Abstract
Cryptococcus neoformans is a human fungal pathogen that exists as three distinct varieties or sibling species: the predominantly opportunistic pathogens C. neoformans var. neoformans (serotype D) and C. neoformans var. grubii (serotype A) and the primary pathogen C. neoformans var. gattii (serotypes B and C). While serotypes A and D are cosmopolitan, serotypes B and C are typically restricted to tropical regions. However, serotype B isolates of C. neoformans var. gattii have recently caused an outbreak on Vancouver Island in Canada, highlighting the threat of this fungus and its capacity to infect immunocompetent individuals. Here we report a large-scale analysis of the mating abilities of serotype B and C isolates from diverse sources and identify unusual strains that mate robustly and are suitable for further genetic analysis. Unlike most isolates, which are of both the a and α mating types but are predominantly sterile, the majority of the Vancouver outbreak strains are exclusively of the α mating type and the majority are fertile. In an effort to enhance mating of these isolates, we identified and disrupted the CRG1 gene encoding the GTPase-activating protein involved in attenuating pheromone response. crg1 mutations dramatically increased mating efficiency and enabled mating with otherwise sterile isolates. Our studies provide a genetic and molecular foundation for further studies of this primary pathogen and reveal that the Vancouver Island outbreak may be attributable to a recent recombination event.
Mating in the microbial world reassorts genomes, increases genetic diversity, and produces strains that can colonize new environmental niches. In several microorganisms, mating is also linked to pathogenicity. In the parasite Toxoplasma gondii, meiosis produces more-pathogenic isolates by recombining the genomes of two less-virulent strains (13, 42). In the plant fungal pathogen Ustilago maydis, mating is required for infection of maize because it is the filamentous dikaryon form that is pathogenic (for a review, see reference 32). In the human fungal pathogen Cryptococcus neoformans, the α mating type has been linked to virulence (25) and sexual recombination produces spores, the suspected infectious propagule (43; J. Cohen, J. R. Perfect, and D. T. Durack, Letter, Lancet i:1301, 1982).
C. neoformans is a haploid, encapsulated yeast that is the most common cause of fungal meningitis, which is fatal if untreated (17, 22, 29). C. neoformans strains can be classified into three divergent varieties and four serotypes based on capsular antigens: C. neoformans var. neoformans (serotype D), C. neoformans var. grubii (serotype A), and C. neoformans var. gattii (serotypes B and C). Serotypes A (the most clinically prevalent) and D are opportunistic pathogens that infect immunocompromised individuals throughout the world, whereas the divergent B and C serotypes are primary pathogens that infect predominantly immunocompetent hosts (40). In contrast to serotypes A and D, which are cosmopolitan and most frequently associated with pigeon guano, the serotype B and C C. neoformans var. gattii is usually restricted to tropical and subtropical regions of the world, where it is found in association with woody debris of eucalyptus trees (10).
Mating among serotype D strains (the least virulent variety) has been well characterized, and a congenic pair has been created to facilitate genetic analysis (16, 25). In contrast, the first accounts of the characterization of mating among serotype A C. neoformans var. grubii strains have only recently been reported (18, 35), an advance that was made possible by the discovery of a isolates long thought to be extinct (30, 45). The sexual system is bipolar (involving a and α cell types) and is dependent on an unusually large mating type locus (28).
Serotype A (C. neoformans var. grubii) strains exist predominantly as clonal isolates of the α mating type throughout the world (12, 23, 47). In contrast, mixed populations of a and α C. neoformans var. gattii strains have been identified colonizing hollows of eucalyptus trees in the Australian environment, where there is a relatively high incidence of cryptococcosis in native animals and indigenous human populations (4, 11, 14, 15). Despite the presence of both mating types, no meiotic recombination has been detected, suggesting that this ecological niche may support only clonal propagation of yeast cells and that sexual development occurs elsewhere if at all (15).
Interest in the primary pathogenic form of C. neoformans has increased due to an outbreak of C. neoformans var. gattii infection on Vancouver Island in Canada. Prior to 1999, an average of two or three cases of cryptococcosis occurred annually in British Columbia and were caused by infections with serotype D. Since 1999, more than 66 human cases in otherwise healthy individuals, with at least four fatalities, have been reported and are attributable to infection with serotype B (M. Fyfe, W. Black, M. Romney, P. Kibsey, L. MacDougall, M. Starr, M. Pearce, C. Stephen, L. Stein, S. Mak, B. Emerson, J. Isaac-Renton, and D. Patrick, 5th Int. Conf. Cryptococcus Cryptococcosis, abstr. P1.1, 2002). There has also been an equally large increase in the rate of infection of animals, including dogs, cats, and porpoises (41). On Vancouver Island, C. neoformans var. gattii has been found in association with a number of native tree species (Douglas fir, alder, maple, and Garry oak) rather than eucalyptus trees, a previously identified environmental source of this variety. This expansion of the serotype B habitat to include nontropical areas increases the human risk of acquiring cryptococcosis and makes it paramount to understand the mechanisms by which C. neoformans var. gattii reproduces sexually.
Our studies address the role of sexual reproduction in the life cycle and virulence of C. neoformans. The majority of C. neoformans var. gattii strains are sterile under most laboratory conditions. Although the perfect state of C. neoformans var. gattii was first reported over 25 years ago (21), these findings have proven difficult to reproduce (14, 26, 31, 34). Here we have recapitulated and extensively analyzed the sexual cycle of C. neoformans var. gattii. First, we describe conditions that support mating of a number of serotype B and C isolates from diverse sources. Second, we show that the vast majority of isolates from the Vancouver Island outbreak are fertile and that all are of the α mating type. Third, we identify a pair of a and α strains that mate robustly to produce viable meiotic progeny, providing a platform for genetic studies. Finally, we perform targeted gene disruptions in this variety and define a conserved role for the regulator of G protein signaling (RGS) protein Crg1 in pheromone sensing by demonstrating that crg1Δ strains mate more vigorously than wild-type strains. Taken together, our studies provide a genetic and molecular foundation for further studies of this primary pathogen and reveal that the Vancouver Island outbreak may be attributable to a recent recombination event in an unusual, fertile clade of C. neoformans.
MATERIALS AND METHODS
Strains, media, and mating assays.
Reference strains used in this study were the congenic serotype D strains JEC20 (a) and JEC21 (α) (16, 25). Strains were grown on yeast extract-peptone-dextrose (YEPD) medium. Mating assays were carried out on V8 medium (24) (5% [vol/vol] V8 juice, 3 mM KH2PO4, 4% [wt/vol] agar) at pH 5 or 7, carrot medium (5% [vol/vol] carrot juice, 3 mM KH2PO4, 4% agar), or synthetic low ammonium dextrose (SLAD) medium. The ability to mate was tested against the serotype D tester strains JEC20 and JEC21, the serotype C strains NIH312 (α) and B4546 (a), and crg1Δ mutant derivatives JF101 (α) and JF109 (a). Strains were pregrown on YEPD agar for 2 days, and a small amount of cells was removed and patched onto solid mating medium either alone or mixed with a reference strain. Plates were incubated at room temperature in the dark for 9 days and assessed by light microscopy for filament and basidiospore formation.
Molecular techniques.
Standard methods were performed as described by Sambrook et al. (37). Serotyping was done using the Crypto Check serotyping kit from Iatron Laboratories (Tokyo). C. neoformans genomic DNA for Southern blot analysis was prepared as described by Pitkin et al. (36). C. neoformans biolistic transformation was performed using the technique for serotype D as described by Davidson et al. (7).
Primers.
Sequences of primers are as follows: STE12αU, CAATCTCAAAGCGGGGACAG; STE12αL, CTTTGTTTCGGTCCTAATACAGCC; JOHE6013,GCAACCAATCACTATGAA; JOHE6014, TGTGATCTGGTTCATGTA; JOHE8733, GCTGCGAGGATGTGAGCTGGA; JOHE8734, TCCATCTTGTTCAATCATAGACATGTTGGGCGAGTT; JOHE8735, AACTCGCCCAACATGTCTATGATTGAACAAGATGGA; JOHE8736, ACCCCTTACCGCCTTCACTCAGAAGAACTCGTCAAG; JOHE8737, CTTGACGAGTTCTTCTGAGTGAAGGCGGTAAGGGGT; JOHE8738, GGTTTATCTGTATTAACACGG; JOHE8896, CCCCTTCAACACAGCTCTTTA; JOHE8897, CAAGCCACCGGTCCTTATCCC; JOHE8987, AAGTTCGGACTGATTTATTCA; JOHE8988, CGGAGGATAGAAGCTGTAAGCCAAAGTAAGGGGTAAGTTGGCAGA; JOHE8989, CCGTGTTAATACAGATAAACCGTCTTCAGAATACGAAAAGGACGA; JOHE8990, AAGCAAGACGGGACTTATTGT; JOHE9365, CAGAAACGGTAACGGGAGCAC; JOHE9366, GCCCCTTCTTTCCTTATTCCT; JOHE9421, ATCCGCCCTCGAGTCAAA; JOHE9422, TGGCGACCGACTGTAGAT; JOHE9779, CCTTACTCCATAACCCGCTAC; JOHE9780, AATTTCATAACGCTTTGGGAA; JOHE9787, ACACCGCCTGTTACAATGGAC; JOHE9788, CAGCGTTTGAAGATGGACTTT; JOHE9574, ATTCGCCACCGGTTTATCTGTATTAAC; JOHE9575, ATTCGCCCTTTAAACGAGGATGTGAGC; JOHE9592, GAGTTCAGGCTTTTTCATAGACATGTTGGGCGAGTT; JOHE9593, AACTCGCCCAACATGTCTATGAAAAAGCCTGAACTC; JOHE9594, ACCCCTTACCGCCTTCACCTATTCCTTTGCCCTCGG; JOHE9595, CCGAGGGCAAAGGAATAGGTGAAGGCGGTAAGGGGT.
Determination of mating type.
Mating type was established by PCR using genomic DNA prepared with the Camgen yeast genomic DNA extraction kit (Whatman BioSciences Ltd., Cambridge, United Kingdom) with primers specific to the STE20a (JOHE9421-JOHE9422), STE12α (STE12αU-STE12αL), MFa (JOHE9787-JOHE9788), and MFα (JOHE9779-JOHE9780) genes. PCR was repeated twice.
Plasmids.
The Neor marker cassette clone pJAF1 was generated by combining the serotype A strain H99 ACT1 promoter (JOHE8733-JOHE8734), the transposon Tn5 Neor gene (JOHE8735-JOHE8736), and the H99 TRP1 terminator (JOHE8737-JOHE8738) via overlap PCR using primer pair JOHE8733-JOHE8738 and cloned into plasmid pCR-TOPO2.1. The Hygr marker cassette clone pJAF15 was created in an equivalent fashion by combining the ACT1 promoter (JOHE8733-JOHE9592), the Hygr gene (JOHE9593-JOHE9594), and the TRP1 terminator (JOHE9595-JOHE8738) via overlap PCR using primer pair JOHE8733-JOHE8738 and cloned into plasmid pCR-TOPO2.1. Bacterial artificial chromosomes (BACs) containing the serotype B CRG1 gene (Australian isolates WM276 BAC 2B19 and E566 BAC 2B13) were identified by hybridization with a 1.2-kb CRG1 PCR fragment (JOHE6013-JOHE6014) from the serotype A strain H99. Based on results of Southern blot analysis, the CRG1 gene was subcloned as a 6.5-kb XhoI fragment into pBluescript SK− SalI to yield plasmids pJAF2 (WM276) and pJAF3 (E566). Sequencing of the 6.5-kb subclones revealed only one single nucleotide polymorphism between the two isolates. The CRG1 reconstitution plasmid was created by amplifying the pCH233 Natr cassette (JOHE9574-JOHE9575), digesting it with AgeI/DraI, and using it to replace the NgoMIV/BsaA1 f1 origin fragment of pBluescript SK− to give the Natr cloning plasmid pJAF13. The WM276 CRG1 gene was then subcloned from pJAF2 into pJAF13 as a 6.6-kb ApaI/PstI fragment, yielding pJAF14.
Disruption of the CRG1 gene.
crg1 deletion alleles for the Australian isolates were generated by replacing the 3-kb Eco47III/SpeI fragment of the CRG1 gene in plasmid pJAF2 with the Natr cassette from plasmid pCH233 and the Neor cassette from plasmid pJAF1 as an EcoRV/XbaI fragment to yield plasmids pJAF4 and pJAF6, respectively.
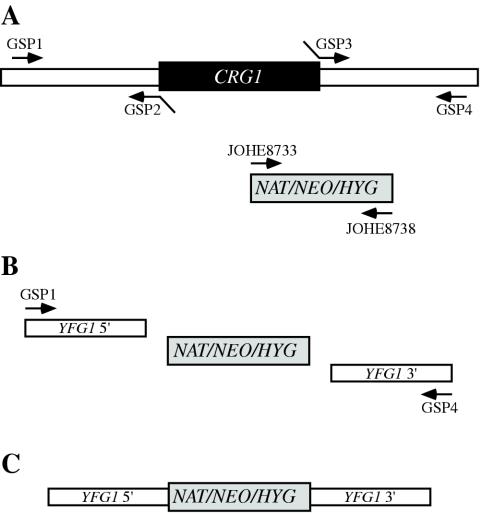
With the use of the CRG1 gene sequence from strain WM276, overlap primers were designed to allow the construction of deletion alleles in strains NIH312, B4546, and R265 without the need to subclone and sequence the CRG1 gene. Building upon the technique of deletion allele generation by overlap PCR (6), we found that the process could be simplified by avoiding the use of chimeric primers for the marker segment, reducing the number of gene-specific primers required from six to four (Fig. 1). Furthermore, the design of the Neor and Hygr cassettes allows the same primer sets to be used for either a Neor, Natr, or Hygr disruption cassette. PCR was used to generate CRG1 5′ (JOHE8987-JOHE9094), 3′ (JOHE8989-JOHE8990), and Neor (JOHE8733-JOHE8738) products, which were combined in a PCR overlap reaction (JOHE8987-JOHE8990) to yield strain-specific CRG1 deletion alleles. We used biolistic transformation for each strain of interest, introducing the appropriate circular plasmid (in strains WM276 and E566) or overlap PCR product (in strains B4546, NIH312, and R265) and selecting for resistance to G418 (200 μg/ml) or nourseothricin (100 μg/ml). crg1Δ transformants were identified by Southern blotting using the 6.5-kb WM276 CRG1 gene fragment as a probe (data not shown). Homologous integration frequencies were higher with overlap PCR products (7 of 60 for B4546, 6 of 70 for NIH312, and 15 of 32 for R265) than with circular plasmid-borne crg1 deletion alleles (2 of 86 and 2 of 65 for the E566 Natr and Neor alleles, respectively, and 2 of 17 for the WM276 Neor allele).
FIG. 1.
Simplification of the creation of knockout constructs by using overlap PCR. We found that only four gene-specific primers (GSP), as opposed to six, were required to create knockout constructs. The flanking regions of the gene to be deleted were amplified (A) using one normal (GSP1-GSP4) and one chimeric (GSP2-GSP3) primer for each side. The marker cassette was amplified using the oligonucleotides JOHE8733 and JOHE8738, which work with the Natr (NAT), Neor (NEO), and Hygr (HYG) cassettes. Like the products of normal overlap PCR, the three products were combined with the primers GSP1 and GSP4 (B) and amplified to yield the final knockout construct (C). For disruption of CRG1, the primers JOHE8987 (GSP1), JOHE9094 (GSP2), JOHE8989 (GSP3), and JOHE8990 (GSP4) were used.
Restriction fragment length polymorphism (RFLP) analysis.
Fragments of the PLB1 (27) and SOD1 (3) genes were amplified from strains NIH312 and B4546 by PCR using the JOHE8896-JOHE8897 and JOHE9365-JOHE9366 primers pairs, respectively, and sequenced. Comparison revealed unique restriction sites in each gene—a BglI restriction site present only in the PLB1 gene of strain NIH312 and a BsaJI restriction site present only in the SOD1 gene of strain B4546. For analysis of meiotic progeny and diploids, gene fragments were PCR amplified as described above and DNA was incubated with 1 U of the appropriate restriction enzyme and buffer for 2 h and separated on a 1.5% agarose gel.
Slide matings.
Glass microscope slides were sterilized, placed at a 20° angle in a plastic petri dish by using a sterile microfuge tube, and coated with a thin layer of molten V8 agar, with 5 ml of liquid V8 medium added to the bottom of each. The strains to be mated were resuspended in distilled H2O, and 10 μl was pipetted at the apex of the slide and allowed to drain down the slide, creating a mating patch 3 to 4 cm in length and 0.5 cm wide. Slides were incubated at room temperature for 3 to 5 days in the dark.
Microscopy and staining.
For filament staining, slides were transferred to a fresh petri dish, washed three times with 10 ml of phosphate-buffered saline (PBS), fixed in 10 ml of 70% ethanol for 20 min, washed three times with PBS, permeabilized with 1% Triton X-100 in PBS for 10 min, and washed three times with PBS prior to staining. To visualize septa and DNA, filaments from matings were incubated in a 10-ml preparation of calcofluor white (fluorescent brightener 28 F-3397; Sigma) at 20 μg/ml and a 5,000× dilution of a 5 mM solution of Sytox Green (Molecular Probes) in PBS for 20 min. Agar sections containing filaments were mounted on fresh microscope slides with 5 μl of antifade (1 mg of p-phenylenediamine/ml in 50% glycerol).
Bright field, differential interference microscopy (DIC), and fluorescent images were captured with a Zeiss Axioskop 2 Plus fluorescent microscope equipped with an AxioCam MRM digital camera and corresponding software.
Environmental scanning electron microscopy (ESEM) was performed using a Philips XL30 ESEM TMP (FEI Company, Portland, Oreg.). Mating reactions were carried out on V8 plates, fixed by exposure to osmium tetroxide vapor for 30 min, and excised blocks of agar were adhered to prechilled stubs by using double-sided tape. The samples were viewed with a secondary electron detector at pressures of 5.2 to 5.5 torr and temperatures of 2.5 to 4.4°C and with a beam of 15 to 18 kV. Working distance ranged from 8.7 to 11 mm.
Nucleotide sequence accession numbers.
The sequence for the CRG1 genes of strains WM276 and E566 has been deposited in GenBank under accession number AY327612. The accession numbers for the partial sequences of the SOD1 and PLB1 genes are AY327613 and AY327615 for NIH312 and AY327614 and AY327616 for B4546, respectively.
RESULTS
Intervarietal mating of C. neoformans var. gattii strains.
Previous analyses of C. neoformans var. gattii mating involved intervarietal mating between serotype D tester strains and serotype B or C strains of the opposite mating type (21). We employed a similar approach and systematically analyzed the ability of a collection of 145 serotype B and C strains to mate with congenic serotype D a (JEC20) and α (JEC21) strains on V8 pH 7 medium (see Table S1 in the supplemental material). Our collection consisted of three groups: laboratory serotype B (20) and C (14) isolates from diverse sources, Vancouver Island outbreak serotype B isolates (83), and Australian serotype B isolates (28).
Analysis of the lab isolates revealed that 18 of 34 (53%) were sterile when mated with the appropriate serotype D tester strain. Of the 47% that did mate, mating was exclusively with partners of a single mating type (a or α). Accordingly, PCR analysis of these isolates with primers for α-specific and a-specific genes revealed that each isolate was of a single mating type (see Table S1 in the supplemental material).
The Australian isolates exhibited a different pattern. Of the clinical isolates, three of six (50%) mated with the tester strains. In contrast, 100% (22 of 22) of the environmental isolates were sterile. Overall, only 10% of isolates obtained from Australia were fertile.
Importantly, the majority of the Vancouver Island outbreak isolates were fertile (70%, or 58 of 83). In contrast to isolates from Australia and elsewhere, which were both a and α, all of the Vancouver isolates were of the α mating type. Additionally, the mating abilities of the Vancouver Island outbreak isolates were not correlated with their clinical or environmental origins.
A C. neoformans var. gattii pair can mate robustly.
Most previous attempts to recapitulate mating of C. neoformans var. gattii used the type cultures NIH444 (serotype B and mating type α) and NIH191 (serotype C and mating type a). We found that both of these isolates mated with the appropriate serotype D tester strains, but mating to each other was much more difficult to detect and produced only a very weak mating reaction that was restricted to V8 pH 5 or carrot medium and required incubation for 6 weeks (data not shown).
Further analysis of our collection revealed an alternative pair of serotype C isolates (NIH312, α, and B4546, a) (Fig. 2) that mated with serotype D tester strains and robustly with each other. These isolates also mated with the type strains NIH444 and NIH191. We retested our collection of 146 isolates with this new mating pair. All of the strains that were mating competent with the serotype D testers also mated with these serotype C tester strains (see Table S1 in the supplemental material; Fig. 2). Additionally, we detected mating with 18 additional isolates from Vancouver Island (nine clinical or veterinary and nine environmental) that failed to mate with the serotype D strains, raising the percentage of mating-competent outbreak isolates from 70 to 91%.
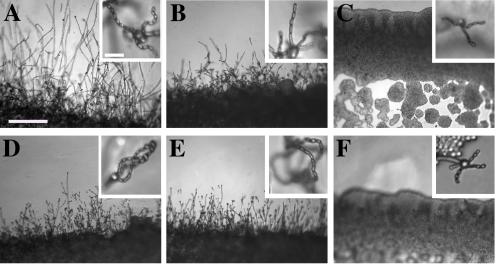
FIG. 2.
Morphology of mating C. neoformans var. gattii pairs. C. neoformans isolates were mated on V8 pH 7 medium on glass slides for 3 days, and morphology was observed. DIC microscopy allowed comparison of the mating ability and morphology of the congenic serotype D pair (JEC20 [serotype D and mating type a] × JEC21 [serotype D and mating type α] [crosses are hereafter indicated in the form JEC20 Da × JEC21 Dα]) (A) with those of the robustly mating serotype C pair (B4546 Ca × NIH312 Cα) (B) and a serotype B-serotype C pair including one of the Vancouver isolates (B4546 Ca × R265 Bα) (C). All three of these C. neoformans var. gattii isolates underwent intervarietal mating with the serotype D congenic pair. (D) B4546 Ca × JEC21 Dα. (E) JEC20 Da × NIH312 Cα. (F) JEC20 Da × R265 Bα. A comparison of morphologies of mating pairs (scale bar, 200 μm) and basidial spore chain morphologies (inset) (scale bar, 20 μm) is given.
Morphology of mating serotype C strains.
Based on examination by light microscopy and ESEM, filaments produced by mating of the serotype C a and α strains B4546 and NIH312 exhibit morphological features characteristic of those produced by mating of basidiomycetes, including segregation of two haploid nuclei per filament cell and the production of fused clamp connections that enable faithful segregation of these nuclei. In contrast to serotype D crosses, which produce long peripheral hyphae, the intervarietal and serotype C intravarietal crosses produce abundant, shorter filaments primarily above the mating reaction. Calcofluor white staining revealed the presence of fused clamp connections (Fig. 3). Also unlike mating serotype D strains, mating serotype C strains form a protrusion from the subapical cell to which the clamp fuses, reminiscent of the peg structures observed in Coprinus cinereus (Fig. 3A) (20). This structure was not observed in serotype D in intervarietal crosses with serotype C (data not shown). Sytox Green staining revealed that each filament cell was dikaryotic and contained paired, comigrating nuclei (Fig. 3).
FIG.3.
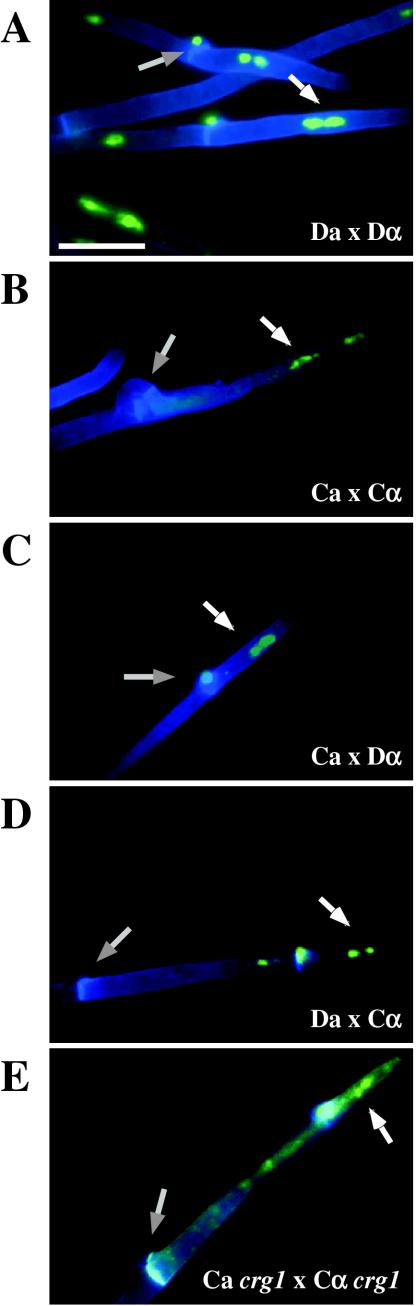
Serotype C strains form dikaryon filaments and fused clamps during mating. Staining filaments from V8 pH 7 medium slide matings with calcofluor white and Sytox Green revealed that, as with serotype D (JEC20 Da × JEC21 Dα [Da × Dα]) (A), both serotype C (B4546 Ca × NIH312 Cα [Ca × Cα]) (B) and intervarietal serotype C-serotype D (B4546 Ca × JEC21 Dα [Ca × Dα] and JEC20 Da × NIH312 Cα [Da × Cα]) (C and D, respectively) crosses form dikaryotic filaments with paired nuclei (white arrows) and fused clamp connections (grey arrows). These same structures could also be observed in a crg1Δ bilateral cross (JF109 Ca × JF101 Cα [Ca crg1 × Cα crg1]) (E), supporting the observation that these strains undergo enhanced mating, as opposed to haploid fruiting which produces unfused clamps and uninucleate filaments. Scale bar, 10 μm.
ESEM was used to more closely examine the filaments produced during mating. In both serotype D and serotype C mating reactions, abundant filaments form, which in turn develop immature basidia. Four evenly spaced buds form simultaneously on the surface of the basidium and develop into basipetal chains of spores through consecutive rounds of budding (Fig. 4B). The serotype C rod-shaped spores are approximately twice as long as their lemon-shaped elliptical serotype D counterparts (Fig. 4B).
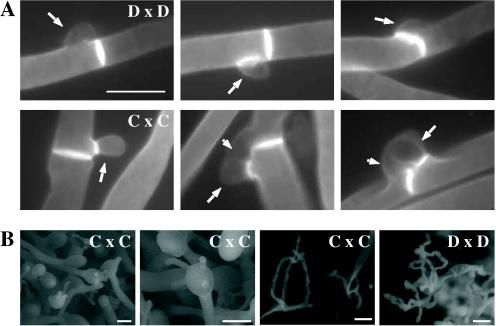
FIG. 4.
Clamp cell formation and ESEM of serotype C basidia. (A) Staining with calcofluor white to analyze clamp cell formation revealed that the morphologies of mating pairs differ in serotype D and serotype C matings. In a serotype D cross (JEC20 Da × JEC21 Dα [D × D]) (top row, left to right), the clamp cell (large arrows) forms and fuses to the subapical filament cell. In a serotype C cross (B4546 Ca × NIH312 Cα [C × C]) (bottom row, left to right), clamp cell fusion involves the formation of a peglike protrusion (small arrows) from the filament subapical to the clamp cell (large arrows), to which the clamp fuses. Scale bar, 5 μm. (B) ESEM of mating serotype C strains (B4546 Ca × NIH312 Cα [C × C]) shows the formation of four evenly spaced buds on the immature basidia, which develop into chains of smooth, rod-shaped spores. In contrast, mating serotype D strains (JEC20 Da × JEC21 Dα [D × D]) form more-spherical, rough basidiospore chains. Scale bar, 10 μm.
The morphology of the mating Vancouver isolates differed from that of the serotype C isolates. While the serotype C mating pair produced intermediate-length aerial filaments over the vegetative mass, the Vancouver isolates produced basidia on stunted filaments close to the surface of the colony. However, the morphologies of the basidia and spores were the same as those of serotype C.
Dominant selectable markers in C. neoformans var. gattii.
To facilitate molecular studies of spore formation in C. neoformans var. gattii, we investigated the efficacy of several dominant markers in serotypes B and C. We confirmed that biolistic transformation of strains NIH312, B4546, R265, WM276, and E566 with the Natr cassette (33) plasmid pCH233 conferred resistance to nourseothricin (100 μg/ml). Firstly, this verified that we could use biolistic transformation to transform a variety of serotype B and C strains, and secondly it demonstrated that the serotype A strain H99 ACT1 promoter that drives the cassette can function in all four serotypes.
Building upon the design of the Natr cassette, we generated two derivatives by overlap PCR, replacing the Natr coding region with the Tn5 Neor gene (pJAF1) or the Escherichia coli Hygr gene (pJAF15). Biolistic transformation of serotype A strain H99, serotype B strains R265, WM276, and E566, serotype C strains NIH312 and B4546, and the serotype D strain JEC21 with these plasmids confirmed that pJAF1 and pJAF15 were sufficient to confer resistance to G418 (200 μg/ml) and hygromycin B (200 μg/ml), respectively, in all four serotypes of C. neoformans. Previous marker cassettes designed to confer resistance to hygromycin B had not functioned in serotypes B and C (5).
Differentially marked serotype C strains mate to produce recombinant spores.
The morphology of C. neoformans var. gattii is distinct from those of C. neoformans var. neoformans and C. neoformans var. grubii, as C. neoformans var. gattii forms both round and elongated cells during infection (44) and elongated spores during mating (21). Elongated vegetative cells also form during growth on V8 medium irrespective of the mating partner and are difficult to distinguish from basidiospores during microdissection. Therefore, to select meiotic progeny, we differentially marked the serotype C mating pair NIH312 and B4546 by biolistic transformation with the Natr and Neor cassettes. These marked strains (MATα Natr [JF65] and MATa G418r ura5 [JF66]) were mated for 6 days and plated onto YEPD medium containing nourseothricin and G418. Similar to results with serotype D, stable Natr G418r diploids were obtained at 37°C that were thermally dimorphic, growing as yeast at 37°C but self-filamentously on V8 medium at 25°C (38). By selecting at 30°C, we could identify Natr G418r recombinant isolates that were not thermally dimorphic. To validate that these were bona fide meiotic progeny, we analyzed the segregation of four markers in a subset of the nonfilamentous isolates and observed recombinant progeny (Fig. 5). The markers included ura5 (from the a parent), RFLPs in the PLB1 and SOD1 genes, and the MAT locus (a and α). This analysis demonstrates that a random spore analysis approach can be used to isolate mating-competent meiotic progeny of serotype C.
FIG. 5.
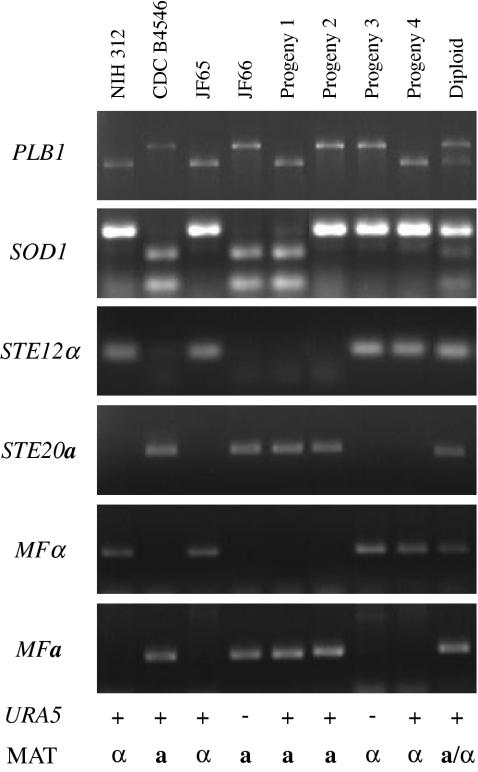
Analysis of meiotic progeny of a serotype C cross. The mating mixture of two marked strains—MATα Natr (JF65) and MATa G418r ura5 (JF66)—was plated onto YEPD medium containing both nourseothricin and G418 to select for recombinants. The parental strains and four progeny, in addition to diploid controls, were analyzed by PCR analysis for the segregation of RFLPs in the PLB1 and SOD1 genes and the mating type loci by using primers for the STE12α, STE20a, MFα, and MFa genes. Segregation of the ability to grow in the absence of uracil supplementation (URA5) and the ability to mate with the parental strains B4546 (serotype C and mating type a) and NIH312 (serotype C and mating type α) was also tested.
Mutation of the RGS protein Crg1.
In Saccharomyces cerevisiae, mating is controlled by the pheromone response pathway, which is conserved in C. neoformans. Detection of pheromones by a heterotrimeric G protein-coupled receptor initiates a conserved mitogen-activated protein kinase cascade to activate transcriptional regulators (for a review, see reference 9). In yeast, this pathway is negatively regulated by the RGS protein Sst2, which accelerates the intrinsic GTPase activity of the Gα subunit to silence signaling (1, 8). The C. neoformans SST2 homologue is the CRG1 gene, and crg1 mutations enhance mating of serotype A (35; P. Wang, personal communication). Here we show that a CRG1-targeted gene disruption enhances mating of serotypes B and C.
The first targeted gene disruption in C. neoformans var. gattii, that of the serotype B SOD1 gene with the use of the URA5 gene, was recently reported (34). We disrupted the CRG1 gene in the serotype B strain WM276 by using dominant selectable markers that confer resistance to nourseothricin or G418. We also isolated crg1::Neor mutations in the serotype C mating pair NIH312 (α) and B4546 (a) and the Vancouver Island clinical isolate R265 (α).
When cocultured with a strain of the opposite mating type, the crg1 mutant derivatives of NIH312, B4546, and R265 underwent much more rapid and proliferative mating reactions than the wild types, producing more hyphae and basidia (Fig. 6). This was also observed in intervarietal matings with serotype D strains. The enhanced mating of the crg1 mutant strains was restored to the wild-type level when the wild-type CRG1 gene was reintroduced (data not shown). Comparison of unilateral mutant crosses to bilateral mutant crosses revealed that the effect of the crg1 mutation is additive. Deletion of CRG1 was sufficient to allow mating on YEPD, a medium that usually strongly represses mating, in the case of a unilateral or bilateral cross (data not shown), indicating that this mutation is sufficient to bypass the nutritional limitation and desiccation signals normally required for C. neoformans mating.
FIG. 6.
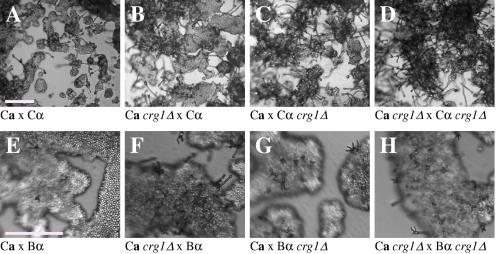
The crg1Δ mutation accelerates mating. Mating C. neoformans isolates and crg1Δ mutant strains were incubated on V8 pH 7 medium on glass slides for 3 days, and their morphology was observed by using DIC microscopy. A comparison of the effects of the crg1Δ mutation on mating ability was performed on V8 pH 7 medium. Compared to the serotype C wild-type crosses (B4546 Ca × NIH312 Cα [Ca × Cα] and B4546 Ca × R265 Bα [Ca × Bα]) (A and E, respectively), introducing a crg1Δ mutant in unilateral crosses (JF109 crg1Δ Ca × NIH312 Cα [Ca crg1Δ × Cα], B4546 Ca × JF101 crg1Δ Cα [Ca × Cα crg1Δ], JF109 crg1Δ Ca × R265 Bα [Ca crg1Δ × Bα], and B4546 Ca × JF117 crg1Δ Bα [Ca × Bα crg1Δ]) (B, C, F, and G, respectively) increased the amount of filamentation and basidia production. This effect was even greater in bilateral crosses (JF109 crg1Δ Ca × JF101 crg1Δ Cα [Ca crg1Δ × Cα crg1Δ] and JF109 crg1Δ Ca × JF117 crg1Δ Bα [Ca crg1Δ × Bα crg1Δ]) (D and H, respectively). Scale bar, 200 μm.
Mutation of CRG1 enables mating with a subset of previously sterile strains.
Unlike the crg1 serotype C mating pair and Vancouver isolates, neither of the Australian crg1Δ strains exhibited any detectable mutant phenotype, nor could the CRG1 wild-type Australian isolates WM276 (serotype B and mating type α) or E566 (serotype B and mating type a) mate with crg1Δ strains. Also, attempts to identify fusion of the B4546 crg1::Neor strain JF109 and the E566 crg1::Natr strain JF9 by selecting for diploids at 37°C on YEPD medium containing nourseothricin and G418 failed to identify Natr G418r strains. The failure of the Australian isolate E566 to produce mating structures is therefore due to a defect prior to fusion. This suggests that mutation of the CRG1 gene increases existing mating compatibility, leading to the production of more mating filaments. To test this hypothesis, we repeated our mating analysis of the original collection of 145 isolates by using NIH312 and B4546 crg1Δ mutants as tester strains. In all instances in which strains previously mated with NIH312 or B4546, this reaction was enhanced in a crg1Δ unilateral cross (see Table S1 in the supplemental material). In addition, one more serotype C strain (a total of 10 of 14, or 71%) and three more Vancouver Island environmental isolates (a total of 56 of 59, or 95%) mated. The crg1Δ strains are therefore useful tools for analyzing mating of C. neoformans var. gattii, allowing mating to be more readily observed and in some cases enabling otherwise sterile strains to mate.
DISCUSSION
Recapitulation of mating of C. neoformans var. gattii.
The role of a sexual cycle in virulence of the human fungal pathogen C. neoformans is of great interest, as its importance in virulence is twofold. First, the vast majority of clinical cases are caused by strains of the α mating type, and in serotype D, α strains have been shown to be more pathogenic than a strains in a murine model (25). Second, the basidiospore is thought to be the infectious propagule. The sexual cycle and the components of the mating signaling cascade of C. neoformans var. neoformans have been studied intensely over the last decade, and recently this research has been extended to include serotype A C. neoformans var. grubii, in which a isolates and a complete sexual cycle have only recently been identified (18, 30, 35, 45). In contrast, the sexual cycle of C. neoformans var. gattii was first reported almost 30 years ago (21). Here we have recapitulated the original data on mating of C. neoformans var. gattii and extended these original studies by identifying strains that mate robustly with one another and with a large number of clinical and environmental isolates. Independently, Tscharke and Kwon-Chung have also identified serotype B strains capable of mating (R. L. Tscharke and K. J. Kwon-Chung, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. F-063, 2003). The recapitulation of data on mating and the discovery of robustly mating strains provide a platform for the generation of congenic serotype C strains and serve as an important foundation for genetic studies of this variety.
As we have previously observed for serotype A (35), some C. neoformans var. gattii isolates are sterile, some are fertile, and others are capable of mating only with certain tester strains. This preference for mating partners can potentially be explained by three possible mechanisms. First, cryptic speciation events may be occurring that restrict mating between some members of the population, as has occurred in several other fungi, such as Coccidioides immitis and Coccidioides posadasii (19). Second, a spontaneous loss of mating ability may be occurring in nature or during laboratory passage, as has recently been reported for C. neoformans serotype D strains (46). Third, alterations in the mating type locus may lead to a loss or restriction of mating capacity, and we are currently investigating this possibility by sequencing the a and α alleles of the MAT locus from strains WM276 and E566.
Targeted gene disruption using dominant selectable markers in serotypes B and C.
In addition to the availability of genetic analysis, the ability to perform targeted disruptions is an essential attribute with regard to a model organism. Our studies presented here demonstrate that the Natr, Hygr, and Neor cassettes can be used as dominant selectable markers in C. neoformans var. gattii to readily disrupt genes by biolistic transformation and homologous recombination, providing valuable tools for facilitating molecular manipulation of the genome. The ability to transform a range of clinical and environmental isolates by using plasmids that can function in other varieties not only provides a tool for performing targeted disruptions in C. neoformans var. gattii but also allows the option of utilizing marker constructs and gene regulatory sequences designed for use in serotype A or D.
The Vancouver Island outbreak is attributable to an unusually fertile set of isolates.
Attention has recently focused on the primary pathogenic form of C. neoformans due to an outbreak on Vancouver Island, Canada. The presence of C. neoformans var. gattii on Vancouver Island indicates that this primary pathogen is not restricted to tropical regions. Our study reveals that, unlike isolates originating from elsewhere in the world, the strains from the Vancouver Island outbreak are exclusively of the α mating type. Furthermore, the vast majority of the outbreak isolates are fertile. This observation gives rise to several possible models. In the first, mating and meiotic recombination in a fertile clade may have given rise to an isolate with increased virulence, expanded host range, or an altered environmental niche. In the second, the fertility of the isolates may increase the production of the infectious propagule, increasing the incidence of infection. Third, the outbreak may be attributable to a so far unobserved ability of these isolates to undergo haploid fruiting, which perhaps explains the presence of exclusively α isolates. Finally, the relationship between the outbreak and the fertility of these isolates may be coincidental.
Australian environmental isolates are sterile.
In contrast to the fertility of isolates from Vancouver Island and elsewhere in the world is the infertility of the Australian environmental isolates, as we were unable to identify mating of any of these isolates. This finding supports previous reports that failed to identify recombining populations in association with eucalyptus trees (15). In contrast, 50% (three of six) of clinical isolates were fertile. These isolates may have originated elsewhere or may be due to the production of spores from rare environmental isolates. It is still possible that the conditions for mating of this subset of isolates differ from those for mating of the fertile strains identified thus far and that given appropriate conditions or mating partners these strains may be capable of undergoing sexual recombination. This represents an important avenue of study, as the C. neoformans var. gattii genome project that is currently under way uses a sterile Australian environmental isolate (WM276) (J. Kronstad, personal communication). One possibility is that these geographically isolated strains no longer have functional mating type loci. To investigate this prospect, we are currently sequencing the mating type loci of both α (WM276) and a (E566) isolates. Molecular analysis of the mating type loci of these environmental isolates may reveal the reason for this infertility, providing not only the means to render the platform sequence reference strain WM276 fertile but also insight into the fertility of strains from elsewhere in the world.
Disruption of the RGS protein-encoding gene CRG1 enhances mating of C. neoformans var. gattii.
In C. neoformans var. grubii, loss of the RGS protein Crg1 required for pheromone desensitization enhances mating. Disruption of the CRG1 gene greatly enhanced mating of both serotype C isolates and an isolate from the Vancouver Island outbreak and conferred the ability to mate with a subset of partners that appeared to be sterile when crossed with a wild-type strain. Furthermore, the crg1Δ mutation overrides the nutritional and desiccation signals normally required for mating. In direct contrast to strains from Vancouver Island, environmental isolates originating from Australia had no identifiable sexual cycle despite deletion of the CRG1 gene in either or both strains in crosses.
It is striking that the RGS protein mutations exert related phenotypes in the two divergent organisms S. cerevisiae and C. neoformans, enhancing pheromone responsiveness in both and enhancing mating of C. neoformans. This said, there are also interesting distinctions between the phenotypes of an S. cerevisiae sst2 mutant and the crg1 mutant of C. neoformans. In contrast to the alteration in cellular morphology and the reduced mating ability of an sst2Δ mutant (9, 39), crg1Δ strains show no alteration in growth morphology in the absence of a partner and undergo increased mating. Comparison of fungal RGS proteins reveals that all of them contain two N-terminal DEP (Dishevelled, Egl-10, Pleckstrin) domains, except the C. neoformans Crg1 protein, which appears to contain only one. In S. cerevisiae, these domains have been implicated in signaling the stress response pathway (2), and this function may have been lost in C. neoformans, perhaps explaining why crg1Δ mutant strains do not display any detrimental phenotypes. crg1Δ mutant strains therefore represent a valuable tool in the further analysis of mating and subsequent production of the infectious propagule by this primary human pathogen.
In summary, these studies provide insight into a factor that may have contributed to the outbreak on Vancouver Island and a starting point for the generation of congenic pairs of this variety. Additionally, comparison of serotype B and C, Vancouver, and Australian isolates to the opportunistic serotype A and D pathogens provides a model to address the central question of how an opportunistic organism escapes immune control and evolves into a primary pathogen.
Supplementary Material
Acknowledgments
We thank Jim Kronstad, Wiley Schell, June Kwon-Chung, and Wieland Meyer for strains and Leslie Eibest for valuable assistance in performing ESEM (NSF award no. DBI-0098534). The Natr cassette subclone pCH233 was generously provided by Christina Hull. We thank Christina Hull, Ping Wang, Tom Mitchell, John Perfect, and Jim Kronstad for advice and comments on the manuscript.
This work was supported in part by NIAID R01 grant AI50113 to Joseph Heitman and NIAID PO1 program project grant AI44975 to the Duke University Mycology Research Unit. Joseph Heitman is a Burroughs Wellcome Scholar in Molecular Pathogenic Mycology and an associate investigator of the Howard Hughes Medical Institute.
Footnotes
The supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Apanovitch, D. M., K. C. Slep, P. B. Sigler, and H. G. Dohlman. 1998. Sst2 is a GTPase-activating protein for Gpa1: purification and characterization of a cognate RGS-Gα protein pair in yeast. Biochemistry 37:4815-4822. [DOI] [PubMed] [Google Scholar]
- 2.Burchett, S. A., P. Flanary, C. Aston, L. Jiang, K. H. Young, P. Uetz, S. Fields, and H. G. Dohlman. 2002. Regulation of stress response signaling by the N-terminal dishevelled/EGL-10/pleckstrin domain of Sst2, a regulator of G protein signaling in Saccharomyces cerevisiae. J. Biol. Chem. 277:22156-22167. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi, S., A. J. Hamilton, P. Hobby, G. Zhu, C. V. Lowry, and V. Chaturvedi. 2001. Molecular cloning, phylogenetic analysis and three-dimensional modeling of Cu, Zn superoxide dismutase (CnSOD1) from three varieties of Cryptococcus neoformans. Gene 268:41-51. [DOI] [PubMed] [Google Scholar]
- 4.Chen, S., T. Sorrell, G. Nimmo, B. Speed, B. Currie, D. Ellis, D. Marriott, T. Pfeiffer, D. Parr, K. Byth, et al. 2000. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Clin. Infect. Dis. 31:499-508. [DOI] [PubMed] [Google Scholar]
- 5.Cox, G. M., D. L. Toffaletti, and J. R. Perfect. 1996. Dominant selection system for use in Cryptococcus neoformans. J. Med. Vet. Mycol. 34:385-391. [PubMed] [Google Scholar]
- 6.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 7.Davidson, R. C., M. C. Cruz, R. A. Sia, B. Allen, J. A. Alspaugh, and J. Heitman. 2000. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 29:38-48. [DOI] [PubMed] [Google Scholar]
- 8.Dohlman, H. G., D. Apaniesk, Y. Chen, J. Song, and D. Nusskern. 1995. Inhibition of G-protein signaling by dominant gain-of-function mutations in Sst2p, a pheromone desensitization factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:3635-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703-754. [DOI] [PubMed] [Google Scholar]
- 10.Ellis, D. H., and T. J. Pfeiffer. 1990. Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 28:1642-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, D., J. Burrow, D. Lo, and B. Currie. 1993. Cryptococcus neoformans in tropical northern Australia: predominantly variant gattii with good outcomes. Aust. N. Z. J. Med. 23:678-682. [DOI] [PubMed] [Google Scholar]
- 12.Franzot, S. P., J. S. Hamdan, B. P. Currie, and A. Casadevall. 1997. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J. Clin. Microbiol. 35:2243-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grigg, M. E., S. Bonnefoy, A. B. Hehl, Y. Suzuki, and J. C. Boothroyd. 2001. Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science 294:161-165. [DOI] [PubMed] [Google Scholar]
- 14.Halliday, C. L., T. Bui, M. Krockenberger, R. Malik, D. H. Ellis, and D. A. Carter. 1999. Presence of α and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. J. Clin. Microbiol. 37:2920-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halliday, C. L., and D. A. Carter. 2003. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J. Clin. Microbiol. 41:703-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heitman, J., B. Allen, J. A. Alspaugh, and K. J. Kwon-Chung. 1999. On the origins of congenic MATα and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet. Biol. 28:1-5. [DOI] [PubMed] [Google Scholar]
- 17.Hull, C. M., and J. Heitman. 2002. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36:557-615. [DOI] [PubMed] [Google Scholar]
- 18.Keller, S. M., M. A. Viviani, M. C. Esposto, M. Cogliati, and B. L. Wickes. 2003. Molecular and genetic characterization of a serotype A MATa Cryptococcus neoformans isolate. Microbiology 149:131-142. [DOI] [PubMed] [Google Scholar]
- 19.Koufopanou, V., A. Burt, and J. W. Taylor. 1997. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. USA 94:5478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kues, U. 2000. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 64:316-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon-Chung, K. J. 1976. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68:943-946. [PubMed] [Google Scholar]
- 22.Kwon-Chung, K. J., and J. E. Bennett. 1992. Cryptococcosis, p. 396-446. In K. J. Kwon-Chung and J. E. Bennett (ed.), Medical mycology. Lea & Febiger, Philadelphia, Pa.
- 23.Kwon-Chung, K. J., and J. E. Bennett. 1978. Distribution of alpha and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108:337-340. [DOI] [PubMed] [Google Scholar]
- 24.Kwon-Chung, K. J., J. E. Bennett, and J. C. Rhodes. 1982. Taxonomic studies on Filobasidiella species and their anamorphs. Antonie Leeuwenhoek 48:25-38. [DOI] [PubMed] [Google Scholar]
- 25.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon-Chung, K. J., B. L. Wickes, L. Stockman, G. D. Roberts, D. Ellis, and D. H. Howard. 1992. Virulence, serotype, and molecular characteristics of environmental strains of Cryptococcus neoformans var. gattii. Infect. Immun. 60:1869-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latouche, G. N., T. C. Sorrell, and W. Meyer. 2002. Isolation and characterisation of the phospholipase B gene of Cryptococcus neoformans var. gattii. FEMS Yeast Res. 2:551-561. [DOI] [PubMed] [Google Scholar]
- 28.Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester, F. S. Dietrich, and J. Heitman. 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1:704-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lengeler, K. B., and J. Heitman. 2002. Cryptococcus neoformans as a model fungal pathogen, p. 513-557. In H. D. Osiewacz (ed.), Molecular biology of fungal development. Marcel Dekker, Inc., New York, N.Y.
- 30.Lengeler, K. B., P. Wang, G. M. Cox, J. R. Perfect, and J. Heitman. 2000. Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proc. Natl. Acad. Sci. USA 97:14455-14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madrenys, N., C. De Vroey, C. Raes-Wuytack, and J. M. Torres-Rodriguez. 1993. Identification of the perfect state of Cryptococcus neoformans from 195 clinical isolates including 84 from AIDS patients. Mycopathologia 123:65-68. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Espinoza, A. D., M. D. Garcia-Pedrajas, and S. E. Gold. 2002. The Ustilaginales as plant pests and model systems. Fungal Genet. Biol. 35:1-20. [DOI] [PubMed] [Google Scholar]
- 33.McDade, H. C., and G. M. Cox. 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39:151-154. [DOI] [PubMed] [Google Scholar]
- 34.Narasipura, S. D., J. G. Ault, M. J. Behr, V. Chaturvedi, and S. Chaturvedi. 2003. Characterization of Cu, Zn superoxide dismutase (SOD1) gene knock-out mutant of Cryptococcus neoformans var. gattii: role in biology and virulence. Mol. Microbiol. 47:1681-1694. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen, K., G. M. Cox, P. Wang, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 2003. The sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pitkin, J. W., D. G. Panaccione, and J. D. Walton. 1996. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142:1557-1565. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1-3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Sia, R. A., K. B. Lengeler, and J. Heitman. 2000. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet. Biol. 29:153-163. [DOI] [PubMed] [Google Scholar]
- 39.Siekhaus, D. E., and D. G. Drubin. 2003. Spontaneous receptor-independent heterotrimeric G-protein signalling in an RGS mutant. Nat. Cell Biol. 5:231-235. [DOI] [PubMed] [Google Scholar]
- 40.Speed, B., and D. Dunt. 1995. Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin. Infect. Dis. 21:28-34. [DOI] [PubMed] [Google Scholar]
- 41.Stephen, C., S. Lester, W. Black, M. Fyfe, and S. Raverty. 2002. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can. Vet. J. 43:792-794. [PMC free article] [PubMed] [Google Scholar]
- 42.Su, C., D. Evans, R. H. Cole, J. C. Kissinger, J. W. Ajioka, and L. D. Sibley. 2003. Recent expansion of Toxoplasma through enhanced oral transmission. Science 299:414-416. [DOI] [PubMed] [Google Scholar]
- 43.Sukroongreung, S., K. Kitiniyom, C. Nilakul, and S. Tantimavanich. 1998. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Med. Mycol. 36:419-424. [PubMed] [Google Scholar]
- 44.Vanbreuseghem, R., and M. Takashio. 1970. An atypical strain of Cryptococcus neoformans (San Felice) Vuillemin 1894. II. Cryptococcus neoformans var. gattii var. nov. Ann. Soc. Belge Med. Trop. 50:695-702. [PubMed] [Google Scholar]
- 45.Viviani, M. A., M. C. Esposto, M. Cogliati, M. T. Montagna, and B. L. Wickes. 2001. Isolation of a Cryptococcus neoformans serotype A MATa strain from the Italian environment. Med. Mycol. 39:383-386. [DOI] [PubMed] [Google Scholar]
- 46.Xu, J. 2002. Estimating the spontaneous mutation rate of loss of sex in the human pathogenic fungus Cryptococcus neoformans. Genetics 162:1157-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, J., R. Vilgalys, and T. G. Mitchell. 2000. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9:1471-1481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.