Abstract
FcγRIIB is a potent lupus susceptibility gene as demonstrated by the observation that mice deficient in this molecule develop spontaneous antinuclear antibodies (ANA) and fatal glomerulonephritis when on the C57BL/6 background. To determine the mechanisms underlying the epistasis displayed by this gene we have constructed hybrids between FcγRIIB−/− and the systemic lupus erythematosus (SLE) modifiers yaa and lpr and the susceptibility locus Sle1. Sle1 and B6.RIIB−/− are both physically and functionally coupled; compound heterozygotes of Sle1 and B6.RIIB−/− develop significant disease, while single heterozygotes display no evidence of autoimmunity or disease, indicating that these genes lie on the same genetic pathway resulting in the loss of tolerance to nuclear antigens. However, the generation of ANA in itself is insufficient to account for the severity of autoimmune disease in this model, as demonstrated by analysis of yaa and lpr hybrids. Thus, B6.RIIB−/−/lpr mice are protected from disease progression, despite equivalent titers of ANA. In contrast, B6.RIIB−/−/yaa mice have significantly enhanced disease despite reduced ANA titers. Yaa modifies the specificity and thus the pathogenicity of the B6. RIIB−/− ANA, by converting them to antinucleolar antibodies. In addition to these known modifier pathways, we have discovered two novel, recessive loci contributed by the C57BL/6 genome that are required for the ANA phenotype, further indicating the epistatic properties of this SLE model.
Keywords: lpr, yaa, Sle1, autoantibodies, glomerulonephritis
Introduction
The development of systemic autoimmunity in diseases like systemic lupus erythematosus (SLE)* require the interaction of multiple genetic loci with environmental factors to result in the loss of tolerance, the development of an autoreactive repertoire, and the amplification of autoreactive cells to produce pathogenic autoantibodies (1–4). The deposition of these autoantibodies as immune complexes and the activation of effector mechanisms to trigger tissue pathology require still additional genetic interactions to result in the systemic pathology typical of these diseases. Recent studies in both rodents and humans have begun to clarify some of the genetic interactions that underlie autoimmune disease susceptibility and progression (for a review, see reference 5). Two types of studies have been undertaken to identify these genetic components – classic linkage analysis in multiplex SLE families and lupus-prone inbred mouse strains; and candidate gene analysis in human patient populations and animal models via genomic manipulation.
Genetic studies in susceptible human or murine populations have demonstrated that disease susceptibility is multifactorial, involving complex interactions among several genes together with poorly defined environmental factors (4). While the contribution of the MHC to lupus susceptibility has been well documented by numerous studies (6, 7), the importance of non-MHC loci that either increase or suppress susceptibility to lupus has recently been appreciated. A total of 31 susceptibility loci have thus been defined, distributed among 21 nonoverlapping genomic intervals, further indicating the genetic complexity involved in autoimmune diseases like lupus (4). Syntenic loci have been identified in the mouse and human for lupus susceptibility on mouse chromosomes 1, 5, 6, 7, and 18. The distal region of chromosome 1 is perhaps the best characterized with the identification of a cluster of loci including Sle1, Nba2, Bxs3, and Lbw7 involved in antinuclear antibody production (8–10). Sle1, for example, was derived from the autoimmune strain NZM 2410 as a 37 cM region of chromosome 1 from NZW (11). C57BL/6 mice congenic for Sle1 develop antichromatin autoantibodies and will progress to autoimmune disease when combined with other NZM-derived loci. Sle1 has been further subdivided into four distinct, although functionally related regions, designated Sle1a, b, c, and d (12). Although each of these genes will express an autoimmune phenotype when isolated from the others, their autoimmune phenotype is strongly enhanced when they are expressed in combination, suggesting that they may impact a common pathway leading to the loss of tolerance to nuclear antigens (13).
In a second type of study, candidate genes have been modified in unaffected mouse strains to determine their contribution to disease susceptibility. A common theme has emerged from these studies highlighting the central role of inhibitory molecules in maintaining tolerance to nuclear antigens. For example, deletion of the inhibitory surface molecules CD22, cytotoxic T lymphocyte antigen 4, PD-1, or FcγRIIB result in animals with autoimmune phenotypes of differing degrees of severity; references 14–17). Similarly, deletion of the inhibitory signaling molecules src homology 1, cbl-b, or lyn also results in autoimmunity and disease (18–21). These studies further support the threshold nature of autoimmunity and emphasize the importance of preventing inappropriate lymphocyte stimulation at subthreshold levels of antigen.
The central role of autoantibodies and immune complexes in the pathophysiology of autoimmune diseases like lupus has focused attention on the role of cellular receptors for these pathogenic ligands. The Fc receptors for IgG, FcγRs, by transducing signals from the IgG immune complex to APCs, B cells, and effector cells, are responsible for much of the immune responses triggered by these ligands (22). Activation FcγRs, like FcγRIII, are responsible for triggering effector cell responses to cytotoxic IgGs or immune complexes; deletion of this receptor protects mice from autoimmune disease initiated by cytotoxic IgG antibodies or immune complex deposition (23). Conversely, the inhibitory FcγR, FcγRIIB, prevents inappropriate activation of effector responses; its deletion renders animals hyperresponsive to sub-threshold levels of cytotoxic antibodies and immune complexes (24, 25). Expression of FcγRIIB on B cells and APCs plays a critical role in the maintenance of peripheral tolerance. Deletion of FcγRIIB results in autoantibody production in animals presented with potentially cross reactive antigens, like collagens type II or IV or when modified by specific genetic backgrounds, like C57BL/6 (17, 26, 27). This epistatic property of the FcγRIIB deficiency model of SLE mimics the multigenic nature of human SLE.
To investigate the mechanisms that contribute to the loss of tolerance and disease progression by FcγRIIB deficiency, we have pursued genetic studies aimed at dissecting the interactions that are responsible for these phenotypes. In this study, we have constructed hybrids between B6.RIIB−/− and the Sle1 susceptibility locus or the SLE modifiers yaa and lpr and analyzed autoantibody production and disease progression. Sle1 and B6.RIIB−/− lie on a common genetic pathway that results in the loss of tolerance to nuclear antigens. The pathogenicity of these autoantibodies leading to disease progression is determined by loci such as yaa and lpr, independent of autoantibody titer. Yaa enhances disease by changing the specificity of the autoantibodies generated, while lpr uncouples autoantibody production from autoimmune disease thus preventing disease progression. The importance of epistasis is further emphasized by the identification of two novel, recessive loci on B6 that are required for antinuclear antibody production by FcγRIIB. These studies demonstrate the relevance of the B6.RIIB−/− model to the genetics of human SLE and reveal some of the mechanisms required for the manifestation of SLE.
Materials and Methods
Mice.
Mice were maintained in SPF conditions at the Rockefeller University Animal Facility. The production and characterization of FcγRIIB−/− and B6.Sle1 mice has been described previously (11, 12, 17, 35). B6 yaa and B6 lpr/lpr mice were purchased from the Jackson ImmunoResearch Laboratories and then crossed into FcγRIIB−/− and B6.Sle1 mice to produce B6.R2 −/− yaa, R2 −/− lpr/lpr, and B6.Sle1yaa mice. For the F2 analysis, B6.R2−/− and BALB.R2−/− mice were bred to produce 21 F1 mice, which were bred among themselves to obtain 210 F2 mice. Nine F2 mice died at an age <4 mo and they were discarded from the study. All the new strains were maintained for 10 mo with monthly bleeding to check for antinuclear antibodies (ANA) and proteinuria. Spleen weight was measured postmortem at 10 mo of age. Moribund animals (>500 mg/dL proteinuria and <40% hematocrit) were killed before the 10 mo deadline and were counted as deceased. H2 congenic animals were purchased from Jackson ImmunoResearch Laboratories.
Phenotypic Analysis.
ANA was tested using 12-well microscope slides covered with human Hep-2 cells. Slides were blocked for 10 min with PBS containing 5% goat serum (Sigma-Aldrich) and then incubated with mouse serum at the indicated dilutions. After two washes of 5 min with PBS, FITC-labeled anti–mouse IgG was added at 1 μg/ml for 15 min and washed away with PBS for 5 min at least three times. ANA scored was measured as follows: 0, no staining at 1:100 dilution; 1, faint staining at 1:100; 2, bright staining at 1:100; 3, bright staining at 1:1,000; and 4, bright staining at 1:10,000. Proteinuria was measured using urinalysis dip-sticks (Merck) and graded as 0, <50 mg/dL; 1, 50 mg/dL; 2, 100 mg/dL; 3, 500 mg/dL; and 4, >500 mg/dL. Histological analysis of the kidneys of 80 animals was performed and indicated a strong correlation between proteinuria and glomerulonephritis.
Genetic Analysis.
50 (B6.R2 − / − × BALB/c.R2 −/−)F2 mice that scored >3 for ANA, and 44 ANA-negative mice for comparison, were genotyped at 81 microsatellite markers distributed on the 19 autosomes. Linkage groups were developed using MAPMAKER-EXP (34) as described previously (36). Statistical analysis of linkage was performed using MAPMAKER-QTL. Those chromosomes that contained loci showing a suggestive linkage (1, 4, 7, 9, 12, and 17) were selected for a second more thorough screen that included 186 mice and 40 markers (including markers for the H2 complex), spanning the six selected chromosomes.
Results
The Autoimmunity Induced in B6 Mice by FcγRIIB Deficiency Is Functionally Linked to the Sle1 Autoimmunity-promoting Gene.
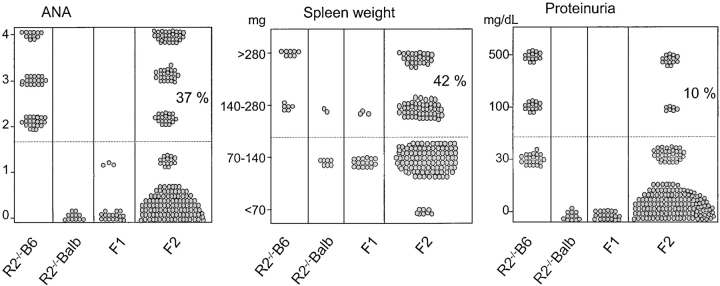
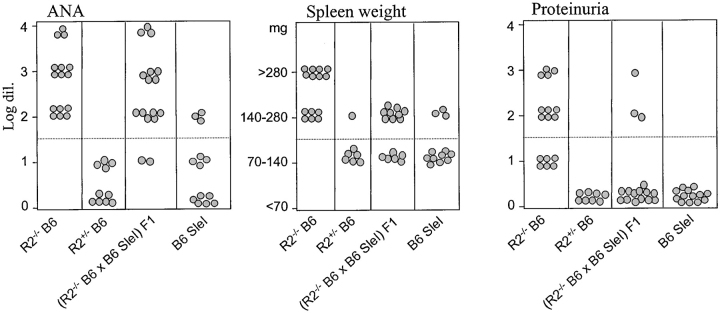
The murine FcγRIIB gene is located on the distal part of chromosome 1, at the same location where genetic studies in humans and various mouse models have identified several autoimmune susceptibility loci (Fig. 1 ; reference 4). FcγRIIB−/− B6 mice develop a spontaneous SLE-like disease, raising the possibility that these susceptibility loci on chromosome 1 and the FcγRIIB gene are genetically or functionally linked. One of these identified susceptibility loci, the Sle1 gene cluster from the NZW strain, leads to ANA production in the B6 background (11). To test for a genetic interaction between Sle1 and FcγRIIB, (B6.Sle1 × B6.R2 − / −) F1 hybrids were produced and analyzed for an autoimmune phenotype. As shown in Fig. 2 , both parental strains demonstrate their previously reported phenotypes: severe lupus disease in B6.R2 − / − animals (100% of the mice with ANA and enlarged spleen, 50% with glomerulonephritis leading to proteinuria) and mild disease in B6.SleI mice (30% ANA and enlarged spleen, 0% proteinuria). Both B6.R2 − / − and B6.Sle1 have predominantly recessive phenotypes with low but detectable levels of ANA production and minimum splenomegaly or proteinuria as heterozygotes. (B6.Sle1 × B6.R2 − / −) F1 mice, which should be equivalent to a double heterozygote for the FcγRII mutation and Sle1, displayed a stronger phenotype than either heterozygote alone or SleI homozygote. As shown in Fig. 2, their autoimmune phenotype was equivalent in disease severity to the phenotype of homozygous B6.R2 − / − mice (Fig. 2 and Table I). 90% of the (B6.Sle1 × B6.R2 − / −) F1 mice produced significant ANA levels (detectable at 102 serum dilution) and >10% further displayed symptoms of severe glomerulonephritis (at least 100 mg/dL protein in urine). These data, together with the fine mapping of the Sle1 region (unpublished data), placing FcγRIIB outside of the four Sle1 susceptibility genes (Fig. 1) and the observation that FcγRIIB levels are normal in both NZW and B6.Sle 1 (data not shown), suggests that Sle 1 and FcγRIIB are nonallelic and noncomplementing and interact synergistically in a dosage-dependent manner to maintain tolerance. These studies also suggest that Sle1, like FcγRIIB, is a negative regulator of B cell responsiveness and that the products of these loci interact within a common pathway. A likely candidate for this pathway is the src homology 2 domain–containing inositol polyphosphate 5′-phosphatase inhibitory signaling pathway, which is known to mediate the FcγRIIB signal. This might occur if the products of the Sle 1 locus modulated either FcγRIIB expression or src homology 2 domain–containing inositol polyphosphate 5′-phosphatase signaling thus changing the responsiveness of B cells to antigen stimulation.
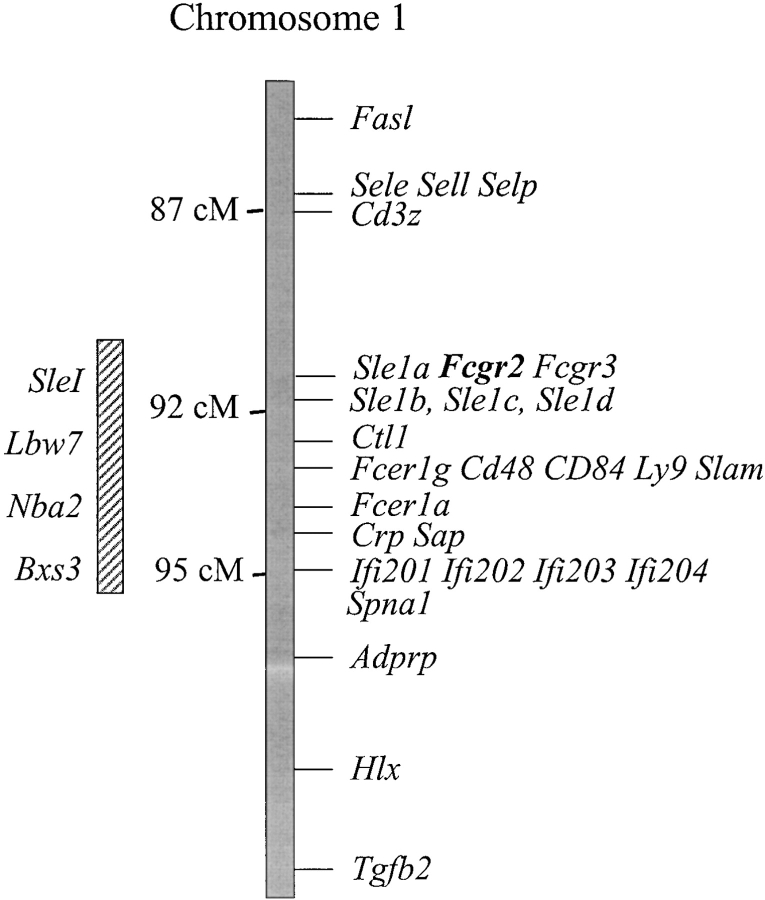
Figure 1.
FcγRIIB maps to the Sle1 locus on chromosome 1. A composite map of chromosome 1 is shown, indicating the Sle1 interval and other lupus susceptibility loci. The FcγRIIB gene (Fcgr2) is located between Sle1a and Sle1b (unpublished data).
Figure 2.
Phenotypic analysis of the F1 cross of (B6.R2−/− × B6.Sle1) for ANA, spleen weight, and proteinuria. The parental phenotypes are indicated, as well as the phenotype of the heterozygous B6.R2 +/−. Dotted lines indicate the level at which animals are considered to be positive for a particular phenotype. ANA > 1/100 dilution, splenomegaly >140 mg, and proteinuria >100 mg/dL.
Table I.
Autoimmune Phenotypes of FcγRIIB-deficient Mice in Different Genetic Backgrounds
| n | ANA (0–4) | Spleen size | Proteinuria | |
|---|---|---|---|---|
| (mg) | ||||
| RII−/−B6 | 16 | 2.75 ± 0.84 | 324 ± 120 | 12/16 (75%) |
| RII+/−B6 | 8 | 0.40 ± 0.52 | 123 ± 32 | 0/8 (0%) |
| B6 SleI homo | 12 | 0.77 ± 0.83 | 142 ± 62 | 0/12 (0%) |
| (B6 SleI × RII−/−B6) F1 | 17 | 2.56 ± 0.96 | 150 ± 43 | 4/17 (23.5%) |
| RII−/−B6 yaaa | 10 | 0.63 ± 0.51 | 484 ± 271 | 9/10 (90%) |
| B6.yaa | 8 | 0.13 ± 0.35 | 296 ± 128 | 0/8 (0%) |
| RII−/−Balb | 16 | 0.00 ± 0.00 | 120 ± 17 | 0/16 (0%) |
| (RII-/-B6 × RII−/−BALB) F1 | 22 | 0.14 ± 0.35 | 111 ± 20 | 0/22 (0%) |
| (RII-/-B6 × RII−/−BALB) F2 | 186 | 0.83 ± 1.04 | 148 ± 97 | 19/186 (10.2%) |
Tested at 5–6 mo of age because of early mortality.
The yaa Gene Enhances the Severity of the Autoimmune Phenotype of B6.R2−/− Mice by Changing Autoantibody Specificity.
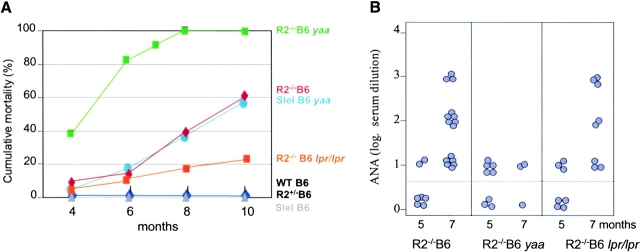
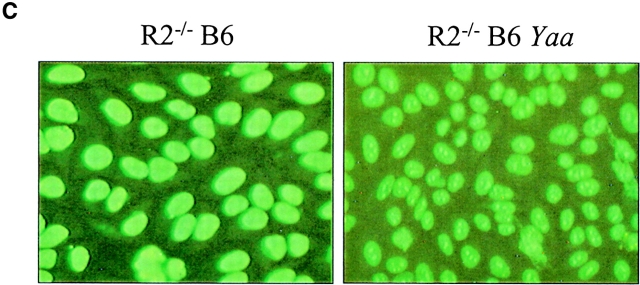
Having determined that Sle1 and B6.RIIB−/− interact epistatically to lead to the loss of tolerance to nuclear antigens and to the development of autoantibodies, we next sought to address the role of autoantibodies in disease progression by characterizing the mechanism which genetic modifiers, such as yaa and lpr influenced autoantibody titers and pathogenicity. The yaa (Y-chromosome autoimmune accelerator) gene can accelerate disease in males of the BXSB, B6.CD22 + / −, and B6.Sle1 strains (13, 28, 29). Similarly, when the yaa gene is transferred to males of the B6.R2 − / − strain, we observe an acceleration of autoimmune disease, resulting in animals that display a disease as severe as observed in BXSB yaa males. B6.R2 − / −/yaa male mice have a median survival of 4.5 mo, compared with the 9-mo median survival of B6.R2 − / − mice (Fig. 3 A). 90% of the B6.R2 − / −/yaa males develop massive proteinuria (>100 mg/dL) with striking splenomegaly (Table I). The yaa mutation has been shown to enhance the autoimmune phenotype of the Sle1 allele in a similar fashion (reference 13, and Fig. 3 A) further pointing to a functional linkage between FcγRIIB and Sle1. One possible cause of this acceleration in disease in B6.R2 − / − animals was suggested by analysis of the autoantibodies produced in the B6.R2 − / − /yaa mice. The ANA produced in these mice were altered as compared with the B6.R2 − / −parental strain. Both the ANA titers and the nuclear staining patterns were different in these mice, as compared with B6.R2 − / − animals (Fig. 3 B). In place of the homogenous nuclear staining seen in B6.R2 − / − (and B6.Sle1 animals, data not shown), the yaa-modified animals displayed a nucleolar staining pattern, typically associated with the presence of anti-RNP autoantibodies (Fig. 3 C). This shift in autoantibody specificity may account for the enhanced disease severity seen in these animals, despite the reduced ANA titers. Yaa, therefore, appears to modify B6.R2 − / − by changing the specificity of the autoantibody repertoire from antichromatin to antiribonuclearprotein with greater pathogenic potential. The identity of the yaa gene product and the mechanism by which yaa modifies autoantibody specificity has not been determined. This gene most likely influences regulatory pathways in B cell, DCs or macrophages, which might affect both cell proliferation, antibody production and antigen presentation. A B cell intrinsic defect in yaa mice has been supported by cell transfer experiments, but a macrophage defect can also be detected as monocytosis from early age (28).
Figure 3.
Yaa and lpr modify the B6.R2 −/− phenotype. (A) Cumulative mortality curves for B6.R2 −/−, B6.Sle1, B6, and crosses to yaa or lpr. (B) ANA titers for B6.R2 −/− crossed to yaa or lpr at 5 and 7 mo of age. (C) ANA staining patterns on Hep2 human epithelial cells for serum derived from B6.R2 −/− and B6.R2 −/− yaa at 1:200 dilution. See Table I for quantitation of these traits.
The lpr Gene Protects B6.RIIB−/− from Autoimmune Disease without Altering ANA Titers.
In contrast to the enhanced disease displayed by B6.R2 − / −/yaa, the lpr lymphoproliferative and autoimmune accelerator factor paradoxically reduced disease severity in the B6.R2 − / − model. While this mutation in the Fas receptor gene is known to cause a very severe autoimmune disease in Mrl mice (30, 31), its addition to B6.R2 − / − had the opposite effect. The resulting mice had longer life spans than B6.R2 − / − mice with little evidence of proteinuria, despite ANA titers comparable to the B6.R2 − / − parental strain (Fig. 3). This uncoupling of autoantibody production from autoimmune disease progression is suggestive of either an lpr-induced defect in effector responses to autoantibodies or a protective effect of the lpr mutation on glomeruli. The lpr mutation may attenuate immune complex deposition or effector cell responses to these immune complexes, thus reducing disease progression. This type of protection has been observed in FcγRIII-deficient animals, where, for example, (NZB × NZW) F1 animals lacking FcγRIII have normal life spans and essentially normal kidney function despite the continuing production and deposition of immune complexes in their glomeruli (23). Alternatively, by modifying the apoptotic pathway, the lpr mutation may protect glomeruli from effector cell mediated inflammation, thus sparing the animals from autoimmune disease. A similar case of a protective role of the lpr mutation over some pathogenic effects of autoimmunity has also been observed in the NOD model for autoimmune diabetes in which the pancreatic expression of FasR has been proposed to be required for the pathology seen in this model (32). Similarly, both lpr and gld mutations have been shown to inhibit autoantibody production in pristine-induced lupus (33).
Recessive B6 Genes Interact with FcγRIIB to Confer Susceptibility to Autoimmune Disease.
To further define the epistatic mechanisms operating to result in SLE in the B6.RIIB−/− model, we next sought to define the B6 loci contributing to the development of disease. We have previously shown that B6.R2 − / − mice develop spontaneous SLE while BALB.R2 − / − do not (17). Neither B6 nor BALB/c wild-type mice develop noticeable spontaneous disease. To study the impact of these background genetic modifiers in the FcγRIIB−/− disease model, a cross between B6.R2 − / − and BALB.R2 − / − mice was performed and the resulting F1 and F2 generations evaluated for autoimmune phenotypes. Three different autoimmune traits were measured in these mice: ANA at 7 mo and spleen weight and proteinuria at 10 mo (Fig. 4 , Table I). ANA measurements were scored 0 to 4 as the log of the highest dilution that gives a positive result in an ANA test. As we have observed in previous studies, all B6.R2 − / − mice were positive at or above the 102 dilution, while all BALB.R2 − / − mice were negative even at the lowest dilution. (B6.R2 − / − × BALB.R2 − / −) F1 mice were protected from disease in all three traits measured. None of the F1 animals had ANA titers at or above the 102 dilution in the ANA score. Spleen weight was normal and no proteinuria was detected. Based on this result, the most influential susceptibility genes from B6 were expected to be recessive, or conversely, some BALB/c resistant alleles were expected to be dominant. To further define these B6 or BALB-derived loci, 186 F2 mice were evaluated for these same three phenotypic traits. 37% of the animals scored positive for ANA and 10% for proteinuria at 100 mg/dL or more. The number of ANA positive F2 mice was more than what would be expected from a single recessive susceptible locus coming from the B6 genome. It is possible that several combinations of susceptibility genes can produce the ANA positive phenotype, or that the BALB/c genome contains susceptibility genes that in BALB/c mice are neutralized by resistant genes. The small number of F2 mice with high levels of proteinuria suggested that this phenotypic trait resulted from the contribution of a larger number of susceptibility genes.
Figure 4.
Phenotypic analysis of the F2 cross (B6.R2 −/− × BALB.R2 −/−) for ANA, spleen weight, and proteinuria. Parental and F1 phenotypes are shown for comparison and summarized in Table I. Valves were measured as in Fig. 1. 56 of the 74 ANA positive animals displayed splenomegaly.
Localization of Autoimmune Susceptibility Loci in B6 and BALB/c Backgrounds.
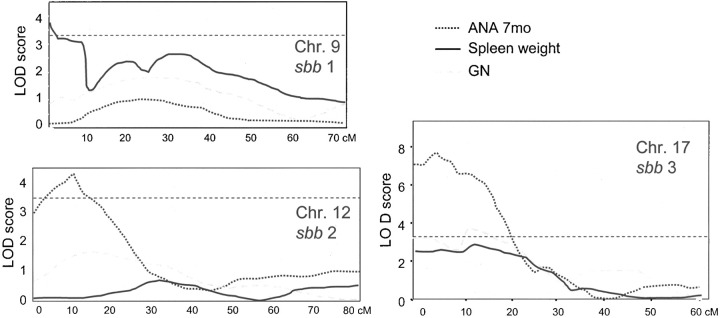
94 F2 mice with extreme scores in the ANA test (44 with 0 and 50 with 3–4 score) were selected for an initial linkage analysis. These 94 genomes were mapped with 81 microsatellite markers that span the entire mouse genome at a coverage of 20 cM. Analysis of these data by MAPMAKER/QTL (34), indicated that chromosomes 1, 4, 7, 9, 12, and 17 were likely to contain regions with positive linkage for ANA. These chromosomes were selected for more refined analysis in which all 186 F2 mice, 40 microsatellite markers and all three phenotypic traits defined in the previous section were evaluated. Table II shows a selection of these results, emphasizing the three regions on chromosomes 9, 12, and 17 that gave LOD scores of >3.4, which can be considered statistically significant according to reference 37 (see Table II and Fig. 5) . The strongest effect came from a proximal region on chromosome 17 defined by the marker D17Mit198, yielding scores 7.58 for ANA, 3.62 for proteinuria, and 2.68 for spleen size. In this locus, the B6 allele provides the susceptibility in a recessive form for all three traits. The minimum interval that contains the susceptibility gene/s was calculated to be 18 cM (5.5–23.5 cM distance from the telomere) for ANA and 10 cM for proteinuria (5.5–15.5 cM) (Table III). These intervals were called sbb3 (for ANA) and sbb3a (for proteinuria). sbb3 contains the mouse H2 complex, which was already shown to be critical in other mouse models of lupus. However, proteinuria, a severe disease trait, mapped at least 4 cM centromeric to the H2 complex. To determine the contribution of the BALB/c or B6 H2 complex on this model of lupus, we performed a locus swap between B6.R2 − / − and BALB.R2 − / − mice to get a better sense of the requirement for a specific H2 haplotype in this disease model. When H2 d (BALB/c carries H2 d) was bred onto B6.R2 − / − mice in homozygous form, ANA production was reduced, but proteinuria could not be detected (data not shown). Conversely, when the reverse H2 swap was generated (BALB. R2 − / − H2 b mice), the autoimmunity was undetectable with all three phenotypes still below the threshold of significant values (data not shown). Thus, the presence of the H2 d haplotype provides some protection for severe autoimmune disease but the H2 b haplotype is insufficient to initiate autoimmunity in BALB.R2 − / −, supporting our mapping of the chromosome 17 locus responsible for severe disease to lie outside of the H2 locus.
Table II.
Loci Associated with Autoimmune Phenotypes by a Genome Scan of (R2−/−B6 × R2−/−BALB) F2 Progeny (188 Mice)
| (Balb) | (Het) | (B6) | Lod | Lod | Lod | Lod | Fix c17 | % Gen | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| cM | Marker | AAa | AB | BB | Free | Dom. | Rec. | Add. | Lod free | Variance | |
| ANA (0–4) | 66.7 | D1Mit90 | 0.65 | 1.19 | 1.38 | 2.24 | 2.01 | 0.81 | 1.98 | 2.30 | 5.8 |
| 10.9 | D7Mit117 | 1.28 | 0.83 | 0.61 | 1.98 | 1.64 | 0.90 | 1.86 | 2.57 | 4.8 | |
| 13.1 | D9Mit67 | 1.10 | 0.81 | 068. | 0.84 | 0.73 | 0.33 | 0.79 | ND | 2.1 | |
| 33.9 | D9Mit32 | 1.24 | 0.68 | 0.82 | 1.78 | 1.69 | 0.04 | 0.79 | 3.20 | 4.5 | |
| 5.5 | D12Mit12 | 0.48 | 0.79 | 1.37 | 3.87 | 1.80 | 3.19 | 3.67 | 4.32 | 9.1 | |
| 5.5 | D17Mit198 | 0.8 | 0.54 | 1.58 | 7.58 | 0.04 | 7.09 | 3.03 | - | 17.1 | |
| Spleen weight (mg) | 66.7 | D1Mit90 | 123.30 | 144.08 | 192.96 | 0.81 | 0.22 | 0.74 | 0.087 | ND | 4.2 |
| 10.9 | D7Mit117 | 142.55 | 145.07 | 148.85 | 0.02 | 0.01 | 0.02 | 0.03 | ND | 0.1 | |
| 13.1 | D9Mit67 | 201.91 | 129.52 | 137.91 | 3.59 | 3.54 | 0.12 | 1.81 | 3.96 | 9.0 | |
| 33.9 | D9Mit32 | 209.94 | 135.07 | 131.90 | 2.43 | 2.42 | 0.81 | 1.69 | ND | 11.3 | |
| 5.5 | D12Mit12 | 122.47 | 114.47 | 139.24 | 0.14 | 0.09 | 0.09 | 0.14 | ND | 0.4 | |
| 5.5 | D17Mit198 | 130.94 | 138.62 | 196.16 | 2.68 | 0.46 | 2.64 | 2.01 | - | 6.8 | |
| Proteinuria (0–4) | 10.9 | D7Mit117 | 0.70 | 0.16 | 0.20 | 1.90 | 1.88 | 0.16 | 0.95 | ND | 6.8 |
| 13.1 | D9Mit67 | 0.26 | 0.31 | 0.34 | 0.15 | 0.00 | 0.13 | 0.04 | ND | 0.4 | |
| 33.9 | D9Mit32 | 0.23 | 0.11 | 0.43 | 1.02 | 0.00 | 0.90 | 0.36 | ND | 2.6 | |
| 5.5 | D12Mit12 | 0.18 | 0.24 | 0.56 | 1.44 | 0.17 | 1.44 | 0.94 | ND | 3.5 | |
| 5.5 | D17Mit198 | 0.21 | 0.35 | 0.86 | 3.62 | 0.92 | 3.43 | 3.05 | - | 8.6 |
Average value of the trait for BALB homozygous at that locus.
Figure 5.
Linkage analysis for autoimmune susceptibility loci in the B6 and BALB/c backgrounds. Mapmaker QTL analysis performed on the F2 cross (B6.R2 −/− × BALB.R2 −/−) gave significant LOD scores in three chromosomes, shown as charts in this Figure. Three loci, named sbb1, sbb2, and sbb3 have been identified which yielded LOD scores >3.4 and are displayed as horizontal patterned bars. sbb1 is linked to splenomegaly in this cross, while sbb2 and sbb3 displayed linkage for ANA at 7 mo. Tables II and III summarize the relevant markers, LOD scores, and inheritance pattern for these loci.
Table III.
New Autoimmune Susceptibility B6 × BALB (sbb) Loci
| Trait | Locus | Chromosome | Interval | Lodfree | Susceptibleallele |
|---|---|---|---|---|---|
| (cM) | |||||
| ANA (0–4) | sbb2 | Chr.12 | 2.5–8.5 | 3.87 | B6 |
| sbb3 | Chr.17 | 5.5–23.5 | 7.58 | B6 | |
| Spleen weight (mg) | sbb1 | Chr.9 | 0.0–14.0 | 3.59 | BALB/c |
| Proteinuria (0–4) | sbb3 | Chr.17 | 5.5–15.5 | 3.62 | B6 |
A second strong linkage (lod 3.87) was observed on chromosome 12 for the ANA phenotype (Table II). As in the case of the chromosome 17 locus, susceptibility is mediated by an allele derived from the B6 background. Analysis of the inheritance of this susceptibility among F2 progeny suggests that it contributes to susceptibility in an allele dose manner, (intermediate for heterozygotes, complete for homozygotes). The interval containing this allele was calculated to be 6 cM (2.5–8.5 cM) and was named sbb2 (Table III). A third significant genetic linkage was found on chromosome 9 (lod 3.62) for the splenomegaly trait. A 14-cM interval on the distal portion of chromosome 9 was found to include this susceptibility allele, which is derived from the BALB/c background in recessive form and has been named sbb1. Other loci with minor linkage for ANA were found on chromosome 1 (D1Mit60) and 7 (D7Mit117) (Table I). So far no obvious candidate genes have been found in any of these newly identified genomic intervals, thus increasing the likelihood that fine mapping of these loci will lead to the identification of novel genes that modify susceptibility of autoimmune disease.
Discussion
The sequence of events that result in the lupus phenotype are the result of distinct genetic interactions between susceptibility loci and their modifiers. We have dissected the mechanisms of this epistasis in the B6.RIIB−/− model of lupus, a model that mimics many aspects of the genetics of the human disease. Loss of tolerance to nuclear antigens in this model involves interactions of genes lying on the same pathway, a pathway that includes RIIB and Sle1, resulting in the appearance of ANA. The pathogenicity of these antibodies, in turn, is influenced by genes such as yaa and lpr, yaa enhances disease by altering the specificity of the autoantibodies despite a reduction in total titer. In contrast, the lpr mutation protects animals from the potentially pathogenic effects of B6.R2 − / − induced autoantibodies, without affecting ANA titers, thus acting like a suppressor locus in this disease model. The results indicate that modulation of FcγRIIB expression or signaling is a critical step in regulating B cell responsiveness. Thus, the inappropriate activation of B cells, which occur when RIIB expression is reduced or signaling is impaired will result in the loss of tolerance and development of autoimmunity.
Acknowledgments
We would like to thank Dr. David Scott for valuable suggestions and Vicki Lappi and Devon White for technical assistance.
This work was supported by grants from the National Institutes of Health (NIH) and Alliance for Lupus Research (to J.V. Ravetch), the Lupus Research Institute and Women & Science (to S. Bolland and E.K. Wakeland).
S. Bolland's current address is Laboratory of Immunogenetics, NIAID/NIH, 12441 Parklawn Dr., Rockville, MD 20852.
Footnotes
Abbreviations used in this paper: ANA, antinuclear antibodies; SLE, systemic lupus erythematosus.
References
- 1.Kotzin, B.L. 1996. Systemic lupus erythematosus. Cell. 85:303–306. [DOI] [PubMed] [Google Scholar]
- 2.Theofilopoulos, A.N. 1995. The basis of autoimmunity: part II. Genetic predisposition. Immunol. Today. 16:150–159. [DOI] [PubMed] [Google Scholar]
- 3.Vyse, T.J., and J.A. Todd. 1996. Genetic analysis of autoimmune disease. Cell. 85:311–318. [DOI] [PubMed] [Google Scholar]
- 4.Wanstrat, A., and E. Wakeland. 2001. The genetics of complex autoimmune diseases: non-MHC susceptibility genes. Nat. Immunol. 2:802–809. [DOI] [PubMed] [Google Scholar]
- 5.Wakeland, E.K., K. Liu, R.R. Graham, and T.W. Behrens. 2001. Delineating the genetic basis of systemic lupus erythematosus. Immunity. 15:397–408. [DOI] [PubMed] [Google Scholar]
- 6.McDevitt, H.O. 1998. The role of MHC class II molecules in susceptibility and resistance to autoimmunity. Curr. Opin. Immunol. 10:677–681. [DOI] [PubMed] [Google Scholar]
- 7.Ridgway, W.M., M. Fasso, and C.G. Fathman. 1999. A new look at MHC and autoimmune disease. Science. 284:749–751. [DOI] [PubMed] [Google Scholar]
- 8.Drake, C.G., S.J. Rozzo, T.J. Vyse, E. Palmer, and B.L. Kotzin. 1995. Genetic contributions to lupus-like disease in (NZB × NZW)F1 mice. Immunol. Rev. 144:51–74. [DOI] [PubMed] [Google Scholar]
- 9.Rozzo, S.J., J.D. Allard, D. Choubey, T.J. Vyse, S. Izui, G. Peltz, and B.L. Kotzin. 2001. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 15:435–443. [DOI] [PubMed] [Google Scholar]
- 10.Hogarth, M.B., J.H. Slingsby, P.J. Allen, E.M. Thompson, P. Chandler, K.A. Davies, E. Simpson, B.J. Morley, and M.J. Walport. 1998. Multiple lupus susceptibility loci map to chromosome 1 in BXSB mice. J. Immunol. 161:2753–2761. [PubMed] [Google Scholar]
- 11.Morel, L., C. Mohan, Y. Yu, B.P. Croker, N. Tian, A. Deng, and E.K. Wakeland. 1997. Functional dissection of systemic lupus erythematosus using congenic mouse strains. J. Immunol. 158:6019–6028. [PubMed] [Google Scholar]
- 12.Morel, L., K.R. Blenman, B.P. Croker, and E.K. Wakeland. 2001. The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc. Natl. Acad. Sci. USA. 98:1787–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morel, L., B.P. Croker, K.R. Blenman, C. Mohan, G. Huang, G. Gilkeson, and E.K. Wakeland. 2000. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc. Natl. Acad. Sci. USA. 97:6670–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tivol, E.A., F. Borriello, A.N. Schweitzer, W.P. Lynch, J.A. Bluestone, and A.H. Sharpe. 1995. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity. 3:541–547. [DOI] [PubMed] [Google Scholar]
- 15.O'Keefe, T.L., G.T. Williams, F.D. Batista, and M.S. Neuberger. 1999. Deficiency in CD22, a B cell-specific inhibitory receptor, is sufficient to predispose to development of high affinity autoantibodies. J. Exp. Med. 189:1307–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimura, H., M. Nose, H. Hiai, N. Minato, and T. Honjo. 1999. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 11:141–151. [DOI] [PubMed] [Google Scholar]
- 17.Bolland, S., and J.V. Ravetch. 2000. Spontaneous autoimmune disease in FcγRII deficient mice results from strain-specific epistasis. Immunity. 13:277–285. [DOI] [PubMed] [Google Scholar]
- 18.Chiang, Y.J., H.K. Kole, K. Brown, M. Naramura, S. Fukuhara, R.J. Hu, I.K. Jang, J.S. Gutkind, E. Shevach, and H. Gu. 2000. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 403:216–220. [DOI] [PubMed] [Google Scholar]
- 19.Bachmaier, K., C. Krawczyk, I. Kozieradzki, Y.Y. Kong, T. Sasaki, A. Oliveira-dos-Santos, S. Mariathasan, D. Bouchard, A. Wakeham, A. Itie, et al. 2000. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 403:211–216. [DOI] [PubMed] [Google Scholar]
- 20.Hibbs, M.L., D.M. Tarlinton, J. Armes, D. Grail, G. Hodgson, R. Maglitto, S.A. Stacker, and A.R. Dunn. 1995. Multiple defects in the immune system of Lyn-deficient mice, culminating in autoimmune disease. Cell. 83:301–311. [DOI] [PubMed] [Google Scholar]
- 21.Tsui, F.W., and H.W. Tsui. 1994. Molecular basis of the motheaten phenotype. Immunol. Rev. 138:185–206. [DOI] [PubMed] [Google Scholar]
- 22.Ravetch, J.V., and S. Bolland. 2001. IgG Fc receptors. Annu. Rev. Immunol. 19:275–290. [DOI] [PubMed] [Google Scholar]
- 23.Clynes, R., C. Dumitru, and J.V. Ravetch. 1998. Uncoupling of immune complex formation and kidney damage in autoimmune glomerulonephritis. Science. 279:1052–1054. [DOI] [PubMed] [Google Scholar]
- 24.Clynes, R., J.S. Maizes, R. Guinamard, M. Ono, T. Takai, and J.V. Ravetch. 1999. Modulation of immune complex-induced inflammation in vivo by the coordinate expression of activation and inhibitory Fc receptors. J. Exp. Med. 189:179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clynes, R.A., T.L. Towers, L.G. Presta, and J.V. Ravetch. 2000. Inhibitory Fc receptors modulate in vivo cytoxicity against tumor targets. Nat. Med. 6:443–446. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura, A., T. Yuasa, A. Ujike, M. Ono, T. Nukiwa, J.V. Ravetch, and T. Takai. 2000. Fcγ receptor IIB-deficient mice develop Goodpasture's syndrome upon immunization with type IV collagen: a novel murine model for autoimmune glomerular basement membrane disease. J. Exp. Med. 191:899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuasa, T., S. Kubo, T. Yoshino, A. Ujike, K. Matsumura, M. Ono, J.V. Ravetch, and T. Takai. 1999. Deletion of Fcγ receptor IIB renders H-2(b) mice susceptible to collagen-induced arthritis. J. Exp. Med. 189:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izui, S., M. Iwamoto, L. Fossati, R. Merino, S. Takahashi, and N. Ibnou-Zekri. 1995. The Yaa gene model of systemic lupus erythematosus. Immunol. Rev. 144:137–156. [DOI] [PubMed] [Google Scholar]
- 29.Mary, C., C. Laporte, D. Parzy, M.L. Santiago, F. Stefani, F. Lajaunias, R.M. Parkhouse, T.L. O'Keefe, M.S. Neuberger, S. Izui, and L. Reininger. 2000. Dysregulated expression of the Cd22 gene as a result of a short interspersed nucleotide element insertion in Cd22a lupus-prone mice. J. Immunol. 165:2987–2996. [DOI] [PubMed] [Google Scholar]
- 30.Vidal, S., D.H. Kono, and A.N. Theofilopoulos. 1998. Loci predisposing to autoimmunity in MRL-Fas lpr and C57BL/6-Faslpr mice. J. Clin. Invest. 101:696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singer, G.G., A.C. Carrera, A. Marshak-Rothstein, C. Martinez, and A.K. Abbas. 1994. Apoptosis, Fas and systemic autoimmunity: the MRL-lpr/lpr model. Curr. Opin. Immunol. 6:913–920. [DOI] [PubMed] [Google Scholar]
- 32.Chervonsky, A.V., Y. Wang, F.S. Wong, I. Visintin, R.A. Flavell, C.A. Janeway, Jr., and L.A. Matis. 1997. The role of Fas in autoimmune diabetes. Cell. 89:17–24. [DOI] [PubMed] [Google Scholar]
- 33.Satoh, M., J.P. Weintraub, H. Yoshida, V.M. Shaheen, H.B. Richards, M. Shaw, and W.H. Reeves. 2000. Fas and Fas ligand mutations inhibit autoantibody production in pristane-induced lupus. J. Immunol. 165:1036–1043. [DOI] [PubMed] [Google Scholar]
- 34.Lander, E.S., P. Green, J. Abrahamson, A. Barlow, M.J. Daly, S.E. Lincoln, and L. Newburg. 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1:174–181. [DOI] [PubMed] [Google Scholar]
- 35.Takai, T., M. Ono, M. Hikida, H. Ohmori, and J.V. Ravetch. 1996. Augmented humoral and anaphylactic responses in Fcγ RII-deficient mice. Nature. 379:346–349. [DOI] [PubMed] [Google Scholar]
- 36.Morel, L., U.H. Rudofsky, J.A. Longmate, J. Schiffenbauer, and E.K. Wakeland. 1994. Polygenic control of susceptibility to murine systemic lupus erythematosus. Immunity. 1:219–229. [DOI] [PubMed] [Google Scholar]
- 37.Lander, E., and L. Kruglyak. 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11:241–247. [DOI] [PubMed] [Google Scholar]