Abstract
CD22, a negative regulator of B cell antigen receptor signaling, binds glycoconjugates terminating in α2, 6 sialic acid. The physiological ligand(s) for CD22 remain unknown. We asked whether the sialic acid binding domains are necessary for CD22 to function as a negative regulator. We generated two mutants that lack sialic acid binding activity and expressed them in a novel CD22−/− murine B cell line. Anti-IgM activated B cells expressing either CD22 mutant had greater Ca2+ responses than cells expressing wild-type CD22. Each variant also had reduced CD22 tyrosine phosphorylation and Src homology 2 domain–containing protein tyrosine phosphatase-1 association. These data suggest that the α2, 6 sialic acid ligand binding activity of CD22 is critical for its negative regulatory functions.
Keywords: signal transduction, SHP-1, calcium, siglec, tyrosine phosphorylation
Introduction
CD22 is a type I transmembrane glycoprotein expressed on mature B cells that specifically recognizes α2, 6 sialic acid. It has seven Ig-like extracellular domains and three cytoplasmic immunoreceptor tyrosine-based inhibitory motifs (ITIMs). CD22 is a member of the rapidly growing family of Siglec (sialic acid–binding Ig domain–containing lectin) proteins (for a review, see reference 1). Siglecs, such as sialoadhesin (Siglec-1), CD22 (Siglec-2), CD33 (Siglec-3), myelin-associated glycoprotein (Siglec-4), Siglec-5, Siglec-6, Siglec7, Siglec-8, Siglec-9, Siglec-10, mSiglec-E, MIS, and S2V, are type 1 transmembrane proteins containing multiple extracellular Ig-like domains that recognize different types and linkages of sialic acid (1). Most Siglecs, including CD22, have one or more ITIMs in their cytoplasmic domains, which suggests they inhibit cellular activity (1). Each Siglec has its own unique tissue distribution, with CD22 being the only α2, 6 sialic acid binding member that is expressed exclusively on B lymphocytes (1).
CD22 is a well-characterized negative regulator of B cell antigen receptor (BCR) signaling (2). Upon BCR cross-linking, its cytoplasmic portion is rapidly tyrosyl phosphorylated and recruits the Src homology 2 domain–containing protein tyrosine phosphatase (SHP)-1 (3, 4), which is believed to mediate inhibitory signaling by CD22 (5). Consistent with a role for CD22 in negative regulation of BCR signaling, CD22−/− mice exhibit an increased and prolonged Ca2+ response after BCR stimulation, an expanded peritoneal B-1 cell population, and increased levels of autoantibodies (6–9).
Although the role of the CD22 cytoplasmic domain is fairly clear, the function of its highly conserved extracellular domain is poorly understood. Previous work established that the most NH2-terminal Ig domains, Ig1 and Ig2, mediate α2, 6 sialic acid–binding activity (10). Mutagenesis was used to identify residues within the Ig domains essential for binding (10, 11). However, the physiological ligand(s) for CD22 remain unknown. IgM and CD45 have been proposed to be major CD22 ligands, but the physiological relevance of these interactions has remained controversial and other molecules also may bind (10, 12–14).
Interestingly, in resting B cells CD22 is bound to an endogenous ligand(s) and is unavailable for binding to exogenous molecules (15). This “masking” of CD22 can be relieved by treatment with sialidase or a combination of anti-IgM and anti-CD40 (15). The significance of CD22 binding to its endogenous ligands for B cell function remains unknown. Thus, it was of interest to explore the relationship between the binding of CD22 to its endogenous ligand(s) and its negative regulatory function in B cells. To address this issue, we generated two CD22 mutants that lack sialic acid binding activity and introduced them into J2–44, a novel immortalized B cell line derived from a CD22−/− mouse.
Materials and Methods
Cell Line and Culture.
The CD22−/− J2–44 mouse cell line was generated by immortalizing splenic B cells from (B6 × 129)F2 CD22−/− mice (6) with the J2 virus (16). Details of the generation and characterization of these cells will be published elsewhere. Cells were cultured in RPMI 1640 (Life Technologies) supplemented with 20% heat-inactivated FCS, 20 mM Hepes, pH 7.4, 1 mM sodium pyruvate, 1 mM nonessential amino acids (GIBCO BRL), 50 μM 2-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate.
Flow Cytometry.
Immunostaining was performed as described previously (15). Antibodies used included biotinylated anti-B220/CD45R antibody (BD PharMingen), biotinylated-B7.6 anti-IgM, and biotinylated-NIM-R6 anti-CD22 mAb. Biotinylated-Sambucus Nigra Agglutinin was obtained from Vector Laboratories. Flow cytometric data were obtained and analyzed using a FACSCalibur™ and WinMDI software.
Retrovirus Generation and B Cell Infections.
Previously modified murine CD22.2 cDNA (17) was cloned into the NotI and EcoRI sites of the modified MSCV 2.2 vector, which expresses inserted genes under the control of the MCSV long terminal repeats and GFP by means of an internal ribosome entry sequence (18). Mutations in CD22 were introduced in this MSCV CD22.2 construct by overlap-extension PCR (19). The two common primers used were the forward primer MSCV-forward: 5′CCTACATCGTGACCTGGGAAG3′ and the reverse primer 5′CD22-R2: 5′ GCCACGGTCCTGAGTTTCG 3′. The primers used to generate the DEL mutant were the reverse primer Ig12DR: 5′CTTGATCTCGAGCTTCGGGGTGTACCGAGCCGAAGCCAC 3′ and the forward primer Ig12DF: 5′ ACCCCGAAGCTCGAGATCAAGGTC3′. The primers used to generate the P2 mutant were reverse primer: MCD22R 5′ TTCAGTCCCTGCGTCATCGCCAACCCCAGATTCCC 3′ and the forward primer MCD22F: 5′ATGACCGCAGGGACTGAAGAATGGATGGAGCCCATTCA3′ (mutations underlined). The product of the PCR reactions was digested with EcoRI and HpaI. The digested fragment was used to replace the WT sequence. DNA sequencing confirmed that only the desired mutations were present.
Retroviruses were generated by transfection of 293T cells with plasmid DNA (10 μg) as described previously (20, 21). Retrovirus-containing 293T cell supernatants were collected 48 h after transfection and used to infect J2–44 cells. The retrovirus-infected cell (GFP+) populations were purified by FACS® and cultured as described above. Sorted, GFP+, cell populations were used within a few weeks after their generation. This approach avoided clonal variation and generated lines that expressed CD22 and mIgM for several months. Each experiment was repeated on three to five independently infected cell populations.
Measurement of Ca2+ Mobilization.
Ca2+ experiments were conducted as described previously (17). Briefly, cells were loaded with 2 μM Fura-2AM/0.01% Pluronic 127 (Molecular Probes) for 35 min at 37°C in Fura-2 buffer (135 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM glucose, 10 mM Hepes, pH 7.4, and 0.1% high purity BSA; Roche Laboratories), washed, and warmed at 37°C for 10 min before analysis. Assays were performed using a SC-500 spectrofluorometer (Photon Technologies). Cells were activated with 1 μg/ml F(ab′)2 anti-IgM (Jackson ImmunoResearch Laboratories). The emission values at 510 nm after excitation at 340 and 380 nm were collected and analyzed using FELIX software (Photon Technologies). Data was analyzed and graphs generated with Deltagraph 4.0 software (SPSS).
Immunoprecipitation and Immunoblotting.
Immunoprecipitations were performed as described previously (17). CD22 was immunoprecipitated with rabbit antibody raised against the 17 COOH-terminal amino acids of murine CD22 (details to be published elsewhere). SHP-1 was immunoprecipitated with rabbit anti–mouse SHP-1 antibody (C-19) (Santa Cruz Biotechnology, Inc.). Immunoprecipitates were collected on protein G Sepharose beads (Amersham Pharmacia Biotech), which were then washed three times in lysis buffer (1% Nonidet P-40, 150 mM sodium chloride, 5 mM sodium fluoride, 2 mM sodium o-vanadate, 25 mM Hepes, pH 7.4, and 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin, 1 μg/ml antipain, and 10 μg/ml PMSF), boiled in Laemmli buffer, and resolved by a 10% SDS-PAGE.
Phosphotyrosine was detected on immunoblots with mAb 4G10 (1 μg/ml) (UBI) while CD22 and SHP-1 were detected with anti-CD22 COOH-terminal peptide Ab (1 μg/ml) or anti–mouse SHP-1 (1 μg/ml) (C-19; Santa Cruz Biotechnology, Inc.), respectively, followed by the appropriate HRP-goat anti-IgG antibody (Chemicon). Immunoblots were developed using an enhanced chemiluminescence (ECL) kit (NEN Life Science Products) and exposed to BioMax MR film (Kodak).
Results
Expression of Wild-Type and Mutant CD22 in CD22−/− Cells.
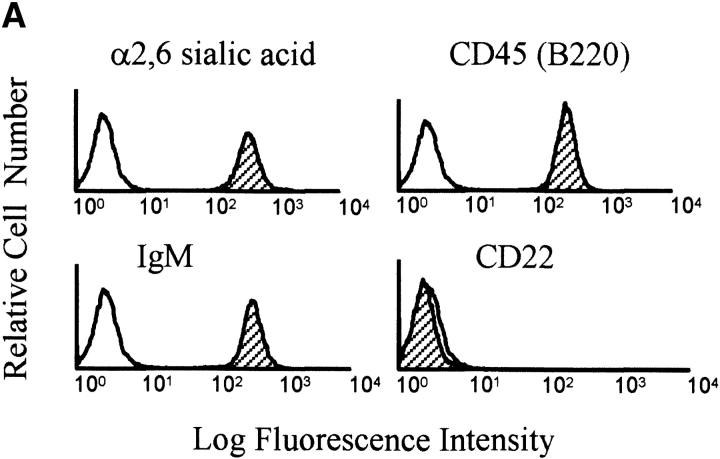
To explore the function of wild-type (WT) CD22 and CD22 extracellular domain mutants, we made use of a novel J2 virus-immortalized B cell line, J2–44, generated from CD22−/− mice. This cell retains high levels of membrane IgM and CD45(B220), expresses surface α2, 6 sialic acids (Fig. 1 A) and responds to BCR cross-linking. A full description of this line and its properties will be published elsewhere.
Figure 1.
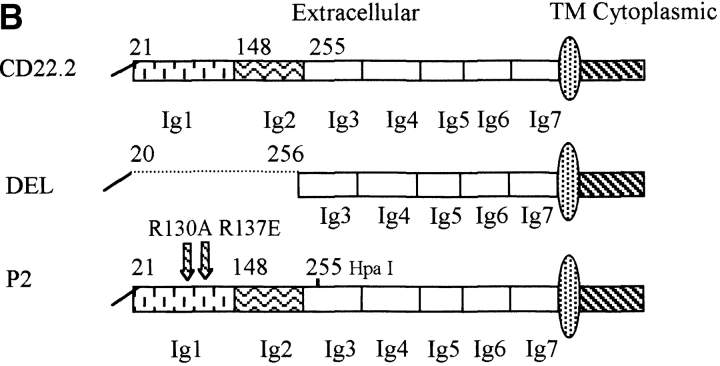
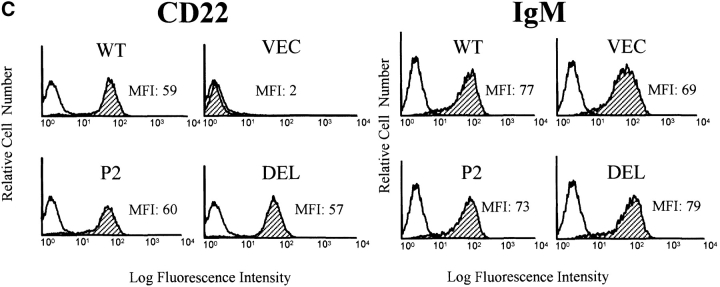
Generation of B cell lines expressing WTCD22 or CD22 ectodomain mutants. (A) Surface phenotype of parental J2–44 cells. Cells were stained with Sambuca nigra lectin or the indicated Abs (filled curve) and isotype control antibody (unfilled curve). One of three similar experimental results. Note that J2–44 cells are IgM+, CD45(B220)+, α2, 6 sialic acid-positive and CD22-negative. (B) Structure of the CD22 mutants used in this study. The first 20 amino acids are the signal peptide. Extracellular domains are assigned as described previously (reference 11). Hpa I lies within the third Ig domain and is used to clone the PCR product. TM, transmembrane domain. (C) Cells infected with WT and mutant CD22 containing retroviruses have similar surface IgM and CD22 expression. Cells were infected with the indicated retrovirus and stained with the indicated Abs (filled curve) or isotype control antibodies (unfilled curve). Median fluorescence intensity (MFI) of each stain is shown. DEL, del mutant-infected cells; P2, P2 mutant-infected cells; Vector, vector-infected J2–44 cells; WT, WT CD22-infected cells. Similar staining was observed in three independently infected cell lines.
We expressed WT or extracellular domain mutant CD22 by using the dual expression retrovirus MSCV-IRES-GFP, which coexpresses the gene of interest and GFP (see Materials and Methods). As a control, J2–44 cells were infected with the parental GFP-containing MSCV vector. Two different ectodomain mutants of CD22 were tested. The DEL mutant is an internal deletion that fuses the first twenty NH2-terminal amino acids of CD22 to amino acid 256; this mutant lacks Ig domains 1 and 2 (Fig. 1 B), which are necessary for CD22 sialic acid binding activity (10). CD22 P2 has two point mutations, R130A and R137E, in Ig domain 1 (Fig. 1 B). These point mutations individually prevent CD22 from binding to CD45 (11).
Infected cells (GFP+) were purified by MoFlo and tested for surface CD22 and IgM expression by flow cytometry using the rat anti–mouse CD22 mAb NIM-R6, which recognizes both the DEL and P2 forms of CD22 (10) The four infected cell pools had similar levels of surface IgM, and the DEL, P2 and WT CD22−/− infected cell populations expressed similar levels of surface CD22 (Fig. 1 C).
Increased Ca2+ Flux in DEL and P2 CD22 Cells.
Previous studies have shown that loss of CD22 expression on B cells causes increased Ca2+ mobilization after BCR cross-linking (6–9). We were interested in whether mutation of the sialic acid binding domains of CD22 alters Ca2+ mobilization.
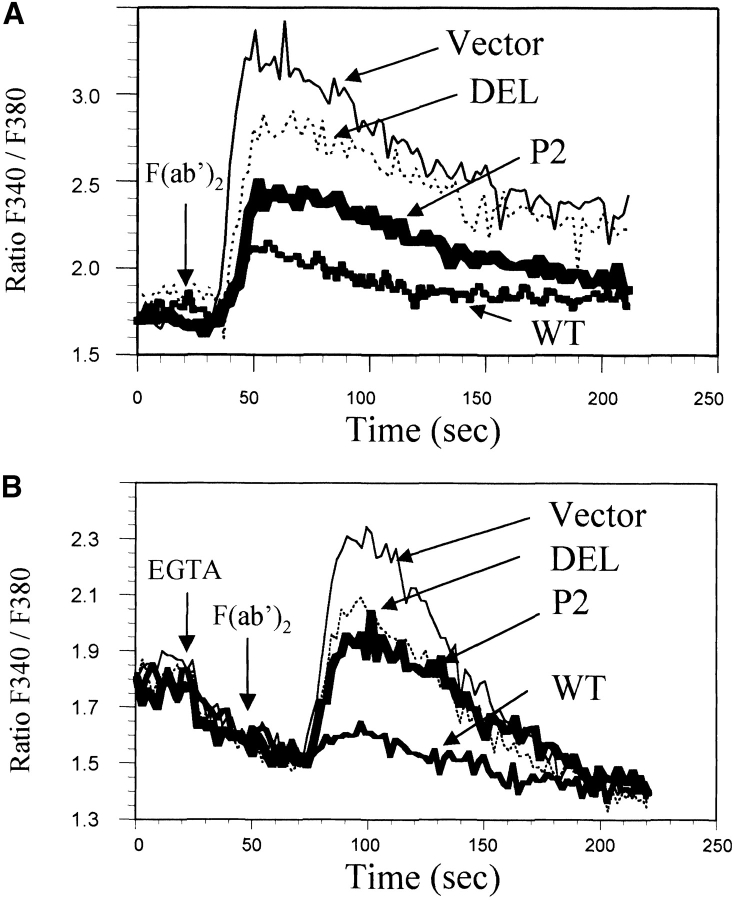
J2–44 cells infected with WT CD22, the DEL or P2 mutants, or vector alone were stimulated with F(ab′)2 goat anti–mouse μ and Ca2+ transients recorded. Previous studies showed that differences in Ca2+ responses of WT CD22 and CD22-deficient mice were more obvious when a low dose of anti-IgM was used (8). We had similar observations with CD22-deficient variants of a transformed cell line (WEHI 231) (17). We titrated the anti-IgM dose response in our J2–44 cells. When we used a high dose anti-IgM (16 μg/ml) to activate, we were unable to see any Ca2+ response difference between WT CD22 and CD22-deficient cells (data not shown). In contrast, differences in Ca2+ responses were obvious when we activated cells with a low dose of anti-IgM, such as 1 μg/ml F(ab′)2 of goat anti–mouse IgM. Thus, we decided to use 1 μg/ml F(ab′)2 anti-IgM for experiments with CD22 variants.
As expected, vector-infected J2–44 cells, which do not express CD22, had a significant Ca2+ response to BCR cross-linking. However, restoring WT CD22 expression dramatically reduced the BCR-evoked calcium response (Fig. 2 A). These data support previous work that indicates that CD22 is a cell autonomous negative regulator of the BCR calcium response (17). Interestingly, neither the DEL nor the P2 mutants of CD22 were as effective as WT CD22 in diminishing the calcium response (Fig. 2 A). This indicated that to fully repress BCR activation, CD22 needs its sialic acid binding domains. Neither mutant totally restored CD22 inhibitory function.
Figure 2.
DEL and P2 mutant cells have increased Ca2+ transients. Shown are representative real time Fura-2 ratios (340:380) for cells activated with 1 μg/ml F(ab′)2 anti-IgM. (A) Cells expressing the indicated CD22 construct or vector alone were activated by BCR cross-linking and total calcium flux was monitored. (B) Ca2+ release was measured by activating cells in the presence of 2 mM EGTA. Representative of three or more experiments with independently infected cells.
Ca2+ transients in response to BCR activation consist of both release from intracellular stores and the subsequent influx of extracellular Ca2+ from the medium. We asked whether the mutations in CD22 affected calcium release from internal stores. Infected cells were subjected to BCR cross-linking in the presence of EGTA (Fig. 2 B). Cells expressing CD22 DEL or P2 exhibited increased intracellular Ca2+ release compared with cells expressing WT CD22. Thus, mutations in the CD22 sialic acid binding domains diminished the ability of CD22 to inhibit Ca2+ release from internal stores.
CD22 Tyrosyl Phosphorylation and SHP-1 Association Require the CD22 Ectodomain.
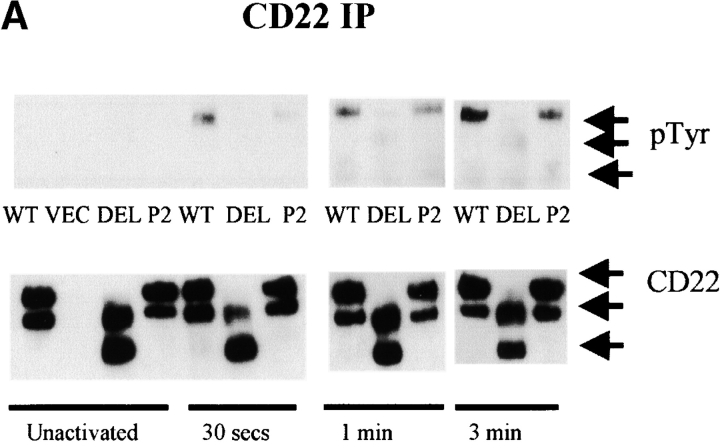
CD22 has six cytoplasmic tyrosines, three of which (Y783, Y843, and Y863) are within ITIMs. Upon BCR cross-linking, some, if not all of these tyrosines are rapidly phosphorylated and recruit SHP-1 (3, 4), which is believed to mediate negative signaling. To explore the mechanisms of ectodomain involvement in CD22 function, we assessed the tyrosyl phosphorylation status of the WT and mutant proteins. Anti-phosphotyrosine immunoblots of CD22 immunoprecipitates showed diminished tyrosine phosphorylation on the DEL and P2 mutant forms of CD22 after BCR cross-linking (Fig. 3 A and C). In contrast to wild-type, there was no detectable tyrosyl phosphorylation on the DEL and little on the P2 mutant 30 s after BCR cross-linking. However, after 1 min (P2 mutant) or 3 min (DEL mutant), tyrosyl phosphorylation was readily detected, but was never at levels seen in WT CD22. This is consistent with the decrease in inhibition of Ca2+ transients seen in cells expressing these mutants. Two isoforms were seen in the CD22 immunoblots. They can be immunoprecipitated by any of three different anti–mouse CD22 antibodies, NIM-R6, CY34, and our own anti-COOH terminal peptide (data not shown). Two isoforms have been observed previously and the suggestion made that they represent different posttranslational modifications (22). A cytoplasmic isoform has been found in transformed B cells (23). We have not addressed this issue.
Figure 3.
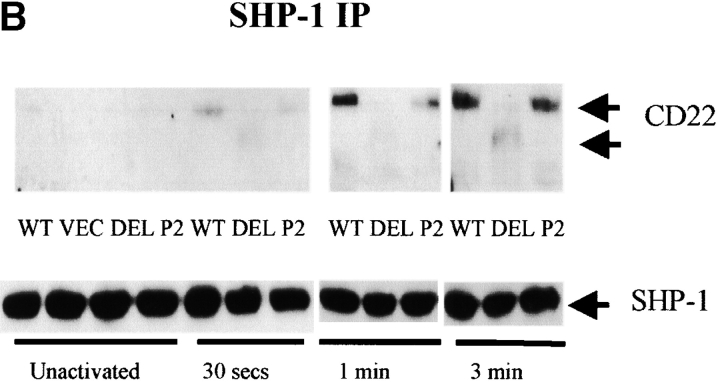
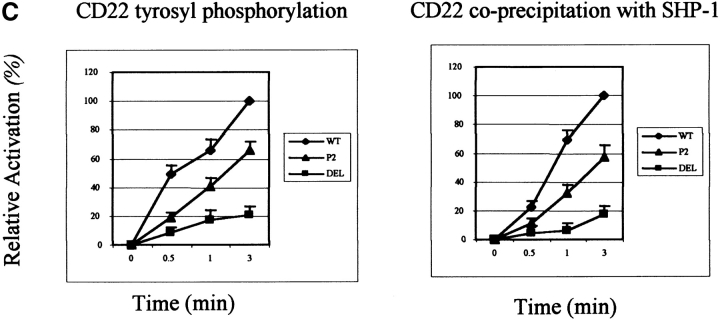
Decreased CD22 tyrosyl phosphorylation and SHP-1 association in CD22 ectodomain mutants (n > 3). (A) J2–44 cells expressing WT or mutant CD22 or vector alone were assayed in the absence of stimulation or at the indicated times after BCR cross-linking. CD22 was immunoprecipitated with rabbit anti–mouse CD22 COOH-terminal peptide. Cells were assayed at different times after activation. The blot was first probed with 4G10 mouse anti-phosphotyrosine Ab, stripped and reprobed with anti- CD22 COOH-terminal peptide Ab. (B) SHP-1 was immunoprecipitated with rabbit anti–mouse SHP-1 Ab. Cells were assayed at different times after activation. The blot was first probed with anti SHP-1 Ab, stripped, and re-probed with anti–CD22 COOH-terminal peptide. (C) Quantitative analysis of the results shown in a and b. The intensities of the bands were quantified by densitometry. The intensities of the P-tyr and CD22 bands were normalized by reference to a loading control. The densities of bands in the WT lysates at 3 min was set as 100%. The percentage of activation was calculated by comparing the corrected intensity of the bands with the 100% control (the activation of WT at 3 min). Mean results from five experiments are shown (±SD), P < 0.05 by Student's t test.
To assess CD22/SHP-1 complex formation, SHP-1 was immunoprecipitated from cells, resolved by SDS-PAGE and immunoblotted with anti-CD22 Ab. Consistent with the decreased tyrosyl phosphorylation of the CD22 ectodomain mutants, there was indeed less CD22 associated with SHP-1 in DEL and P2 mutant expressing cells (Fig. 3 B and C). Again, the SHP-1 association increased with the time after BCR cross-linking and the P2 mutant had more associated SHP-1 than the DEL mutant.
Discussion
Here we have demonstrated that deletion of a sialic acid binding domain or introduction of two point mutations, (R130A and R137E), diminishes the negative regulatory function of CD22. This means that the ectodomains of CD22 are required for full function.
We favor the hypothesis that this loss of function is directly due to the loss of binding to a sialic acid containing ligand. The R130 residue is highly conserved within the Siglec family of sialic acid binding proteins. Previously, it was shown that either the R130A or R137E substitution decreased CD22 binding to the α2, 6 sialic acid of CD45 to <5% of wild-type (11). Therefore, when both mutations are present, ligand binding should be disrupted. As pointed out previously, the R130A and R137E mutations are unlikely to disrupt the overall structure of CD22 (10, 11). Indeed, we found that the P2 mutant can still bind two CD22 mAbs, CY34 (which binds to the Ig-like 1 and 2 ectodomains) and NIM-R6. The DEL mutant, which contains a deletion of the Ig-like domains, can still be recognized by mAb NIM-R6, which suggests that the remaining five Ig-like domains retain their native conformation. However, we cannot rule out that these mutations result in altered protein–protein binding mediated by unknown mechanisms.
As B cells are the only cells present in our system, it is apparent that they must express the relevant ligands. While the physiological ligand(s) for CD22 have not been established, CD45 and IgM were proposed to be major endogenous ligands (10, 12–14). CD45 is the predominant sialylated glycoprotein on lymphocytes (24). CD22 can bind CD45 in vitro (11, 12); however, it also binds cells lacking CD45, which indicates that CD45 cannot be the sole ligand (10, 12).
IgM is a proposed physiological ligand for CD22 (10, 12–14). Previously, it was found that a small percentage (<2%) of membrane IgM is associated with CD22 (13). Recently, quantitative confocal microscopy was used to assess the location of CD22 in relation to IgM. CD22 was found to constitutively associate with the BCR complex even after mIgM ligation by soluble ligand (25). This suggests a model in which ligation of mIg induces phosphorylation of CD22, which can then recruit and activate SHP-1, thereby inhibiting signaling. Therefore, the sequestration of CD22 away from mIgM relieves its ability to function as a negative inhibitor (3). Perhaps, in our case, mutation of the CD22 ligand-binding domain prevented binding of IgM. As a result, when activated, CD22 may not be in a location critical for phosphorylation of its tyrosines.
Previous studies suggested that individual Ig-like domains of mouse CD22 might be responsible for interacting with different glycoproteins (10). Soluble recombinant proteins containing different murine CD22 Ig-like domains immunoprecipitated different glycoproteins from B cell lines (10). Therefore, it is possible that the DEL mutation disrupted CD22 binding to more ligands than did the P2 mutations. As a result, the negative regulatory function of CD22 might be more diminished by the DEL mutation.
SHP-1 is the major negative effector known to associate with CD22 after BCR cross-linking. However, SHIP also associates with CD22, regulating calcium transients (22). Since the mutants phosphorylated poorly, it is possible that their association with other negative effectors, including SHIP, is also affected.
It appears that CD22 binding to endogenous α2, 6 sialic acid–containing ligands contributes to the inhibition of B cell signaling. Relief of this binding in vivo may lower the threshold for B cell activation. While most resting splenic and mesenteric lymph node B cells showed masked CD22, 2–5% of IgD+ murine B cells had unmasked CD22 (15, 26). It is possible that within specific lymphoid microenvironments the threshold for activation for some B cells will be lower. One possibility is that CD22 binding to ligands on adjacent cells (trans binding) might relieve negative regulation that requires self (cis) binding. Moreover, it was shown that a combination of IgM and CD40 mediated-activation, as might be expected to occur in the secondary lymphoid organs, could unmask CD22 (15). Therefore, in the secondary lymphoid organs, previously activated B cells might be relieved of CD22 suppression and be more sensitive to stimulation by antigen.
The question remains how the disruption of CD22 binding to endogenous ligands alters its inhibitory affect on BCR signaling. The observed decrease in tyrosine phosphorylation of mutant CD22 is likely to be key, as SHP-1 association and negative regulation of Ca2+ are believed to be downstream of CD22 phosphorylation. Yet to be explained is how CD22 phosphorylation can be affected by the disruption of binding to endogenous ligand(s). One possibility is that the disruption of CD22–sialic acid binding domains alters the placement of CD22 within the cell so that the critical kinase is less able to access CD22. Another possibility is that the activity of the kinase responsible for CD22 phosphorylation is altered by the disruption of CD22 ligand-binding domains. Lyn is a likely candidate. It can phosphorylate CD22 cytoplasmic peptides in vitro (22), and its loss in Lyn−/− mice causes decreased phosphorylation of CD22 (27, 28). Furthermore, phosphorylation of murine CD22 ectopically expressed in the transformed chicken B cell line DT40 is Syk and Lyn dependent (29). Interestingly CD45, a putative CD22 ligand on B cells, is a regulator of Lyn (30, 31). Lyn has two critical tyrosines: a stimulatory autophosphorylation site and an inhibitory COOH-terminal phosphorylation site. While CD45's role in negative or positive regulation of Lyn is still under discussion (30, 31), CD45 can dephosphorylate either tyrosine. Thus, it is possible that the disruption of CD22 binding to its ligand(s), perhaps including CD45 itself, somehow affects CD45's ability to dephosphorylate the critical tyrosines of Lyn. As a result, the kinase activity of Lyn would be affected, thereby altering phosphorylation of CD22. However, we have not detected altered phosphorylation of Lyn at either site after activation of cells expressing mutant CD22 (unpublished data).
B cells from CD22-deficient mice have decreased cell proliferation and increased apoptosis upon anti-IgM cross-linking (6–9). As yet, we have not generated convincing data to show that cell proliferation and apoptosis of these transformed cells is altered after F(ab′)2 activation (data not shown).
Our results indicate there is a connection between two crucial events regarding CD22: the phosphorylation of CD22 and the ligand binding of its ectodomains. Future work will be directed at understanding the mechanisms by which binding of CD22 to endogenous ligand(s) affects its phosphorylation status. Like CD22, many Siglecs have Ig-like binding ectodomains and ITIM motifs in their cytoplasmic tails. They recognize different types and linkages of sialic acid and have different tissue distributions. Therefore, our finding that the sialic acid binding domains of CD22 are required for negative regulation may have broad implications.
Acknowledgments
We thank Dr. Keming Zhang for providing J2-44 cells, Allen Parmelee for assistance in cell sorting, Dr. Robert Berland for helpful discussion, and Albert Tai for help with early experiments.
This work was supported by National Institutes of Health grants AR43773 and AI15803 (to H.H. Wortis) and R01CA66600 and PO1DK50654 (to B.G. Neel).
References
- 1.Crocker, P.R., and A. Varki. 2001. Siglecs, sialic acids and innate immunity. Trends Immunol. 22:337–342. [DOI] [PubMed] [Google Scholar]
- 2.Cyster, J.G., and C.C. Goodnow. 1997. Tuning antigen receptor signaling by CD22: integrating cues from antigens and the microenvironment. Immunity. 6:509–517. [DOI] [PubMed] [Google Scholar]
- 3.Doody, G.M., L.B. Justement, C.C. Delibrias, R.J. Matthews, J. Lin, M.L. Thomas, and D.T. Fearon. 1995. A role in B cell activation for CD22 and the protein tyrosine phosphatase SHP. Science. 269:242–244. [DOI] [PubMed] [Google Scholar]
- 4.Schulte, R.J., M.A. Campbell, W.H. Fischer, and B.M. Sefton. 1992. Tyrosine phosphorylation of CD22 during B cell activation. Science. 258:1001–1004. [DOI] [PubMed] [Google Scholar]
- 5.Ono, M., H. Okada, S. Bolland, S. Yanagi, T. Kurosaki, and J.V. Ravetch. 1997. Deletion of SHIP or SHP-1 reveals two distinct pathways for inhibitory signaling. Cell. 90:293–301. [DOI] [PubMed] [Google Scholar]
- 6.Sato, S., A.S. Miller, M. Inaoki, C.B. Bock, P.J. Jansen, M.L. Tang, and T.F. Tedder. 1996. CD22 is both a positive and negative regulator of B lymphocyte antigen receptor signal transduction: altered signaling in CD22-deficient mice. Immunity. 5:551–562. [DOI] [PubMed] [Google Scholar]
- 7.Otipoby, K.L., K.B. Andersson, K.E. Draves, S.J. Klaus, A.G. Farr, J.D. Kerner, R.M. Perlmutter, C.L. Law, and E.A. Clark. 1996. CD22 regulates thymus-independent responses and the lifespan of B cells. Nature. 384:634–637. [DOI] [PubMed] [Google Scholar]
- 8.O'Keefe, T.L., G.T. Williams, S.L. Davies, and M.S. Neuberger. 1996. Hyperresponsive B cells in CD22-deficient mice. Science. 274:798–801. [DOI] [PubMed] [Google Scholar]
- 9.Nitschke, L., R. Carsetti, B. Ocker, G. Kohler, and M.C. Lamers. 1997. CD22 is a negative regulator of B-cell receptor signalling. Curr. Biol. 7:133–143. [DOI] [PubMed] [Google Scholar]
- 10.Law, C.L., A. Aruffo, K.A. Chandran, R.T. Doty, and E.A. Clark. 1995. Ig domains 1 and 2 of murine CD22 constitute the ligand-binding domain and bind multiple sialylated ligands expressed on B and T cells. J. Immunol. 155:3368–3376. [PubMed] [Google Scholar]
- 11.van der Merwe, P.A., P.R. Crocker, M. Vinson, A.N. Barclay, R. Schauer, and S. Kelm. 1996. Localization of the putative sialic acid-binding site on the immunoglobulin superfamily cell-surface molecule CD22. J. Biol. Chem. 271:9273–9280. [PubMed] [Google Scholar]
- 12.Engel, P., Y. Nojima, D. Rothstein, L.J. Zhou, G.L. Wilson, J.H. Kehrl, and T.F. Tedder. 1993. The same epitope on CD22 of B lymphocytes mediates the adhesion of erythrocytes, T and B lymphocytes, neutrophils, and monocytes. J. Immunol. 150:4719–4732. [PubMed] [Google Scholar]
- 13.Leprince, C., K.E. Draves, R.L. Geahlen, J.A. Ledbetter, and E.A. Clark. 1993. CD22 associates with the human surface IgM-B-cell antigen receptor complex. Proc. Natl. Acad. Sci. USA. 90:3236–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanasaki, K., L.D. Powell, and A. Varki. 1995. Binding of human plasma sialoglycoproteins by the B cell-specific lectin CD22. Selective recognition of immunoglobulin M and haptoglobin. J. Biol. Chem. 270:7543–7550. [DOI] [PubMed] [Google Scholar]
- 15.Razi, N., and A. Varki. 1998. Masking and unmasking of the sialic acid-binding lectin activity of CD22 (Siglec-2) on B lymphocytes. Proc. Natl. Acad. Sci. USA. 95:7469–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rapp, U.R., J.L. Cleveland, T.N. Fredrickson, K.L. Holmes, H.C. Morse III, H.W. Jansen, T. Patschinsky, and K. Bister. 1985. Rapid induction of hemopoietic neoplasms in newborn mice by a raf(mil)/myc recombinant murine retrovirus. J. Virol. 55:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nadler, M.J., P.A. McLean, B.G. Neel, and H.H. Wortis. 1997. B cell antigen receptor-evoked calcium influx is enhanced in CD22-deficient B cell lines. J. Immunol. 159:4233–4243. [PubMed] [Google Scholar]
- 18.Van Parijs, L., Y. Refaeli, J.D. Lord, B.H. Nelson, A.K. Abbas, and D. Baltimore. 1999. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity. 11:281–288. [DOI] [PubMed] [Google Scholar]
- 19.Cormack, B. 2000. Directed mutagenesis using the polymerase chain reaction Current Protocols In Molecular Biology. V.B. Chanda, editor. John Willey & Sons, Inc., Boston, MA. pp. 1–10. [DOI] [PubMed]
- 20.Pear, W.S., G.P. Nolan, M.L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA. 90:8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller, A.J., J.C. Young, A.M. Pendergast, M. Pondel, N.R. Landau, D.R. Littman, and O.N. Witte. 1991. BCR first exon sequences specifically activate the BCR/ABL tyrosine kinase oncogene of Philadelphia chromosome-positive human leukemias. Mol. Cell. Biol. 11:1785–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poe, J.C., M. Fujimoto, P.J. Jansen, A.S. Miller, and T.F. Tedder. 2000. CD22 forms a quaternary complex with SHIP, Grb2, and Shc. A pathway for regulation of B lymphocyte antigen receptor-induced calcium flux. J. Biol. Chem. 275:17420–17427. [DOI] [PubMed] [Google Scholar]
- 23.Sherbina, N.V., P.S. Linsley, S. Myrdal, L.S. Grosmaire, and J.A. Ledbetter, and G.L. Schieven. 1996. Intracellular CD22 rapidly moves to the cell surface in a tyrosine kinase-dependent manner following antigen receptor stimulation. J. Immunol. 157:4390–4398. [PubMed] [Google Scholar]
- 24.Whiteheart, S.W., J.C. McLenithan, and G.W. Hart. 1990. Surfaces of murine lymphocyte subsets differ in sialylation states and antigen distribution of a major N-linked penultimate saccharide structure. Cell. Immunol. 125:337–353. [DOI] [PubMed] [Google Scholar]
- 25.Phee, H., W. Rodgers, and K.M. Coggeshall. 2001. Visualization of negative signaling in B cells by quantitative confocal microscopy. Mol. Cell. Biol. 21:8615–8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Floyd, H., L. Nitschke, and P.R. Crocker. 2000. A novel subset of murine B cells that expresses unmasked forms of CD22 is enriched in the bone marrow: implications for B-cell homing to the bone marrow. Immunology. 101:342–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cornall, R.J., J.G. Cyster, M.L. Hibbs, A.R. Dunn, K.L. Otipoby, E.A. Clark, and C.C. Goodnow. 1998. Polygenic autoimmune traits: Lyn, CD22, and SHP-1 are limiting elements of a biochemical pathway regulating BCR signaling and selection. Immunity. 8:497–508. [DOI] [PubMed] [Google Scholar]
- 28.Smith, K.G., D.M. Tarlinton, G.M. Doody, M.L. Hibbs, and D.T. Fearon. 1998. Inhibition of the B cell by CD22: a requirement for Lyn. J. Exp. Med. 187:807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otipoby, K.L., K.E. Draves, and E.A. Clark. 2001. CD22 regulates B cell receptor-mediated signals via two domains that independently recruit Grb2 and SHP-1. J. Biol. Chem. 276:44315–44322. [DOI] [PubMed] [Google Scholar]
- 30.Yanagi, S., H. Sugawara, M. Kurosaki, H. Sabe, H. Yamamura, and T. Kurosaki. 1996. CD45 modulates phosphorylation of both autophosphorylation and negative regulatory tyrosines of Lyn in B cells. J. Biol. Chem. 271:30487–30492. [DOI] [PubMed] [Google Scholar]
- 31.Katagiri, T., M. Ogimoto, K. Hasegawa, Y. Arimura, K. Mitomo, M. Okada, M.R. Clark, K. Mizuno, and H. Yakura. 1999. CD45 negatively regulates lyn activity by dephosphorylating both positive and negative regulatory tyrosine residues in immature B cells. J. Immunol. 163:1321–1326. [PubMed] [Google Scholar]