Abstract
Saccharomyces cerevisiae strains lacking the Ppz1 protein phosphatase are salt tolerant and display increased expression of the ENA1 Na+-ATPase gene, a major determinant for sodium extrusion, while cells devoid of the similar Ppz2 protein do not show these phenotypes. However, a ppz1 ppz2 mutant displays higher levels of ENA1 expression than the ppz1 strain. We show here that the increased activity of the ENA1 promoter in a ppz1 ppz2 mutant maps to two regions: one region located at −751 to −667, containing a calcineurin-dependent response element (CDRE), and one downstream region (−573 to −490) whose activity responds to intracellular alkalinization. In contrast, the increased ENA1 expression in a ppz1 mutant is mediated solely by an intact calcineurin/Crz1 signaling pathway, on the basis that (i) this effect maps to a single region that contains the CDRE and (ii) it is blocked by the calcineurin inhibitor FK506, as well as by deletion of the CNB1 or CRZ1 gene. The calcineurin dependence of the increased ENA1 expression of a ppz1 mutant would suggest that Ppz1 could negatively regulate calcineurin activity. In agreement with this notion, a ppz1 strain is calcium sensitive, and this mutation does not result in a decrease in the calcium hypertolerance of a cnb1 mutant. It has been shown that ENA1 can be induced by alkalinization of the medium and that a ppz1 ppz2 strain has a higher intracellular pH. However, we present several lines of evidence that show that the gene expression profile of a ppz1 mutant does not involve an alkalinization effect. In conclusion, we have identified a novel role for calcineurin, but not alkalinization, in the control of ENA1 expression in ppz1 mutants.
The Saccharomyces cerevisiae Ppz phosphatases are encoded by the PPZ1 and PPZ2 genes (21, 37, 38), and they are characterized by a catalytic carboxyl-terminal half related to type 1 phosphatases and an amino-terminal extension. The catalytic moieties of the two proteins are very similar (92%). In contrast, the sequences of the amino-terminal halves are much less related. These phosphatases are involved in a variety of cell processes, including regulation of salt tolerance (39), maintenance of cell wall integrity (26, 37), and regulation of the cell cycle at the G1/S transition (5, 6). In all cases, the function of Ppz1 appears to be more important than that of Ppz2. For instance, a ppz1 deletion mutant is tolerant to high concentrations of sodium or lithium cations, while a ppz2 mutant is not. However, the ppz1 ppz2 double mutant is more tolerant than the ppz1 single mutant (39). Genetic and biochemical evidence has identified the halotolerant determinant Hal3, also known as Sis2 (12, 13), as a negative regulatory subunit of Ppz1 (11). Therefore, overexpression of Hal3 results in increased salt tolerance. Hal3 binds to the carboxy-terminal catalytic moiety of Ppz1, thus modulating the diverse physiological functions of the phosphatase. No evidence of Hal3 interaction with Ppz2 has been reported so far.
In S. cerevisiae, the ENA1 gene plays an important role in salt tolerance. This gene encodes a P-type Na+-ATPase that represents the most important element for the efflux of Na+ and Li+. Consequently, an ena1 mutant is highly sensitive even to low concentrations of Na+ or Li+ (14). The ENA1 gene is barely expressed under standard growth conditions, but it is strongly induced by exposure to high salt concentrations and to an alkaline pH. This transcriptional response of ENA1 is based on a complex regulation of its promoter (28). Expression of ENA1 is repressed by the presence of glucose in the medium (2), through a mechanism that involves the general repressor complex Mig1-Ssn6-Tup1 (40). Saline induction is mediated by two pathways: the Hog1 mitogen-activated protein kinase pathway and the calcineurin pathway. The Hog1 pathway responds to increased osmolarity and acts through the Sko1 transcriptional inhibitor (40), which binds to a cyclic AMP response element present in the ENA1 promoter.
The Ca+-calmodulin-activated protein phosphatase calcineurin is structurally and functionally conserved from yeast to humans. The enzyme is a heterodimer composed of a catalytic A subunit and a regulatory B subunit. In S. cerevisiae, the catalytic subunit is encoded by two genes (CNA1 and CNA2), while a single gene (CNB1) codes for the regulatory subunit (9, 10, 24, 27, 48). Calcineurin regulates ENA1 transcription in response to sodium and lithium toxicity by dephosphorylating and activating the transcriptional activator Crz1/Tcn1/Hal8 (29, 31, 44), which binds specific DNA sequences known as calcineurin-dependent response elements (CDRE). Two such elements are present in the ENA1 promoter, at positions −813 to −821 and −727 to −719; the downstream element is more relevant for the transcriptional response under stress conditions (30). The function of calcineurin can be regulated by a conserved family of proteins termed calcipressins, which are represented in S. cerevisiae by a single gene known as RCN1. Interestingly, although it seems clear that Rcn1 binds and inhibits calcineurin, it is not evident whether this protein acts as a regulator of calcineurin or as a downstream effector of the phosphatase (17, 23). Recently, a role for the TOR (target of rapamycin) pathway in the regulation of ENA1 expression through the Gln3 and Gat1 transcription factors has been documented (7).
The transcriptional response of ENA1 to an alkaline pH is also rather complex and not yet fully understood. Although the effect of a high pH on ENA1 induction was described about 10 years ago (14, 33), only recently has a report (43) addressed the molecular basis of this effect and identified two responsive regions (ARR1 and ARR2) in the ENA1 promoter. ARR1 maps to the downstream CDRE and is responsible for the calcineurin-dependent component of the response. ARR2 spans nucleotides −573 to −490 and integrates Rim101-dependent and -independent inputs. A very recent report has suggested that Rim101 may inhibit the function of the Nrg1 transcription factor, which in turn would negatively regulate the ENA1 promoter (25).
The early observation that a ppz1 ppz2 mutant showed increased expression of the ENA1 gene (39) provided a reasonable explanation for the observed salt-tolerant phenotype. However, it has been shown very recently that a ppz1 ppz2 mutant strain displays an augmented intracellular pH and, under NaCl stress conditions, a higher-than-normal intracellular potassium content. This is likely the result of an inhibitory role of the Ppz phosphatases on the Trk1/Trk2 potassium transporters, and it may explain most of the phenotypes observed in cells lacking both phosphatase genes (49). Despite the evident link between lack of Ppz phosphatases and Trk function, a number of important issues remain to be resolved. For instance, there was evidence that deletion of both PPZ phosphatases in a trk1 trk2 background still produced a detectable increase in salt tolerance (49), suggesting that Trk-independent mechanisms may exist. In addition, since the saline tolerances of ppz1 and ppz2 mutants are not equivalent, it was important to evaluate the relative contributions of the two gene products to this phenotype.
The fact that the ENA1 ATPase gene is a major determinant for salt tolerance prompted us to investigate how the absence of Ppz1, Ppz2, or both phosphatases affects ENA1 expression. In this report we present a detailed dissection of the relative contributions of the Ppz phosphatases to the regulation of ENA1, providing evidence that Ppz1 negatively controls ENA1 expression through mechanisms that involve calcineurin but not intracellular alkalinization.
MATERIALS AND METHODS
Yeast strains and growth conditions.
Yeast cells were grown at 28°C in YPD medium (containing, per liter, 10 g of yeast extract, 20 g of peptone, and 20 g of dextrose) or, when indicated, in synthetic minimal (SD) or complete minimal (CM) medium (1). The relevant genotypes of the strains described in this work are given in Table 1. Sensitivity to LiCl and NaCl was monitored essentially as in reference 39.
TABLE 1.
S. cerevisiae strains used in this work
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| DBY746 | MATα ura3-52 leu2-3, 112 his3-Δ1 trp1-Δ239 | D. Botstein |
| EDN2 | DBY746 ppz1::TRP1 | This work |
| EDN85 | DBY746 ppz1::TRP1 ppz2::KAN | This work |
| MAR55 | DBY746 ppz2::TRP1 | This work |
| MAR15 | DBY746 cnb1::KAN | 43 |
| EDN92 | DBY746 crz1::KAN | 43 |
| MAR18 | DBY746 crz1::KAN ppz1::URA3 | This work |
| MAR19 | DBY746 cnb1::KAN ppz1::URA3 | This work |
| EDN308 | DBY746 crz1::KAN ppz1::TRP1 | This work |
| MAR40 | DBY746 cnb1::KAN ppz1::TRP1 | This work |
| ESV212 | DBY746 trk1::LEU2 trk2::HIS3 | This work |
| MAR37 | DBY746 ppz1::TRP1 trk1::LEU2 trk2::HIS3 | This work |
| RSC25 | DBY746 rcn1::KAN | This work |
| MAR39 | DBY746 ppz1::TRP1 rcn1::KAN | This work |
| MAR43 | DBY746 mck1::KAN | This work |
| MAR46 | DBY746 ppz1::TRP1 mck1::KAN | This work |
| JA100 | MATaura3-52 leu2-3, 112 his4 trp1-1 can-1r | 11 |
| EDN75 | JA100 ppz1::KAN | 11 |
| JA-103 | JA100 ppz2::TRP1 | 6 |
| EDN76 | JA100 ppz1::KAN ppz2::TRP1 | This work |
Gene disruptions and plasmids.
Disruption of the PPZ1 gene with the URA3 marker was carried out as in reference 38. Deletion of this gene with the TRP1 marker was performed as follows. Plasmid pYC2Z1 (6) was digested with BamHI/HindIII, and the 1.6-kbp insert was cloned into the same sites of plasmid pGEM-3Z (Promega). This construct was then digested with SalI/StuI, and the DNA fragment released was replaced with a 0.85-kbp TRP1 marker, previously released from plasmid YDp-W (4) by digestion with SalI/SmaI. The disruption cassette was then digested with SacI, and the 1.3-kbp fragment was used for yeast transformation. This strategy removes most of the Ppz1 protein (from residue 1 to residue 547). Deletion of PPZ2 with a TRP1 marker was performed as described by Clotet et al. (6). To disrupt the PPZ2 gene with the KanMX marker, a 2.2-kbp BamHI/BamHI fragment of the gene was cloned into pUC18 and then digested with HpaI and BglII to remove approximately 1.5 kbp corresponding to a small fragment of the 5′ untranslated region plus the first 460 codons. This fragment was replaced by the KanMX marker, obtained by digestion of plasmid pFA6-kanMX4 (45) with BglII and EcoRV. The final construct was then cleaved by BamHI, and the insert was used to transform yeast cells. Disruptions of CNB1 and CRZ1 with the KAN marker have been described previously (43). The TRK1 and TRK2 genes were disrupted with the LEU2 and HIS3 markers, respectively, by using deletion cassettes described previously (20). Strains carrying deletions of the RCN1 and MCK1 genes were generated as follows. Deletion cassettes were amplified by PCR from genomic DNA prepared from rcn1::KanMX and mck1::KanMX deletion mutants in the BY4741 background (46). The oligonucleotides used spanned nucleotides −842 to +746 (RCN1) and −467 to +1782 (MCK1). Positions are relative to the initiating ATG codon. The amplification fragments were purified and used to transform the desired strains.
For high-copy expression of HAL3, the gene was recovered from plasmid YEp351-HAL3 (13) by digestion with EcoRI/HindIII and cloned into these sites of plasmid YEplac181 (16).
β-Galactosidase reporter constructs and assays.
Plasmid pKC201, containing the entire promoter of ENA1, has been described previously (2, 8). The pMP and pMR series, containing defined fragments of the ENA1 promoter, have also been described previously (40, 43). pMRK211 was constructed similarly to pMRK212 and pMRK213 (43). Plasmid pLA, a gift from I. Fernandez de Larrinoa (Universidad del Pais Vasco, San Sebastián, Spain), contains the −732-to-−711 sequence of the ENA1 promoter, corresponding to the high-affinity Crz1-binding element (30), and derives from plasmid pLGΔ312s (19). Plasmid pAMS366 contains four copies in tandem of the wild-type CDRE present in the FKS2 promoter, while pAMS364 contains a mutated version that cannot bind Crz1 (44). The construction of pPHO84-LacZ, pPHO89-LacZ, and pPHO12-LacZ has been described previously (43). Plasmids pFRE1-LacZ, pTRR1-LacZ, and pFIT2-LacZ were constructed by PCR amplification of the −792-to-+73, −758-to-+82, and −695-to-+82 regions of the respective genes, which were cloned into the EcoRI/HindIII sites of plasmid YEp357 (34). Plasmid pSIT1-LacZ was constructed by cloning the −786-to-+52 region of SIT1 into the EcoRI/SalI site of YEp357. In all cases, cloning resulted in translational fusions with the β-galactosidase gene.
The levels of expression driven by the various promoters tested (as well as by specific CDRE) in different mutant backgrounds were determined by using β-galactosidase reporter constructs as follows. Yeast cells were transformed with the desired plasmids, and positive clones were grown in selective medium (lacking uracil) to an A660 of 1.5 to 2.0. These cultures were used to inoculate YPD medium (2.5 ml), and growth was resumed until an A660 of 0.6 to 0.8 was reached (approximately 5 h). When sensitivity to FK506 was tested, YPD medium was made 1.5 μg/ml FK506 from a 10-mg/ml stock solution of the drug (in 90% ethanol-10% Tween P-20 [vol/vol]). Control cells were exposed only to the vehicle. Cells were recovered by centrifugation, and β-galactosidase assays were performed as described previously (41). The sensitivity to ambient pH of the response of the ARR1 and ARR2 elements of the ENA1 promoter was determined essentially as described previously (43).
In vitro binding experiments.
Interaction experiments were performed essentially as described previously (11). The construction of the glutathione S-transferase (GST) fusion protein containing the catalytic moiety (amino acids 345 to 692) of Ppz1 (Ppz1CAT-GST) has been documented previously (6). The catalytic domain (amino acids 379 to 710) of Ppz2 (Ppz2CAT) was cloned by PCR from genomic DNA using primers 5′ ATATCTAGATAGGACTAATACTATGGTCGA and 3′ ATAATAGTCGACTCAGCGATTGGCTAATTTAC. The resulting PCR product was then digested with XbaI and SalI and was subcloned into the pGEX-KG vector (18). The yeast strain (LY127) used as a source of Hal3 has been described previously (49).
Intracellular cation measurements.
Intracellular sodium and potassium concentrations were determined essentially as described previously (3). Briefly, YPD medium containing 0.3 M sodium chloride was inoculated with the different strains, and growth was resumed until an optical density at 660 nm (OD660) of 0.9 to 1.0 was reached. Cells were recovered by filtration and washed with a 0.6 M sorbitol solution containing 10 mM magnesium chloride, and extracts were prepared by boiling for 30 min. Extracts were centrifuged for 1 min at 10,000 × g, and the concentrations of the cations were measured in the supernatants by flame spectroscopy.
RESULTS
The Hal3-dependent regulation of ENA1 expression is mediated by both Ppz1 and Ppz2 phosphatases.
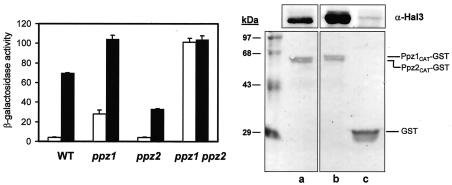
Hal3 was identified some time ago as a negative regulatory subunit of Ppz1. To characterize the relative influences of Ppz1 and Ppz2 phosphatases on ENA1 expression, we first compared the effects of high-copy expression of Hal3 on the expression of the ATPase gene in wild-type cells and cells deficient in Ppz1, Ppz2, or both phosphatases. As shown in Fig. 1, overexpression of Hal3 in wild-type cells resulted in increased ENA1 expression. When Hal3 was overexpressed in ppz1 cells, the activity of the ENA1 promoter reached virtually the same level as that observed in a ppz1 ppz2 strain carrying an empty plasmid. Interestingly, expression of ENA1 did not increase further when Hal3 was overexpressed in the ppz1 ppz2 double mutant. A ppz2 mutant did not show altered ENA1 expression, and in this background, overexpression of Hal3 produced a level of activity of the ENA1 promoter almost identical to that observed in a ppz1 strain bearing an empty plasmid. These results indicated that Hal3 influences both Ppz1 and Ppz2 functions. To confirm the possibility of Ppz2 being a target for Hal3, the catalytic domains of both Ppz1 and Ppz2 were purified from Escherichia coli as GST fusion proteins and used as an affinity system to purify Hal3 from crude yeast extracts. As shown in Fig. 1, Hal3 was retained in similar amounts by Ppz1 and Ppz2, proving that Hal3 is able to interact in vitro with both phosphatases.
FIG. 1.
Effects of overexpression of Hal3 on ENA1 expression in different Ppz backgrounds. (Left) Strains JA100 (wild type [WT]), EDN75 (ppz1), JA103 (ppz2), and EDN76 (ppz1 ppz2) bearing plasmid pKC201 (2), which encodes the β-galactosidase reporter gene fused to the ENA1 promoter, were transformed with the high-copy-number plasmid YEplac181 carrying no insert (open bars) or the same plasmid carrying the entire HAL3 gene. Cells were grown as described in Materials and Methods, and β-galactosidase activity was measured. Data are means ± standard errors of the means from three to nine independent clones. (Right) The catalytic domains of both Ppz1 and Ppz2 were purified from E. coli as GST fusion proteins. Approximately 3 μg of purified protein was then used as an affinity resin to copurify Hal3 from crude yeast extracts. The material retained by the resin after extensive washing was then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and probed with antibodies specific for Hal3. The bottom panel shows the Ponceau S staining of the membrane, as a control for protein loading. The upper panel shows the result of Western blot analysis with Hal3-specific antibodies. Lane a, Hal3 purified by using the catalytic domain of Ppz2; lane b, Hal3 purified by using the catalytic domain of Ppz1). No Hal3 was detected by using GST alone as an affinity resin (lane c).
The increased ENA1 expression of a ppz1 mutant functionally maps to a CDRE and depends on an intact calcineurin-signaling pathway.
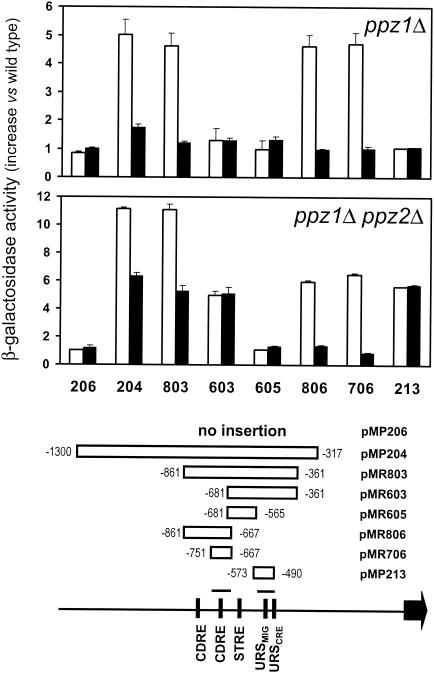
The ENA1 promoter is rather complex and is able to respond to different stimuli. We have analyzed the levels of expression in wild-type, ppz1, and ppz1 ppz2 cells by using a number of reporter constructs that allow functional mapping of this promoter. As shown in Fig. 2, the increased ENA1 expression characteristic of a ppz1 mutant could be detected in all constructs including the −751-to-−667 region, which contains the downstream CDRE defined for this promoter, but not in those lacking this element. In addition, incubation of the cells with the calcineurin inhibitor FK506 reduced the activity of the promoter in the ppz1 mutant to the levels observed in wild-type cells. Interestingly, when the same experiments were performed with a ppz1 ppz2 mutant, expression levels from the full promoter increased twofold and were reduced only partially after addition of FK506. As observed, the FK506-inhibitable expression fully maps to the −751-to-−667 region. In contrast, the non-FK506-inhibitable component maps to a downstream region, delimited by nucleotides −573 and −490. The level of expression driven from this region was not increased at all in a ppz2 mutant.
FIG. 2.
Mapping the activity of the ENA1 promoter in ppz1 and ppz1 ppz2 mutants. Strains JA100 (wild type), EDN75 (ppz1), and EDN76 (ppz1 ppz2) were transformed with the indicated constructs. (Bottom) Relevant regulatory elements described in the main text are schematically depicted. Rectangles represent the fragments of the ENA1 promoter contained in the constructs, and numbers indicate their relative nucleotide positions (from the initiating Met codon). (Top) Data represent the ratio of β-galactosidase activity between each mutant and the isogenic wild-type strain in the absence (open bars) or presence (solid bars) of 1.5 μg of FK506/ml. Each data point corresponds to the mean ± standard error of the mean from three to nine independent clones.
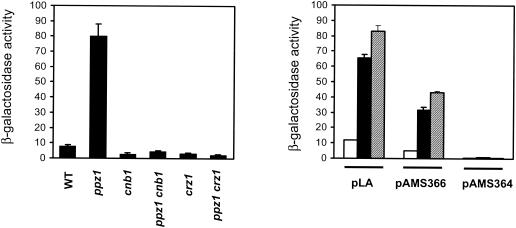
The results described above suggested that the increased ENA1 expression of a ppz1 mutant was fully dependent on the existence of a functional calcineurin pathway. Consequently, we show (Fig. 3, left) that this increase in expression was completely abolished in a strain lacking the CNB1 gene, which encodes the regulatory subunit of calcineurin. It is accepted that most, if not all, of the transcriptional response triggered by activation of calcineurin appears to be mediated by the transcriptional factor Crz1. As shown, lack of Ppz1 did not result in increased ENA1 expression in a crz1 mutant, providing additional support to the idea that the Ppz1-dependent effect on ENA1 promoter activity requires an intact calcineurin/Crz1-signaling pathway. Furthermore, expression driven exclusively by a synthetic sequence reproducing the CDRE found at positions −732 to −711 of the ENA1 promoter was also increased in a ppz1 strain (Fig. 3, right), and this increase was fully dependent on the presence of Crz1 (data not shown). Finally, we found that expression from the pAMS366 construct, which contains four tandem copies of the CDRE found in the calcium-responsive gene FKS2, was also increased in a ppz1 mutant. This increase, however, was not detected when a mutated version of the FKS2 CDRE (pAMS364), unable to bind Crz1 (44), was used (Fig. 3, right). The levels of expression driven from these chimeric promoters were only slightly increased by further deletion of the PPZ2 phosphatase gene in a ppz1 background.
FIG. 3.
Regulation of ENA1 expression by Ppz1 requires the integrity of the calcineurin-Crz1 pathway. (Left) Strain DBY746 (wild type [WT]) and the indicated isogenic derivatives were transformed with plasmid pKC201, and β-galactosidase activity was measured. Data are means ± standard errors of the means from 6 to 12 independent clones. (Right) Strains DBY746 (wild type) (open bars), EDN2 (ppz1) (filled bars), and EDN85 (ppz1 ppz2) (hatched bars) were transformed with plasmid pLA (containing the −732-to-−711 CDRE found in the ENA1 promoter as the only regulatory element), pAMS366 (bearing the wild-type CDRE found in the FKS2 promoter), or pAMS364 (bearing a mutated, nonfunctional version of the CDRE in pAMS366). Data are means ± standard errors of the means of β-galactosidase activities measured from three to nine independent clones.
Cation tolerance and ENA1 gene expression analysis of strains lacking Ppz phosphatases and components of the calcineurin pathway.
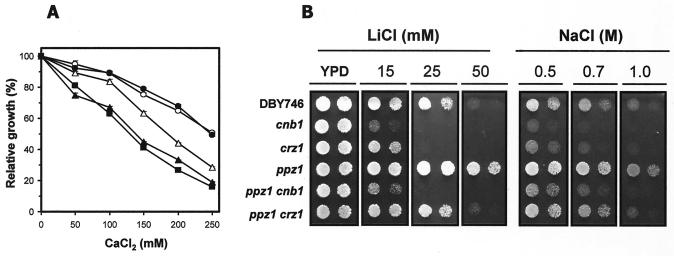
These results suggested that a ppz1 strain may have an activated calcineurin pathway, and they were compatible with the notion of Ppz1 acting as a negative regulator of calcineurin. Accordingly, we observed that ppz1 and ppz1 ppz2 mutants were more sensitive to addition of exogenous CaCl2 than the wild-type strain (Fig. 4A). Because deletion of CNB1 in an otherwise wild-type background has been shown to confer calcium tolerance (47), we tested whether lack of ppz1 affected this tolerance. We observed that a strain lacking Ppz1 and Cnb1 displayed a tolerance identical to that of a cnb1 single mutant, demonstrating that Ppz1 is not required for this tolerance and lending further support to the idea that Ppz1 may act upstream of calcineurin. We next examined the salt tolerance phenotypes of strains lacking the Ppz1 phosphatase and components of the calcineurin pathway. As shown in Fig. 4B, we observed a decrease in the salt tolerance of the ppz1 mutant upon further removal of either CNB1 or its downstream transcription factor, CRZ1. Mutation of the CNB1 gene in a ppz1 background has more pronounced effects in terms of salt tolerance, as expected, since calcineurin is known to have cellular effects independent of Crz1.
FIG. 4.
Effects on tolerance to calcium, lithium, and sodium ions of mutation of CNB1 and CRZ1 in a ppz1 background. (A) Strains DBY746 (wild type) (▵), EDN2 (ppz1) (▴), EDN85 (ppz1 ppz2) (▪), MAR15 (cnb1) (○), and MAR19 (ppz1 cnb1) (•) were tested for growth in liquid cultures at the indicated concentrations of CaCl2. Data are expressed as the percentage of growth relative to that in cultures without added salt. Each data point is the mean ± standard error of the mean from four independent cultures. (B) Cultures (OD660, 0.05 and 0.01) of the indicated strains were spotted onto YPD plates containing the indicated concentrations of LiCl or NaCl. Growth was monitored after incubation at 28°C for 60 h.
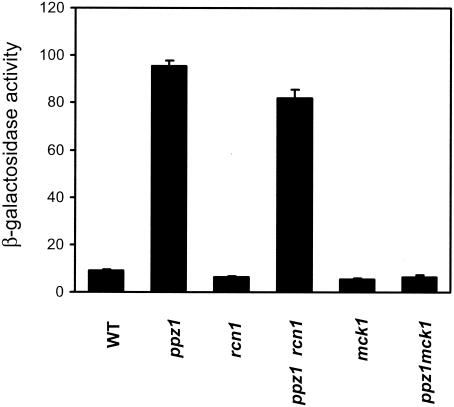
Because Rcn1 has been reported as a protein that is able to interact with calcineurin and regulate its function, we were interested in evaluating whether the lack of Rcn1 could affect the level of expression of ENA1 in a ppz1 mutant. To this end, the RCN1 gene was disrupted in wild-type and ppz1 backgrounds, and the resulting strains were transformed with the ENA1-LacZ reporter pKC201. As shown in Fig. 5, in the absence of Rcn1, expression from the ENA1 promoter was almost unaffected. We also tested the effect of disrupting MCK1, a gene encoding one of the four putative yeast homologs of glycogen synthase kinase-3, which has recently been postulated to be an upstream activator of calcineurin (K. Cunningham, personal communication). Interestingly, mutation of MCK1 fully abolished the increased ENA1 expression of the ppz1 mutant (Fig. 5). The mck1 mutant displayed a tolerance to high calcium concentrations virtually identical to that of a cnb1 strain, and this tolerance was not decreased by deletion of PPZ1 (data not shown).
FIG. 5.
Lack of Ppz1 does not result in increased ENA1 expression in an Mck1 kinase-deficient background. The indicated strains were transformed with plasmid pKC201, and β-galactosidase activity was measured. Data are means ± standard errors of the means from six independent clones. WT, wild type.
The increased ENA1 expression of a ppz1 mutant is likely not due to intracellular alkalinization.
Recent evidence (49) has shown that the absence of both Ppz1 and Ppz2 results in an increased intracellular pH and that this increase is quantitatively similar to the transient rise in the intracellular pH observed after shifting of a standard yeast culture to pH 5.8 to 8.5. Because it is known that expression from the ENA1 promoter is substantially enhanced by exposure of cells to a high pH, it can be postulated that the increase in ENA1 expression found in the ppz1 ppz2 double mutant is due to intracellular alkalinization. In this context, it was reasonable to ask whether the higher ENA1 expression characteristic of a ppz1 mutant, which we have now observed to be calcineurin dependent, could also be influenced by intracellular pH.
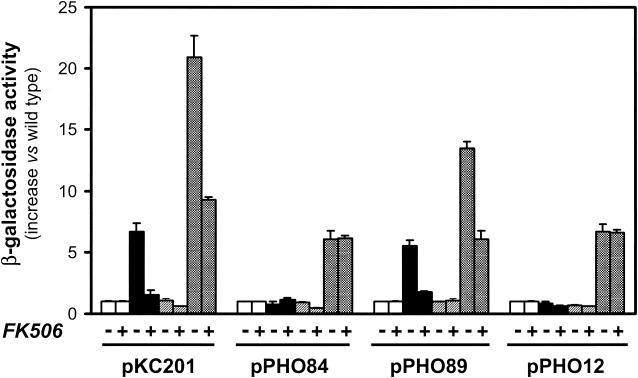
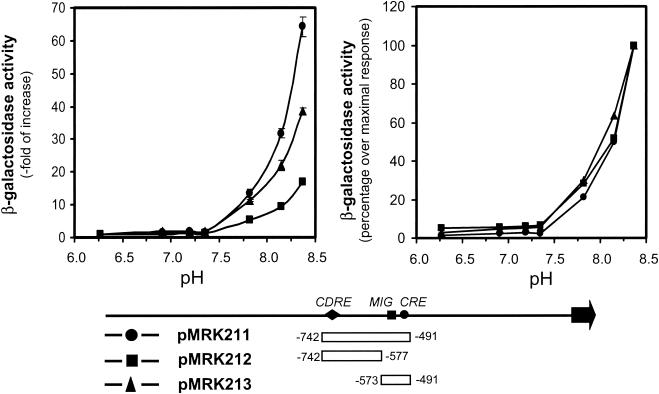
To test this hypothesis, we first compared the expression of four different genes known to be induced by exposure of cells to an alkaline environment (ENA1, PHO84, PHO89, and PHO12) in wild-type, ppz1, ppz2, and ppz1 ppz2 strains, both in the presence and in the absence of the calcineurin inhibitor FK506. Interestingly, as documented in Fig. 6, while ENA1 promoter activity (plasmid pKC201) was increased in ppz1 and ppz1 ppz2 cells, expression of PHO84 and PHO12 was increased only in the ppz1 ppz2 double mutant and was not affected by the presence of the drug FK506. A remarkable observation was that the expression of PHO89 was also increased in a single ppz1 mutant (but not in a ppz2 strain), and this increase was fully abolished by exposure of the cells to FK506. The level of PHO89 expression in the ppz1 ppz2 double mutant was higher than that observed in the ppz1 single mutant, and in this case, a substantial part of the response was blocked by the calcineurin inhibitor. We extended the study to four additional genes whose expression has been found to increase under alkaline stress: FRE1, SIT1, TRR1, and FIT2. The appropriate reporter constructs were transformed into wild-type, ppz1, ppz2, and ppz1 ppz2 cells in two different genetic backgrounds, and the level of expression of each construct was monitored. As shown in Table 2, none of these promoters showed increased activity in ppz1 or ppz2 cells, while all of them were more active in the double mutant. It is worth noting that all four genes respond to an alkaline pH essentially in a calcineurin-independent manner (L. Viladevall and J. Ariño, unpublished data). We considered then the possibility that the lack of ppz1 could result in a relatively light increase in pH and that the response of the calcineurin-responsive region could be particularly sensitive to small increases in pH. To test this, we determined the responses of reporter constructs containing the −742-to-−490 (pMRK211), −742-to-−577 (pMRK212), or −573-to-−490 (pMRK213) region of the ENA1 promoter when cells were grown at different ambient pHs (Fig. 7). As can be observed, the profile of the response was essentially the same when expression was driven from the region containing the CDRE or from the downstream region to which the FK506-insensitive increase in ENA1 expression of a ppz1 ppz2 strain was mapped (note that the region of the ENA1 promoter in plasmid pMRK213 is the same as that in plasmid pMP213). Therefore, our results do not support the hypothesis that the region of the ENA1 promoter containing the CDRE is more sensitive to an increase in pH than the downstream responsive region.
FIG. 6.
Effects of calcineurin inhibition on the expression levels of different alkali-inducible genes in wild-type and Ppz-deficient backgrounds. Strains JA100 (wild type) (open bars), EDN75 (ppz1) (filled bars), JA103 (ppz2) (hatched bars), and EDN76 (ppz1 ppz2) (stippled bars) were transformed with plasmid pKC201, pPHO84, pPHO89, or pPHO12. Cultures were incubated in the presence (1.5 μg/ml) or absence of FK506 as described in Materials and Methods, and β-galactosidase activity was measured. Data are means ± standard errors of the means from 3 to 12 independent clones.
TABLE 2.
Effects of the absence of Ppz1 and Ppz2 functions on the activities of the FRE1, FIT2, SIT1, and TRR1 promoters
| Reporter and background | β-Galactosidase activity (fold induction)a in the following mutant:
|
|||
|---|---|---|---|---|
| Wild type | ppz1 | ppz2 | ppz1 ppz2 | |
| pFRE1 | ||||
| JA100 | 4.4 ± 0.3 (1.0) | 4.1 ± 0.8 (0.9 ± 0.2) | 3.0 ± 0.4 (0.7 ± 0.1) | 14.8 ± 1.6 (3.4 ± 0.4) |
| DBY746 | 1.7 ± 0.1 (1.0) | 1.3 ± 0.2 (0.8 ± 0.1) | 1.3 ± 0.2 (0.7 ± 0.1) | 27.9 ± 8.7 (16.6 ± 2.2) |
| pFIT2 | ||||
| JA100 | 4.2 ± 0.5 (1.0) | 3.2 ± 0.2 (0.8 ± 0.0) | 2.1 ± 0.2 (0.8 ± 0.1) | 41.3 ± 3.0 (9.9 ± 0.7) |
| DBY746 | 0.6 ± 0.1 (1.0) | 1.1 ± 0.3 (1.9 ± 0.5) | 1.0 ± 0.1 (1.8 ± 0.2) | 47.8 ± 5.1 (83.1 ± 8.8) |
| pSIT1 | ||||
| JA100 | 166.6 ± 9.9 (1.0) | 128.3 ± 11.8 (0.8 ± 0.1) | 140.0 ± 6.0 (0.8 ± 0.0) | 350.4 ± 25.4 (2.1 ± 0.2) |
| DBY746 | 174.4 ± 6.9 (1.0) | 185.4 ± 7.4 (1.1 ± 0.0) | 179.0 ± 5.3 (1.0 ± 0.0) | 627.1 ± 35.7 (3.6 ± 0.2) |
| pTRR1 | ||||
| JA100 | 111.1 ± 9.9 (1.0) | 132.7 ± 9.1 (1.2 ± 0.1) | 118.8 ± 2.6 (1.1 ± 0.0) | 424.2 ± 39.6 (3.8 ± 0.4) |
| DBY746 | 164.5 ± 6.0 (1.0) | 148.6 ± 9.2 (0.9 ± 0.1) | 151.7 ± 3.9 (0.9 ± 0.0) | 336.0 ± 21.1 (2.0 ± 0.1) |
β-Galactosidase activity is expressed in Miller units. Fold induction is given relative to activity in the isogenic wild-type strain.
FIG. 7.
Sensitivities of Ppz-regulatable regions of the ENA1 promoter to different levels of alkalinization. The wild-type strain DBY746 was transformed with the indicated constructs bearing the regions of the ENA1 promoter depicted schematically at the bottom. Cells were exposed to the indicated pH for 60 min and then processed for β-galactosidase activity measurement. The responses of each construct are expressed as the ratio of expression between each pH tested and the lowest pH used in the experiment (6.3) (left graph) and as the percentage of the maximal response (right graph). Data are means ± standard errors of the means from three independent clones.
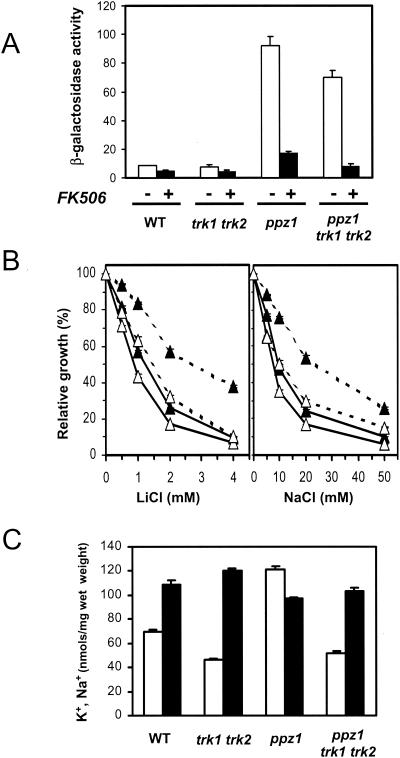
Marked increases in intracellular concentrations of potassium can lead to alkalinization of the cytoplasm. Because the Trk1/Trk2 transporters represent by far the most important K+ uptake system, we considered it important to evaluate whether the increase in ENA1 expression derived from the absence of Ppz1 was dependent on intact Trk1/Trk2-mediated transport. As shown in Fig. 8A, cells carrying deletions of both the TRK1 and TRK2 genes retain most of the increased ENA1 expression characteristic of a ppz1 mutant, and under these circumstances, the expression is still completely blocked by incubation of the cells with FK506. When the sensitivities of these strains to sodium and lithium cations were tested, we observed (Fig. 8B) that, as was observed with the ppz1 ppz2 mutant (49), the absence of Ppz1 increased tolerance to these cations in the hypersensitive trk1 trk2 background. Interestingly, this increase was abolished in the presence of the calcineurin inhibitor FK506. Addition of the drug slightly increased the sensitivity of the trk1 trk2 mutant. Finally, the intracellular concentrations of sodium and potassium ions were determined in wild-type, trk1 trk2, ppz1, and trk1 trk2 ppz1 cells grown in the presence of 0.3 M NaCl (Fig. 8C). As previously reported for the ppz1 ppz2 mutant, disruption of PPZ1 clearly increased the intracellular potassium ion concentration and slightly reduced the content of sodium, while lack of the Trk system decreased the potassium and increased the sodium ion concentration. Deletion of PPZ1 in the trk-deficient background showed a slight decrease in the intracellular Na+ concentration relative to that in the trk1 trk2 mutant. Equivalent results were obtained when cells were grown in the presence of a higher NaCl concentration (0.5 M). This decrease in Na+ accumulation in the ppz1 trk1 trk2 strain is consistent with the continued induction of ENA1 in this strain and likely explains the relative tolerance to NaCl and LiCl.
FIG. 8.
Effects of the absence of Ppz1 on the expression of ENA1 and on salt tolerance in a Trk-deficient background. (A) The wild-type (WT) strain DBY746 and its derivatives EDN2 (ppz1), ESV212 (trk1 trk2), and MAR37 (ppz1 trk1 trk2) were transformed with plasmid pKC201 and cultured in the presence or absence of FK506. β-Galactosidase activity was measured, and data are presented as means ± standard errors of the means from six independent clones. (B) Strains ESV212 (solid lines) and MAR37 (dashed lines) were tested for growth in liquid cultures at the indicated concentrations of LiCl or NaCl in the absence (solid symbols) or presence (open symbols) of 1.5 μg of FK506/ml. Data, expressed as percentages of the growth of cultures without added salt, are means ± standard errors of the means from three independent cultures. (C) The indicated strains were grown in the presence of 0.3 M NaCl, extracts were prepared as described in Materials and Methods, and the concentrations of potassium (open bars) and sodium (solid bars) were measured. Data, expressed as nanomoles per milligram of cells (fresh weight), are means ± standard errors of the means from two independent experiments, performed in duplicate.
DISCUSSION
It has been known for some time that the Ppz1 and Ppz2 phosphatases are not functionally equivalent. For instance, the absence of Ppz1 results in clear-cut phenotypes, while cells lacking Ppz2 are indistinguishable from wild-type cells. Although the recent finding that the Ppz phosphatases are negative effectors of potassium influx, probably by regulating the Trk1/Trk2 potassium transporters (49), has provided a very fruitful framework for understanding the diverse phenotypes associated with the absence or the overexpression of these proteins, we considered it necessary to initiate a detailed evaluation of the function and regulation of these proteins independently, in order to better understand their individual physiological roles. Because it was known that Hal3 was able to interact with and inhibit Ppz1, we first raised the question of whether Hal3 could be considered fully specific for Ppz1. We show here that Hal3 can bind Ppz2 in vitro with roughly the same efficiency as that for Ppz1 binding. Furthermore, high-copy expression of Hal3 in the ppz1 mutant results in a level of ENA1 expression virtually identical to that observed in the ppz1 ppz2 double mutant, suggesting that high levels of Hal3 could fully inhibit Ppz2 function. The observation that high-copy expression of Hal3 fails to increase expression of the ENA1 promoter in a ppz1 ppz2 mutant is in agreement with the notion that the regulation of the expression of the ATPase gene by Hal3 is fully mediated by both the Ppz1 and Ppz2 phosphatases. Consistent with these results, overexpression of Hal3 also fails to increase salt tolerance in a ppz1 ppz2 double mutant (data not shown). In conclusion, Hal3 appears to regulate both the Ppz1 and Ppz2 phosphatases. A practical consequence of these observations would be that high-copy expression of Hal3 could not be considered a suitable tool for specific inhibition of Ppz1.
Since the absence of Ppz1 and Ppz2 affects ENA1 expression, we considered that a detailed functional analysis of the ENA1 promoter in ppz1 and ppz1 ppz2 mutants might provide useful information on the mechanism of action of these phosphatases. Here we present evidence that a region containing the downstream CDRE of ENA1 (30, 43) is necessary and sufficient to mimic the increased expression of the full promoter in a ppz1 mutant. It should be noted that this region also contains one of the two possible binding sites for Nrg1, a transcription factor which, very recently, has been proposed to negatively regulate ENA1 transcription under the influence of Rim101 (25). However, we observed that the increase in ENA1 expression characteristic of ppz1 mutants was completely blocked by incubation of the cells with the calcineurin inhibitor FK506; by mutation of the CNB1 gene, encoding the regulatory subunit of calcineurin; or by the absence of the calcineurin-activated Crz1/Tcn1 transcription factor (Fig. 3), which is responsible for most, if not all, of the transcriptional effects of calcineurin (50). Furthermore, we show (Fig. 3, right) that a reporter construct containing the CDRE sequence found in FKS2, which does not contain the CCCCT or CCCTC sequences proposed as Nrg1 binding sites (35), was also overexpressed in a ppz1 strain. These findings are consistent with the hypothesis that the effect of Ppz1 on the regulation of ENA1 expression is not related to Nrg1 function. Mapping of the ENA1 promoter activity in a ppz1 ppz2 mutant revealed an additional functionally relevant target region, designated ARR2 by Serrano et al. (43). Transcription from this region is not blocked by FK506, indicating that it is not influenced by calcineurin activity. This region was identified recently as being responsible for a substantial, calcineurin-insensitive component of the alkaline response of ENA1 (43). Therefore, the complete absence of Ppz activity results in increased ENA1 expression by modulation of the activity of at least two different sites in its promoter.
The results presented in this paper suggest that Ppz1 might act as an upstream negative regulator of the calcineurin pathway (see Fig. 9 for a schematic depiction). The observations that a ppz1 mutant is sensitive to calcium ions, as would be expected for a strain with higher-than-normal calcineurin activity, and that this sensitivity is fully abolished by deletion of cnb1 are in agreement with this model. It should be noted that a previous report failed to identify a ppz1 mutant as calcium sensitive (42). These authors based their work in a genetic background (W303-1A-derived strains) different from those used in this work. We have also tested calcium tolerance for ppz1 and ppz1 ppz2 mutants in the W303-1A background, and we were able to observe a slight calcium-sensitive phenotype in the ppz1 mutant that was exacerbated in the double mutant (data not shown).
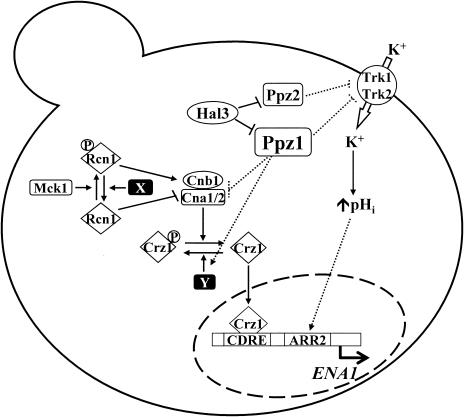
FIG. 9.
Schematic depiction of the relationship of Hal3, Ppz1, and Ppz2 with the calcineurin-dependent pathway and the transcriptional regulation of the ENA1 gene. “X” represents the protein phosphatase(s) that dephosphorylates Rcn1, and “Y” represents the protein kinase that phosphorylates Crz1. Discontinuous lines indicate functional relationships whose mechanisms have yet to be clarified.
It has recently been shown that calcineurin function can be modulated by a conserved family of proteins termed calcipressins, represented in budding yeast by a single gene, RCN1 (17, 23). It is not evident from the literature how Rcn1 acts on calcineurin. For instance, overproduction has been shown to inhibit calcineurin function, and bacterially expressed Rcn1 inhibits calcineurin in vitro (23). In contrast, a null mutant exhibits reduced calcineurin activity and, like calcineurin mutants, presents increased sensitivity to sodium and lithium cations (15, 17, 22, 23). Recently, K. Cunningham's laboratory (John Hopkins University) has suggested a model in which Rcn1 stimulates calcineurin when phosphorylated at a given site, while a nonphosphorylated form of Rcn1 would have a potent inhibitory activity. These authors have also identified the Mck1 protein kinase, a member of the glycogen synthase kinase-3 family, as the major Rcn1 kinase activity in yeast (K. Cunningham, personal communication).
We have found that expression from the ENA1 promoter is somewhat lower in an rcn1 mutant than in a wild-type strain (6.5 ± 0.4 versus 8.9 ± 0.4 Miller units) and that lack of Rcn1 in a ppz1 mutant only marginally decreases the activity of the promoter (Fig. 5). In contrast, deletion of MCK1 in a ppz1 background decreases the activity of the promoter to wild-type levels. We consider that these results provide support for the model of Mck1/Rcn1-mediated regulation of calcineurin activity mentioned above. Lack of Rcn1 would represent the absence of an inhibitor (when dephosphorylated) but also that of a possible activator of calcineurin (when phosphorylated by Mck1). This might have little influence on ENA1 expression in a ppz1 mutant, in which calcineurin activity would already be abnormally increased due to the absence of the Ppz1 phosphatase. However, lack of Mck1 would result in the incapacity to phosphorylate Rcn1, and this unphosphorylated form of the protein would strongly inhibit the activity of calcineurin, possibly overriding the positive effect due to the lack of Ppz1. It should be noted that our results rule out the possibility of Ppz1 acting as an Rcn1 phosphatase (which would account for the calcineurin-dependent activation of the ENA1 promoter in the ppz1 mutant), as in this scenario the absence of Ppz1 should not result in increased ENA1 promoter activity in an rcn1 strain. A role for Mck1 in sodium tolerance was postulated some time ago (36). These authors observed that an mck1 mutant was more sensitive to high sodium concentrations and that expression of ENA1 was reduced under 0.5 M NaCl stress. In addition, overexpression of MCK1 was able to somewhat improve the growth of a strain lacking both catalytic subunits of calcineurin, which would suggest a calcineurin-independent mechanism. However, in this experiment the MCK1 gene was expressed from the strong ADH1 promoter, and therefore, conditions were far from physiological.
Recent work (49) has shown that the ppz1 ppz2 mutant has a more alkaline cytosolic pH, and it was postulated that the increased basal activity of the ENA1 promoter may be the result of this alkalinization. In this study, we show that the induction of the ENA1 promoter in the double mutant maps to two different regions; a pH-responsive region (−573 to −490) and the calcineurin-dependent CDRE (−751 to −667). Interestingly, several lines of evidence demonstrate that, in a single ppz1 mutant, the CDRE is necessary and sufficient for the observed ENA1 induction and that alterations in intracellular pH are not involved in the changes in gene expression observed in this mutant. First, only promoters of calcineurin-responsive genes, such as ENA1 or PHO89, show increased activity in a ppz1 mutant, while several other alkaline pH-responsive genes do not. Second, the CDRE-containing region of ENA1 is not particularly sensitive to alkaline pHs. Thus, the possibility that lack of Ppz1 might result in a slight increase in pH that would be to drive transcription from this element seems unlikely. In fact, the increase in expression of ENA1 in a ppz1 mutant is quantitatively similar at different ambient pHs (from pH 5.8 to 7.2 [data not shown]). Third, the increased activity of the ENA1 promoter due to the lack of Ppz1 is largely conserved in a trk1 trk2 mutant (Fig. 8), ruling out the possibility of intracellular alkalinization as a result of enhanced Trk activity in the absence of Ppz1. In addition, we confirm and extend here the previous observation (49) that the absence of both the Ppz1 and Ppz2 phosphatases results in increased expression of alkali-inducible genes. This increased expression is not at all affected by calcineurin inhibition in some cases, while in other cases it displays a significant calcineurin-dependent component, suggesting that the absence of both phosphatases triggers regulatory mechanisms that are not elicited by the simple loss of Ppz1 function.
An interesting question is how much the increased expression of ENA1 contributes to the salt-tolerant phenotype of a ppz1 mutant. As shown in Fig. 4B, a crz1 mutant is less sensitive to lithium or sodium than a cnb1 mutant, supporting the notion that calcineurin plays a role(s) in salt tolerance that does not involve Crz1-mediated transcriptional regulation. Interestingly, deletion of CRZ1 in a ppz1 background reduces tolerance to lithium and sodium cations, although the effect is less potent than that observed upon deletion of CNB1. As we show that increased ENA1 expression observed in the absence of Ppz1 is dependent on the presence of Crz1, it could be hypothesized that the difference in tolerance between a ppz1 and a ppz1 crz1 mutant would be due to the incapacity of the latter to increase ENA1 expression levels. Similarly, a higher ENA1 expression would explain the FK506-sensitive increase in sodium and lithium tolerance provided by disruption of PPZ1 in a trk1 trk2 strain (Fig. 8), suggesting a role for calcineurin as a downstream effector of Ppz1. In addition to regulating ENA1 expression, it is known that, in the presence of high NaCl concentrations, calcineurin is able to regulate the Trk potassium transporters, positively influencing the ability of Trk to convert to the high-affinity state which allows for better discrimination of potassium from sodium (32, 33). Therefore, a likely possibility that would contribute to the marked salt tolerance phenotype of a ppz1 mutant would be that the absence of Ppz1 would activate calcineurin and consequently influence the cation selectivity of the Trk transporters. Of course, this model does not rule out an alternative, calcineurin-independent regulation of Trk function.
Acknowledgments
We thank F. Pérez-Bermejo and A. Friedrich (Fujisawa Co.) for kindly supplying the calcineurin inhibitor FK506 and R. Haro, R. Serrano, E. Simón, D. Bernal, and J. L. Revuelta for strains and constructs. The contribution of F. Posas in the earliest steps of this work and the excellent technical assistance of Anna Vilalta, Manuel Clemente, and Jacobo Molins are acknowledged. We are particularly grateful to Kyle Cunningham (John Hopkins University, Baltimore, Md.) for generously sharing unpublished data.
This work was supported by a research grant from the “Fundación Ramón Areces” to J.A. and by grants BMC2002-04011-C05-04, BMC2002-04011-C05-02, and GEN2001-4707-C08-03 (Ministerio de Ciencia y Tecnología, Government of Spain, and Fondo Europeo de Desarrollo Regional). A. Ruiz is the recipient of a fellowship from the CIRIT (Generalitat de Catalunya). L. Yenush is supported by the Ramón y Cajal Program (Ministerio de Ciencia y Tecnología).
REFERENCES
- 1.Adams, A., D. E. Gottschlings, C. A. Kaiser, and T. Stearns. 1998. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Alepuz, P. M., K. W. Cunningham, and F. Estruch. 1997. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol. Microbiol. 26:91-98. [DOI] [PubMed] [Google Scholar]
- 3.Ariño, J., F. Posas, and J. Clotet. 1998. The search for the biological function of novel yeast Ser/Thr phosphatases. Methods Mol. Biol. 93:305-313. [DOI] [PubMed] [Google Scholar]
- 4.Berben, G., J. Dumont, V. Gilliquet, P. A. Bolle, and F. Hilger. 1991. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7:475-477. [DOI] [PubMed] [Google Scholar]
- 5.Clotet, J., E. Garí, M. Aldea, and J. Ariño. 1999. The yeast Ser/Thr phosphatases Sit4 and Ppz1 play opposite roles in regulation of the cell cycle. Mol. Cell. Biol. 19:2408-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clotet, J., F. Posas, E. de Nadal, and J. Ariño. 1996. The NH2-terminal extension of protein phosphatase PPZ1 has an essential functional role. J. Biol. Chem. 271:26349-26355. [DOI] [PubMed] [Google Scholar]
- 7.Crespo, J. L., K. Daicho, T. Ushimaru, and M. N. Hall. 2001. The GATA transcription factors GLN3 and GAT1 link TOR to salt stress in Saccharomyces cerevisiae. J. Biol. Chem. 276:34441-34444. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham, K. W., and G. R. Fink. 1996. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2226-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cyert, M. S., and J. Thorner. 1992. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 12:3460-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cyert, M. S., R. Kunisawa, D. Kaim, and J. Thorner. 1991. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc. Natl. Acad. Sci. USA 88:7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Nadal, E., J. Clotet, F. Posas, R. Serrano, N. Gomez, and J. Ariño. 1998. The yeast halotolerance determinant Hal3p is an inhibitory subunit of the Ppz1p Ser/Thr protein phosphatase. Proc. Natl. Acad. Sci. USA 95:7357-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Como, C. J., R. Bose, and K. T. Arndt. 1995. Overexpression of SIS2, which contains an extremely acidic region, increases the expression of SWI4, CLN1 and CLN2 in sit4 mutants. Genetics 139:95-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrando, A., S. J. Kron, G. Rios, G. R. Fink, and R. Serrano. 1995. Regulation of cation transport in Saccharomyces cerevisiae by the salt tolerance gene HAL3. Mol. Cell. Biol. 15:5470-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garciadeblás, B., F. Rubio, F. J. Quintero, M. A. Banuelos, R. Haro, and A. Rodríguez-Navarro. 1993. Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol. Gen. Genet. 236:363-368. [DOI] [PubMed] [Google Scholar]
- 15.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 16.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 17.Gorlach, J., D. S. Fox, N. S. Cutler, G. M. Cox, J. R. Perfect, and J. Heitman. 2000. Identification and characterization of a highly conserved calcineurin binding protein, CBP1/calcipressin, in Cryptococcus neoformans. EMBO J. 19:3618-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:1262-1267. [DOI] [PubMed] [Google Scholar]
- 19.Guarente, L., and T. Mason. 1983. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell 32:1279-1286. [DOI] [PubMed] [Google Scholar]
- 20.Haro, R., L. Sainz, F. Rubio, and A. Rodríguez-Navarro. 1999. Cloning of two genes encoding potassium transporters in Neurospora crassa and expression of the corresponding cDNAs in Saccharomyces cerevisiae. Mol. Microbiol. 31:511-520. [DOI] [PubMed] [Google Scholar]
- 21.Hughes, V., A. Muller, M. J. Stark, and P. T. Cohen. 1993. Both isoforms of protein phosphatase Z are essential for the maintenance of cell size and integrity in Saccharomyces cerevisiae in response to osmotic stress. Eur. J. Biochem. 216:269-279. [DOI] [PubMed] [Google Scholar]
- 22.Jesus Ferreira, M. C., X. Bao, V. Laize, and S. Hohmann. 2001. Transposon mutagenesis reveals novel loci affecting tolerance to salt stress and growth at low temperature. Curr. Genet. 40:27-39. [DOI] [PubMed] [Google Scholar]
- 23.Kingsbury, T. J., and K. W. Cunningham. 2000. A conserved family of calcineurin regulators. Genes Dev. 14:1595-1604. [PMC free article] [PubMed] [Google Scholar]
- 24.Kuno, T., H. Tanaka, H. Mukai, C. D. Chang, K. Hiraga, T. Miyakawa, and C. Tanaka. 1991. cDNA cloning of a calcineurin B homolog in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 180:1159-1163. [DOI] [PubMed] [Google Scholar]
- 25.Lamb, T. M., and A. P. Mitchell. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, K. S., L. K. Hines, and D. E. Levin. 1993. A pair of functionally redundant yeast genes (PPZ1 and PPZ2) encoding type 1-related protein phosphatases function within the PKC1-mediated pathway. Mol. Cell. Biol. 13:5843-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, Y., S. Ishii, M. Tokai, H. Tsutsumi, O. Ohki, R. Akada, K. Tanaka, E. Tsuchiya, S. Fukui, and T. Miyakawa. 1991. The Saccharomyces cerevisiae genes (CMP1 and CMP2) encoding calmodulin-binding proteins homologous to the catalytic subunit of mammalian protein phosphatase 2B. Mol. Gen. Genet. 227:52-59. [DOI] [PubMed] [Google Scholar]
- 28.Márquez, J. A., and R. Serrano. 1996. Multiple transduction pathways regulate the sodium-extrusion gene PMR2/ENA1 during salt stress in yeast. FEBS Lett. 382:89-92. [DOI] [PubMed] [Google Scholar]
- 29.Matheos, D. P., T. J. Kingsbury, U. S. Ahsan, and K. W. Cunningham. 1997. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 11:3445-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendizabal, I., A. Pascual-Ahuir, R. Serrano, and I. F. de Larrinoa. 2001. Promoter sequences regulated by the calcineurin-activated transcription factor Crz1 in the yeast ENA1 gene. Mol. Genet. Genomics 265:801-811. [DOI] [PubMed] [Google Scholar]
- 31.Mendizabal, I., G. Rios, J. M. Mulet, R. Serrano, and I. F. de Larrinoa. 1998. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 425:323-328. [DOI] [PubMed] [Google Scholar]
- 32.Mendoza, I., F. J. Quintero, R. A. Bressan, P. M. Hasegawa, and J. M. Pardo. 1996. Activated calcineurin confers high tolerance to ion stress and alters the budding pattern and cell morphology of yeast cells. J. Biol. Chem. 271:23061-23067. [DOI] [PubMed] [Google Scholar]
- 33.Mendoza, I., F. Rubio, A. Rodríguez-Navarro, and J. M. Pardo. 1994. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J. Biol. Chem. 269:8792-8796. [PubMed] [Google Scholar]
- 34.Myers, A. M., A. Tzagoloff, D. M. Kinney, and C. J. Lusty. 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299-310. [DOI] [PubMed] [Google Scholar]
- 35.Park, S. H., S. S. Koh, J. H. Chun, H. J. Hwang, and H. S. Kang. 1999. Nrg1 is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piao, H. L., K. T. Pih, J. H. Lim, S. G. Kang, J. B. Jin, S. H. Kim, and I. Hwang. 1999. An Arabidopsis GSK3/shaggy-like gene that complements yeast salt stress-sensitive mutants is induced by NaCl and abscisic acid. Plant Physiol. 119:1527-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Posas, F., A. Casamayor, and J. Ariño. 1993. The PPZ protein phosphatases are involved in the maintenance of osmotic stability of yeast cells. FEBS Lett. 318:282-286. [DOI] [PubMed] [Google Scholar]
- 38.Posas, F., A. Casamayor, N. Morral, and J. Ariño. 1992. Molecular cloning and analysis of a yeast protein phosphatase with an unusual amino-terminal region. J. Biol. Chem. 267:11734-11740. [PubMed] [Google Scholar]
- 39.Posas, F., M. Camps, and J. Ariño. 1995. The PPZ protein phosphatases are important determinants of salt tolerance in yeast cells. J. Biol. Chem. 270:13036-13041. [DOI] [PubMed] [Google Scholar]
- 40.Proft, M., and R. Serrano. 1999. Repressors and upstream repressing sequences of the stress-regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol. Cell. Biol. 19:537-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds, A., V. Lundblad, D. Dorris, and M. Keaveney. 1997. Yeast vectors and assays for expression of cloned genes, p. 13.6.1-13.6.6. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, N.Y. [DOI] [PubMed]
- 42.Sakumoto, N., I. Matsuoka, Y. Mukai, N. Ogawa, Y. Kaneko, and S. Harashima. 2002. A series of double disruptants for protein phosphatase genes in Saccharomyces cerevisiae and their phenotypic analysis. Yeast 19:587-599. [DOI] [PubMed] [Google Scholar]
- 43.Serrano, R., A. Ruiz, D. Bernal, J. R. Chambers, and J. Ariño. 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 46:1319-1333. [DOI] [PubMed] [Google Scholar]
- 44.Stathopoulos, A. M., and M. S. Cyert. 1997. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11:3432-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 46.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 47.Withee, J. L., J. Mulholland, R. Jeng, and M. S. Cyert. 1997. An essential role of the yeast pheromone-induced Ca2+ signal is to activate calcineurin. Mol. Biol. Cell 8:263-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye, R. R., and A. Bretscher. 1992. Identification and molecular characterization of the calmodulin-binding subunit gene (CMP1) of protein phosphatase 2B from Saccharomyces cerevisiae. An α-factor inducible gene. Eur. J. Biochem. 204:1713-1723. [DOI] [PubMed] [Google Scholar]
- 49.Yenush, L., J. M. Mulet, J. Ariño, and R. Serrano. 2002. The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: implications for salt tolerance, cell wall integrity and cell cycle progression. EMBO J. 21:920-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshimoto, H., K. Saltsman, A. P. Gasch, H. X. Li, N. Ogawa, D. Botstein, P. O. Brown, and M. S. Cyert. 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277:31079-31088. [DOI] [PubMed] [Google Scholar]