Abstract
Minor histocompatibility antigens (minor H antigens) are targets of graft-versus-host disease and graft-versus-leukemia responses after allogeneic human leukocyte antigen identical hematopoietic stem cell transplantation. Only a few human minor H antigens have been molecularly characterized and in all cases, amino acid differences between homologous donor and recipient proteins due to nucleotide polymorphisms in the respective genes were responsible for immunogenicity. Here, we have used cDNA expression cloning to identify a novel human minor H antigen encoded by UGT2B17, an autosomal gene in the multigene UDP-glycosyltransferase 2 family that is selectively expressed in liver, intestine, and antigen-presenting cells. In contrast to previously defined human minor H antigens, UGT2B17 is immunogenic because of differential expression of the protein in donor and recipient cells as a consequence of a homozygous gene deletion in the donor. Deletion of individual members of large gene families is a common form of genetic variation in the population and our results provide the first evidence that differential protein expression as a consequence of gene deletion is a mechanism for generating minor H antigens in humans.
Keywords: hematopoietic stem cell transplantation, graft-versus-host disease, cytotoxic T lymphocyte, UDP glycosyltransferase 2B family, cDNA expression cloning
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT)* from a HLA identical donor is curative for a variety of hematologic malignancies but GVHD mediated by T cells transplanted with the stem cell graft remains a major complication (1). Minor histocompatibility antigens (minor H antigens) are the targets of GVHD and consist of HLA bound peptides, which are derived from cellular proteins that differ in amino acid sequence between donor and recipient due to polymorphisms in the genome (2). Because minor H antigens are also targets of the graft-versus-leukemia (GVL) effect, defining the polymorphisms in the genome that are responsible for generating minor H antigens and the tissues in which individual determinants are expressed may lead to novel strategies to reduce GVHD and/or augment GVL activity after HSCT (3).
The structure, genetics, and tissue distribution have been characterized for only a few human minor H antigens, although extrapolation from studies in mice suggests a very large number of minor H antigens are likely to be important in human transplantation (4). T cell clones that are specific for minor H antigens have been derived from allogeneic HSCT recipients and provide reagents for discovery of the genes that encode these antigens using either peptide elution and mass spectrometry (5), cDNA expression cloning (6), or genetic linkage analysis (7). Human minor H antigens that have been molecularly characterized include those encoded by the Y chromosome genes SMCY, UTY, DFFRY, and DBY, which are polymorphic with homologues on the X chromosome and are recognized by donor T cells after sex mismatched HSCT (5, 8–13). Additionally, four minor H antigens encoded by autosomal genes have been described and designated HA-1, HA-2, HA-8, and HB-1 (6, 14–17). The immunogenicity of all of the previously discovered human minor H antigens results from polymorphism in the coding sequences of homologous donor and recipient genes that give rise to unique peptides, which are displayed at the surface of recipient cells bound to class I MHC and recognized by donor T cells.
Here, we have used cDNA expression cloning to identify a new autosomal human minor H antigen encoded by the UDP glycosyltransferase 2 family, polypeptide B17 (UGT2B17) gene. This minor H antigen is presented by HLA-A*2902 and recognized by CD8+ T cells that were isolated from a patient with GVHD involving the gastrointestinal tract, liver, and skin. In contrast to previously defined human minor H antigens, the immunogenicity of UGT2B17 was not due to a polymorphism in the coding sequence between homologous donor and recipient genes but resulted from absent expression of UGT2B17 in donor cells due to a homozygous deletion of the UGT2B17 gene. Analysis of 36 unrelated normal donors demonstrated that four (11%) were also deficient in UGT2B17 demonstrating that this polymorphism is not uncommon in the human population. These results provide the first evidence in humans that minor H antigens can result from discordance in gene expression as a consequence of homozygous deletion of individual members of multigene families.
Materials and Methods
Cell Culture.
The CD8+ clone PL8 CTL was isolated from a blood sample obtained post-transplant from an allogeneic HLA identical HSCT recipient as described previously (18). HLA typing was performed by standard serologic methods and revealed that both the recipient and his sibling donor were HLA-A2, A29, B44, Cw5, DR3, DR11. PBMCs were obtained from the transplant recipient after engraftment at the onset of GVHD and were stimulated in vitro with aliquots of γ-irradiated PBMCs that had been obtained from the recipient pretransplant and cryopreserved. After three weekly stimulations, the PL8 CTL clone was isolated from the polyclonal T cell culture by limiting dilution cloning. The PL8 CTL clone was propagated by stimulation every 14 d with 30 ng/ml of OKT3 monoclonal antibody (Ortho Biotech), using unrelated allogeneic γ irradiated (35Gy) PBMCs and γ irradiated (50 Gy) Epstein Barr virus transformed lymphoblastoid cells (B-LCL) as feeder cells. The culture media consisted of RPMI-HEPES (GIBCO BRL) containing 10% pooled, heat-inactivated human serum, and IL-2 (50 U/ml; Chiron Corp.). The T cell clone was used in cytotoxicity and epitope reconstitution assays either 12–16 d after stimulation (19), or 1 d after thawing a frozen aliquot. B-LCL were maintained in RPMI-HEPES with 10% FBS (ATLAS Biologicals). COS cells were obtained from the American Type Culture Collection and maintained in DMEM (GIBCO BRL) with 10% FBS.
cDNA Library Construction.
A cDNA library consisting of 40,000 clones was constructed using the Superscript Choice System (GIBCO BRL). Total RNA was isolated from B-LCL and poly (A)+ mRNA was prepared by Straight A's mRNA Isolation (Novagen). mRNA was converted into cDNA using an oligo-dT primer that contains a NotI site at its 3′ end and Thermoscript reverse transcriptase (GIBCO BRL). The cDNA was ligated to BstXI adaptors, digested with NotI, size fractionated by column chromotography, and ligated into BstXI and NotI sites of the pEAK10 expression vector (EdgeBioSystems). E. coli DH10B (GIBCO BRL) were electroporated with the recombinant plasmids and transformed clones were selected with ampicillin. cDNA pools, each comprising ∼50 bacterial clones were amplified in liquid culture for 20 h, following which plasmid DNA was extracted using the MultiScreen filtration system (Millipore) and stored in 96-well plates.
Transfection of COS Cells and CTL Stimulation Assay.
The cDNA expression cloning methodology for identifying genes encoding antigens recognized by CD8+ CTL was performed as described by Boon et al. with modifications (20). In brief, 5 × 103 COS cells were plated in individual wells of 96-well plates, cultured for 24 h, and then transfected with 80 ng of plasmid DNA from each pool of the cDNA library and 40 ng of a plasmid encoding HLA-A*2902 using the FuGENE transfection reagent (Roche). 2 × 104 CD8+ CTL were added to each well of COS cells 48 h after transfection, and after a further 24 h of coculture at 37°C, IFN-γ was measured in the supernatant by ELISA (Endogen).
cDNA Constructs of UGT2B17.
Constructs containing either full-length or a defined portion of the UGT2B17 gene were generated from the 4A2 cDNA by PCR and cloned into pEAK10. Sense and antisense primers contained the recognition sequence (underlined in the primer sequences) for EcoRV and NotI, respectively, to facilitate cloning of the PCR product into pEAK10. The following sequences were used: full-length construct–5′-ATCGGATATCATGTCTCTGAAATGGATGTCAGT-3′ (primer A) and 5′-ATCGGCGGCCGCCTAATCCCTTTTCTTCTTCTTTCCT-3′; construct I–primer A and 5′-ATCGGCGGCCGCTTAAACGGCATCTGCCAGAAGGA-3′; construct II–5′-ATCGGATATCATGGTCCTTCTGGCAGATGCCGTTAA-3′ (primer B) and 5′-ATCGGCGGCCGCTTAGATCATCGACCCCAGAGAAAAC-3′; construct III–primer B and 5′-ATCGGCGGCCGCTTAGTACAGAAAGGGTATGTTAAGTAGC-3′. An additional construct UGT493–561 was generated using primer B and 5′-ATCGGCGGCCGCTTACAGAAAGGGTATGTTAAGTAGCTC-3′.
Transfection of B-LCL with UGT2B17 cDNA.
Donor B-LCL (5 × 106) were electroporated (220 V, 500 μFD) in 200 μl K-PBS with 15 μg of plasmid DNA encoding the full-length UGT2B17 cDNA and with truncated versions of UGT2B17. The transfected B-LCL were placed in culture media, selected with 0.6 μg/ml of puromycin (EdgeBioSystems) beginning 48 h after transfection, and used as targets in cytotoxicity assays 3 d after selection.
Chromium Release Assay.
B-LCL were labeled for 1 h with 51Cr, washed twice, dispensed at 2 × 103 cells/well into triplicate cultures in 96-well plates, and incubated for 4 h at 37°C with PL8 CTL at various E:T ratios. In peptide recognition assays, B-LCL were preincubated with various concentrations of peptide for 2 h at 37°C before labeling with 51Cr. These cells were then washed and aliquoted with PL8 CTL as described above.
Peptides.
Peptides were synthesized with a free COOH terminus using standard FMOC chemistry. The synthetic peptides were dissolved in dimethyl sulfoxide and stored at −20°C.
Northern Blot Analysis.
Northern blot analysis was performed by standard methodology. In brief, 20 μg of total RNA was extracted from B-LCL using RNeasy Minikit (QIAGEN), electrophoresed on a 1.2% agarose gel, and transferred to a Nytran SuperCharge Membrane (Schleicher & Schuell). The membrane was hybridized for 2 h at 65°C in ExpressHyb Solution (CLONTECH Laboratories, Inc.) with salmon sperm DNA (GIBCO BRL) and for 16 h at 68°C in the solution with a 32P-labeled probe from corresponding to nt 400–751 of the UGT2B17 gene. After washing, the membrane was exposed for 2 d at room temperature on Storage Phosphor Screen (Molecular Dynamics). A glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probe (CLONTECH Laboratories, Inc.) was used as a control for RNA integrity and loading. Expression of UGT2B17 in various tissues was analyzed using a dot-blot array of poly (A)+ RNA (MTE; CLONTECH Laboratories, Inc.).
Sequence-specific Primer (SSP)-PCR for UGT2B17 in Genomic DNA and cDNA.
The sense and antisense sequence-specific primers for PCR to detect exon 1 of the UGT2B17 gene were (exon 1a) 5′-TGTTGGGAATATTCTGACTATAA-3′ and 5′-CCCACTTCTTCAGATCATATGC-3′: (exon 1b) 5′-AAATGACAGAAAGAAACAA-3′ and 5′-GCATCTTCACAGAGCTTATAT-3′. The sequence specific primers for exon 6 were 5′-GAATTCATCATGATCAACCG-3′ and 5′-ACAGGCAACATTTTGTGATC-3′. The primers for the region 5′ to UGT2B17 were 5′-GGCAGTATCTTGCCAATGT-3′ and 5′-AGACTCCAAGTGCCAGTT-3′. 38 cycles of amplification were performed on 0.5 μl genomic DNA prepared from B-LCL using QIAamp DNA Blood Minikit (QIAGEN), or cDNA synthesized from total RNA using Superscript II (GIBCO BRL). Each reaction contained 0.4 μl of Advantage 2 Polymerase Mix (CLONTECH Laboratories, Inc.), 0.2 mmol/L of each of the four deoxyribonucleotides, 10 pmol of each primer, and PCR buffer in a volume of 20 μl. Each cycle consisted of denaturation (94°C; 30 s), annealing (68°C; 20 s), and elongation (72°C; 30 s). 10 μl of the PCR product was analyzed by electrophoresis on a 1.5% agarose gel. The primers for PCR to detect the GAPDH gene were 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ and 5′-CATGTGGGCCATGAGGTCCACCAC-3′ and 22 cycles of amplification were used. Expression of UGT2B17 in various tissues was analyzed using a panel of normalized first strand cDNA prepared from poly (A)+ RNA (MTC; CLONTECH Laboratories, Inc.) as a template for PCR with the sequence specific primers for exon 1 of UGT2B17 and with the primers for GAPDH described above.
Generation of Dendritic Cells and B Cells.
Immature dendritic cells (DCs) were generated from cryopreserved PBMC by culturing for 6 d in AIM-V (GIBCO BRL) with 800 U/ml of recombinant human GM-CSF (Immunex) and 500 U/ml of IL-4 (R&D Systems) as described (21). Mature DCs were generated from immature DCs by culturing for 2 additional days in AIM-V with recombinant human GM-CSF (800 U/ml), IL-4 (500 U/ml), IL-6 (1,000 U/ml; R&D Systems), IL-1β (10 ng/ml; R&D Systems), tumor necrosis factor α (10 ng/ml; R&D Systems), and prostaglandin E2 (1 μg/ml; Sigma-Aldrich) as described previously (22). Activated B cells were generated by culturing PBMCs for 15 d on γ-irradiated (96 Gy) human CD40L-transfected NIH3T3 cell (gift from J. Schultze, Dana-Farber Cancer Institute, Boston, MA) in IMDM (GIBCO BRL) containing IL-4 (100 U/ml) as described previously (23). First strand cDNA was prepared from total RNA extracted from each subset of APCs using RNeasy Minikit and reverse transcribed using Superscript II. PCR to detect UGT2B17 or GAPDH in cDNA was performed as described above. For other subsets of peripheral blood, a cDNA panel (Human blood fractions MTC; CLONTECH Laboratories, Inc.) was used as a template for PCR.
Results
Isolation of a cDNA Encoding the Minor H Antigen Recognized by PL8 CTL.
The PL8 CTL clone was isolated from blood obtained from an allogeneic HLA identical HSCT recipient at the onset of acute GVHD involving the gastrointestinal tract, liver, and skin. PL8 CTL lysed recipient B-LCL and 21 of 24 B-LCL lines from unrelated individuals that shared HLA-A29 with the recipient, but failed to lyse B-LCL from the donor and 10 unrelated individuals that shared class I HLA molecules other than HLA-A29 (unpublished data). This data indicated that PL8 CTL recognizes a minor H antigen that is frequent in the population (∼88%) and presented by HLA-A29.
To identify the minor H antigen recognized by PL8 CTL, we constructed a cDNA library from B-LCL that expressed the antigen and cotransfected COS cells with plasmid pools containing ∼50 cDNAs from the library, and with a plasmid encoding HLA-A*2902. The transfected COS cells were cocultured with PL8 CTL and the production of IFN-γ measured in the supernatant. COS cells transfected with two of 384 cDNA pools stimulated IFN-γ production (Fig. 1 A). These two positive pools were then subcloned, and individual plasmids were rescreened by COS cell transfection. Two plasmids containing inserts of 2,896 bp and 2,958 bp, respectively, induced HLA-A29–dependent IFN-γ production by PL8 CTL after transfection into COS cells (Fig. 1 B). The cDNA inserts of each plasmid were sequenced and a search of DNA sequence databases revealed that both cDNAs had greater than 99% identity with the UGT2B17 gene (GI: 4507820).
Figure 1.
Identification of a cDNA encoding the minor H antigen recognized by PL8 CTL. (A) Pools of a cDNA library containing ∼50 cDNA clones were transiently transfected into COS cells in individual wells of a 96-well plate together with a plasmid encoding HLA-A*2902. COS transfectants were cocultured with PL8 CTL and IFN-γ production was measured in supernatant collected after 24 h. Two pools (indicated by arrows) stimulated IFN-γ production by PL8 CTL. (B) Isolation of a cDNA that stimulates IFN-γ production by PL8 CTL. The two pools of cDNA were subcloned in E. coli, and individual plasmids were transfected into COS cells alone or with HLA-A*2902. Two clones, designated 2H9 and 4A2 stimulated IFN-γ production by PL8 CTL only when transfected with HLA-A*2902. Data is shown as the mean of triplicate determinations.
Identification of the Antigenic Epitope Encoded by UGT2B17.
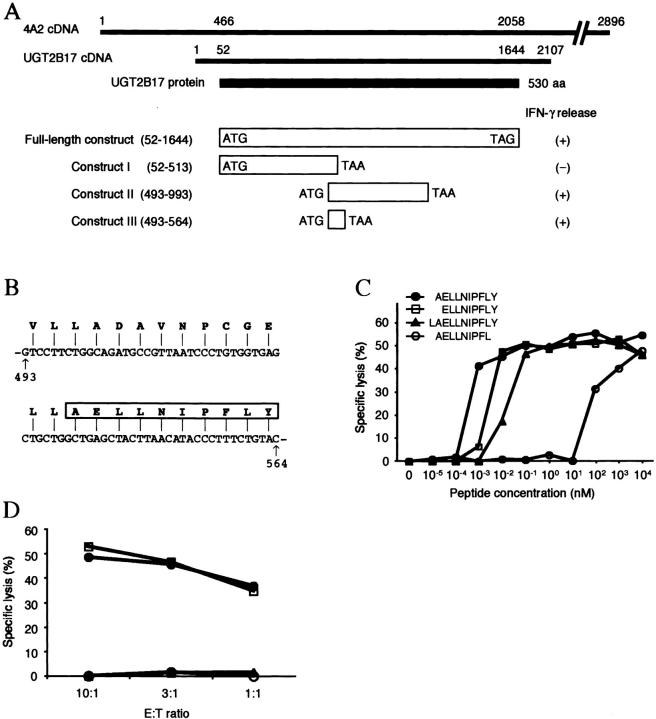
The UGT2B17 sequence that encoded the epitope recognized by PL8 CTL was localized by generating truncated UGT2B17 constructs containing selected portions of the gene, transfecting each construct into COS cells with the plasmid encoding HLA-A29, and screening the COS transfectants for recognition by PL8 CTL. Construct I included nt 52–513 of the primary open reading frame (ORF) of UGT2B17 and was generated with a TAA codon at the 3′ end to terminate transcription. Constructs II and III included nt 493–993 and nt 493–564 of UGT2B17, respectively, and were engineered to have ATG and TAA codons at the 5′ and 3′ ends of the primary ORF, respectively (Fig. 2 A). COS cells transfected with either construct II or III, but not with construct I, stimulated IFN-γ production by PL8 CTL demonstrating that the antigenic epitope recognized was derived from the 24 amino acid polypeptide encoded by nt 493–564 of UGT2B17 (Fig. 2 A).
Figure 2.
Identification of the antigenic epitope encoded by UGT2B17. (A) Location of the epitope-containing region by transfection of truncated UGT2B17 constructs. Alignment of 4A2 cDNA with the UGT2B17 cDNA (NM_001077), the nucleotide numbering of the constructs used for transfection corresponds to that provided for the UGT2B17 GenBank sequence. Constructs I, II, and III contained the indicated UGT2B17 sequences and were transfected with HLA-A*2902 into COS cells. IFN-γ production by PL8 CTL was measured after coculture with COS transfectants and is indicated by (+) or (−) in the right-hand column. (B) UGT2B17 493–564 encodes a decamer peptide with anchor residues for HLA-A*2902. The amino acid sequence encoded by UGT2B17 493–564 is shown and a putative epitope for PL8 CTL is boxed. (C) CTL recognition of donor B-LCL cultured with synthetic peptides corresponding to UGT2B17 sequences. The concentration of peptide that elicited half-maximal lysis was ∼0.7 pM for AELLNIPFLY, ∼6 pM for ELLNIPFLY, ∼50 pM for LAELLNIPFLY, and ∼80 nM for AELLNIPFL. Specific lysis is shown as the mean of triplicate cultures at an E:T ratio of 5:1. (D) Transfection of minigene constructs define a requirement for tyrosine at the COOH terminus of the naturally processed UGT2B17 epitope. Donor B-LCL were transfected by electroporation with either UGT2B17 493–564 that encodes the 24 amino acid polypeptide described in B or UGT2B17 493–561 that encodes 23 amino acids with the COOH-terminal tyrosine deleted. Transfected B-LCL were selected for 3 d with puromycin (0.6 μg/ml). The lysis of UGT2B17 493–564-transfected donor B-LCL (solid circles), UGT2B17 493–561-transfected donor B-LCL (triangles), recipient B-LCL (squares), and untransfected donor B-LCL (open circles) is shown as the mean of triplicate cultures at various E:T ratios.
Peptides that bind to HLA-A*2902 frequently contain a glutamic acid at P2 and tyrosine at the COOH terminus as preferred anchor residues (24). We examined the amino acid sequence encoded by nt 493–564 of UGT2B17 for candidate peptides that might bind to HLA-A*2902 and identified the 10 mer peptide, AELLNIPFLY, which contained the appropriate anchor residues (Fig. 2 B). Donor B-LCL were pulsed with various concentrations of synthetic AELLNIPFLY and tested as targets for PL8 CTL in a cytotoxicity assay. PL8 CTL lysed donor B-LCL pulsed with AELLNIPFLY and half-maximal lysis occurred at peptide concentrations of 100 fM–1 pM (Fig. 2 C). Synthetic peptides which have 1 amino acid deleted (ELLNIPFLY) or added (LAELLNIPFLY) to the NH2 terminus, respectively, also sensitized donor B-LCL for lysis by PL8 CTL, but at a one to two log higher concentration. We also synthesized the nonamer peptide AELLNIPFL with the COOH-terminal tyrosine deleted because the computer program PAProC (www.paproc.de) that identifies likely COOH-terminal proteosomal cleavage sites predicted AELLNIPFL, but not AELLNIPFLY, as a cleavage product of UGT2B17. Leucine also is a preferred residue at the COOH terminus for binding to HLA A*2902 (24). The synthetic AELLNIPFL sensitized donor B-LCL for recognition by PL8 CTL but at a 5 log higher concentration than AELLNIPFLY (Fig. 2 C). Therefore, to further define the COOH terminus of the naturally processed epitope, we transfected donor B-LCL with construct III (UGT2B17493–564) encoding a 24 amino acid peptide ending with tyrosine at the COOH terminus, or with a construct (UGT2B17493–561) that encoded a 23 amino acid polypeptide with the COOH-terminal tyrosine deleted, and analyzed recognition by PL8 CTL. The donor B-LCL transfected with UGT2B17493–564 were lysed as well as recipient B-LCL, but donor B-LCL transfected with UGT2B17493–561 were not recognized at all by PL8 CTL (Fig. 2 D). These results indicate that the naturally processed peptide contains tyrosine and not leucine at the COOH terminus and suggest the optimal peptide for PL8 CTL is AELLNIPFLY.
The Epitope of UGT2B17 Differs in Amino Acid Sequence with All Other UGT2B Family Members.
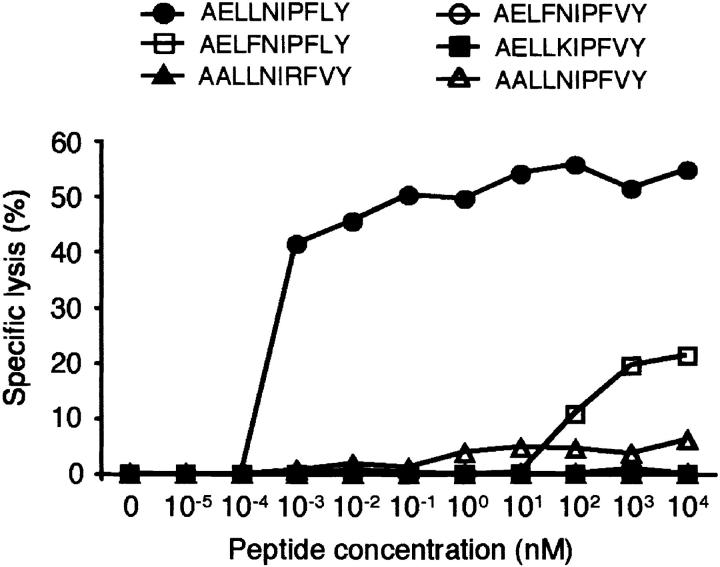
UGT2B17 is a member of the UGT2B multigene family, which presently consists of 7 genes and 5 pseudogenes and maps to chromosome 4q13. We compared the sequence of the recipient UGT2B17 cDNA with the canonical sequences of other UGT2B family members obtained from the GenBank DNA sequence database (25–31). Although there was a high degree of homology between UGT2B17 and other family members, all family members had one or more amino acid differences in the peptide sequence corresponding to the epitope identified in UGT2B17 (Table I). The most closely related sequence in this region was that encoded by UGT2B15, which contained only a single amino acid difference, a phenylalanine in place of leucine at P4. The synthetic peptide (AELFNIPFLY) corresponding to the UGT2B15 sequence failed to efficiently sensitize donor B-LCL for recognition by PL8 CTL, although a low level of lysis was observed at μM concentrations of peptide (Fig. 3) . The amino acid sequences of UGT2B10 and UGT2B7 in the epitope region were identical with each other but differed from UGT2B17 at P4 with a phenylalanine instead of leucine and at P9 with a valine instead of leucine. The sequences encoded by UGT2B11, UGT2B4, and UGT2B28 also contained the valine substitution at P9 and either an alanine in place of glutamic acid at P2 (UGT2B11 and UGT2B28), an arginine in place of proline at P7 (UGT2B11), or a lysine in place of asparagine at P5 (UGT2B4). Synthetic peptides corresponding to the sequence of each of these other UGT2B17 family members also failed to sensitize donor B-LCL for recognition by PL8 CTL (Fig. 3). Taken together, these data identify an important role for leucine at P4, asparagine at P5, and leucine at P9 in the UGT2B17 epitope for recognition by PL8 CTL, either directly as T cell receptor contact residues or as secondary anchor residues for binding, or indirectly by altering the conformation of T cell receptor contact or MHC binding residues in the epitope (32).
Table I.
Homology between UGT2B17 and Other UGT2B Family Members
| Members | GI numbers | Homologya | Peptidesb |
|---|---|---|---|
| UGT2B15 | 475758 | 97.6 (1555/1593) | AELFNIPFLY |
| UGT2B11 | 4507822 | 85.9 (1368/1593) | AALLNIRFVY |
| UGT2B10 | 4507816 | 85.2 (1358/1593) | AELFNIPFVY |
| UGT2B7 | 4507824 | 85.6 (1363/1593) | AELFNIPFVY |
| UGT2B4 | 10863940 | 85.2 (1357/1593) | AELLKIPFVY |
| UGT2B28 | 16596679 | 84.2 (1342/1593) | AALLNIPFVY |
| UGT2B29P c | 6979424 | 82.8 (565/682) | Not available |
| UGT2B27P | 6979423 | 83.9 (559/666) | Not available |
| UGT2B26P | 6979422 | 82.9 (539/650) | Not available |
| UGT2B25P | 6979421 | 82.6 (598/724) | Not available |
| UGT2B24P | 6979420 | 82.3 (596/724) | Not available |
Homology between recipient UGT2B17 cDNA and canonical sequences of each UGT2B family member is presented as the percentage and actual number (parentheses) of identical nucleotides.
Amino acid sequence of the peptides encoded by other family members and corresponding to AELLNIPFLY encoded by UGT2B17. Amino acids that differ from AELLNIPFLY are shown in bold.
P indicates a pseudogene.
Figure 3.
Donor cells pulsed with synthetic peptides corresponding to homologous sequences of other UGT2B family members are not recognized by PL8 CTL. Donor B-LCL were pulsed with various concentrations of the following synthetic peptides: AELLNIPFLY (encoded by UGT2B17), AELFNIPFLY (UGT2B15), AALLNIRFVY (UGT2B11), AELFNIPFVY (UGT2B10 and UGT2B7), AELLKIPFVY (UGT2B4), or AALLNIPFVY (UGT2B28), and tested as targets for PL8 CTL in a cytotoxicity assay. Specific lysis is shown as the mean of triplicate cultures at an E:T ratio of 5:1.
Immunogenicity of UGT2B17 Is Due to Differential Transcription.
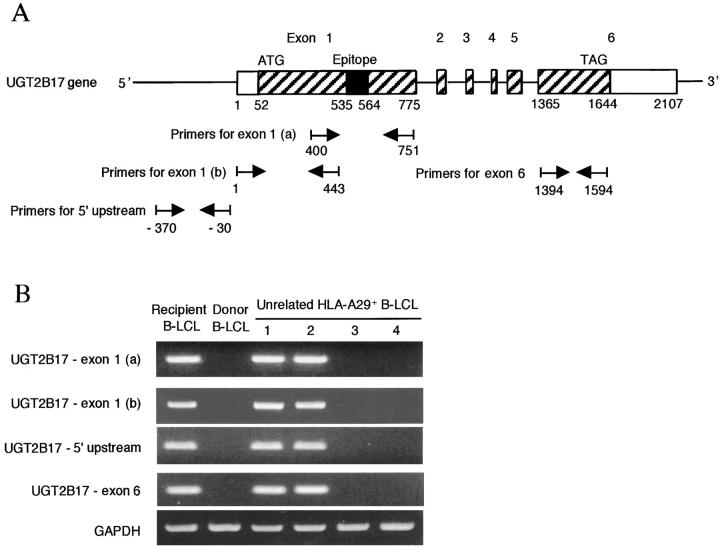
All previously defined human minor H antigens contain amino acid substitutions that result from nucleotide polymorphism between homologous recipient and donor genes. We initially presumed that the sequence of the donor and recipient UGT2B17 genes would contain a polymorphism in the epitope region. However, a search of the human genome database of single nucleotide polymorphisms failed to uncover any previously described polymorphisms in this region of UGT2B17 encoding the epitope. Therefore, we attempted to isolate UGT2B17 cDNA from donor cells for sequencing to identify a possible polymorphic UGT2B17 allele. However, a PCR product was not obtained from donor cells despite trying several primer pairs designed specifically to amplify UGT2B17 (unpublished data). We next determined if the levels of UGT2B17 gene expression were comparable in total RNA prepared from recipient and donor cells using a probe that overlapped the exon 1 sequence encoding the antigenic peptide and was polymorphic with other UGT family members to minimize crosshybridization. UGT2B17 transcripts were easily detected in recipient B-LCL and in unrelated HLA-A29+ B-LCL that were lysed by PL8 CTL but not in donor B-LCL or unrelated HLA-A29+ B-LCL that were not lysed by PL8 CTL (Fig. 4, A and B) . To determine if lack of recognition by PL8 CTL was solely attributable to the absence of UGT2B17 gene expression, the full-length UGT2B17 cDNA in a plasmid containing the puromycin resistance gene was transfected into donor B-LCL and cells were selected in puromycin. Donor B-LCL transfected with the UGT2B17 cDNA were lysed equally well as recipient B-LCL, demonstrating the lack of recognition of donor cells by PL8 CTL resulted entirely from differential transcription of this gene in recipient versus donor cells (Fig. 4 C).
Figure 4.
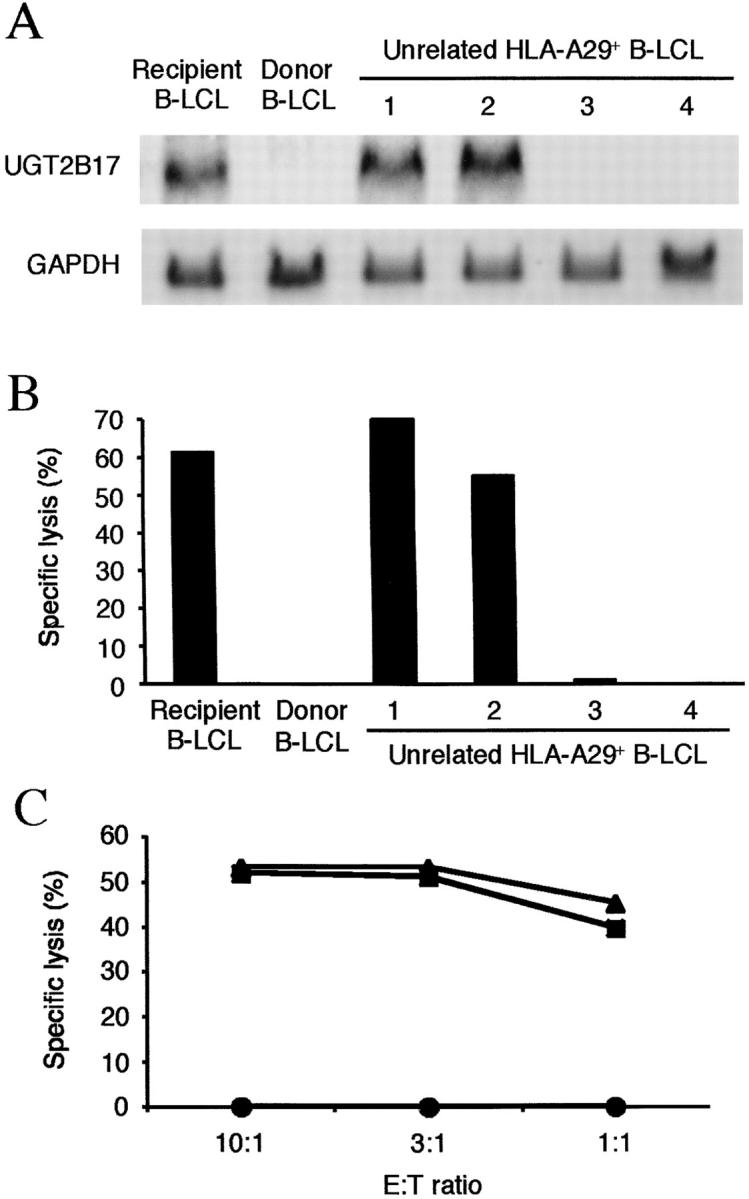
UGT2B17 mRNA is transcribed in minor H antigen positive but not minor H antigen negative cells. (A) Northern blot analysis of total RNA from recipient and donor B-LCL, and from HLA-A29+ B-LCLs from 4 unrelated donors (designated 1–4). The blot was hybridized with a 32P-labeled UGT2B17 probe for 16 h at 68°C. Detection of GAPDH mRNA was performed as a control. (B) Cytotoxicity assay for B-LCL by PL8 CTL. The B-LCL from unrelated donors (1–4) are the same lines used for analysis of gene expression in Fig. 3 A. Specific lysis is shown as the mean of triplicate cultures at an E:T ratio of 5:1. (C) Transfection of UGT2B17 cDNA into donor B-LCL restores recognition by PL8 CTL. Donor B-LCL were transfected by electroporation with the full-length construct of UGT2B17, selected for 3 d with puromycin (0.6 μg/ml), and assayed as targets for PL8 CTL. The lysis of UGT2B17-transfected donor B-LCL (triangles), untransfected donor B-LCL (circles), and recipient B-LCL (squares) is shown at various E:T ratios.
Lack of Expression of UGT2B17 in Donor Cells Is Due to a Gene Deletion.
Several possibilities could account for the absence of UGT2B17 transcripts in donor cells, including a homozygous gene deletion, polymorphism in promoter sequences, or a polymorphism in the 3′ portion of the gene that resulted in termination of transcription upstream of the region encoding the epitope and detected by our probe. To determine if all or a portion of the UGT2B17 gene was deleted in donor cells, we analyzed genomic DNA prepared from donor B-LCL using SSP-PCR with primer pairs to detect exon 1 and exon 6 sequences, which represent the first and last exons of the UGT2B17 gene (33), and a primer pair that recognizes sequences 5′ to the initiation codon for UGT2B17. PCR products for exon 1, exon 6, and the region immediately 5′ to exon 1 were not detected by amplification of genomic DNA from donor B-LCL or minor H antigen–negative B-LCL from unrelated individuals but were obtained by amplification of genomic DNA from recipient B-LCL and unrelated minor H antigen–positive B-LCL (Fig. 5) . These data indicated that the failure to detect a UGT2B17 transcript in donor cells was a result of homozygous deletion of a large portion of the UGT2B17 gene including the ATG initiation codon, exon 1, and at least a portion of exon 6. Analysis of B-LCL from 36 randomly selected normal individuals by SSP-PCR identified 4 (11%) that similarly lacked both exon 1 and exon 6 of UGT2B17 demonstrating that homozygous deletion of the UGT2B17 gene was not restricted to this particular stem cell donor but occurs as a variant in the human population.
Figure 5.
The UGT2B17 gene is deleted in donor cells and HLA-A*2902 positive cells from unrelated individuals that are not recognized by PL8 CTL. (A) UGT2B17 consists of six exons encoded over 27 Kb of DNA on chromosome 4. Primer pairs used for PCR to detect exon 1 and exon 6 sequences were selected such that at least one primer of each pair contained nucleotides that were mismatched with all other known UGT family members. The primer pair for the region immediately 5′ to the UGT2B17 start site was selected to amplify nt −370 to −30. (B) SSP-PCR for exon 1 and exon 6 sequences of UGT2B17 and PCR for 5′ sequences upstream of UGT2B17 on genomic DNA prepared from B-LCL. PCR products for exon 1, exon 6 and the 5′ region upstream of UGT2B17 were detected in the recipient B-LCL and unrelated HLA-A29+ B-LCL that were recognized by PL8 CTL. These PCR products were sequenced and found to be identical to UGT2B17 (unpublished data). No PCR products were detected in donor B-LCL or unrelated HLA-A29+ B-LCL that were not lysed by PL8 CTL. The B-LCL from unrelated donors (indicated as 1–4) are the same lines used for analysis of gene expression and cytotoxicity in Fig. 4, A and B. PCR for GAPDH was performed as a control.
Expression of UGT2B17 in Tissues and APCs.
PL8 CTL were isolated from an HSCT recipient with GVHD involving the liver, gastrointestinal tract, and skin. To determine if UGT2B17 was expressed in tissues that were targets of GVHD in this patient, first-strand cDNA prepared from different human tissues pooled from multiple donors was evaluated for sequences corresponding to exon 1 using SSP-PCR. Expression of UGT2B17 was easily detected in liver, colon, small intestine, and pancreas, but absent or barely detectable in other tissues (Fig. 6 A). Similar data was obtained using hybridization of a dot-blot array of poly (A)+ RNA from human tissues. A dominant signal for UGT2B17 mRNA was found in liver, colon, and pituitary gland, with a weaker signal detected in small intestine and spleen (unpublished data).
Figure 6.
Tissue expression of UGT2B17. (A) Expression of UGT2B17 in tissues by RT-PCR. First-strand cDNA prepared from a series of different human tissues pooled from multiple donors was analyzed by SSP-PCR for UGT2B17 using the exon 1(a) primer pair. The highest level of expression of UGT2B17 was observed in cDNA from liver and colon. Detectable bands were also obtained from lung, skeletal muscle, pancreas, spleen, thymus, prostate, testis, ovary, and small intestine. No band was obtained from heart, brain, placenta, and kidney. PCR for GAPDH was performed as a control. (B) Expression of UGT2B17 in subsets of PBMC. First-strand cDNA was isolated from PBMCs or from subsets of PBMCs and analyzed by SSP-PCR using the exon 1(a) primer pair. A PCR product of the correct size was obtained from PBMCs, immature and mature DCs, resting and activated CD19+ B cells and B-LCL. No bands were obtained from resting or activated CD4+ and CD8+ T cells, or from CD14+ cells (monocytes). PCR for GAPDH was performed on all samples as a control. (C) APCs stimulate IFN-γ production by PL8 CTL. PL8 CTL (5 × 103) were cocultured with B-LCL, mature DCs, and activated B cells (0.625 × 103) in triplicate wells of a 96-well plate for 24 h at 37°C. IFN-γ production was measured in the supernatant by ELISA. The mean and SD of triplicate measurements are shown.
The expression profile of UGT2B17 was compatible with it being a target for T cells mediating GVHD in this patient. However, studies in murine models have suggested that expression of minor H antigens by APC is required for initiating CD8+ T cell responses in GVHD (34). Thus, we investigated the expression of UGT2B17 in subsets of hematopoietic cells prepared from cryopreserved pretransplant recipient blood. UGT2B17 was expressed in immature and mature DC, and resting and activated B cells, but not in resting or activated T cells (Fig. 6 B). To determine whether the level of expression was sufficient for mature DCs or activated B cells to present the minor H antigen encoded by UGT2B17 to PL8 CTL, we measured IFN-γ production after coculture of CTL with these APCs. Both activated B cells and mature DCs stimulated IFN-γ production by PL8 CTL (Fig. 6 C). These data are consistent with participation of APC in the induction of T cell responses to minor H antigens that are expressed by epithelial cells and serve as targets for GVHD.
Discussion
T cell responses to minor H antigens have been implicated in both GVHD and GVL responses after allogeneic HLA-identical HSCT, and a more precise understanding of the genetics and tissue expression of these determinants could potentially lead to improvements in transplant outcome. However, the polymorphisms in the genome that encode minor H antigens and the basis for antigenicity have been defined for only a few human determinants. In all cases described so far, human minor H antigens have been derived from genes that are expressed in both donor and recipient cells but have polymorphism in their coding sequences. The resulting changes in amino acid sequence may alter the processing of recipient and donor peptides (17), the ability of the peptide to bind MHC (5, 12), or the ability of T cells to recognize the MHC/peptide complex (6). Here, we have identified a new human minor H antigen encoded by the UGT2B17 gene and demonstrated that differential expression in donor and recipient cells as a consequence of a homozygous gene deletion in the donor is responsible for immunogenicity. This is the first example in humans of differential expression of a protein as the basis for generating a minor H antigen. The murine minor H antigens, H60 and H28, that result in rejection of BALB/B skin grafts by C57Bl/6 recipients, are also immunogenic as a result of differential transcription, although the genetic basis for absent expression of these genes in some strains of mice remains to be elucidated (4, 35).
The optimal peptide of UGT2B17 for recognition by PL8 CTL was AELLNIPFLY, which contains preferred amino acid residues at both of the anchor positions for binding to HLA-A*2902, and sensitized donor B-LCL for lysis at pM concentrations. Peptides with one amino acid deleted or added to the NH2 terminus of AELLNIPFLY, respectively, or with one amino acid deleted from the COOH terminus also sensitized donor B-LCL for recognition by PL8 CTL, although a much higher concentration of peptide was required. Donor B-LCL that were transfected with UGT2B17493–564 encoding AELLNIPFLY but not UGT2B17493–561 encoding AELLNIPFL, were recognized by PL8 CTL suggesting tyrosine is the anchor residue at the COOH terminus of the naturally processed UGT2B17 epitope presented by HLA-A*2902. The elution of peptides from HLA-A29 molecules of recipient cells that reconstitute recognition by PL8 CTL will be required to conclusively define the NH2 terminus of the naturally processed peptide, since the synthetic peptides LAELLNIPFLY and ELLNIPFLY were both able to sensitize target cells for recognition by PL8 CTL, albeit at 1–2 log10 higher concentration than AELLNIPFLY.
The UDP glycosyl transferases are comprised of two subfamilies, UGT1 and UGT2 (36), and serve a major role in the conjugation and subsequent elimination of endogenous compounds including steroid hormones and bilirubin, and potentially toxic exogenous compounds (37). A single gene located on chromosome 2 encodes several UGT1 isoforms that arise by differential splicing of the encoded mRNA. Mutations of UGT1 family members have been described and are responsible for the Crigler-Najjar and Gilbert syndromes (38–40). Individual genes located on chromosome 4 encode the UGT2 family members, but clinical syndromes resulting from mutation or deletion of UGT2 members have not been described. The lack of UGT2B17 in the adult donor in this study was not clinically significant since this individual had a normal physical exam, complete blood count, and serum chemistry at the time of evaluation for donation of hematopoietic stem cells. UGT2B17 has been shown to catalyze the conjugation of the 17β-hydroxy position of dihydrotestosterone (DHT), testosterone, and androstane-3α17β-diol (3α-Diol), and glucuronidates androsterone (33, 41). Other UGT family members, such as UGT2B7 and UGT2B15 also conjugate DHT, testosterone, and 3α-Diol, and UGT2B7 can glucuronidate androsterone (25, 28, 42–44). Thus, redundancy of other UGT2B family members for these substrates may compensate for the deficiency of UGT2B17 in normal individuals and be responsible for the absence of a clinical phenotype associated with homozygous UGT2B17 deletion.
Despite the redundancy in substrate specificity, there is significant variation in the nucleotide and amino acid sequences of UGT2B17 compared with other UGT2B family members. The region of UGT2B17 that encodes the epitope recognized by PL8 CTL contains one or more unique amino acids compared with all other family members and synthetic peptides corresponding to the sequences of other family members failed to sensitize donor cells for recognition by PL8 CTL. We have analyzed other regions of UGT2B17 that also differ in amino acid sequence with all other family members using computer algorithms that identify peptides predicted to bind to class I HLA molecules based on preferred anchor and secondary anchor residues (45). Several peptides were identified that are predicted to bind to common HLA alleles such as HLA-A2 and -B44. Thus, studies in donor/recipient pairs that express these HLA molecules and are discordant for UGT2B17 gene expression are warranted to determine if UGT2B17 encodes additional minor H antigens.
Establishing a causative role for individual human minor H antigens in GVHD after allogeneic HSCT has proven difficult for several reasons. Until recently, few minor H antigens were molecularly characterized and reagents and methods to detect T cells of selected antigen specificities at tissue sites of GVHD were not available. Additionally, other factors such as the intensity of the conditioning regimen, polymorphism in genes that encode cytokines or cytokine receptors, the intensity of posttransplant immunosuppression, and the relative immunodominance of individual minor H antigens may influence the development of clinical GVHD even when disparity for defined minor H antigens is known to exist (46). Studies in an situ model of human skin GVHD have demonstrated that T cells specific for minor H antigens expressed by both epithelial cells and APC induced the histologic features of GVHD whereas T cells specific for minor H antigens that are selectively expressed in APC did not cause GVHD (47). The requirement that the target antigen be expressed in APC for the induction of GVHD by CD8+ CTL is consistent with prior results obtained in vivo in murine models (34). The patient from whom the PL8 CTL clone was isolated had acute GVHD involving the liver and gastrointestinal tract and we found UGT2B17 was highly and preferentially expressed in both of these target organs. An in situ model is not available to address the potential for PL8 CTL to induce GVHD in the liver or intestine, but based on the known function of UGT2B17, it seemed unlikely this gene would fulfill the requirement that it be expressed in APCs. Surprisingly, our data showed that both dendritic cells and activated B cells express sufficient levels of UGT2B17 to stimulate PL8 CTL in vitro. While these results are consistent with the principles suggested by murine studies and in situ models of human GVHD involving the skin, they do not definitively establish UGT2B17 as a target of GVHD. However, the development of the SSP-PCR assay used to demonstrate the homozygous deletion of UGT2B17 in the donor in this study will permit prospective genotyping of HSCT donors and recipients and analysis of the occurrence of GVHD in UGT2B17 positive recipients who undergo HSCT from donors that lack this gene.
The demonstration in this study that differential expression of proteins in donor and recipient cells provides a mechanism for generating minor H antigens in humans has potentially broad implications for understanding T cell responses that mediate GVHD after allogeneic HSCT. Deficiencies of members of other enzyme families have been described in humans. These include deletion or lack of expression of the cytochrome P450 (CYP) family genes CYP2A6*4, CYP2D6*5, CYP2D6*8, and CYP2C19*4, which metabolize foreign chemicals as well as endogenous steroids (48–51), and deletion of the glutathione S-transferase (GST) T1 and GSTM1 genes, which belong to the GST gene family that detoxify mutagenic hydrophobic and electrophilic compounds (52, 53). Deletions of GSTT1 and GSTM1 are especially common, occurring in 38 and 50% of individuals, respectively, and these proteins share only 55% amino acid identity. Both GSTT1 and GSTM1 are expressed in the gastrointestinal tract and liver and could potentially be targets of GVHD involving these organs after HSCT from a donor with a deficiency of one of these enzymes into a recipient who expresses the protein (54). Thus, the novel mechanism responsible for antigenicity of UGT2B17 may apply to other minor H antigens involved in allogeneic reactions after HSCT.
Acknowledgments
We would like to thank Drs. Marc A. Gavin, Tomoki Naoe, Nobuhiko Emi, and Yoshiki Akatsuka for helpful discussion and Wendy Hilliker for assistance in preparation of the manuscript.
This research was supported by a grant from the National Institutes of Health CA 18029 (to S.R. Riddell) and by the Damon Runyon Walter Winchell Clinical Scholar Program (to E.H. Warren).
M. Murata is on leave from Nagoya University Graduate School of Medicine, Nagoya 466 8550, Japan.
Footnotes
Abbreviations used in this paper: B-LCL, Epstein Barr virus transformed lymphoblastoid cells; CYP, cytochrome P450; DC, dendritic cells; GST, glutathione S-transferase; HSCT, hematopoietic stem cell transplantation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GVL, graft-versus-leukemia; minor H antigens, minor histocompatibility antigens; SSP-PCR, sequence specific primer-PCR; UGT2B17, UDP glycosyltransferase 2 family, polypeptide B17.
References
- 1.Storb, R., H.J. Deeg, J. Whitehead, F. Appelbaum, P. Beatty, W. Bensinger, C.D. Buckner, R. Clift, K. Doney, V. Farewell, et al. 1986. Methotrexate and cyclosporine compared with cyclosporine alone for prophylaxis of acute graft versus host disease after marrow transplantation for leukemia. N. Engl. J. Med. 314:729–735. [DOI] [PubMed] [Google Scholar]
- 2.Goulmy, E. 1997. Human minor histocompatibility antigens: new concepts for marrow transplantation and adoptive immunotherapy. Immunol. Rev. 157:125–140. [DOI] [PubMed] [Google Scholar]
- 3.Warren, E.H., M. Gavin, P.D. Greenberg, and S.R. Riddell. 1998. Minor histocompatibility antigens as targets for T-cell therapy after bone marrow transplantation. Curr. Opin. Hematol. 5:429–433. [DOI] [PubMed] [Google Scholar]
- 4.Malarkannan, S., T. Horng, P. Eden, F. Gonzalez, P. Shih, N. Brouwenstijn, H. Klinge, G. Christianson, D. Roopenian, and N. Shastri. 2000. Differences that matter: major cytotoxic T cell-stimulating minor histocompatibility antigens. Immunity. 13:333–344. [DOI] [PubMed] [Google Scholar]
- 5.Wang, W., L.R. Meadows, J.M. den Haan, N.E. Sherman, Y. Chen, E. Blokland, J. Shabanowitz, A.I. Agulnik, R.C. Hendrickson, C.E. Bishop, et al. 1995. Human H-Y: a male-specific histocompatibility antigen derived from the SMCY protein. Science. 269:1588–1590. [DOI] [PubMed] [Google Scholar]
- 6.Dolstra, H., H. Fredrix, F. Maas, P.G. Coulie, F. Brasseur, E. Mensink, G.J. Adema, T.M. de Witte, C.G. Figdor, and E. van de Wiel-van Kemenade. 1999. A human minor histocompatibility antigen specific for B cell acute lymphoblastic leukemia. J. Exp. Med. 189:301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warren, E.H., B.E. Otterud, R.W. Linterman, A.G. Brickner, V.H. Engelhard, M.F. Leppert, P.J. Martin, and S.R. Riddell. 2002. Feasibility of using genetic linkage analysis to identify the genes encoding T cell-defined minor histocompatibility antigens. Tissue Antigens. 59:293–303. [DOI] [PubMed] [Google Scholar]
- 8.Meadows, L., W. Wang, J.M. den Haan, E. Blokland, C. Reinhardus, J.W. Drijfhout, J. Shabanowitz, R. Pierce, A.I. Agulnik, C.E. Bishop, et al. 1997. The HLA-A*0201-restricted H-Y antigen contains a posttranslationally modified cysteine that significantly affects T cell recognition. Immunity. 6:273–281. [DOI] [PubMed] [Google Scholar]
- 9.Vogt, M.H., E. Goulmy, F.M. Kloosterboer, E. Blokland, R.A. de Paus, R. Willemze, and J.H. Falkenburg. 2000. UTY gene codes for an HLA-B60-restricted human male-specific minor histocompatibility antigen involved in stem cell graft rejection: characterization of the critical polymorphic amino acid residues for T-cell recognition. Blood. 96:3126–3132. [PubMed] [Google Scholar]
- 10.Warren, E.H., M.A. Gavin, E. Simpson, P. Chandler, D.C. Page, C. Disteche, K.A. Stankey, P.D. Greenberg, and S.R. Riddell. 2000. The human UTY gene encodes a novel HLA-B8-restricted H-Y antigen. J. Immunol. 164:2807–2814. [DOI] [PubMed] [Google Scholar]
- 11.Vogt, M.H., R.A. de Paus, P.J. Voogt, R. Willemze, and J.H. Falkenburg. 2000. DFFRY codes for a new human male-specific minor transplantation antigen involved in bone marrow graft rejection. Blood. 95:1100–1105. [PubMed] [Google Scholar]
- 12.Pierce, R.A., E.D. Field, J.M. den Haan, J.A. Caldwell, F.M. White, J.A. Marto, W. Wang, L.M. Frost, E. Blokland, C. Reinhardus, et al. 1999. The HLA-A*0101-restricted HY minor histocompatibility antigen originates from DFFRY and contains a cysteinylated cysteine residue as identified by a novel mass spectrometric technique. J. Immunol. 163:6360–6364. [PubMed] [Google Scholar]
- 13.Vogt, M.H.J., J.W. van den Muijsenberg, E. Goulmy, E. Spierings, P. Kluck, M.G. Kester, R.A. van Soest, J.W. Drijfhout, R. Willemze, and J.H. Falkenburg. 2002. The DBY gene codes for an HLA-DQ5-restricted human male-specific minor histocompatibility antigen involved in graft-versus-host disease. Blood. 99:3027–3032. [DOI] [PubMed] [Google Scholar]
- 14.den Haan, J.M., L.M. Meadows, W. Wang, J. Pool, E. Blokland, T.L. Bishop, C. Reinhardus, J. Shabanowitz, R. Offringa, D.F. Hunt, et al. 1998. The minor histocompatibility antigen HA-1: a diallelic gene with a single amino acid polymorphism. Science. 279:1054–1057. [DOI] [PubMed] [Google Scholar]
- 15.den Haan, J.M., N.E. Sherman, E. Blokland, E. Huczko, F. Koning, J.W. Drijfhout, J. Skipper, J. Shabanowitz, D.F. Hunt, V.H. Engelhard, and E. Goulmy. 1995. Identification of a graft versus host disease-associated human minor histocompatibility antigen. Science. 268:1476–1480. [DOI] [PubMed] [Google Scholar]
- 16.Pierce, R.A., E.D. Field, T. Mutis, T.N. Golovina, C. Von Kap-Herr, M. Wilke, J. Pool, J. Shabanowitz, M.J. Pettenati, L.C. Eisenlohr, et al. 2001. The HA-2 minor histocompatibility antigen is derived from a diallelic gene encoding a novel human class I myosin protein. J. Immunol. 167:3223–3230. [DOI] [PubMed] [Google Scholar]
- 17.Brickner, A.G., E.H. Warren, J.A. Caldwell, Y. Akatsuka, T.N. Golovina, A.L. Zarling, J. Shabanowitz, L.C. Eisenlohr, D.F. Hunt, V.H. Engelhard, and S.R. Riddell. 2001. The immunogenicity of a new human minor histocompatibility antigen results from differential antigen processing. J. Exp. Med. 193:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warren, E.H., P.D. Greenberg, and S.R. Riddell. 1998. Cytotoxic T-lymphocyte-defined human minor histocompatibility antigens with a restricted tissue distribution. Blood. 91:2197–2207. [PubMed] [Google Scholar]
- 19.Akatsuka, Y., E. Kondo, H. Taji, Y. Morishima, M. Yazaki, Y. Obata, Y. Kodera, S.R. Riddell, and T. Takahashi. 2002. Targeted cloning of cytotoxic T cells specific for minor histocompatibility antigens restricted by HLA class I molecules of interest. Transplantation. 74:1773–1780. [DOI] [PubMed] [Google Scholar]
- 20.van Pel, A., P. van der Bruggen, P.G. Coulie, V.G. Brichard, B. Lethe, B. van den Eynde, C. Uyttenhove, J.C. Renauld, and T. Boon. 1995. Genes coding for tumor antigens recognized by cytolytic T lymphocytes. Immunol. Rev. 145:229–250. [DOI] [PubMed] [Google Scholar]
- 21.Heiser, A., P. Dahm, D.R. Yancey, M.A. Maurice, D. Boczkowski, S.K. Nair, E. Gilboa, and J. Vieweg. 2000. Human dendritic cells transfected with RNA encoding prostate-specific antigen stimulate prostate-specific CTL responses in vitro. J. Immunol. 164:5508–5514. [DOI] [PubMed] [Google Scholar]
- 22.Jonuleit, H., U. Kuhn, G. Muller, K. Steinbrink, L. Paragnik, E. Schmitt, J. Knop, and A.H. Enk. 1997. Pro-inflammatory cytokines and prostaglandins induce maturation of potent immunostimulatory dendritic cells under fetal calf serum-free conditions. Eur. J. Immunol. 27:3135–3142. [DOI] [PubMed] [Google Scholar]
- 23.Schultze, J.L., A.A. Cardoso, G.J. Freeman, M.J. Seamon, J. Daley, G.S. Pinkus, J.G. Gribben, and L.M. Nadler. 1995. Follicular lymphomas can be induced to present alloantigen efficiently: a conceptual model to improve their tumor immunogenicity. Proc. Natl. Acad. Sci. USA. 92:8200–8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boisgerault, F., I. Khalil, V. Tieng, F. Connan, T. Tabary, J.H. Cohen, J. Choppin, D. Charron, and A. Toubert. 1996. Definition of the HLA-A29 peptide ligand motif allows prediction of potential T-cell epitopes from the retinal soluble antigen, a candidate autoantigen in birdshot retinopathy. Proc. Natl. Acad. Sci. USA. 93:3466–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, F., J.K. Ritter, M.G. Wang, O.W. McBride, R.A. Lubet, and I.S. Owens. 1993. Characterization of a cloned human dihydrotestosterone/androstanediol UDP-glucuronosyltransferase and its comparison to other steroid isoforms. Biochemistry. 32:10648–10657. [DOI] [PubMed] [Google Scholar]
- 26.Beaulieu, M., E. Levesque, D.W. Hum, and A. Belanger. 1998. Isolation and characterization of a human orphan UDP-glucuronosyltransferase, UGT2B11. Biochem. Biophys. Res. Commun. 248:44–50. [DOI] [PubMed] [Google Scholar]
- 27.Jin, C.J., J.O. Miners, K.J. Lillywhite, and P.I. Mackenzie. 1993. cDNA cloning and expression of two new members of the human liver UDP-glucuronosyltransferase 2B subfamily. Biochem. Biophys. Res. Commun. 194:496–503. [DOI] [PubMed] [Google Scholar]
- 28.Ritter, J.K., Y.Y. Sheen, and I.S. Owens. 1990. Cloning and expression of human liver UDP-glucuronosyltransferase in COS-1 cells. 3,4-catechol estrogens and estriol as primary substrates. J. Biol. Chem. 265:7900–7906. [PubMed] [Google Scholar]
- 29.Jackson, M.R., L.R. McCarthy, D. Harding, S. Wilson, M.W. Coughtrie, and B. Burchell. 1987. Cloning of a human liver microsomal UDP-glucuronosyltransferase cDNA. Biochem. J. 242:581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levesque, E., D. Turgeon, J.S. Carrier, V. Montminy, M. Beaulieu, and A. Belanger. 2001. Isolation and characterization of the UGT2B28 cDNA encoding a novel human steroid conjugating UDP-glucuronosyltransferase. Biochemistry. 40:3869–3881. [DOI] [PubMed] [Google Scholar]
- 31.Turgeon, D., J.S. Carrier, E. Levesque, B.G. Beatty, A. Belanger, and D.W. Hum. 2000. Isolation and characterization of the human UGT2B15 gene, localized within a cluster of UGT2B genes and pseudogenes on chromosome 4. J. Mol. Biol. 295:489–504. [DOI] [PubMed] [Google Scholar]
- 32.Sharma, A.K., J.J. Kuhns, S. Yan, R.H. Friedline, B. Long, R. Tisch, and E.J. Collins. 2001. Class I major histocompatibility anchor substitutions alter the conformation of T-cell receptor contacts. J. Biol. Chem. 276:21443–21449. [DOI] [PubMed] [Google Scholar]
- 33.Beaulieu, M., E. Levesque, A. Tchernof, B.G. Beatty, A. Belanger, and D.W. Hum. 1997. Chromosomal localization, structure, and regulation of the UGT2B17 gene, encoding a C19 steroid metabolizing enzyme. DNA Cell Biol. 16:1143–1154. [DOI] [PubMed] [Google Scholar]
- 34.Shlomchik, W.D., M.S. Couzens, C.B. Tang, J. McNiff, M.E. Robert, J. Liu, M.J. Shlomchik, and S.G. Emerson. 1999. Prevention of graft versus host disease by inactivation of host antigen-presenting cells. Science. 285:412–415. [DOI] [PubMed] [Google Scholar]
- 35.Malarkannan, S., P.P. Shih, P.A. Eden, T. Horng, A.R. Zuberi, G. Christianson, D. Roopenian, and N. Shastri. 1998. The molecular and functional characterization of a dominant minor H antigen, H60. J. Immunol. 161:3501–3509. [PubMed] [Google Scholar]
- 36.Mackenzie, P.I., I.S. Owens, B. Burchell, K.W. Bock, A. Bairoch, A. Belanger, S. Fournel-Gigleux, M. Green, D.W. Hum, T. Iyanagi, et al. 1997. The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics. 7:255–269. [DOI] [PubMed] [Google Scholar]
- 37.Dutton, G.J. 1980. Glucuronidation of Drugs and Other Compounds. CRC Press, Inc., Boca Raton, FL. 261 pp.
- 38.Ritter, J.K., M.T. Yeatman, P. Ferreira, and I.S. Owens. 1992. Identification of a genetic alteration in the code for bilirubin UDP-glucuronosyltransferase in the UGT1 gene complex of a Crigler-Najjar type I patient. J. Clin. Invest. 90:150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moghrabi, N., D.J. Clarke, M. Boxer, and B. Burchell. 1993. Identification of an A-to-G missense mutation in exon 2 of the UGT1 gene complex that causes Crigler-Najjar syndrome type 2. Genomics. 18:171–173. [DOI] [PubMed] [Google Scholar]
- 40.Bosma, P.J., J.R. Chowdhury, C. Bakker, S. Gantla, A. de Boer, B.A. Oostra, D. Lindhout, G.N.J. Tytgat, P.L.M. Jansen, R.P.J.O. Elferink, and N.R. Chowdhury. 1995. The genetic basis of the reduced expression of Bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N. Engl. J. Med. 333:1171–1175. [DOI] [PubMed] [Google Scholar]
- 41.Beaulieu, M., E. Levesque, D.W. Hum, and A. Belanger. 1996. Isolation and characterization of a novel cDNA encoding a human UDP-glucuronosyltransferase active on C19 steroids. J. Biol. Chem. 271:22855–22862. [DOI] [PubMed] [Google Scholar]
- 42.Jin, C., J.O. Miners, K.J. Lillywhite, and P.I. Mackenzie. 1993. Complementary deoxyribonucleic acid cloning and expression of a human liver uridine diphosphate-glucuronosyltransferase glucuronidating carboxylic acid-containing drugs. J. Pharmacol. Exp. Ther. 264:475–479. [PubMed] [Google Scholar]
- 43.Levesque, E., M. Beaulieu, M.D. Green, T.R. Tephly, A. Belanger, and D.W. Hum. 1997. Isolation and characterization of UGT2B15 (Y85): a UDP-glucuronosyltransferase encoded by a polymorphic gene. Pharmacogenetics. 7:317–325. [DOI] [PubMed] [Google Scholar]
- 44.Turgeon, D., J.S. Carrier, E. Levesque, D.W. Hum, and A. Belanger. 2001. Relative enzymatic activity, protein stability, and tissue distribution of human steroid-metabolizing UGT2B subfamily members. Endocrinology. 142:778–787. [DOI] [PubMed] [Google Scholar]
- 45.Rammensee, H., J. Bachmann, N.P. Emmerich, O.A. Bachor, and S. Stevanovic. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 50:213–219. [DOI] [PubMed] [Google Scholar]
- 46.Murphy, W.J., and B.R. Blazar. 1999. New strategies for preventing graft-versus-host disease. Curr. Opin. Immunol. 11:509–515. [DOI] [PubMed] [Google Scholar]
- 47.Dickinson, A.M., X.N. Wang, L. Sviland, F.A. Vyth-Dreese, G.H. Jackson, T.N. Schumacher, J.B. Haanen, T. Mutis, and E. Goulmy. 2002. In situ dissection of the graft-versus-host activities of cytotoxic T cells specific for minor histocompatibility antigens. Nat. Med. 8:410–414. [DOI] [PubMed] [Google Scholar]
- 48.Nunoya, K., T. Yokoi, K. Kimura, K. Inoue, T. Kodama, M. Funayama, K. Nagashima, Y. Funae, C. Green, M. Kinoshita, and T. Kamataki. 1998. A new deleted allele in the human cytochrome P450 2A6 (CYP2A6) gene found in individuals showing poor metabolic capacity to coumarin and (+)-cis-3,5-dimethyl-2-(3-pyridyl)thiazolidin-4-one hydrochloride (SM-12502). Pharmacogenetics. 8:239–249. [DOI] [PubMed] [Google Scholar]
- 49.Gaedigk, A., M. Blum, R. Gaedigk, M. Eichelbaum, and U.A. Meyer. 1991. Deletion of the entire cytochrome P450 CYP2D6 gene as a cause of impaired drug metabolism in poor metabolizers of the debrisoquine/sparteine polymorphism. Am. J. Hum. Genet. 48:943–950. [PMC free article] [PubMed] [Google Scholar]
- 50.Broly, F., D. Marez, J.M. Lo Guidice, N. Sabbagh, M. Legrand, P. Boone, and U.A. Meyer. 1995. A nonsense mutation in the cytochrome P450 CYP2D6 gene identified in a Caucasian with an enzyme deficiency. Hum. Genet. 96:601–603. [DOI] [PubMed] [Google Scholar]
- 51.Ferguson, R.J., S.M. De Morais, S. Benhamou, C. Bouchardy, J. Blaisdell, G. Ibeanu, G.R. Wilkinson, T.C. Sarich, J.M. Wright, P. Dayer, and J.A. Goldstein. 1998. A new genetic defect in human CYP2C19: mutation of the initiation codon is responsible for poor metabolism of S-mephenytoin. J. Pharmacol. Exp. Ther. 284:356–361. [PubMed] [Google Scholar]
- 52.Pemble, S., K.R. Schroeder, S.R. Spencer, D.J. Meyer, E. Hallier, H.M. Bolt, B. Ketterer, and J.B. Taylor. 1994. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem. J. 300:271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Board, P.G. 1981. Biochemical genetics of glutathione-S-transferase in man. Am. J. Hum. Genet. 33:36–43. [PMC free article] [PubMed] [Google Scholar]
- 54.Sherratt, P.J., D.J. Pulford, D.J. Harrison, T. Green, and J.D. Hayes. 1997. Evidence that human class Theta glutathione S-transferase T1-1 can catalyse the activation of dichloromethane, a liver and lung carcinogen in the mouse. Comparison of the tissue distribution of GST T1-1 with that of classes Alpha, Mu and Pi GST in human. Biochem. J. 326:837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]