Abstract
Urm1p is a ubiquitin-related protein that serves as a posttranslational modification of other proteins. Urm1p conjugation has been implicated in the budding process and in nutrient sensing. Here, we have identified the first in vivo target for the urmylation pathway as the antioxidant protein Ahp1p. The attachment of Urm1p to Ahp1p requires the E1 for the urmylation pathway, Uba4p. Loss of the urmylation pathway components results in sensitivity to a thiol-specific oxidant, as does loss of Ahp1p, implying that urmylation has a role in an oxidative-stress response. Moreover, treatment of cells with thiol-specific oxidants affects the abundance of Ahp1p-Urm1p conjugates. These results suggest that the conjugation of Urm1p to Ahp1p could regulate the function of Ahp1p in antioxidant stress response in Saccharomyces cerevisiae.
An emerging theme in the regulation of protein function is the covalent attachment of one protein to another. An example of this type of modification is provided by ubiquitin, a 76-residue protein that can be attached to other proteins. In many cases, ubiquitination targets the modified proteins for degradation, but other regulatory effects of ubiquitination are also known. Ubiquitin is the founding member of a family of small modifier proteins present in most eukaryotes and known as ubiquitin-like proteins (Ubls) (20). As in the case of the ubiquitin pathway, the attachment of Ubls to targets requires a series of enzymatic steps that include an E1 (an activating enzyme), an E2 (a conjugating enzyme), and sometimes an E3 (protein ligase). Each Ubl is thought to be attached to other proteins via an isopeptide bond between the C terminus of the modifier and a lysine residue of a target protein (18). In contrast to ubiquitin, some of these Ubls do not appear to promote proteolysis but instead appear to alter the function or localization of the target (26, 31, 33, 36).
Most of the Ubls are attached to many targets and consequently are involved in the regulation of many processes. Probably the best-characterized ubiquitin-like pathway involves the attachment of SUMO to a rapidly growing set of targets. These substrates include RanGAP1, a Ran GTPase-activating protein involved in nucleocytoplasmic transport (29, 36); IκBα, an inflammatory-response regulatory protein (10); PML (promyelocytic leukemia protein), whose fusion to the retinoic acid receptor causes acute promyelocytic leukemia (29); and Cdc3p, Cdc11p, and Shs1p, proteins in the yeast Saccharomyces cerevisiae that are components of the septin ring involved in cytokinesis (25).
In addition to sumoylation, four other ubiquitin-like pathways have been identified in yeast (11, 20). The modifier of one of these protein conjugation systems is Urm1p (12); thus, the pathway is termed urmylation. The function of Urm1p conjugation is only beginning to be understood. The E1 for this pathway is Uba4p (12), but other components of the conjugation pathway, such as an E2 or an E3, have yet to be identified. Although urmylation has been linked to budding and invasion (17), no substrates are known for the urmylation pathway.
As a first step toward understanding the physiological role of the Urm1p conjugation pathway in S. cerevisiae, we were interested in identifying and characterizing substrate proteins that are modified by Urm1p. Here, we show that Urm1p is attached to Ahp1p (alkyl hydroperoxide reductase), a protein implicated in oxidative-stress protection in yeast (41). Furthermore, we identified a potential role for urmylation in oxidative-stress response.
MATERIALS AND METHODS
Strains, growth conditions, and plasmids.
The yeast strains used in this study are listed in Table 1. Yeast transformations were performed as described previously (15). Haploid MATa nonessential yeast deletion strains were purchased from Research Genetics (Huntsville, Ala). Other gene deletions were constructed by PCR (4) using either the pRS (49) or pFA6a (34) plasmid series as templates. In all cases, the entire coding region was replaced with the indicated marker, and successful replacement was confirmed by PCR and phenotype when applicable.
TABLE 1.
Yeast strains used in this study
| Straina | Relevant genotypeb | Source or reference |
|---|---|---|
| SY3357 | MATaleu2-Δ1 ura3-52 his3-Δ200 trpl-Δ63 ade8Δade2-101 mfa2-Δ1::FUS1-lacZ | 39 |
| SY3358 | MATα leu2-Δ1 ura3-52 lys2-801 his3-Δ200 trpl-Δ63 ade8Δade2-101 mfa2-Δ1::FUS1lacZ | 39 |
| SY3362 | SY3357, except cla4Δ::TRP1 (pRS316ADE8CLA4) | 39 |
| SY3754 | SY3358, except cla4Δ::TRP1 ste20Δ::TRP1 (pRS316ADE8CLA4) | 16 |
| SY3805 | SY3358, except uba4Δ::KanMX6 | 16 |
| SY3806 | SY3358, except urmlΔ::KanMX6 | 16 |
| SY4117 | SY3357, except urmIΔ::HIS3 (PGALI pRS315 URMI) | This study |
| SY4139 | SY3357, except urmIΔ::HIS3 | This study |
| SY4118 | SY3357, except urmIΔ::HIS3 (PGALI pRS315 HFURMI) | This study |
| Y258 | pep4-3 his4-580 ura3-52 leu2-3, 112 | 55 |
| SY3972 | pep4-3 his4-580 ura3-52 leu2-3,112 (pEGH PGALI-GST-HisX6-AHPI) | 55 |
| SY4050 | pep4-3 his4-580 ura3-52 leu2-3, 112 (pEGH PGALI-GST-HisX6) | 55 |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| SY3839 | BY4741 urm1Δ::KanMX4 | Research Genetics |
| SY3840 | BY4741 uba4Δ::KanMX4 | Research Genetics |
| SY4119 | BY4741 ahp1Δ::KanMX4 | Research Genetics |
| SY3807 | SY3357, except cla4Δ::TRP1 uba4Δ::HIS3 (pRS316ADE8CLA4) | 16 |
| SY3808 | SY3357, except cla4Δ::TRP1 urm1Δ::KanMX6 (pRS316ADE8CLA4) | 16 |
| SY4120 | SY3357, except cla4Δ::TRP1 ahp1Δ::KanMX6 (pRS316ADE8CLA4) | This study |
| HYL333 | MATα ura3-52 | G. Fink |
| SY3826 | HYL333, except his3Δ ura3 | 16 |
| SY3827 | SY3826, except uba4Δ::KanMX6 | 16 |
| SY3828 | SY3826, except urm1Δ::KanMX6 | 16 |
| SY4121 | SY3826, except ahp1Δ::KanMX6 | This study |
| SY3836 | BY4741, except gln3Δ::KanMX4 | Research Genetics |
| SY4122 | BY4741, except urm1Δ::KanMX4 ahp1Δ::NatMX4 | This study |
| SY4123 | BY4741, except uba4Δ::KanMX4 ahp1Δ::NatMX4 | This study |
| SY4124 | BY4741, except urm1Δ::KanMX4 uba4Δ::KanMX4 | This study |
SY3357 and SY3358 are derivatives of S288C.
Strains containing the designation ura3 were made URA3 at the designated chromosomal locus and then ura3 by selection on 5-fluoroorotic acid as described in Materials and Methods.
Yeast strains were propagated using standard methods (47). Rich yeast medium containing 2% glucose (YPD) and synthetic yeast medium containing either 2% glucose (SD) or 2% galactose (SG) were prepared as described previously (47). Geneticin (Biovectra, Charlottetown, Canada) selection was performed as previously described (34). Strains were cured of URA3-containing plasmids using 5-fluoroorotic acid (Biovectra). Rapamycin (Sigma, St. Louis, Mo.) was added to the medium from a concentrated stock solution in 90% ethanol-10% Tween 20. To test for tert-butyl hydroperoxide (t-BOOH) and diamide sensitivities, cells were grown overnight in YPD medium, spotted on yeast extract-peptone-dextrose (YEPD) plates containing either 0.4 mM t-BOOH (Sigma), 1 mM t-BOOH, 1 mM diamide (Sigma), or 1.3 mM diamide. The plates were monitored after 3 days of incubation at 30°C.
The PGAL1-driven URM1 construct that bears the N-terminal extension MTSHHHHHHMHDYKDDDDKMGS containing His6 and FLAG tags (PGAL1-PRS315 HFURM1) has been described (17). The other plasmids used in this study, YCpHIS3cla4-75, pML40, pML41, pRS316ADE8CLA4, pEGH PGAL1-GST-HisX6, and pEGH PGAL1-GST-HisX6-AHP1, were described previously (9, 35, 39, 55).
Immunoblot analysis of whole-yeast lysates and antibodies.
Immunoblot analysis of yeast lysates was performed as previously described (17). When indicated, strains were treated with 1.5 mM diamide or 1.3 mM t-BOOH for 4 h prior to lysis. Urm1p was detected by using polyclonal antibody to Urm1p (17), a secondary anti-rabbit horseradish peroxidase-conjugated antibody (Bio-Rad Laboratories, Hercules, Calif.), and chemiluminescence (Pierce, Rockford, Ill.). A monoclonal antibody against Dpm1p (a kind gift from T. Stevens, University of Oregon, Eugene, Oreg.) and a secondary anti-mouse horseradish peroxidase-conjugated antibody (Bio-Rad Laboratories) were used to confirm that equal amounts of protein were loaded in each lane.
Isolation of Urm1p-conjugated proteins.
SY4118 containing pRS315 PGAL1-HFURM1 was grown at 30°C in 4 liters of YP containing 2% raffinose to an optical density at 600 nm of 0.75. Expression of PGAL1-HFURM1 was induced by the addition of 2% galactose, and the cells were grown for an additional 4 h at 30°C. The cells were harvested by centrifugation and converted to spheroplasts by incubation with 150 μg of Zymolyase 100T (Seikagaku America, Ijamsville, Md.)/ml in 1.2 M sorbitol-50 mM potassium phosphate buffer (pH 7.5)-1 mM MgCl2 for 45 min at 30°C. The spheroplasts were washed once with 1.2 M sorbitol and lysed in buffer A. The lysates were clarified by centrifugation at 12,000 × g, supplemented to 20 mM imidazole and 10% glycerol, and bound in batch for 1.5 h to 4 ml of Talon resin (Clontech, Palo Alto, Calif.). The Talon resin was loaded into a column, washed with 40 ml of buffer B (buffer A with 10% glycerol), and then eluted with 200 mM imidazole in buffer B. Four milliliters of the eluate was bound in batch for 1 h at 4°C to 0.4 ml of anti-FLAG agarose (Sigma). The anti-FLAG agarose was loaded into a column, washed with 50 ml of buffer B supplemented with 0.2% Trition X = 100, washed with 5 ml of buffer B, and then eluted with 1.2 ml of buffer B supplemented with 0.2 mg of FLAG peptide (Sigma). The eluate was precipitated by the addition of trichloroacetic acid to 5%, resuspended in loading buffer, and applied to a 10% acrylamide gel. Following electrophoresis, protein bands were detected by silver staining and excised. The sample was subjected to in-gel digestion (48), desalted using Millipore μC18 ZipTips, and dried. The sample was then resuspended in 5 μl of water and analyzed by liquid chromatography microspray-mass spectrometry (MS-MS) with a ThermoFinnigan LCQ DECA XP mass spectrometer (FHCRC Proteomics) using the configuration of Gatlin et al. (14). Data were collected in the data-dependent mode in which an MS scan was followed by MS-MS scans of the three most abundant ions from the preceding MS scan. The MS-MS data were searched using SEQUEST software against the translated open reading frames from the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/). The resulting peptide matches were scored by SEQUEST, and protein identifications were considered valid if the identified protein contained at least two peptides with Xcorr scores above 2.0.
Glutathione S-transferase (GST) pull-down assays.
Yeast strains SY4050 and SY3972 were grown at 30°C in 50 ml of YP containing 2% raffinose to an optical density at 600 nm of 0.75. Expression of PGAL1-GST-HisX6-AHP1 and PGAL1-GST-HisX6 was induced by the addition of 2% galactose, and the cells were grown for an additional 4 h at 30°C. The cells were harvested by centrifugation, converted to spheroplasts as described above, and lysed in 500 μl of buffer A. The lysates were clarified by centrifugation at 12,000 × g and bound in batch for 1 h to 100 μl of glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech, Piscataway, N.J.). The beads were collected by centrifugation at 500 × g for 5 min, washed four times in phosphate-buffered saline, eluted in loading buffer, and applied to a 10% acrylamide gel. Following electrophoresis, protein bands were detected by Western blot analysis using a previously described purified anti-Urm1p antibody (17).
Invasive-growth assays.
For the plate-washing assay, strains were patched on YPD plates and incubated at 30°C. After 2 days of incubation, the plates were washed under a stream of running water as the surface was gently rubbed with a finger to remove cells not in the agar, and invasive growth was scored (45).
RESULTS
In an attempt to identify substrates of the urmylation pathway, we constructed a strain that expressed His6- and FLAG-tagged URM1 as its only copy of URM1. Extracts from cells expressing HFURM1 were incubated with Talon resin. Bound proteins were eluted with imidazole and bound to anti-FLAG agarose. When proteins bearing both tags were eluted with loading buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and visualized by Western blotting, the banding pattern was similar to the pattern of Urm1p conjugates recognized by the Urm1p antibody (Fig. 1A; also see Fig. 3) (17). We were able to obtain sufficient amounts of one of these bands (32 kDa) to identify it by direct MS analysis of a protease digestion mixture. Six tryptic peptides sequenced by MS exhibited 100% identity with regions of a protein known as Ahp1p (Fig. 1B). The 32-kDa product is consistent with modification of Ahp1p (19 kDa) by a single Urm1p polypeptide (13 kDa).
FIG. 1.
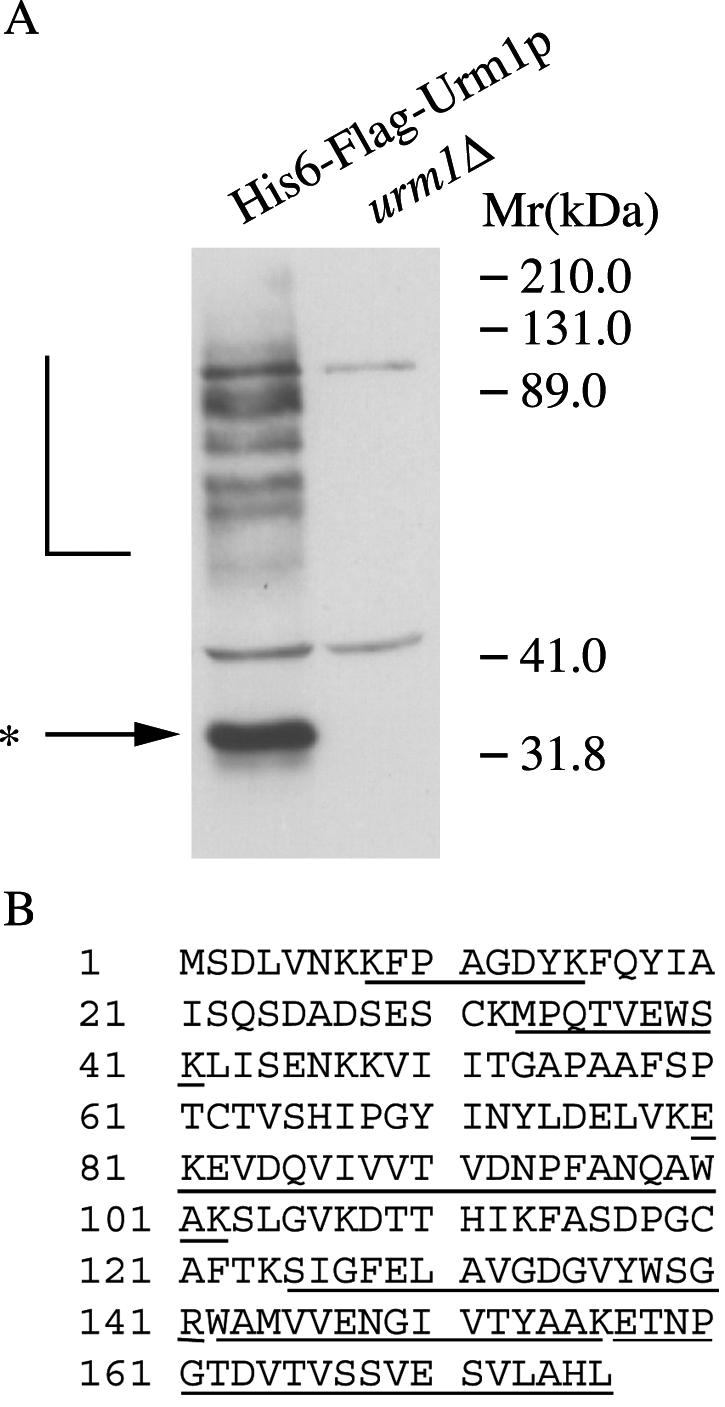
Identification of Ahp1p as a target of the urmylation pathway. (A) Lysates of urm1Δ cells (SY4139) and urm1Δ cells expressing His6- and FLAG epitope-tagged Urm1p (SY4118) were subjected to affinity chromatography on Talon resin. Eluates from the Talon resin were applied to anti-FLAG agarose. The proteins eluted from the FLAG agarose were analyzed by SDS-PAGE and immunoblotting with polyclonal antibodies against Urm1p. The L bracket designates high-molecular-weight Urm1p-modified species, while the asterisk indicates the position of a 32-kDa Urm1p-modified species. (B) BLAST sequence analysis of five peptides (underlined) obtained from the MS-identified Ahp1p.
FIG. 3.
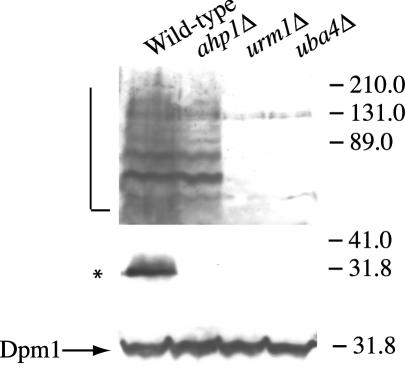
A 32-kDa band disappears in ahp1Δ cells. Strains SY3358 (wild-type), SY3806 (urm1Δ), SY3805 (uba4Δ), and SY3815 (ahp1Δ) were grown to mid-log phase in YEPD at 30°C. The lysates were analyzed by SDS-PAGE and immunoblot analysis using affinity-purified polyclonal anti-Urm1p antibodies and monoclonal antibodies to Dpm1p (to confirm equal protein loading). The L bracket designates high-molecular-weight Urm1p-modified species, while the asterisk designates the position of a 32-kDa Urm1p-modified species.
Recently, Ahp1p was identified as a peroxiredoxin (24, 32, 52). Peroxiredoxins are a class of enzymes, present in most organisms, that promote the elimination of H2O2 and alkyl hydrogen peroxides as a cellular defense against oxidative stress (8, 37, 40). In vitro assays show that Ahp1p preferentially reduces alkyl hydrogen peroxides, such as t-BOOH, while other peroxiredoxins preferentially reduce H2O2 (24, 42). Furthermore, loss of Ahp1p results in sensitivity of cells to the oxidants t-BOOH, an organic peroxide, and diamide, a thiol oxidant (32).
Urm1p is conjugated to Ahp1p.
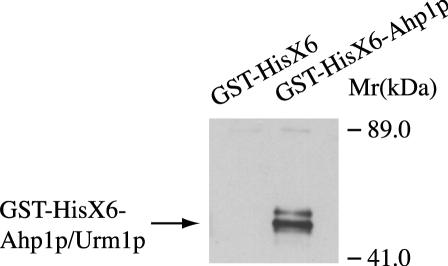
In order to confirm that Urm1p was interacting with Ahp1p, we used a GST-tagged version of Ahp1p. Purification of Ahp1p from wild-type extracts and immunoblotting with Urm1p-specific antibodies indicated that a 32-kDa species of Ahp1p contained Urm1p (Fig. 2, right lane). These bands were not detected in extracts from cells expressing GST alone (Fig. 2, left lane). Intriguingly, either Urm1p itself or Ahp1p may undergo posttranslational modification, since the conjugated product appears as a doublet.
FIG. 2.
Urm1p coimmunoprecipitates with Ahp1p from whole-cell extracts. Exponential cultures of haploid strains SY4050 (wild type) carrying GST-HisX6 and SY3972 (wild type) carrying GST-HisX6-Ahp1p were lysed in the presence of 20 mM N-ethylmaleimide. The lysates were analyzed by SDS-PAGE and immunoblot analysis using affinity-purified polyclonal anti-Urm1p. The bands corresponding to Urm1p conjugated to Ahp1p are indicated.
To further support the idea that Urm1p was conjugated to Ahp1p, we performed Western analysis using the anti-Urm1p antibody on strains lacking URM1, UBA4, and AHP1. Loss of AHP1 resulted in the disappearance of the ∼32-kDa protein, but did not affect the abundance of the ∼75-, 80-, 89-, and 125-kDa polypeptides (Fig. 3, lane ahp1Δ). However, loss of URM1 and UBA4 eliminated the ∼32-kDa protein, as well as many other polypeptides of higher molecular mass (Fig. 3, lanes urm1Δ and uba4Δ). Collectively, these results support the conclusion that Ahp1p is one of the several targets of the urmylation pathway.
Ahp1p is not involved in budding or invasion.
Previously, potential functions for the urmylation pathway in budding and nutrient sensing were identified. It was shown that the simultaneous loss of Urm1p and Cla4p, a p21-activated kinase that functions in budding, is lethal (16). The simultaneous loss of Cla4p and urmylation pathway components caused cells to arrest with a severe defect in budding, suggesting that the urmylation pathway has a role in vegetative growth. To begin to investigate the function of Urm1p conjugation to Ahp1p, we tested whether the loss of Ahp1p was essential in a cla4Δ background. Unlike uba4 and urm1 mutations, mutations in ahp1 were not synthetically lethal with cla4Δ (data not shown).
The other role identified for the urmylation pathway is related to nutrient sensing. When haploid cells are starved for nutrients, the cells become elongated, bud primarily from one pole, and can invade the agar substratum. Previously, we showed that the urmylation pathway has a role in invasive growth (17). In order to determine if Ahp1p is the target of the urmylation pathway during invasion, we tested whether the loss of Ahp1p causes a defect in invasion. While deletion of UBA4 and URM1 rendered cells unable to invade agar under starvation conditions (data not shown), deletion of AHP1 did not affect invasive growth. Therefore, Ahp1p does not appear to play a role in invasion.
Potential role for Ahp1p in TOR signaling.
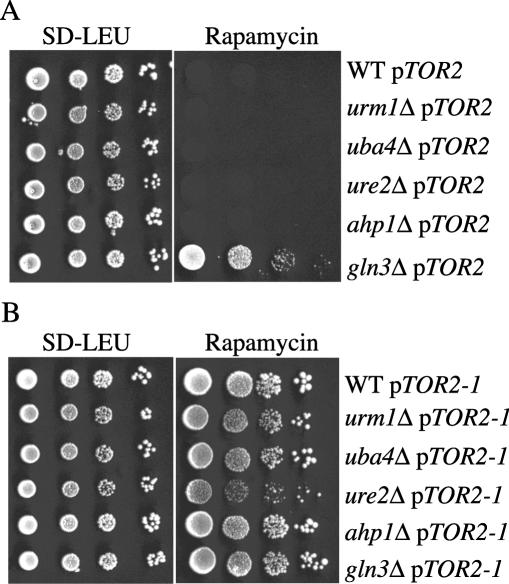
Loss of the urmylation pathway also resulted in hypersensitivity to rapamycin (a macrocyclic antibiotic) (17). Rapamycin inhibits Tor1p and Tor2p, ultimately resulting in cellular responses characteristic of nutrient deprivation through a mechanism involving translational and transcriptional arrest (3, 19, 27, 30). Since Ahp1p appears to be a target of urmylation, we tested whether ahp1 mutants are hypersensitive to rapamycin and if the mutations exhibited genetic interaction with TOR pathway mutations. A gln3Δ mutant, which is defective for a GATA-type transcription factor regulated by the TOR kinases and by the Ure2p repressor (5, 7, 44), served as a rapamycin-resistant control strain. A ure2Δ mutant served as a control for rapamycin sensitivity. We found that ahp1 null mutants were unable to grow after 5 days on medium containing rapamycin (data not shown). Moreover, even if the ahp1 null mutants were carrying a wild-type copy of TOR2 on a plasmid, the cells were still unable to grow after 5 days on medium containing rapamycin (Fig. 4A). The sensitivity to rapamycin was not due to a reduction in protein synthesis or a general drug sensitivity, because the ahp1 null mutants were not sensitive to cycloheximide (data not shown). To verify that the sensitivity of these mutants to rapamycin reflected inactivity of the TOR pathway, we measured the rapamycin sensitivities of ahp1Δ mutants carrying a plasmid-borne TOR2S1972I (TOR2-1) allele (35). This allele of TOR2 has previously been shown to confer rapamycin resistance on many sensitive mutants. We found that all the double mutants were no longer sensitive to rapamycin (Fig. 4B). In contrast, the ahp1Δ mutants carrying a wild-type version of TOR2 on a plasmid were still sensitive to rapamycin (data not shown). These results suggest that, like the urmylation pathway mutants, ahp1 mutants are hypersensitive to rapamycin because of inhibition of TOR pathway signaling.
FIG. 4.
Ahp1p functions in TOR signaling. (A) Strains SY3836 (gln3Δ), BY4741 (wild type [WT]), SY3839 (urm1Δ), SY3840 (uba4Δ), SY3845 (ure2Δ), and SY4121 (ahp1Δ) carrying pTOR2 (pML40) were grown to mid-log phase in YEPD at 30°C. A serial dilution (1/10) was performed starting with 10,000 cells. Cells were spotted onto either SD-LEU (left) or SD-LEU plus 25 nM rapamycin (right) and grown for 3 days at 30°C. (B) Strains SY3836 (gln3Δ), BY4741 (wild-type), SY3839 (urm1Δ), SY3840 (uba4Δ), SY3845 (ure2Δ), and SY4121 (ahp1Δ) carrying pTOR2-1 (pML41) were grown to mid-log phase in YEPD at 30°C. A serial dilution (1/10) was performed starting with 10,000 cells. Cells were spotted onto SD-LEU (left) or SD-LEU plus 25 nM rapamycin (right) and grown for 3 days at 30°C.
Urmylation is involved in oxidative stress.
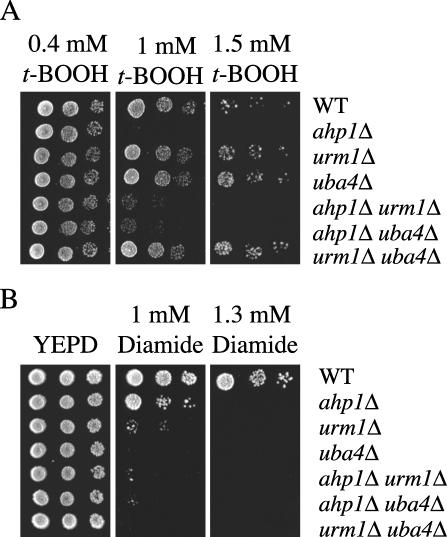
Ahp1p is believed to function as an antioxidant specific for t-BOOH and the thiol oxidant diamide (24, 32, 42). We therefore tested whether the loss of the urmylation pathway results in sensitivity to either t-BOOH or diamide. The urm1 and the uba4 mutant strains were sensitive to diamide (Fig. 5B). In fact, the loss of URM1 or UBA4 caused cells to be more sensitive to diamide than did loss of AHP1. In contrast, although the ahp1 null mutant strain was hypersensitive to t-BOOH compared to the wild-type control, the urm1 and the uba4 mutant strains were not sensitive to t-BOOH (Fig. 5A). Taken together, these results suggest that the urmylation pathway has an antioxidant role specifically toward diamide in vivo. Furthermore, these results also suggest that the conjugation of Urm1p to Ahp1p affects the antioxidant role of Ahp1p toward diamide but not toward t-BOOH.
FIG. 5.
Sensitivities of the urmylation pathway mutants to oxidative stress. (A) Loss of the urmylation pathway does not result in sensitivity to t-BOOH. Strains BY4741 (wild type [WT]), SY4119 (ahp1Δ), SY3839 (urm1Δ), SY3840 (uba4Δ), SY4122 (ahp1Δ urm1Δ), SY4123 (ahp1Δ uba4Δ), and SY4124 (urm1Δ uba4Δ) were grown to mid-log phase in YEPD at 30°C. A serial dilution (1/5) was performed starting with 10,000 cells. Cells were spotted onto either YEPD plus 0.4 mM t-BOOH (left), YEPD plus 1 mM t-BOOH (middle), or 1.5 mM t-BOOH (right) and grown for 3 days at 30°C. (B) Loss of Ahp1p and the urmylation pathway results in diamide sensitivity. Strains BY4741 (wild type), SY4119 (ahp1Δ), SY3839 (urm1Δ), SY3840 (uba4Δ), SY4122 (ahp1Δ urm1Δ), SY4123 (ahp1Δ uba4Δ), and SY4124 (urm1IΔ uba4IΔ) were grown to mid-log phase in YEPD at 30°C. A serial dilution (1/5) was performed starting with 10,000 cells. Cells were spotted onto either YEPD (left), YEPD plus 1 mM diamide (middle), or 1.3 mM diamide (right) and grown for 3 days at 30°C.
Given that Ahp1p is urmylated and controls sensitivity to t-BOOH and to diamide, we asked whether the state of Ahp1p urmylation was modulated by treatment with either of these agents. We observed a modest increase in the Ahp1p-Urm1p conjugate when cells were treated with diamide (Fig. 6). In contrast, the Ahp1p-Urm1p conjugate was absent from cells treated with t-BOOH. Whether urmylation of Ahp1p is blocked by t-BOOH or Ahp1p is degraded under these conditions cannot be determined from these data, though the former possibility seems more likely. Nonetheless, the salient point is that urmylation of Ahp1p is differentially affected by diamide and t-BOOH, concordant with the observation that the loss of urmylation causes a differential effect on sensitivity to these agents.
FIG. 6.
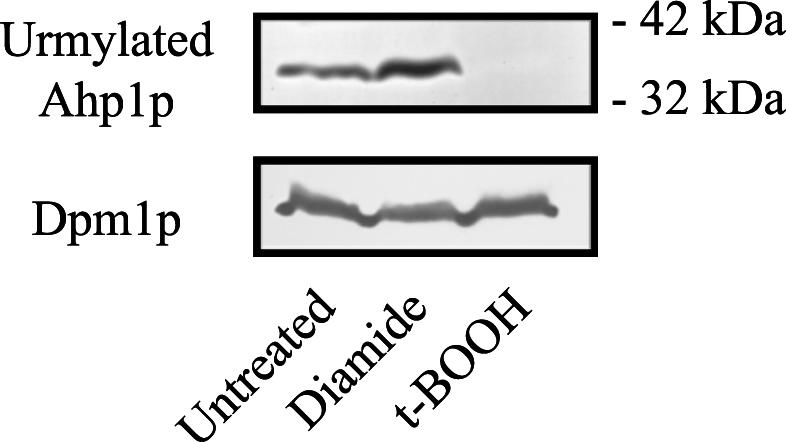
The Ahp1p-Urm1p conjugate is affected by diamide and t-BOOH treatment. Strain SY3357 (wild type) was grown to mid-log phase in YEPD at 30°C. Cells were then grown in YEPD at 30°C in the presence of 1.5 mM diamide or 1.3 mM t-BOOH or without drug treatment for 4 h. The lysates were analyzed by SDS-PAGE and immunoblot analysis using affinity-purified polyclonal anti-Urm1p antibodies and monoclonal antibodies to Dpm1p.
DISCUSSION
In order to investigate the role of the Urm1p conjugation pathway, we sought to identify proteins modified by Urm1p using MS. This effort identified the first target for the urmylation pathway, the antioxidant protein Ahp1p. We showed that the conjugation of Urm1p to Ahp1p requires the E1 of the urmylation pathway, Uba4p. Moreover, we found that the loss of the urmylation pathway components causes cells to be sensitive to diamide. These results suggest a role for the urmylation pathway in antioxidant activity specific for diamide.
Conjugation of Urm1p to Ahp1p.
Only a single copy of Urm1p appears to be attached to each Ahp1p polypeptide. Two species of urmylated GST-HisX6-Ahp1p were detected by Western blotting. One is the expected size for urmylated GST-HisX6-Ahp1p. The second band is not sufficiently larger to contain a second Urm1p moeity. This result suggests that either Ahp1p or Urm1p is posttranslationally modified (Fig. 2). Since our previous results suggest that there is a higher-molecular-weight Urm1p species, it is possible that Urm1p is the protein that is modified (17).
Although we have not identified which of the 14 lysines on Ahp1p is modified by Urm1p, two results support the idea that Ahp1p is a target and not an enzyme of the urmylation pathway. First, although the Ahp1p-Urm1p conjugate is detected only when 20 mM N-ethylmaleimide is present during cell lysis, these samples were resuspended in loading buffer that contained 2-mercaptoethanol (βME) (17)(Fig. 3). Sample buffer containing this reducing agent should disrupt the Urm1 thiol esters of E1 or E2, whereas Urm1p-protein conjugates would be resistant to βME (21). Because the Urm1p-Ahp1p product is present when treated with βME, it seems likely that Ahp1p is a target of the urmylation pathway. Second, loss of AHP1 affects the presence of only the 32-kDa Urm1p-Ahp1p conjugate, not other urmylated targets (Fig. 3). If Ahp1p functioned as an E2 or an E3 for the urmylation pathway, then the loss of Ahp1p would be expected to affect at least some of these other bands. Together, our results suggest that Ahp1p is a target of the urmylation pathway.
Possible functions for Urm1p attachment to Ahp1p.
When cells are exposed to elevated levels of reactive oxygen species, they suffer oxidative stress. Oxidative stress can lead to DNA damage, lipid peroxidation, and protein oxidation (22, 50). The altered diamide tolerance phenotype seen in the ahp1 null mutant is believed to be related to the generation of lipid hydroperoxides upon exposure to the oxidant diamide (32). Similarly, mutations in either URM1 or UBA4 cause cells to become hypersensitive to diamide. Our results therefore suggest a potential role for the urmylation pathway in an antioxidant activity specific for diamide. In keeping with this possibility, treatment of cells with diamide causes an increase, albeit a modest one, in the amount of urmylated Ahp1p. Interestingly, the loss of the urmylation pathway components causes cells to be more sensitive to diamide than does the loss of Ahp1p. One possible explanation for the difference in diamide sensitivity may be that Urm1p interacts with another antioxidant enzyme(s). In support of this possibility, previous results indicated that different peroxiredoxins in yeast might have redundant and nonredundant functions (42, 54).
We also identified a potential interaction between Ahp1p and the TOR pathway. As seen for loss of URM1 or UBA4, the loss of AHP1 confers sensitivity to rapamycin. Since loss of AHP1 confers rapamycin sensitivity, it might be an enhancer of TOR signaling or of a cellular process regulated by TOR. Recent results suggest that the TOR pathway may play a role in oxidative-stress response. In Schizosaccharomyces pombe, tor1+ is required for response to oxidative stress, as well as other stresses (53). Therefore, Urm1p targeting of Ahp1p may provide a link between the TOR pathway and oxidative-stress response in S. cerevisiae.
Implications of urmylation and oxidative-stress response in mammals.
Damage to cells caused by oxidative stress has been implicated in aging (51), neurodegenerative diseases (6), diabetes (28), and cancer (2). In mammals, reactive oxygen species are a well-established signal for regulation of transcription factors, such as NF-κB (nuclear factor κB), Ap1 (activator protein 1), and p53 (tumor suppressor) (1, 13, 23, 38). In higher plants, as well as animals, reactive oxygen species are also established signals in response to wound healing (43, 46). Thus, a growing area of research is focused on trying to understand the mechanisms that protect cells from oxidative stress or promote cellular recovery. Potential homologues for Ahp1p have been identified in plants, animals, fungi, and prokaryotes (52). Sequence similarity with these other potential antioxidants is highest around a putative catalytic active site that may be important for the peroxidase function (cysteine 62 in Ahp1p) (52). Since there also appears to be a homologue to Urm1p in mammals, perhaps urmylation of Ahp1p is important in oxidative-stress response in mammals. Further characterization of Ahp1p and the attachment of Urm1p is necessary to determine if this conjugate is involved in an antioxidant mechanism and/or other functions in S. cerevisiae.
Acknowledgments
April S. Goehring and David M. Rivers contributed equally to this work.
We thank J. Heitman and T. Stevens for providing plasmids and reagents. Thanks are also due to Paul Cullen, Hilary Kemp, Megan Keniry, and Dave Mitchell for helpful comments and suggestions.
This work was supported by research (GM-30027) and training (HD07348 to D.M.R. and GM07759 to A.S.G) grants from the National Institutes of Health.
REFERENCES
- 1.Abate, C., L. Patel, F. J. Rauscher III, and T. Curran. 1990. Redox regulation of fos and jun DNA-binding activity in vitro. Science 249:1157-1161. [DOI] [PubMed] [Google Scholar]
- 2.Ames, B. N., M. K. Shigenaga, and L. S. Gold. 1993. DNA lesions, inducible DNA repair, and cell division: three key factors in mutagenesis and carcinogenesis. Environ. Health Perspect. 101:(Suppl. 5):35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell. 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 6.Benzi, G., and A. Moretti. 1995. Are reactive oxygen species involved in Alzheimer's disease? Neurobiol. Aging 16:661-674. [DOI] [PubMed] [Google Scholar]
- 7.Bertram, P. G., J. H. Choi, J. Carvalho, W. Ai, C. Zeng, T. F. Chan, and X. F. Zheng. 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275:35727-35733. [DOI] [PubMed] [Google Scholar]
- 8.Bryk, R., P. Griffin, and C. Nathan. 2000. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407:211-215. [DOI] [PubMed] [Google Scholar]
- 9.Cvrckova, F., C. De Virgilio, E. Manser, J. R. Pringle, and K. Nasmyth. 1995. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9:1817-1830. [DOI] [PubMed] [Google Scholar]
- 10.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell. 2:233-239. [DOI] [PubMed] [Google Scholar]
- 11.Dittmar, G. A., C. R. Wilkinson, P. T. Jedrzejewski, and D. Finley. 2002. Role of a ubiquitin-like modification in polarized morphogenesis. Science 295:2442-2446. [DOI] [PubMed] [Google Scholar]
- 12.Furukawa, K., N. Mizushima, T. Noda, and Y. Ohsumi. 2000. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 275:7462-7465. [DOI] [PubMed] [Google Scholar]
- 13.Gaiddon, C., N. C. Moorthy, and C. Prives. 1999. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 18:5609-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gatlin, C. L., G. R. Kleemann, L. G. Hays, A. J. Link, and J. R. Yates III. 1998. Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem. 263:93-101. [DOI] [PubMed] [Google Scholar]
- 15.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 16.Goehring, A. S., D. A. Mitchell, A. H. Tong, M. E. Keniry, C. Boone, and G. F. Sprague, Jr. 2003. Synthetic lethal analysis implicates Ste20p, a p21-activated protein kinase, in polarisome activation. Mol. Biol. Cell 14:1501-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goehring, A. S., D. M. Rivers, and G. F. J. Sprague. Urmylation: a ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed]
- 18.Haas, A. L., and T. J. Siepmann. 1997. Pathways of ubiquitin conjugation. FASEB J. 11:1257-1268. [DOI] [PubMed] [Google Scholar]
- 19.Helliwell, S. B., P. Wagner, J. Kunz, M. Deuter-Reinhard, R. Henriquez, and M. N. Hall. 1994. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5:105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochstrasser, M. 2000. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2:E153-E157. [DOI] [PubMed] [Google Scholar]
- 21.Jahngen-Hodge, J., M. S. Obin, X. Gong, F. Shang, T. R. Nowell, Jr., J. Gong, H. Abasi, J. Blumberg, and A. Taylor. 1997. Regulation of ubiquitin-conjugating enzymes by glutathione following oxidative stress. J. Biol. Chem. 272:28218-28226. [DOI] [PubMed] [Google Scholar]
- 22.Jamieson, D. J. 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14:1511-1527. [DOI] [PubMed] [Google Scholar]
- 23.Jayaraman, L., K. G. Murthy, C. Zhu, T. Curran, S. Xanthoudakis, and C. Prives. 1997. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 11:558-570. [DOI] [PubMed] [Google Scholar]
- 24.Jeong, J. S., S. J. Kwon, S. W. Kang, S. G. Rhee, and K. Kim. 1999. Purification and characterization of a second type thioredoxin peroxidase (type II TPx) from Saccharomyces cerevisiae. Biochemistry 38:776-783. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, E. S., and G. Blobel. 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147:981-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawakami, T., T. Chiba, T. Suzuki, K. Iwai, K. Yamanaka, N. Minato, H. Suzuki, N. Shimbara, Y. Hidaka, F. Osaka, M. Omata, and K. Tanaka. 2001. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20:4003-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komeili, A., K. P. Wedaman, E. K. O'Shea, and T. Powers. 2000. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151:863-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowluru, R., T. S. Kern, and R. L. Engerman. 1994. Abnormalities of retinal metabolism in diabetes or galactosemia. II. Comparison of gamma-glutamyl transpeptidase in retina and cerebral cortex, and effects of antioxidant therapy. Curr. Eye Res. 13:891-896. [DOI] [PubMed] [Google Scholar]
- 29.Kretz-Remy, C., and R. M. Tanguay. 1999. SUMO/sentrin: protein modifiers regulating important cellular functions. Biochem. Cell Biol. 77:299-309. [PubMed] [Google Scholar]
- 30.Kunz, J., R. Henriquez, U. Schneider, M. Deuter-Reinhard, N. R. Movva, and M. N. Hall. 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73:585-596. [DOI] [PubMed] [Google Scholar]
- 31.Lammer, D., N. Mathias, J. M. Laplaza, W. Jiang, Y. Liu, J. Callis, M. Goebl, and M. Estelle. 1998. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 12:914-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, J., D. Spector, C. Godon, J. Labarre, and M. B. Toledano. 1999. A new antioxidant with alkyl hydroperoxide defense properties in yeast. J. Biol. Chem. 274:4537-4544. [DOI] [PubMed] [Google Scholar]
- 33.Liakopoulos, D., G. Doenges, K. Matuschewski, and S. Jentsch. 1998. A novel protein modification pathway related to the ubiquitin system. EMBO J. 17:2208-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longtine, M. S., A. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 35.Lorenz, M. C., and J. Heitman. 1995. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J. Biol. Chem. 270:27531-27537. [DOI] [PubMed] [Google Scholar]
- 36.Mahajan, R., L. Gerace, and F. Melchior. 1998. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J. Cell Biol. 140:259-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall, H. E., K. Merchant, and J. S. Stamler. 2000. Nitrosation and oxidation in the regulation of gene expression. FASEB J. 14:1889-1900. [DOI] [PubMed] [Google Scholar]
- 38.Meyer, M., R. Schreck, and P. A. Baeuerle. 1993. H2O2 and antioxidants have opposite effects on activation of NF-κB and AP-1 in intact cells: AP-1 as secondary antioxidant-responsive factor. EMBO J. 12:2005-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell, D. A., and G. F. Sprague. 2001. The phosphotyrosyl phosphatase activator, Ncs1p (Rrd1p), functions with Cla4p to regulate the G(2)/M transition in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:488-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nathan, C., and M. U. Shiloh. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 97:8841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen-Nhu, N., and B. Knoops. 2002. Alkyl hydroperoxide reductase 1 protects Saccharomyces cerevisiae against metal ion toxicity and glutathione depletion. Toxicol. Lett. 135:219-228. [DOI] [PubMed] [Google Scholar]
- 42.Park, S. G., M. K. Cha, W. Jeong, and I. H. Kim. 2000. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem. 275:5723-5732. [DOI] [PubMed] [Google Scholar]
- 43.Rea, G., M. Laurenzi, E. Tranquilli, R. D'Ovidio, R. Federico, and R. Angelini. 1998. Developmentally and wound-regulated expression of the gene encoding a cell wall copper amine oxidase in chickpea seedlings. FEBS Lett. 437:177-182. [DOI] [PubMed] [Google Scholar]
- 44.Rempola, B., A. Kaniak, A. Migdalski, J. Rytka, P. P. Slonimski, and J. P. di Rago. 2000. Functional analysis of RRD1 (YIL153w) and RRD2 (YPL152w), which encode two putative activators of the phosphotyrosyl phosphatase activity of PP2A in Saccharomyces cerevisiae. Mol. Gen. Genet. 262:1081-1092. [DOI] [PubMed] [Google Scholar]
- 45.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 46.Rojkind, M., J. A. Dominguez-Rosales, N. Nieto, and P. Greenwel. 2002. Role of hydrogen peroxide and oxidative stress in healing responses. Cell Mol. Life Sci. 59:1872-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rose, M. D., F. Winston, and P. Hieter (ed.). 1990. Methods in yeast genetics. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 48.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 49.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 51.Taylor, A., and T. Nowell. 1997. Oxidative stress and antioxidant function in relation to risk for cataract. Adv. Pharmacol. 38:515-536. [DOI] [PubMed] [Google Scholar]
- 52.Verdoucq, L., F. Vignols, J. P. Jacquot, Y. Chartier, and Y. Meyer. 1999. In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J. Biol. Chem. 274:19714-19722. [DOI] [PubMed] [Google Scholar]
- 53.Weisman, R., and M. Choder. 2001. The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J. Biol. Chem. 276:7027-7032. [DOI] [PubMed] [Google Scholar]
- 54.Wong, C. M., Y. Zhou, R. W. Ng, H. F. Kung, and D. Y. Jin. 2002. Cooperation of yeast peroxiredoxins Tsa1p and Tsa2p in the cellular defense against oxidative and nitrosative stress. J. Biol. Chem. 277:5385-5394. [DOI] [PubMed] [Google Scholar]
- 55.Zhu, H., M. Bilgin, R. Bangham, D. Hall, A. Casamayor, P. Bertone, N. Lan, R. Jansen, S. Bidlingmaier, T. Houfek, T. Mitchell, P. Miller, R. A. Dean, M. Gerstein, and M. Snyder. 2001. Global analysis of protein activities using proteome chips. Science 293:2101-2105. [DOI] [PubMed] [Google Scholar]