In Saccharomyces cerevisiae, ammonia can be incorporated into the amino group of glutamate, the source of 80% of cellular nitrogen, by two pathways: the reductive amination of 2-ketoglutarate, catalyzed by glutamate dehydrogenase, in which NADPH serves as the source of electrons, or by the ATP-dependent synthesis of glutamine from glutamate and ammonia catalyzed by glutamine synthetase. The latter reaction is followed by the reductive transfer of the amide group of glutamine to 2-ketoglutarate that is catalyzed by glutamate synthase and in which NADH serves as the source of electrons. In cells growing with an excess of glucose as the source of carbon and energy and ammonia as the sole source of nitrogen, the rate of growth is determined by the ability of the cell to use glutamate dehydrogenase to assimilate ammonia and is not affected by mutations resulting in the loss of glutamate synthase. It can, however, be shown that the use of the NADH-linked glutamate synthase in place of the NADPH-linked glutamate dehydrogenase results in a more efficient utilization of glucose, which is a distinct advantage when the availability of glucose limits the rate of growth. The reduction of the intracellular concentration of 2-ketoglutarate during glucose restriction favors the switch from the use of glutamate dehydrogenase to the use of glutamate synthase by the threefold greater affinity of glutamate synthase for 2-ketoglutarate and by the irreversibility of the reaction catalyzed by this enzyme.
Like many other microorganisms (Escherichia coli, for example), the yeast Saccharomyces cerevisiae grows well in a medium containing glucose as the sole source of carbon and energy and ammonia as the sole source of nitrogen. Ammonia is incorporated into the amino group of glutamate, the source of 80% of the cellular nitrogen, and into the amide group of glutamine, the source of the remaining 20% (8, 9).
The reaction responsible for the synthesis of glutamine is catalyzed by glutamine synthetase:
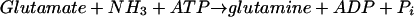 |
(1) |
Both organisms can produce glutamate by the reductive amination of 2-ketoglutarate catalyzed by glutamate dehydrogenase:
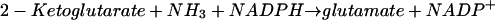 |
(2) |
In addition, both organisms can produce glutamate in a two-step process in which the first step is catalyzed by glutamine synthetase (reaction 1) and the second step is catalyzed by glutamate synthase. In this reaction, glutamine serves as the donor of the amino group, 2-ketoglutarate as its receptor, and NADPH in bacteria or NADH in S. cerevisiae as the reducing agent:
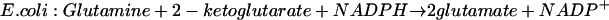 |
(3) |
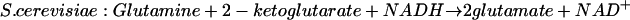 |
(4) |
It is evident that reaction 1 combined with reaction 3 or 4 accomplish together exactly what reaction 2 accomplishes by itself, i.e., the conversion of 2-ketoglutarate and ammonia to glutamate, but in the case of reaction 1 plus reaction 3 or 4, this is done at the cost of the hydrolysis of ATP.
In the bacterial case, the essential pathway to glutamate is via reactions 1 and 3, as shown by the fact that mutants lacking glutamate synthase (reaction 3) fail to grow in a medium containing ammonia at a concentration less than 1 mM or containing serine, a compound which is slowly hydrolyzed to yield ammonia, as the sole source of nitrogen (9). On the other hand, mutants lacking glutamate dehydrogenase (reaction 2) grow as well as the parent strain in these media. The only defect that results from the lack of glutamate dehydrogenase is a slightly lower rate of growth with ammonia as the source of nitrogen when a poor energy source replaces glucose; apparently in this case, the lower cost in ATP favors the synthesis of glutamate by glutamate dehydrogenase (16). The reason that glutamate dehydrogenase (reaction 2) cannot be used by bacteria when the ammonia concentration of the medium is less than 1 mM is the high Km of the enzyme for ammonia. The hydrolysis of ATP in reactions 1 and 3 ensures a favorable equilibrium for the synthesis of glutamate.
By the same reasoning, reactions 1 and 4 should be required for the synthesis of glutamate by S. cerevisiae when the concentration of ammonia in the growth medium is low. However, experiments with mutants of S. cerevisiae have led to a different conclusion: the lack of glutamate synthase (reaction 4) does not prevent growth of the mutant in a medium containing a concentration of ammonia less than 1 mM or when cytosine, a poor source of ammonia, serves as the source of nitrogen. On the other hand, a mutant lacking glutamate dehydrogenase (reaction 2) that depends on reactions 1 and 3 to generate glutamate grows with ammonia as the source of nitrogen at one-half the rate of its parent strain (8, 13).
These observations present us with two problems regarding ammonia assimilation by S. cerevisiae. (i) In spite of its unfavorable equilibrium, how can glutamate dehydrogenase catalyze the formation of glutamate when the concentration of ammonia in the medium is insufficient to allow reaction 2 to proceed? (ii) What is the role of glutamate synthase (reaction 4), and why does this enzyme use NADH rather than the more abundant NADPH, which is the preferred reducing agent for biosynthetic reactions?
One way to use reaction 1 for the synthesis of glutamate when the concentration of ammonia in the growth medium is low is to increase the intracellular concentration of ammonia to a level exceeding that of the growth medium. It has been shown that the ability of S. cerevisiae to grow in a medium containing ammonia at a concentration less than 10 mM depends on the presence of proteins located in the cell membrane that are the products of the MEP genes. The product of MEP1 is a high-affinity ammonia permease and that of MEP3 is a low-affinity, but high-capacity ammonia permease (11, 12). The product of MEP2, another high-affinity ammonia permease, functions as an ammonia sensor in the regulation of pseudohyphal differentiation (7). The energy necessary for the concentrative uptake of ammonia by these permeases is provided by the intracellular hydrolysis of ATP (18).
Accordingly, in contrast to what occurs in enteric bacteria, where glutamine is an intermediate in the synthesis of glutamate (reactions 1 and 3), in S. cerevisiae, glutamine and glutamate are synthesized independently by reactions 1 and 2, respectively. This may explain why in enteric bacteria a single transcription factor, the product of glnG (ntrC), is responsible for the activation of the expression of nitrogen-regulated genes in response to a drop in the intracellular concentration of glutamine (9), while in S. cerevisiae, two transcription factors, the products of GLN3 and NlL1, activate the expression of these genes in response to a drop in the intracellular concentrations of glutamine and glutamate, respectively (10).
To understand the potential role of the NADH-linked glutamate synthase, it is necessary to consider that in studies of ammonia assimilation glucose is used as the source of carbon and energy. In S. cerevisiae, glucose exerts strong catabolite repression on the enzymes required for respiration and on the enzymes of the tricarboxylic acid cycle beyond those required for the synthesis of 2-ketoglutarate (4). Consequently, during the exponential phase of growth, the production of ATP depends entirely on the conversion of glucose to ethanol and CO2. Under these conditions, the rate of growth is determined by the ability of the cell to assimilate ammonia by using the NADPH-linked glutamate dehydrogenase, as shown by the following observations: substitution of glutamine for ammonia increases the rate of growth, the loss of glutamate dehydrogenase decreases the rate of growth, and the loss of glutamate synthase has no effect on the rate of growth (8).
This rapid phase of growth on glucose reduces the glucose concentration to a level which permits only very slow growth and is no longer adequate for catabolite repression. It is during this second phase of growth that the enzymes necessary for respiration and for the operation of the tricarboxylic acid cycle are formed, thus enabling the cell to resume more rapid growth by utilizing the accumulated ethanol as a source of energy and carbon (14).
It is evident that the efficient utilization of glucose as the source of energy and carbon is more important during growth on limiting glucose than when glucose is abundantly available. A comparison of the amount of glucose needed to produce glutamate using glutamate dehydrogenase with that needed to produce glutamate using glutamate synthase shows that the latter reaction permits the more efficient utilization of glucose and should therefore be preferable during glucose limitation.
The synthesis of amino acids in cells using glucose as the source of carbon and ammonia as the source of nitrogen requires the withdrawal of pyruvate from the pathway of glycolysis; the pyruvate then serves as the source of keto acids which are converted to the corresponding amino acids in reactions catalyzed by amino transferases with glutamate as the amino donor. The glutamate consumed in these reactions must then be restored through reductive amination of 2-ketoglutarate by either glutamate dehydrogenase (reaction 2) or glutamine synthetase and glutamate synthase (reactions 1 and 4).
It is apparent that the withdrawal of pyruvate from the pathway of glycolysis prevents the formation of acetaldehyde from pyruvate and consequently the restoration of NAD+ by the reduction of acetaldehyde to ethanol by NADH. Consequently, to continue the synthesis of ATP by glycolytic reactions it is necessary to regenerate NAD+ from NADH by other means. This regeneration is easily achieved in cells using glutamate synthase for the synthesis of glutamate by the use of NADH in the reductive amination of 2-ketoglutarate by glutamine (reactions 1 and 4). On the other hand, in cells using glutamate dehydrogenase, NADPH is the agent responsible for the reductive amination of 2-ketoglutarate by ammonia (reaction 2), and since S. cerevisiae does not possess a transhydrogenase which would allow NADPH to reduce NAD+ to NADH, the regeneration of NAD+ is achieved by the formation of glycerol from glucose (15):
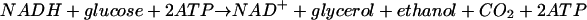 |
(5) |
It is apparent that in this case glucose is consumed to regenerate NAD+ without any gain of ATP.
If we now compare the cost of the synthesis of one molecule of glutamate from 2-ketoglutarate and ammonia by glutamate synthase (reactions 1 and 3) with that by glutamate dehydrogenase (reactions 2 and 5), it is apparent that in both reaction sequences ATP is required: for the formation of glutamine in the case of glutamate synthase and to raise the intracellular concentration of ammonia in the case of glutamate dehydrogenase. The important difference is that the synthesis of one molecule of glutamate by glutamate dehydrogenase instead of by glutamine synthetase-glutamate synthase requires the consumption of an additional molecule of glucose.
To assess the importance of this additional consumption in relation to the resources of the cell, we have to consider that in the absence of respiration the synthesis of 1 g (dry weight) of cell mass requires the conversion of approximately 40 mmol of glucose to ethanol and CO2 (14, 15). This amount of cell mass contains 140 mg of nitrogen, of which 80% is derived from the amino group of glutamate. Thus, the formation of 1 g of cell mass requires the synthesis of 8.0 mmol of glutamate from 2-ketoglutarate and ammonia. Consequently, the use of an additional molecule of glucose for the synthesis of glutamate by the NADPH-linked glutamate dehydrogenase would reduce the amount of glucose available to produce energy for the synthesis of 1 g of cell material from 40 to 32 mmol, a reduction of approximately 20%. A reduction in the efficiency of glucose utilization of this size is not significant for cells growing rapidly with an abundant supply of glucose; however, for cells growing anaerobically on a growth rate-limiting concentration of glucose, it would clearly be advantageous to use glutamate synthase to make 20% more of the glucose available for the synthesis of cell material. This view is supported by the observation that the lack of glutamate synthase reduces the biomass of cells grown in a glucose-limited chemostat but not that of cells grown in a nitrogen-limited chemostat (6).
The preferential utilization of glutamate synthase over glutamate dehydrogenase by glucose-limited cells may be explained by the properties of these enzymes. Glutamate synthase has greater affinity for 2-ketoglutarate than does glutamate dehydrogenase (Kms of 0.14 and 0.36 mM, respectively) and, in contrast to glutamate dehydrogenase, catalyzes an irreversible reaction (5, 17). The low intracellular concentration of 2-ketoglutarate in glucose-limited cells (2) would therefore favor glutamate synthase as the agent responsible for ammonia assimilation.
According to this analysis, glutamate synthase plays its important role during the transition from fermentative to respiratory growth.
It has been reported that the transition from glucose to ethanol as the source of carbon and energy results in the gradual replacement of the NADPH-dependent glutamate dehydrogenase, encoded by the GDH1 gene, by another NADPH-dependent glutamate dehydrogenase, encoded by the GDH3 gene, with even lower affinity for 2-ketoglutarate (Km, 1 mM) (3). This replacement favors the oxidation of 2-ketoglutarate to serve as the source of energy over its reductive amination to glutamate. In addition, any accumulation of glutamate results in the induction of the NAD+-linked glutamate dehydrogenase, encoded by GDH2, which reconverts glutamate to ammonia and 2-ketoglutarate (1, 3).
Thus far, the possible effect of the deletion of GLT1, the structural gene for glutamate synthase, on growth with ethanol as the source of carbon and ammonia as the source of nitrogen has not been examined; however, the observation that deletion of GDH1 and GDH3, which places full responsibility for ammonia assimilation on glutamate synthase, reduces the rate of growth with ethanol as the source of carbon and ammonia as the source of nitrogen by one-half (3) indicates that in this medium, too, NADPH-linked glutamate dehydrogenase has an important role in ammonia assimilation.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Coschigano, P. W., S. M. Miller, and B. Magasanik. 1991. Physiological and genetic analysis of the carbon regulation of the NAD-dependent glutamate dehydrogenase of Saccharomyces cerevisiae. Mol. Cell. Biol. 11:4455-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox, K. H., J. J. Tate, and T. G. Cooper. 2002. Cytoplasmic compartmentation of Gln3 during nitrogen catabolite repression and the mechanism of its nuclear localization during carbon starvation in Saccharomyces cerevisiae. J. Biol. Chem. 277:37559-37566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLuna, A., A. Avenadaño, L. Riego, and A. Gonzalez. 2001. NADP-glutamate dehydrogenase isoenzymes of Saccharomyces cerevisiae. Purification, kinetic properties, and physiological roles. J. Biol. Chem. 276:43775-43783. [DOI] [PubMed] [Google Scholar]
- 4.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holzer, M., and S. Schneider. 1957. Anreicherung und Trennung einer DPN-spezifischen und einer TPN-spezifischen Glutaminsaure-dehydrogenase aus Hefe. Biochem. Z. 329:89-95. [PubMed] [Google Scholar]
- 6.Lacerda, V., A. Marsden, and W. M. Ledingham. 1992. Ammonia utilization in S. cerevisiae under chemostatic growth. Appl. Biochem. Biotechnol. 32:15-21. [DOI] [PubMed] [Google Scholar]
- 7.Lorenz, M. C., and J. Heitman. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magasanik, B. 1992. Regulation of nitrogen utilization, p. 283-317. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces cerevisiae: gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Magasanik, B. 1996. Regulation of nitrogen utilization, p. 1344-1356. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C.
- 10.Magasanik, B., and C. A. Kaiser. 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290:1-18. [DOI] [PubMed] [Google Scholar]
- 11.Marini, A. M., S. Soussi-Boudekou, S. Vissers, and B. Andre. 1997. A family of ammonia transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4282-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marini, A. M., J. Y. Springael, W. B. Frommer, and B. André. 2001. Cross-talk between ammonium transporters in yeast and interference by the soybean SAT1 protein. Mol. Microbiol. 35:378-385. [DOI] [PubMed] [Google Scholar]
- 13.Miller, S. M., and B. Magasanik. 1990. Role of NAD-linked glutamate dehydrogenase in nitrogen metabolism in Saccharomyces cerevisiae. J. Bacteriol. 172:4927-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nissen, T. L., C. W. Hamann, M. C. Kielland-Brandt, J. Nielsen, and J. Villadsen. 2000. Anaerobic and aerobic batch cultivation of Saccharomyces cerevisiae mutants impaired in glycerol synthesis. Yeast 16:463-474. [DOI] [PubMed] [Google Scholar]
- 15.Nissen, T. L., M. Anderlund, J. Nielsen, J. Villadsen, and M. C. Kielland-Brandt. 2001. Expression of cytoplasmic transhydrogenase in Saccharomyces cerevisiae results in formation of 2-oxoglutarate due to depletion of the NADPH pool. Yeast 18:19-32. [DOI] [PubMed] [Google Scholar]
- 16.Reitzer, L., and B. L. Schneider. 2001. Metabolic context and possible physiological themes of δ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65:422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roon, R. Y., L. Eisen, and F. Larimore. 1974. Glutamate synthase: properties of the reduced nicotinamide adenine dinucleotide-dependent enzyme from Saccharomyces cerevisiae. J. Bacteriol. 118:89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soupene, E., R. Ramirez, and S. Kustu. 2001. Evidence that fungal MEP proteins mediate diffusion of the uncharged species NH3 across the cytoplasmic membrane. Mol. Cell. Biol. 21:5733-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]


