidative stress (0.4 mM H2O2) for the times indicated was carried out. The Ask10p and slower-migrating species (Ask10p*) are indicated by arrows. The vector control lane (V) controls for nonspecific cross-reactivity of the HA monoclonal antibody. (B) Ask10p is phosphorylated in response to oxidative stress. The oxidative stress time course depicted in panel A was repeated, and the Ask10p-HA immunoprecipitates were either treated with phosphatase or phosphatase plus inhibitor as indicated. The arrows indicate the phosphorylated and unphosphorylated species of Ask10p.
Ask10p phosphorylation is mediated through an alternative pathway for the Pkc1p MAP kinase cascade.
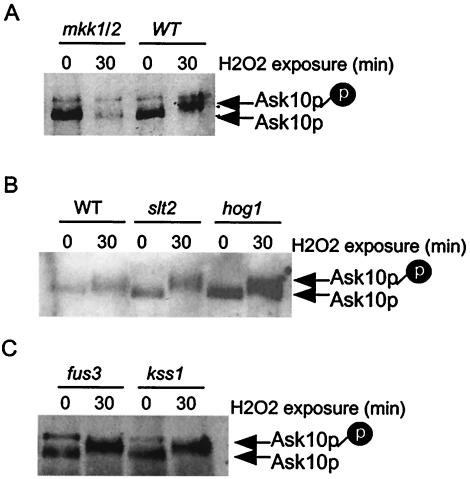
The results presented above indicate that Ask10p is phosphorylated in response to oxidative stress. To identify the kinase responsible for this activity, a candidate approach was initiated. Mutants defective for individual protein kinases that function in environmental sensing (Pho85p [55], Mkk1/2p [22], Rim11p [1], and Rim15p [57]), a developmental switch (Mck1p [41, 49] and Prr1p [5]), and checkpoint/transcription (Bub1p [45] and Ctk1p [30]) were tested. The mutant strains were transformed with a single-copy plasmid harboring an HA epitope-tagged derivative of ASK10 under the control of its own promoter (pAK3). The transformants were grown to mid-log phase and then split in half. One portion was harvested immediately for the T = 0 control, whereas the remaining half was subjected to oxidative stress (0.5 mM H2O2) for 30 min. Protein extracts were prepared from these samples, and Ask10p was visualized by Western analysis of immunoprecipitates. Of the strains tested, only a mutant defective for the redundant pair of MAP kinase kinases (or MEKs), Mkk1p/Mkk2p, displayed a defect in the H2O2-induced phosphorylation of Ask10p (Fig. 6A). Mkk1p and Mkk2p are considered redundant and therefore were always analyzed by using the double mutant. To determine whether their activity was redundant for Ask10p phosphorylation, these experiments were repeated in the individual signal mutants. No effect on oxidative-stress-induced phosphorylation of Ask10p was observed in either single mutant (data not shown), indicating that either Mkk1p or Mkk2p is sufficient for this process.
FIG. 6.
Ask10p phosphorylation requires Mkk1p/Mkk2p. (A, B, and C) Strains with the indicated genotypes containing ASK10-HA expression plasmid (pAK3) were harvested in mid-log phase (T = 0) or 30 min after treatment with H2O2 (0.5 mM). Extracts were prepared, and Ask10p-HA was visualized as described in Materials and Methods. The hyperphosphorylated form of Ask10p is indicated by the shaded “p.”
Mkk1p/Mkk2p are components of a well-characterized MAP kinase cascade (Bck1p→Mkk1p/Mkk2p→Slt2) that is activated by Pkc1p (reviewed in reference 18). This pathway responds to stress, most notably hypo-osmotic conditions and heat shock (25, 32). To determine whether Slt2p is necessary for transducing the oxidative stress signal from Mkk1p/Mkk2p, Ask10p phosphorylation was monitored in an slt2 mutant after treatment with H2O2. Interestingly, Ask10p phosphorylation was not altered in the slt2 mutant strain (Fig. 6B), suggesting that another MAP kinase was the recipient of the signal from Mkk1p/Mkk2p. Since the Hog1p MAP kinase is also stress activated (2, 11), its requirement or Ask10p phosphorylation was also tested. As observed for Slt2p, Hog1p is not necessary for oxidative stress-induced Ask10p phosphorylation (Fig. 6B). This experiment was repeated with mutants lacking the two remaining MAP kinases that function in vegetative cells: Kss1p (9) and Fus3p (13). Surprisingly, these mutants also did not affect the phosphorylation of Ask10p (Fig. 6C). These results indicate that, although Ask10p phosphorylation requires the MEKs Mkk1p/Mkk2p, this reaction is independent of Slt2p or any other known MAP kinase functioning in mitotic cells. These findings suggest that either there are redundancies in MAP kinase activity or that another, yet-unidentified protein kinase is able to transmit the oxidative stress signal (see Discussion).
DISCUSSION
Srb11p-Srb10p C-type cyclin-Cdk represses several genes required for the cellular response to stress. To relieve this repression, Srb11p is destroyed in cells subjected to stress such as heat shock or oxidative stress. We provide evidence here that Ask10p is a direct mediator of the normal ROS-induced destruction of Srb11p. First, Ask10p associates with Srb11p and two additional components of the RNA Pol II holoenzyme in vivo. Second, mutants lacking ASK10 display a threefold reduction in cyclin degradation kinetics after oxidative stress. The physiological relevance of this regulatory circuit is underscored by the ability of srb11 mutations to suppress the H2O2 hypersensitivity displayed in ask10 mutant strains. Interestingly, heat-induced destruction of Srb11p is independent of Ask10p, suggesting that the heat shock and oxidative stress pathways do not converge prior to triggering cyclin degradation. In addition, Ask10p is rapidly phosphorylated in ROS-treated cells but not in cultures after heat shock. Combined with the results of the coimmunoprecipitation studies, these data indicate that Ask10p directly links the ROS signaling pathway and the basal transcription machinery. Finally, the ROS-dependent phosphorylation of Ask10p requires the redundant MEKs Mkk1p/Mkk2p but not their cognate MAP kinase Slt2p or any of the other mitotic MAP kinases. These findings suggest the presence of a novel bifurcation in the Pkc1p MAP kinase cascade that utilizes an alternative pathway for transmitting the oxidative stress signal.
Srb11p was only partially stabilized in the ask10Δ mutant exposed to H2O2 (Fig. 3). Previous studies have identified mutants (plc1 and doa4) that more fully protected Srb11p from destruction under these conditions (7). These observations suggest that Ask10p is not the only pathway able to mediate ROS-induced Srb11p destruction. Sequence analysis identified two additional genes in yeast that displayed homology to ASK10 (Ynl1215p and Ypr115p). However, no difference in Srb11p turnover was observed in strains deleted for either gene after oxidative stress (data not shown). These results suggest that Ask10p is the only member of this gene family that regulates Srb11p turnover in response to oxidative stress. It should be noted that although the single mutants were readily constructed, double mutants of this gene family were not obtained in any pairwise combination, suggesting that these mutations may be synthetically lethal. Taken together, these results suggest that an Ask10p-independent pathway exists that directs Srb11p destruction in response to oxidative stress. However, given that ask10 mutants exhibit a significant growth defect when challenged with H2O2, Ask10p must have an important role to play in the cellular response to ROS. Moreover, since deleting SRB11 is able to rescue the H2O2-induced growth defect of ask10 mutants, this arm of the ROS response is critical for the cell to survive ROS-induced damage.
Ask10p was originally identified as a high-copy enhancer of Skn7p-dependent transcription (42). Skn7p is part of the two-component histidine phosphorylation pathway that provides a sensor to hyperosmotic conditions. The mechanism by which Ask10p enhances Skn7p-dependent transcription is unclear. One possible model is that Ask10p stimulates the regulatory pathway upstream of Skn7p. Alternatively, Ask10p could enhance Skn7p activity at the promoter level. Our finding that Ask10p associates with the RNA Pol II holoenzyme favors the latter model. One simplistic mechanism to explain the data presented here suggests that Skn7p and Srb10p-Srb11p provide antagonistic functions. For example, Ask10p's stimulation of Skn7p activity when overexpressed could be the result of precocious destruction of the cyclin. However, several pieces of data argue against this model. First, several genes induced by Skn7p after oxidative stress (OCH1 [34] and TRX1 and TRR1 [46]) are not derepressed in srb11 mutants (21). Second, overexpression of ASK10 did not induce Srb11p destruction in the absence of stress (data not shown). Therefore, we can find no evidence for a functional interaction between Srb10-Srb11p and Skn7p. We did, however, notice that overexpression of Ask10p induced transcription of the ADH1 promoter (T. J. Cohen and R. Strich, unpublished results). In the initial report of Ask10p function, Skn7p was fused to the lexA DNA-binding domain under the control of the ADH1 promoter (42). Therefore, the enhancement of Skn7p activation function may be indirect and related to increased expression of the ADH1-SKN7 fusion gene itself.
The finding that Ask10p is phosphorylated in response to oxidative stress, but not heat shock, suggests that the two stress signaling pathways are separable but converge at Srb11p. This result is consistent with our earlier results that multiple cis-acting destruction signals are present on Srb11p that appear to respond to different stresses (6). What is the role of Ask10p phosphorylation? One obvious possibility is that modifying Ask10p triggers Srb11p destruction. Phosphorylation of ubiquitin ligases has been shown to be a key step in activation (58). Since Ask10p does not contain any motifs associated with known ubiquitin ligases (e.g., ring fingers [for a review, see reference 56]), its role may be to serve as a docking site for a ligase or to facilitate Srb11p translocation to a different compartment where ubiquitination and/or destruction takes place. Indeed, a green fluorescent protein-Srb11p fusion protein does relocalize to the cytoplasm after stress (E. Krasely and R. Strich, unpublished observations). Similarly, the transcriptional repressor Matα2p is transported to the endoplasmic reticulum, where it is ubiquitinated by Doa10p prior to destruction (54). Further experiments are under way to determine what role, if any, localization plays in mediating Srb11p degradation and to determine whether Ask10p is involved in this process.
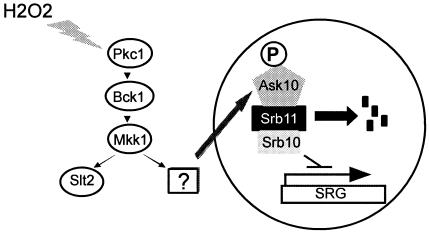
The present study demonstrates that Ask10p is phosphorylated in response to oxidative stress and that this modification requires the redundant MEKs Mkk1p/Mkk2p. Mkk1p/Mkk2p are downstream of a signaling cascade that includes the control kinase Pkc1p and the MEK kinase Bck1p (Fig. 7). This pathway has a well-established role in transducing changes in environmental osmolarity and heat shock (15). However, our data indicate that this pathway, as defined by Ask10p phosphorylation, is activated in cells exposed to oxidative stress but not in cells exposed to heat shock. These findings are consistent with other signal transduction pathways that are able to alter their final target depending on the impute stimulus. However, the finding that Ask10p phosphorylation is independent of Slt2p, or any of the MAP kinases that function during mitotic cell division, was unanticipated. One possibility is that Slt2p and another MAP kinase, perhaps Hog1p, have overlapping activities with respect to signaling oxidative stress. However, a previous report failed to detect the activation of the MAP kinase Slt2p after exposure to oxidative stress (25). These results argue against redundant functions and are more consistent with the existence of a novel bifurcation in the Pkc1p MAP kinase cascade. This model would predict that rather than Mkk1p or Mkk2p activating a MAP kinase, another intermediate is used to transduce the signal to the nucleus (boxed question mark, Fig. 7).
FIG. 7.
Model for oxidative stress-induced destruction of Srb11p. H2O2 activates the Pkc1p-dependent MAP kinase cascade, including the redundant MAP kinase kinases Mkk1/Mkk2. This pathway bifurcates following this step and a new intermediary (boxed question mark) is proposed that transduces the signal to Ask10p. Phosphorylation of Ask10p triggers Srb11p degradation, thus relieving Srb10p-dependent repression of stress response genes (SRG).
An alternative explanation to the unknown intermediary kinase is that either Mkk1p or Mkk2p is directly phosphorylating Ask10p. The possibility is unlikely for two reasons. First, MEKs target a specific domain on MAP kinases termed the T-loop that contains the modified resides Thr-X-Tyr (where X is any amino acid). No other substrate recognition site has been reported to date. Since Ask10p does not contain a T-loop motif, the direct modification of Ask10p by Mkk1p/Mkk2p would dictate the unlikely involvement of a novel noncanonical recognition site. Second, although transient MEK shuttling from the cytoplasm through the nucleus has been reported in mammalian cells (23), this phenomenon has not been clearly demonstrated in yeast. Therefore, we feel it is most likely that Mkk1p/Mkk2p work through another protein kinase (or kinases) that ultimately modifies Ask10p. Identifying this kinase and its regulation may provide important new insight into how signal transduction cascades can affect gene expression through direct modification of the basal transcription machinery.
Acknowledgments
We thank E. Winter, E. Golemis, and J. Jaehning for strains, plasmids, and antibodies. We also thank K. F. Cooper and J. Chernoff for critical reading of the manuscript and helpful suggestions.
This work was supported by the National Institutes of Health (GM57842) to R.S. and NHSA to L.H.R. Institutional support was also provided by the National Cancer Center (Comprehensive Cancer Center core grant CA 06927) and an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Bowdish, K. S., H. E. Yuan, and A. P. Mitchell. 1994. Analysis of RIM11, a yeast protein kinase that phosphorylates the meiotic activator IME1. Mol. Cell. Biol. 14:7909-7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osomsensing signal transduction pathway in yeast. Science 259:1760-1763. [DOI] [PubMed] [Google Scholar]
- 3.Brown, J. L., H. Bussey, and R. C. Stewart. 1994. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 13:5186-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, J. L., S. North, and H. Bussey. 1993. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall beta-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J. Bacteriol. 175:6908-6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burchett, S. A., A. Scott, B. Errede, and H. G. Dohlman. 2001. Identification of novel pheromone-response regulators through systematic overexpression of 120 protein kinases in yeast. J. Biol. Chem. 276:26472-26478. [DOI] [PubMed] [Google Scholar]
- 6.Cooper, K. F., M. J. Mallory, J. S. Smith, and R. Strich. 1997. Stress and developmental regulation of the yeast C-type cyclin UME3 (SRB11/SSN8). EMBO J. 16:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper, K. F., M. J. Mallory, and R. Strich. 1999. Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires the Phosphatidylinositol-specific phospholipase C and the 26S proteasome. Mol. Cell. Biol. 19:3338-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper, K. F., and R. Strich. 1999. Functional analysis of the yeast C-type cyclin Ume3p/Srb11p-RNA polymerase II holoenzyme interaction. Gene Exp. 8:43-57. [PMC free article] [PubMed] [Google Scholar]
- 9.Courchesne, W. E., R. Kunisawa, and J. Thorner. 1989. A putative protein kinase overcomes pheromone-induced arrest of cell cycling in Saccharomyces cerevisiae. Cell 58:1107-1119. [DOI] [PubMed] [Google Scholar]
- 10.Craig, E. A. 1993. The heat-shock response of Saccharomyces cerevisiae, p. 501-538. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast saccharomyces, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.Davenport, K. R., M. Sohaskey, Y. Kamada, D. E. Levin, and M. C. Gustin. 1995. A second osmosensing signal transduction pathway in yeast. Hypotonic shock activates the PKC1 protein kinase-regulated cell integrity pathway. J. Biol. Chem. 270:30157-30161. [DOI] [PubMed] [Google Scholar]
- 12.Dodou, E., and R. Treisman. 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:1848-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elion, E. A., P. L. Grisafi, and G. R. Fink. 1990. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell 60:649-664. [DOI] [PubMed] [Google Scholar]
- 14.Flick, J. S., and J. Thorner. 1993. Genetic and biochemical characterization of a phosphatidylinositol-specific phospholipase C in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:5861-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrington, T. P., and G. L. Johnson. 1999. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr. Opin. Cell Biol. 11:211-218. [DOI] [PubMed] [Google Scholar]
- 16.Gorner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schuller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenblat, J. 1997. RNA polymerase II holoenzyme and trancriptional regulation. Curr. Opin. Cell Biol. 9:310-319. [DOI] [PubMed] [Google Scholar]
- 18.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gyuris, J., E. A. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 20.Hengartner, C. J., V. E. Myer, S.-M. Liao, C. J. Wilson, S. S. Koh, and R. A. Young. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2:43-53. [DOI] [PubMed] [Google Scholar]
- 21.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 22.Irie, K., M. Takase, K. S. Lee, D. E. Levin, H. Araki, K. Matsumoto, and Y. Oshima. 1993. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol. Cell. Biol. 13:3076-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaaro, H., H. Rubinfeld, T. Hanoch, and R. Seger. 1997. Nuclear translocation of mitogen-activated protein kinase kinase (MEK1) in response to mitogenic stimulation. Proc. Natl. Acad. Sci. USA 94:3742-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung, U. S., A. K. Sobering, M. J. Romeo, and D. E. Levin. 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46:781-789. [DOI] [PubMed] [Google Scholar]
- 25.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9:1559-1571. [DOI] [PubMed] [Google Scholar]
- 26.Kofoid, E. C., and J. S. Parkinson. 1988. Transmitter and receiver modules in bacterial signaling proteins. Proc. Natl. Acad. Sci. USA 85:4981-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krems, B., C. Charizanis, and K. D. Entian. 1996. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet. 29:327-334. [DOI] [PubMed] [Google Scholar]
- 28.Kuchin, S., I. Treich, and M. Carlson. 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 97:7916-7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274:16040-16046. [DOI] [PubMed] [Google Scholar]
- 30.Lee, J. M., and A. L. Greenleaf. 1991. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1:149-167. [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, K. S., K. Irie, Y. Gotoh, Y. Watanabe, H. Araki, E. Nishida, K. Matsumoto, and D. E. Levin. 1993. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol. Cell. Biol. 13:3067-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin, D. E., F. O. Fields, R. Kunisawa, J. M. Bishop, and J. Thorner. 1990. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell 62:213-224. [DOI] [PubMed] [Google Scholar]
- 33.Li, S., A. Ault, C. L. Malone, D. Raitt, S. Dean, L. H. Johnston, R. J. Deschenes, and J. S. Fassler. 1998. The yeast histidine protein kinase, sln1p, mediates phosphotransfer to two response regulators, ssk1p and skn7p. EMBO J. 17:6952-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, S., S. Dean, Z. Li, J. Horecka, R. J. Deschenes, and J. S. Fassler. 2002. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol. Biol. Cell 13:412-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao, S.-M., J. Zhang, D. A. Jeffery, A. J. Koleske, C. M. Thompson, D. M. Chao, M. Viljoen, H. J. J. van Vuuren, and R. A. Young. 1995. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374:193-196. [DOI] [PubMed] [Google Scholar]
- 36.Longtine, M. S., A. r. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 37.Madden, K., Y. J. Sheu, K. Baetz, B. Andrews, and M. Snyder. 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275:1781-1784. [DOI] [PubMed] [Google Scholar]
- 38.Martinez-Pastor, M. T., G. Marchler, C. Schuller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15:2227-2235. [PMC free article] [PubMed] [Google Scholar]
- 39.Moradas-Ferreira, P., V. Costa, P. Piper, and W. Mager. 1996. The molecular defenses against reactive oxygen species in yeast. Mol. Microbiol. 19:651-658. [DOI] [PubMed] [Google Scholar]
- 40.Morgan, B. A., G. R. Banks, W. M. Toone, D. Raitt, S. Kuge, and L. H. Johnston. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neigeborn, L., and A. P. Mitchell. 1991. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 5:533-548. [DOI] [PubMed] [Google Scholar]
- 42.Page, N., J. Sheraton, J. L. Brown, R. C. Stewart, and H. Bussey. 1996. Identification of ASK10 as a multicopy activator of Skn7p-dependent transcription of a HIS3 reporter gene. Yeast 12:267-272. [DOI] [PubMed] [Google Scholar]
- 43.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 44.Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865-875. [DOI] [PubMed] [Google Scholar]
- 45.Roberts, B. T., K. A. Farr, and M. A. Hoyt. 1994. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol. Cell. Biol. 14:8282-8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross, S. J., V. J. Findlay, P. Malakasi, and B. A. Morgan. 2000. Thioredoxin peroxidase is required for the transcriptional response to oxidative stress in budding yeast. Mol. Biol. Cell 11:2631-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santoro, N., and D. J. Thiele. 1997. Yeast stress responses, p. 171-203. In S. Hohmann and W. H. Mager (ed.), Molecular biology intelligence unit. R. G. Landes Co., Austin, Tex.
- 48.Serebriiskii, I., V. Khazak, and E. A. Golemis. 1999. A two-hybrid dual bait system to discriminate specificity of protein interactions. J. Biol. Chem. 274:17080-17087. [DOI] [PubMed] [Google Scholar]
- 49.Shero, J. H., and P. Hieter. 1991. A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1). Genes Dev. 5:549-560. [DOI] [PubMed] [Google Scholar]
- 50.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh, K. K. 2000. The Saccharomyces cerevisiae Sln1p-Ssk1p two-component system mediates response to oxidative stress and in an oxidant-specific fashion. Free Radic. Biol. Med. 29:1043-1050. [DOI] [PubMed] [Google Scholar]
- 52.Stephen, D., S. Rivers, and D. Jamieson. 1995. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol. Microbiol. 16:415-423. [DOI] [PubMed] [Google Scholar]
- 53.Surosky, R. T., R. Strich, and R. E. Esposito. 1994. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol. Cell. Biol. 14:3446-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swanson, R., M. Locher, and M. Hochstrasser. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev. 15:2660-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toh-e, A., K. Tanaka, Y. Uesono, and R. B. Wickner. 1988. PHO85, a negative regulator of the PHO system, is a homolog of the protein kinase gene, CDC28, of Saccharomyces cerevisiae. Mol. Gen. Genet. 214:162-164. [DOI] [PubMed] [Google Scholar]
- 56.Tyers, M., and P. Jorgensen. 2000. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr. Opin. Genet. Dev. 10:54-64. [DOI] [PubMed] [Google Scholar]
- 57.Vidan, S., and A. P. Mitchell. 1997. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol. Cell. Biol. 17:2688-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zachariae, W., and K. Nasmyth. 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13:2039-2058. [DOI] [PubMed] [Google Scholar]