Abstract
Celiac disease is caused by a selective lack of T cell tolerance for gluten. It is known that the enzyme tissue transglutaminase (tTG) is involved in the generation of T cell stimulatory gluten peptides through deamidation of glutamine, the most abundant amino acid in gluten. Only particular glutamine residues, however, are modified by tTG. Here we provide evidence that the spacing between glutamine and proline, the second most abundant amino acid in gluten, plays an essential role in the specificity of deamidation. On the basis of this, algorithms were designed and used to successfully predict novel T cell stimulatory peptides in gluten. Strikingly, these algorithms identified many similar peptides in the gluten-like hordeins from barley and secalins from rye but not in the avenins from oats. The avenins contain significantly lower percentages of proline residues, which offers a likely explanation for the lack of toxicity of oats. Thus, the unique amino acid composition of gluten and related proteins in barley and rye favors the generation of toxic T cell stimulatory gluten peptides by tTG. This provides a rationale for the observation that celiac disease patients are intolerant to these cereal proteins but not to other common food proteins.
Keywords: HLA-DQ2, deamidation, T cell, cereals, algorithm
Introduction
Celiac disease (CD) is a permanent intolerance to gluten (1). Gluten is a complex mixture of storage proteins found in wheat. Similar proteins are present in other grains like barley, rye, and oats. Typical disease symptoms include chronic diarrhea, fatigue, and failure to thrive. These symptoms are associated with a lesion in the upper small intestine, characterized by a (sub) total villous atrophy, an increased number of intraepithelial lymphocytes, and a chronic inflammatory response of the lamina propria lymphocytes (1). A strong indication for CD is the presence of antibodies specific for the enzyme tissue transglutaminase (tTG) in patients (2). Moreover, recent studies demonstrated the involvement of this enzyme in the generation of T cell stimulatory peptides (3, 4). CD is strongly associated with HLA-DQ2 (A1*0501, B1*0201) and HLA-DQ8 (A1*0301, B1*0302) (5). In addition, HLA-DQ2 and/or HLA-DQ8 restricted, gluten-specific CD4+ T lymphocytes can be isolated from small intestinal biopsies of CD patients, and these are thought to cause disease (6, 7). The peptide-binding motif of HLA-DQ2 and -DQ8 predicts a preference for negative charges at anchor positions in the bound peptides (8–10). However, gluten molecules contain few negative charges. This discrepancy was solved by the finding that tTG can convert glutamine residues into glutamic acid (a process termed deamidation), and thus introduces negative charges in gluten peptides. Indeed it was found that tTG is required for, or enhances the gluten-specific response of T cell clones and T cell lines derived from the majority of CD patients (3, 4, 11, 12). Considering the abundance of glutamine residues in gluten (∼30–40%) tTG has many potential target sites in gluten. Mass spectral analysis of the deamidated T cell stimulatory gluten peptides, however, showed a highly restricted pattern of deamidation (3, 4) that often coincides with the positions where negative charges are preferred in the HLA-binding motif, thus favoring the binding of gluten peptides to HLA-DQ2 and/or –DQ8. The specificity of gluten deamidation by tTG, therefore, is a crucial factor in the generation of toxic gluten peptides but it is not known why only particular glutamine residues are targeted by tTG. This has been investigated in this study.
Materials and Methods
Peptides and tTG Treatment.
Peptides were synthesized by standard Fmoc chemistry on a multiple peptide synthesizer (SyroII, MultiSynTech GmbH). Integrity of synthetic peptides was checked by rpHPLC and mass spectrometry. tTG treatment was performed by incubating the peptides (500 μg/ml) with the enzyme (100 μg/ml, T-5398; Sigma-Aldrich) at 37°C for 4 h minimum, in 50 mM triethylamine-acetate, pH 6.5, 2 mM CaCl2. Throughout the manuscript the one letter code for amino acids is used.
Mass Spectrometry.
Electrospray ionization mass spectrometry was performed on the synthetic gluten peptides before and after tTG treatment, using a Q-TOF hybrid mass spectrometer (Micromass). Precursor ions were selected with the quadruple window set to 3 Da and fragments were collected with high efficiency with the orthogonal time of flight mass spectrometer. The collision gas applied was argon (pressure 4 × 10−5 mbar) and the collision voltage ∼30 V. Deamidation of glutamine residues in synthetic gluten peptides was determined by mass spectrometric analysis as described previously (4). Deamidation of a glutamine residue results in an increase of 1 Da. These conversions were assigned to particular glutamine residues by comparison of the fragmentation spectra of tTG treated and nontreated peptides (4).
Database Searching.
The program PeptideSearch was used for pattern searches (13). The patterns were composed on the basis of the newly identified specificity of tTG in combination with the previously identified HLA-DQ2 peptide-binding motif (8, 9). For database searching selected subsets of gluten, hordein, secalin, and avenin proteins were compiled from the Swiss Prot databank, SPTREMBL and PIR.
T Cell Clones and T Cell Lines.
Gluten-specific T cell lines were generated from small intestinal biopsies of CD patients as described previously (4, 7, 14). All patients gave informed consent to the study, which was approved by the hospital ethics committee.
T Cell Proliferation Assays.
Proliferation assays were performed in duplicate in 150 μl RPMI 1640 (GIBCO BRL) supplemented with 10% human serum in 96-well, flat-bottomed plates (Falcon) using 104 T cells stimulated with 105 irradiated PBMCs (3,000 rad) in the presence or absence of antigen (1–10 μg/ml). After 48 h at 37°C, cultures were pulsed with 0.5 μCi of 3[H]thymidine and harvested 18 h thereafter as described previously (7).
Results
The Specificity of tTG.
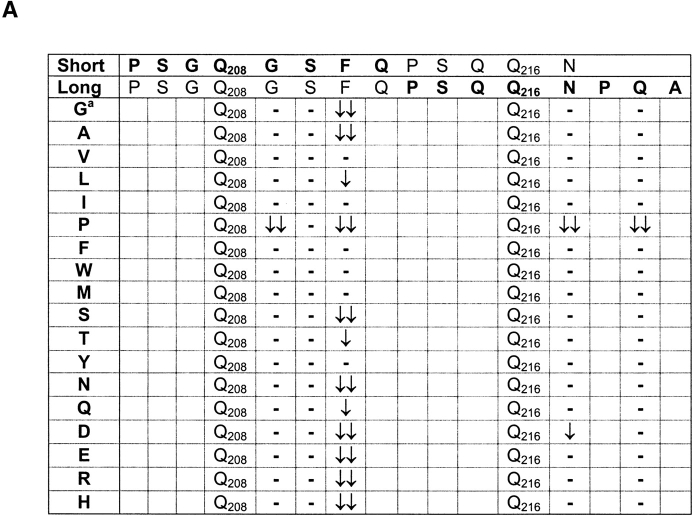
Previously we have characterized an HLA-DQ8–restricted, T cell stimulatory gliadin peptide (4, 7). In the core of this gliadin peptide (sequence PSGQ208GSFQPSQQ216NPQA) the Q208 and Q216 are deamidated by tTG, whereas positions Q212, Q215, and Q219 are not (4). Preliminary studies indicated that the nature of COOH-terminal flanking amino acids influenced deamidation of glutamine residues (data not shown). We have now investigated this systematically by analyzing the deamidation in amino acid substitution analogs of a shorter version of the gliadin peptide (sequence: PSGQ208GSFQPSQQ216N). In this peptide only the Q208 is deamidated by tTG treatment (Fig. 1 A). Mass spectrometric analysis demonstrated that at only two positions in the peptide substitutions had effect on deamidation of Q208 (Fig. 1 A, and data not shown). First, the replacement of Q209 by a P abolished deamidation of Q208. Second, several amino acid substitutions at F211 had a strong negative effect on deamidation of Q208, in particular replacements of F211 for amino acids with significantly smaller or charged side chains or replacement by proline (Fig. 1 A).
Figure 1.
The influence of amino acid substitutions on deamidation of Q208 and Q216 in an HLA-DQ8–restricted gliadin peptide. (A) Overview of the influence of COOH-terminal amino acid substitutions on deamidation of Q208 in the short version of the gliadin peptide and Q216 in the long version of the peptide. aThe amino acids G209, S210, F211, N217, and Q219 were substituted for all amino acids except lysine and cysteine, because these residues could affect deamidation by formation of disulphide bridges and tTG driven cross-linking, respectively. The effect on deamidation was determined by mass spectrometry. Substitutions that resulted in major differences in deamidation are designated with arrows: ↓↓ decrease of 70–100%; ↓ decrease of 30–69%. Dash indicates no effect of the substitution; substitution at other positions in the peptide had only minor effects on deamidation of the Q208 and Q216 residues (data not shown). (B) Patterns for deamidation in the HLA-DQ8–restricted gliadin peptide. X stands for any amino acid.
Next, we performed a similar analysis to determine the influence of amino acid substitutions on the deamidation of Q216 in a longer version of the peptide (Fig. 1 A). The results demonstrate that the substitution of N217 with a P abolishes deamidation of Q216, a finding that underscores the observation that in a QP sequence the Q is not a target for tTG. Moreover, the substitution of Q219 with a P had a negative effect on deamidation of Q216, confirming the negative influence of the P in the sequence QXXP (X stands for any amino acid). Other amino acid substitutions at Q216+3, however, had no effect on deamidation of Q216, a result that is in contrast to the strong influence of amino acid substitutions at position Q208+3 on deamidation of Q208. A marked difference between the sequence Q208GSF and Q216NPQ is the presence of a P in the latter sequence at Q+2 suggesting that in the sequence QXPX the Q is a good target for deamidation and the nature of the amino acid at Q+3 is much less important (see below).
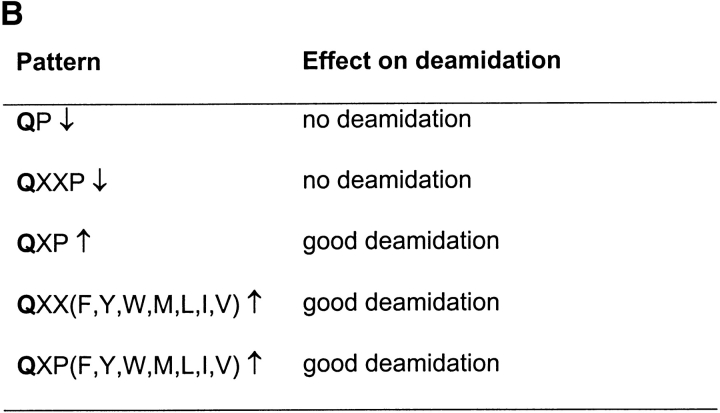
Together the results indicate that in the sequences QP and QXXP the Q is not a target for tTG. In contrast the sequences QXP, QXXF(Y, W, M, L, I, or V) and QXPF(Y, W, M, L, I, or V) favor deamidation of the Q (Fig. 1 B). To verify these “patterns” for deamidation by tTG, a large panel of gluten peptides was synthesized, treated with tTG and analyzed by mass spectrometry (Table I). Without exception we found that a Q preceding a P was not deamidated by tTG while in the majority of cases a P at Q+3 also inhibited deamidation. In contrast, a bulky/hydrophobic residue at Q+3, and P at Q+2 allowed good deamidation. In the case that two opposite rules coincide, as is found in peptide 20 (LQQPQQ6PQFQPQQQF), the negative effect of QP↓ overruled the positive effect of QXXF↑ at position Q6. Together, these results confirmed the defined patterns. Proline thus plays a crucial role in the deamidation of gluten peptides.
Table I.
Validation of the Deamidation Patterns
| Pattern
|
||||
|---|---|---|---|---|
| Peptides | QP ↓ | QXXF ↑ | QXXP ↓ | QXP ↑ |
| 1 VPVPQLQPQNPSQQQPQEQVPL | 100% | 100% | 100% | 66% |
| 2 PLVQQQQFLGQQQPFPPQ | 100% | 100% | 100% | 100% |
| 3 YYPTSPQQSGQG | - | - | - | - |
| 4 QGQQGYYPTSPQ | - | 50% | - | - |
| 5 SSQVSFQPSQLN | 100% | 100% | - | - |
| 6 FPQTQQPQQLFPQSQ | 100% | 0% | 100% | 100% |
| 7 GQQGYYPTSVQQSGQ | - | 50% | - | - |
| 8 GSVQPQQQLPQFEIR | 100% | 100% | 100% | 100% |
| 9 FPQHCNYQQQPQTFPQPF | 100% | - | 100% | 100% |
| 10 QQPIQPQQFPQQQFF | 100% | 50% | 100% | 50% |
| 11 QSGQYQQQPQMQTT | 100% | - | 100% | 100% |
| 12 ESQQSQQDEPQPF | 100% | - | 100% | - |
| 13 FPQPEDQQSQQSE | 100% | - | - | - |
| 14 LQQVQQGPQQQPFPQPQPF | 100% | 100% | 66% | 100% |
| 15 QVQWPQQQPFPQPQQPF | 100% | 50% | 100% | 33% |
| 16 PLLPQQPFPSQQEQPQF | 100% | 50%a | 100% | 100% |
| 17 WQQQPPFSQEQPIL | 100% | 0%a | 33% | 100% |
| 18 QSNLPQPAQQPFQPQVPQQP | 100% | 100% | - | 66% |
| 19 WFQPSQLNPQAQQDQPQ | 100% | - | 100% | - |
| 20 LQQPQQPQFQPQQQF | 100% | 50%a | 100% | 50% |
| 21 VQQQIPVVQPSIL | 100% | 33%a | 100% | 100% |
The observed deamidation patterns in a set of 21 gluten peptides were compared with those predicted according to the deamidation patterns given in Fig. 1. The deamidation patterns are represented in the 4 columns. The percentages shown indicate whether the pattern is fully or partially applicable to the specific deamidation of Q residues in the listed gluten peptides. A bar (-) indicates the absence of that sequence in the peptide. Deamidated glutamine residues are shown in bold and underlined.
QP inhibition of deamidation overrules the positive influence of QXXF.
Prediction of Novel Gluten Epitopes.
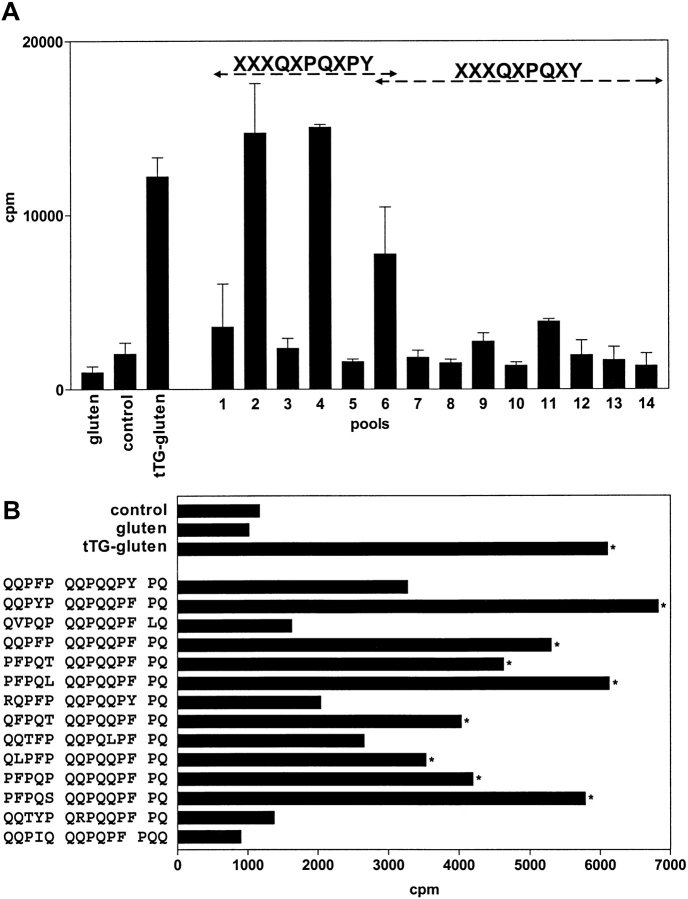
The peptide-binding motifs of the disease-associated molecules HLA-DQ2 and HLA-DQ8 display preferential binding of negative charges at anchor positions (8–10). HLA-DQ2 has a strong preference for a negative charge at p4 and p7 (8, 9). Strikingly, at p6 a proline is one of the preferred amino acids in the HLA-DQ2 peptide binding motif that could thus target specific deamidation at p4 (Table II). Therefore, we combined the HLA-DQ2 peptide-binding motif with the specificity of tTG to construct a search algorithm that could be used to predict T cell stimulatory gluten peptides from the gluten database (Table II). In this algorithm glutamine residues were introduced at the putative DQ2 binding positions p4 and p7. These serve as potential target sites for tTG to allow introduction of a negative charge at these positions. Next, proline was selected for p6 to promote deamidation at Q4, which is also consistent with the requirements for peptide binding at this position (Table II). For the p9 position two options were designed. In the first option bulky/hydrophobic residues were selected for p9, according to the peptide-binding motif. In the second option, a proline residue was placed at p9, followed by bulky/hydrophobic at p10. This option does not agree with the peptide-binding motif but should allow optimal deamidation at Q7 according to our QXPF pattern. The remaining positions p1, p2, p3, p5, and p8 were not defined (Table II). These two algorithms were used to search in the gluten database. A total of 98 matches were obtained, which corresponded to 27 unique gluten sequences. 13 of these were predicted by algorithm 1, the others by algorithm 2. These 27 peptides were synthesized and deamidated by tTG. Subsequent mass spectral analysis confirmed deamidation of the predicted Q4 and Q7 residues (data not shown). The native and deamidated peptides were pooled in pools of five peptides each and tested in T cell proliferation assays with three gluten-specific T cell lines isolated from small intestinal biopsies of CD patients. Two of these T cell lines responded to stimulation by several of the deamidated peptide pools (Fig. 2 A, results shown for one of the T cell lines). Strong T cell responses were only found against pools that contained peptides predicted by the second algorithm, X X X Q4 X P6 Q7 X P9 (Y, F, W, I, L)10 (Fig. 2 A and B). Subsequently, the T cell line was tested against the individual peptides of the positive pools, and found to respond to eight out of 13 peptides tested (Fig. 2 B).
Table II.
Design of Search Algorithms Based on the HLA-DQ2 Peptide-binding Motif and the tTG Specificity
| DQ2 peptide-binding motif | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Residue | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Preferred | F, W, Y, I, L, V | D, E, V, L, I | P, A, E | D, E | F, W, Y, I, L, V, M | - | ||||
| DQ2 epitope search algorithms | ||||||||||
| Algorithm 1 | X | X | X | Q | X | P | Q | X | F, Y, W, I, L | X |
| Algorithm 2 | X | X | X | Q | X | P | Q | X | P | F, Y, W, I, L |
X stands for any amino acid.
Figure 2.
Stimulation of gluten-specific T cell lines by predicted peptides. (A) A gluten-specific T cell line isolated from a small intestinal biopsy of a CD patient was tested against gluten, tTG-treated gluten, and against the predicted peptides that were present in pools of five peptides each (pools 1–14). The odd numbers represent pools that contain the native peptides, whereas the even numbers represent corresponding pools that were treated with tTG. Strong reactivity of the T cell line was only observed with tTG-treated gluten and tTG-treated pools of peptides predicted by the XXXQXPQXPY algorithm. Cpm indicates 3[H]thymidine incorporation. (B) Reactivity of the T cell line against the individual peptides from the T cell stimulatory pools. Bars indicated with an asterisk are considered positive (Stimulation Index > 3).
In addition we performed searches with a less strict variant of algorithm 2, X X X X4 X P6 Q7 X P9 (Y,F,W,I,L)10, which predicts the deamidation of only one Q residue. This search yielded 261 matches in the gluten database (Table III). 18 of these matches corresponded to a known T cell stimulatory α-gliadin peptide (sequence QLQPFPQPQLPY) (11, 12).
Table III.
Database Searches with the Search Algorithms
| Number of matchesin databases
|
|||||
|---|---|---|---|---|---|
| Search algorithm | Gluten | Hordein | Secalin | Avenin | |
| Algorithm 1 | Q4XPQ7X(YFWIL) | 52 | 48 | 8 | - |
| Algorithm 1 – Q4 | XXPQ7X(YFWIL) | 334 | 91 | 14 | 6 |
| Algorithm 1 – Q7 | Q4XPXX(YFWIL) | 286 | 142 | 60 | 11 |
| Algorithm 2 | Q4XPQ7XP(YFWIL) | 46 | 60 | 33 | - |
| Algorithm 2 – Q4 a | XXPQ7XP(YFWIL) | 262 | 196 | 89 | - |
| Algorithm 2 – Q7 | Q4XPXXP(YFWIL) | 51 | 68 | 33 | - |
| Short algorithm | QXP(YFWIL) X ≠ Pb | >500 | 276 | 105 | 7 |
| Predicted epitope | QQPFPQQPQQPFPQ | 6 | 12 | 2 | - |
The search algorithms reveal many matches in the gluten, hordeins, and secalin sequences but very few in the avenin sequences. The predictive algorithm 2 (QXPQXP[YFWIL]) and the less strict variants of that algorithm have no matches in the avenin database. The novel T cells stimulatory gluten peptide (QQPFPQQPQQPFPQ) is also found in the hordeins and secalin but not in the avenins. X stands for any amino acid.
Algorithm 2 without the specification of a Q at position 4 predicts the known gliadin α9 epitope.
A proline at p2 in this algorithm inhibits deamidation of the glutamine.
Presence of T Cell Stimulatory Sequences in Rye, Barley, and Oats.
Next we used the three algorithms to perform database searches in the gluten-like hordeins from barley, secalins from rye, and avenins from oats. While the first two cereals are toxic for CD patients, the latter is not (15, 16). Strikingly, all three search algorithms yielded many matches in the available hordein and secalin sequences but none in the avenin sequences (Table III). Consequently, the predicted T cell stimulatory gluten peptide QQPFPQQPQQPFPQ (Fig. 2) is present in hordeins and secalins but not in avenins (Table III). Additional database searches with less strict algorithms revealed a general lack of matches in the avenin database as compared with the gluten, hordein, and secalin databases (Table III). Notably, no matches were found in the avenins with three variants of the predictive algorithm 2.
Discussion
We have investigated the deamidation patterns of gluten peptides by tTG. The results demonstrate clear and selective deamidation patterns that to a large extent can be explained by strong effects of the spacing between the target glutamine residue and COOH-terminal proline residues. It has been described previously that a glutamine between proline residues is not modified by tTG (17). An important general role for proline in guiding tTG specificity, however, has not been appreciated. Proline, after glutamine, is the second most abundant amino acid in gluten (18) and thus the dominant factor in the selective deamidation of gluten peptides observed. In addition, the presence of a large hydrophobic amino acid three positions COOH terminally from the target glutamine has a strong positive influence on deamidation. This is highly significant since the sequence QXPF(Y) is one of the most frequently found sequences in gluten molecules and is predicted to be invariably deamidated by tTG. Moreover, cleavage by the enzyme pepsin is known to occur after phenylalanine and to a lesser extent after tyrosine, leucine, and isoleucine, and will thus generate many gluten peptides with the tTG substrate sequence QXPF(Y) at the COOH terminus. The sequential activity of pepsin and tTG, therefore, favors the generation of gluten peptides with an appropriate p7 anchor for binding to HLA-DQ2. We also demonstrate that by combining the HLA-DQ2 peptide-binding motif with two deamidation patterns an algorithm is obtained that predicts novel T cell stimulatory peptides in gluten. Strikingly, in this predictive algorithm (X X X Q4 X P6 Q7 X P9 [Y,F,W,I,L]10) we had incorporated a deamidation pattern that results in peptides that do not optimally fit the HLA-DQ2 peptide-binding motif, e.g., they lack an amino acid with a large hydrophobic side chain at position 9 in the peptide. The lack of this anchor is probably compensated for by the introduction of two negative charged anchor residues in the peptide at p4 and p7 as the result of deamidation. Thus, the T cell stimulatory activity of these peptides appears to depend more on optimal deamidation than on strict adherence to the exact HLA-DQ2 peptide-binding motif.
A less stringent search algorithm, combining only one deamidation rule with the HLA-DQ2 peptide-binding motif, proved equally successful since it predicted a previously identified T cell stimulatory α-gliadin peptide (11, 12). Importantly, the first predictive algorithm identified 13 peptides, eight out of which stimulated T cells. The second, less stringent algorithm predicted 261 peptides. 18 of those (1 in 15) matched a known T cell stimulatory gliadin peptide. These algorithms, therefore, have a high predictive value. Therefore, it is striking that these algorithms identified very similar and sometimes identical peptides in the barley- and rye-derived hordeins and secalins but not in the oats-derived avenins. Oats is considered safe for CD patients (15, 16). While gliadins, hordeins, and secalins contain ∼36% glutamine and 20% proline, avenins contain a similar percentage of glutamine (34%) but half the amount of proline (10%) (18). Deamidation of avenins is thus likely to occur in a much more random fashion. Indeed, the lack of proline residues in Q-rich regions leads to nonselective deamidation of the glutamine residues in such regions (data not shown). Collectively, our results indicate that due to nonselective deamidation no T cell stimulatory neoepitopes can be generated from the avenins and thus offer an explanation for the clinical observation that oats is not toxic for CD patients. Moreover, our results indicate that the replacement of particular proline residues in gluten by other amino acids would yield gluten molecules with considerably less toxicity for CD patients but that would retain many of the chemical and physical properties desired in gluten.
tTG is a ubiquitous enzyme and could thus be involved in the modification of proteins at other sites in the body. Therefore, it will be of interest to investigate if the defined deamidation patterns and search algorithms can be used to identify novel T cell stimulatory peptides that are relevant for autoimmune diseases.
Acknowledgments
We thank Drs. F.H.J. Claas, R.R.P. de Vries, and R. Offringa for critical reading of the manuscript.
This study was supported by grants from the European Community (BHM4-CT98-3087 and QLK1-2000-00657) and the Dutch Digestive Disease Foundation (WS 98-24).
References
- 1.Marsh, M.N. 1992. Gluten, major histocompatibility complex, and the small intestine. A molecular and immunobiologic approach to the spectrum of gluten sensitivity (‘celiac sprue’). Gastroenterology. 102:330–354. [PubMed] [Google Scholar]
- 2.Dieterich, W., T. Ehnis, M. Bauer, P. Donner, U. Volta, E.O. Riecken, and D. Schuppan. 1997. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 3:797–801. [DOI] [PubMed] [Google Scholar]
- 3.Molberg, O., S.N. McAdam, R. Korner, H. Quarsten, C. Kristiansen, L. Madsen, L. Fugger, H. Scott, O. Noren, P. Roepstorff, K.E. Lundin, H. Sjostrom, and L.M. Sollid. 1998. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 4:713–717. [DOI] [PubMed] [Google Scholar]
- 4.van de Wal, Y., Y. Kooy, P. van Veelen, S. Pena, L. Mearin, G. Papadopoulos, and F. Koning. 1998. Selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J. Immunol. 161:1585–1588. [PubMed] [Google Scholar]
- 5.Spurkland, A., G. Ingvarsson, E.S. Falk, I. Knutsen, L.M. Sollid, and E. Thorsby. 1997. Dermatitis herpetiformis and celiac disease are both primarily associated with the HLA-DQ (α1*0501, β1*02) or the HLA-DQ (α1*03, β1*0302) heterodimers. Tiss. Anttigens. 49:29–34. [DOI] [PubMed] [Google Scholar]
- 6.Lundin, K.E., H. Scott, T. Hansen, G. Paulsen, T.S. Halstensen, O. Fausa, E. Thorsby, and L.M. Sollid. 1993. Gliadin-specific, HLA-DQ(α1*0501, β1*0201) restricted T cells isolated from the small intestinal mucosa of celiac disease patients. J. Exp. Med. 178:187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Wal, Y., Y.M. Kooy, P.A. van Veelen, S.A. Pena, L.M. Mearin, O. Molberg, K.E. Lundin, L.M. Sollid, T. Mutis, W.E. Benckhuijsen, J.W. Drijfhout, and F. Koning. 1998. Small intestinal T cells of celiac disease patients recognize a natural pepsin fragment of gliadin. Proc. Natl. Acad. Sci. USA. 95:10050–10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansen, B.H., T. Jensen, C.J. Thorpe, F. Vartdal, E. Thorsby, and L.M. Sollid. 1996. Both α and β chain polymorphisms determine the specificity of the disease-associated HLA-DQ2 molecules, with β chain residues being most influential. Immunogenetics. 45:142–150. [DOI] [PubMed] [Google Scholar]
- 9.van de Wal, Y., Y.M.C. Kooy, J.W. Drijfhout, R. Amons, and F. Koning. 1996. Peptide binding characteristics of the coeliac disease-associated DQ(α1*0501, β1*0201) molecule. Immunogenetics. 44:246–253. [DOI] [PubMed] [Google Scholar]
- 10.Kwok, W.W., M.L. Domeier, F.C. Raymond, P. Byers, and G.T. Nepom. 1996. Allele-specific motifs characterize HLA-DQ interactions with a diabetes-associated peptide derived from glutamic acid decarboxylase. J. Immunol. 156:2171–2177. [PubMed] [Google Scholar]
- 11.Arentz-Hansen, H., R. Korner, O. Molberg, H. Quarsten, W. Vader, Y.M. Kooy, K.E. Lundin, F. Koning, P. Roepstorff, L.M. Sollid, and S.N. McAdam. 2000. The intestinal T cell response to α-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J. Exp. Med. 191:603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson, R.P., P. Degano, A.J. Godkin, D.P. Jewell, and A.V. Hill. 2000. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat. Med. 6:337–342. [DOI] [PubMed] [Google Scholar]
- 13.Mann, M., and M. Wilm. 1994. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal.Chem. 66:4390–4399. [DOI] [PubMed] [Google Scholar]
- 14.van de Wal, Y., Y.M. Kooy, P. van Veelen, W. Vader, S.A. August, J.W. Drijfhout, S.A. Pena, and F. Koning. 1999. Glutenin is involved in the gluten-driven mucosal T cell response. Eur. J. Immunol. 29:3133–3139. [DOI] [PubMed] [Google Scholar]
- 15.Janatuinen, E.K., P.H. Pikkarainen, T.A. Kemppainen, V.M. Kosma, R.M. Jarvinen, M.I. Uusitupa, and R.J. Julkunen. 1995. A comparison of diets with and without oats in adults with celiac disease. N. Engl. J. Med. 333:1033–1037. [DOI] [PubMed] [Google Scholar]
- 16.Janatuinen, E.K., T.A. Kemppainen, P.H. Pikkarainen, K.H. Holm, V.M. Kosma, M.I. Uusitupa, M. Maki, and R.J. Julkunen. 2000. Lack of cellular and humoral immunological responses to oats in adults with coeliac disease. Gut. 46:327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pastor, M.T., A. Diez, E. Perez-Paya, and C. Abad. 1999. Addressing substrate glutamine requirements for tissue transglutaminase using substance P analogues. FEBS Lett. 451:231–234. [DOI] [PubMed] [Google Scholar]
- 18.Wieser, H., W. Seilmeier, M. Eggert, and H.D. Belitz. 1983. Tryptophan content of cereal proteins. Z. Lebensm. Unters. Forsch. 177:457–460. [DOI] [PubMed] [Google Scholar]