Abstract
Experimental allergic encephalomyelitis (EAE) is an animal model for multiple sclerosis induced by stimulating myelin basic protein (MBP)-specific T cells. The MBP-specific repertoire in B10.PL mice is shaped by tolerance mechanisms that eliminate MBP121–150–specific T cells. In contrast, MBPAc1–11–specific T cells escape tolerance and constitute the encephalitogenic repertoire. To determine if this differential tolerance is caused by differences in the abundance of MBP epitopes generated by processing, MBP peptides were eluted from I-Au complexes and analyzed by mass spectrometry. Peptides were identified from both the NH2-terminal and MBP121–150 regions. Unexpectedly, MBPAc1–18 and Ac1–17, which contain the MBPAc1–11 epitope, were much more abundant than MBP121–150 peptides. The results demonstrate that competition between two I-Au binding registers, a low affinity register defined by MBPAc1–11 and a high affinity register defined by MBP5–16, prevents most of the NH2-terminal naturally processed peptides from binding in the MBPAc1–11 register. The small fraction of MBPAc1–18 bound in the MBPAc1–11 register is not sufficient to induce tolerance but provides a ligand for MBPAc1–11–specific T cells during disease. These results provide a basis for both the lack of tolerance to MBPAc1–11 and the ability of this epitope to become a target during autoimmunity.
Keywords: experimental allergic encephalomyelitis, antigen processing, T cell tolerance, MHC class II, MBP epitopes
Introduction
The TCR repertoire generated by random rearrangement of V, D, and J gene segments includes TCRs specific for self-antigens. Most self-reactive T cells are eliminated in the thymus by negative selection mediated by high avidity interactions with peptide/MHC ligands (1). Peripheral tolerance mechanisms also eliminate or render nonresponsive T cells that recognize self-antigens under noninflammatory conditions (2). Nevertheless, some T cells escape both central and peripheral tolerance and cause autoimmune disease when activated under inflammatory conditions. This phenomenon is the basis for the induction of experimental allergic encephalomyelitis (EAE), an animal model of multiple sclerosis (MS). EAE is induced by activating self-reactive T cells via immunization with myelin antigens in complete Freund's adjuvant or by adoptive transfer of myelin-specific T cells (3). In the B10.PL strain, almost all T cells primed by immunization with myelin basic protein (MBP) recognize a single I-Au–restricted epitope contained within MBPAc1–11 (4). Initially, this observation was interpreted to mean that MBPAc1–11 was the only epitope processed and presented by I-Au molecules and recognized by T cells. However, our previous studies showed that the MBP-specific repertoire in B10.PL mice is significantly influenced by immune tolerance mechanisms. In B10.PL MBP-deficient shiverer (MBP−/−) mice, immunization with MBP elicits strong T cell responses directed toward MBP121–150 while the MBPAc1–11–specific T cell response is a minor component of the total MBP response (5). Tolerance mechanisms in MBP+/+ wild-type mice eliminate MBP121–150-specific but not MBPAc1–11–specific T cells.
We proposed that differences in the stability of peptide/MHC ligands caused the differential tolerance induction of MBPAc1–11 and MBP121–150–specific T cells. Two synthetic peptides, MBP125–135 and MBP136–146, stimulated T cells from MBP-primed mice and both peptides exhibit high affinity for I-Au (6). In contrast, MBPAc1–11 binds I-Au poorly (7). These data suggested that one or both of the MBP125–135 and 136–146 epitopes are presented on the cell surface in greater abundance than MBPAc1–11 and therefore mediate tolerance more effectively. Analyses of TCR transgenic (Tg) mice supported this notion. MBP121–150-specific Tg T cells undergo central tolerance mediated by bone marrow–derived cells. MBP121–150 epitopes are presented constitutively in the periphery in sufficient quantity to activate naive MBP121–150-specific Tg T cells adoptively transferred into wild-type mice (8). However, insufficient ligand is presented in vivo to induce central tolerance of MBPAc1–11–specific T cells (9) or trigger their activation in the periphery after adoptive transfer (8).
To provide direct support for this model of differential epitope display, we analyzed peptides eluted from I-Au purified from MBP-pulsed APCs. We expected to detect predominantly MBP121–150 peptides and few or no peptides containing MBPAc1–11. Surprisingly, the predominant MBP peptides eluted from I-Au were MBPAc1–17 and Ac1–18. A nested set of peptides containing MBP125–135 was also detected, but the abundance was 20-fold less than MBPAc1–17 and Ac1–18. We demonstrate that the abundance of MBPAc1–17 and Ac1–18 results from the presence of a high-affinity binding register, MBP5–16, that was previously described as “absolutely cryptic” (10). The results show that most, but not all, NH2-terminal peptides are bound in the MBP5–16 register and are not recognized by MBPAc1–11–specific T cells. Our data define the naturally processed MBP peptides and identify the type of APC that presents tolerogenic MBP121–150 epitopes in vivo. The results provide an explanation for how MBPAc1–11–specific T cells escape tolerance but mediate disease under inflammatory conditions.
Materials and Methods
Mice.
B10.PL(73 NS)/Sn were obtained from The Jackson Laboratory. MBP−/− B10.PL mice, MBP1–11- and MBP121–150-specific TCR transgenic mice have been described previously (8, 9, 11).
Sample Preparation for Mass Spectrometry.
MBP was isolated using a modified protocol of Martenson et al. (12). Briefly, frozen mouse brains (Pel-Freez Biologicals) were pulverized and lipids extracted in chloroform/methanol (2:1). Dried residue was homogenized in 0.3 M HCl and the supernatant was clarified by centrifugation, dialyzed against 10 mM acetic acid, and lyophilized. Single cell suspensions were prepared from the spleen and T cell–depleted using anti-TCR biotin (BD Biosciences) and streptavidin-labeled magnetic beads (Dynal). T cell–depleted splenocytes (2 × 106 cells/ml in RPMI) were incubated for 16 h at 37°C with or without MBP (95 μg/ml). I-Au was isolated and peptides eluted according to Luckey (13) using the anti–I-Au 10–3.6.2 antibody (∼1 mg/108 cells) bound to Protein A beads. Peptides were eluted with 10% acetic acid through a 10 kD cutoff spinfilter (Millipore).
Mass Spectrometry.
Approximately 6 × 106 cell equivalents (ce) of sample spiked with 2 fmol angiotensin I was gradient-eluted (C18 microcapillary, acetonitrile:water with 0.1 mM acetic acid, 0–70% in 40 min at 57 nL/min) directly into a home-built FT ion cyclotron resonance mass spectrometer (FT-ICR MS) as described previously (14). Full-scan mass spectra (300 ≤ m/z ≤ 5,000) were collected at one scan per second, with typical resolution of 5,000–10,000 amu. Subtractive analysis software aligned the data based on the shapes of their total ion chromatograms (TIC) and the similarity of masses found in scans at TIC peaks. Multiple charged ions were deconvoluted and compared among MBP pulsed and nonpulsed data sets. For peptide identity verification, ∼6 × 107 ce of the MBP-pulsed sample were gradient eluted into a ThermoFinnigan LCQ Deca ion trap mass spectrometer (ThermoFinnigan) to collect ions with m/z of 300–2,000 amu and predetermined precursor ions were subjected to collision-activated dissociation. MS/MS spectra were manually interpreted and compared with the synthetic peptide MS/MS fragmentation patterns. Coelution studies were performed by adding 60 fmol of synthetic peptide to a 6 × 107 ce sample. Peptide concentrations were estimated by normalizing the ion intensities to the angiotensin internal standard.
Peptide Binding Assays.
Peptides were synthesized using standard Fmoc chemistry on an Applied Biosystems 433A peptide synthesizer (Foster City) and labeled with N-hydroxysuccinimidyl ester of 5(6)-carboxyfluorescein before cleavage. Labeling was on either the NH2 terminus or, for N-acetylated peptides, on a COOH-terminal lysine selectively deprotected with 2% hydrazine. Peptides were high performance liquid chromatography-purified and their identity confirmed by mass spectrometry. Detergent soluble I-Au was prepared as described (6). A solution of I-Au (50–100 nM in 0.2 mM dodecyl maltoside/PBS) was incubated with excess peptide at pH 5.3 at 37°C. Unbound peptide was removed by size exclusion (Sephadex G50-SF) at 4°C, pH 7.4. The amount of peptide bound to I-Au was assessed by high performance size exclusion chromatography (300 mm × 7.8 mm, Biosep-SEC-S 3000; Phenomenex) as the amount of fluorescence associated with the protein peak. Loss in fluorescence as a function of time was fit to an exponential function to obtain dissociation half-times.
Detection of MBP-presenting APCs.
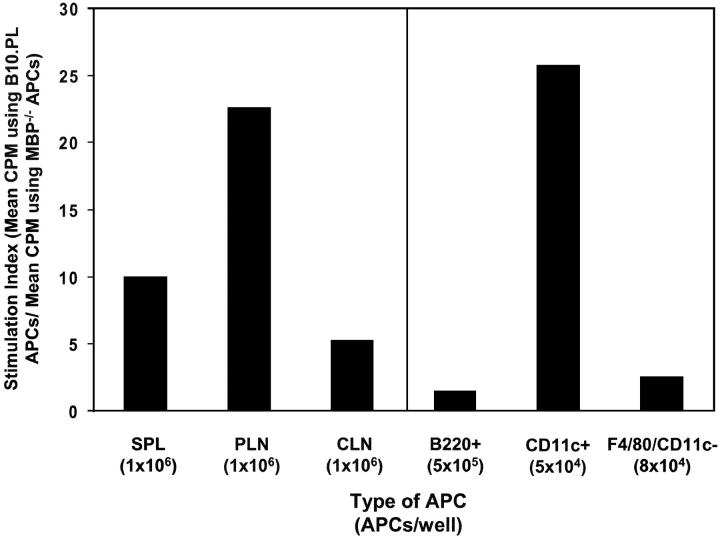
All antibodies were from BD Biosciences except those indicated. T cells from MBP−/− MBP121–150-specific TCR Tg mice were purified using biotinylated anti-Vα2.3 and AutoMACS (Miltenyi Biotec) cell separation. B220+ B cells were purified using biotinylated anti-B220 and AutoMACS cell separation. Dendritic cells (CD11c+) and macrophages (F4/80+/ CD11c−) were isolated from splenocytes depleted of TCR+ and B220+ cells by AutoMACS cell separation and sorted based on F4/80-PE (Caltag) and CD11c-FITC using a FACS Vantage™ (Becton Dickinson) cell sorter. Purity of all populations was greater than 86%. APC subsets (APC numbers/well are indicated in Fig. 3) were cultured with 5 × 104 purified MBP121–150–specific T cells in 96-well round bottom plates in complete RPMI. Cultures were incubated 48 h, pulsed with [H3]-thymidine, and harvested 18 h later. Stimulation index is calculated as the mean CPM of T cells incubated with wild-type APCs divided by the mean CPM of T cells incubated with MBP−/− APCs.
Figure 3.
MBP121–150–specific Tg T cells respond to dendritic cells presenting endogenous MBP directly ex vivo. Stimulation index (SI) was calculated as described in methods. APCs: whole splenocytes (SPL), peripheral lymph node cells (PLN), or superficial cervical lymph nodes cells (CLN), B cells (B220+), dendritic cells (CD11c+), and macrophages (F4/80+/CD11c−). Numbers of APCs/well are indicated in parentheses. Data on the left are responses from an individual mouse representative of 22 mice over seven experiments. Variation in SIs for different tissues was seen between individual mice. Mean, standard deviation, and median of SIs and the fraction of mice with SIs > 2 (respectively) are as follows: SPL (3.6, 2.9, 3.0, 13/22); PLN (19.1, 26.8, 7.7, 19/22); and CLN (6.7, 9.1, 4.5, 19/22). The data shown on the right are representative from two experiments.
T Cell Proliferation Assays Using Fixed APCs.
Assays were performed as described above except T cells from MBPAc1–11-specific TCR Tg mice were purified by AutoMACs separation using anti-Vα2.3 or Vβ8.1/8.2 antibodies and then sorted to remove contaminating I-Au–expressing cells using either anti-Vα2.3 or Vβ8.1/8.2 and anti-I-Ab (clone AF6–120.1, cross-reactive to I-Au). Sorted T cells were plated at 2 × 105/well with 50 uM peptide or 95 μg/ml whole MBP and 106 irradiated live or fixed (0.1% paraformaldehyde for 5 min at room temperature) splenocytes.
Results
Processing of MBP Generates Two Distinct Nested Sets of Peptides.
The large differences in affinity for I-Au observed between peptides within MBP121–150 versus MBPAc1–11 suggested that differential T cell tolerance induction reflected differences in the abundance of peptide/MHC ligands derived from these regions. A caveat to these studies is that the synthetic peptides may not represent the actual epitopes processed from MBP under noninflammatory, tolerizing conditions in vivo. Residues flanking a core epitope can influence binding to MHC by interacting with MHC residues outside the binding groove (15). Additionally, processed peptides may contain multiple epitopes that compete for MHC binding (16). These concerns motivated us to identify the peptides bound to I-Au after processing of whole MBP.
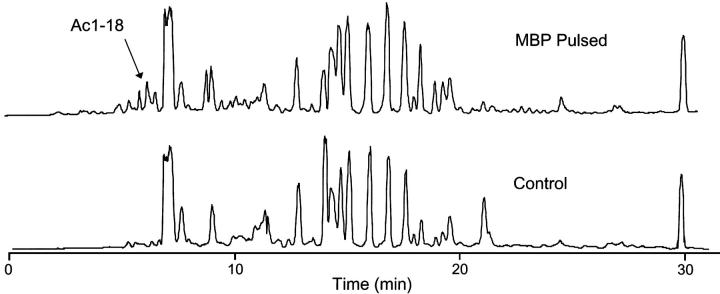
We used differential analysis to identify the MBP peptides bound to I-Au. B10.PL splenocytes were incubated overnight with whole MBP and this positive sample was compared with MBP−/− B10.PL splenocytes incubated without MBP. I-Au molecules were purified and peptides were eluted from both the MBP positive and negative samples. Peptide masses were analyzed using Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR-MS). Total ion chromatograms for each sample are shown in Fig. 1 . Peptides present in the positive (MBP pulsed) and absent in the negative FT-ICR-MS datasets were identified using subtractive analysis software.
Figure 1.
Total ion chromatograms (TIC) from the FT-ICR MS analysis of peptide from MBP-pulsed and unpulsed samples. Mass spectra acquisitions were recorded at 1 s intervals. The majority of the peptides eluted over a ∼28 min period during the gradient.
We identified two nested sets of MBP peptides. The identity of multiple peptides from each set was confirmed by collision-activated dissociation analyses (CAD) and coelution (Table I). One peptide set corresponded to the NH2 terminus of MBP and the other set was derived from the MBP121–150 region. The most abundant MBP peptides were MBPAc1–17 and Ac1–18, which were ∼20 times more abundant than peptides from the MBP121–150 region. Within the MBP121–150 region, only peptides containing the predicted core epitope MBP125–135 and not MBP136–146 were identified (detection limit estimated at one copy/cell).
Table I.
MBP Peptides Eluted from I-Au Identified by Mass Spectrometry
| MBP peptidea | Observed mass | Calculated mass | Scan no. | Abundance | Copy/cellb | CAD | Coelution |
|---|---|---|---|---|---|---|---|
| 3-16 | 545.307 | 545.309 | 153 | 11.7 | 2.4 | ||
| 3-17 | 430.994 | 430.992c | 204 | 51.9 | 10.4 | ||
| 2-16 | 430.994 | 430.992c | 204 | 51.9 | 10.5 | ||
| 4-17 | 531.634 | 531.634 | 190 | 181.6 | 36.3 | Yes | Yes |
| Ac1-16 | 612.010 | 612.003 | 321 | 90.8 | 18.1 | ||
| Ac1-17 | 641.015 | 641.013 | 311 | 2,410.9 | 484.0 | Yes | Yes |
| Ac1-18 | 674.696 | 674.696 | 328 | 3,112.8 | 622.0 | Yes | Yes |
| Ac1-19 | 718.738 | 718.718 | 438 | 33.4 | 6.7 | ||
| Ac1-20 | 756.738 | 756.718 | 439 | 122.1 | 24.4 | ||
| Ac1-22 | 619.823 | 619.814c | 428 | 28.4 | 5.7 | ||
| 124-137 | 499.583 | 499.580 | 183 | 46.2 | 9.2 | Yes | |
| 124-138 | 548.609 | 548.602 | 319 | 32.5 | 6.3 | Yes | |
| 123-138 | 567.616 | 567.610 | 317 | 31.4 | 6.5 | Yes | |
| 123-136 | 499.583 | 499.580 | 183 | 46.2 | 9.2 | Yes | |
| 122-136 | 548.609 | 548.602 | 319 | 32.5 | 6.3 | Yes | Yes |
| 121-136 | 567.616 | 567.610 | 317 | 31.4 | 6.5 | Yes | |
| 122-137 | 567.616 | 567.610 | 317 | 31.4 | 6.5 | ||
| 120-137 | 450.227 | 450.222c | 363 | 16.8 | 3.4 |
Peptide identity assigned by matching observed and calculated masses, CAD column indicates confirmed identities. Numbering is based on the sequence of the 18.5 kD MBP isoform.
Based on copy number/cell of MBPAc1-18 which was determined by comparison to angiotensin control.
Observed masses are from +4 charge state peptides, all other masses are from +3 charge state peptides.
The MBP5–16 Binding Register Accounts for the Abundance of NH2-terminal Peptides.
The paradoxical observation that MBPAc1–17 and Ac1–18 are the most abundant peptides generated by processing MBP despite the low affinity of MBPAc1–11 for I-Au could be due to the presence of a second more stable binding register within these peptides. This idea was initially suggested by the observation that rat MBPAc5–20 and MBPAc9–20 inhibited EAE induced by immunization with rat MBPAc1–11 by competition for I-Au binding (17). Fairchild et al. (10) investigated this binding register in mouse MBP and implicated the tyrosine at MBP12 as the anchor residue binding in the p6 pocket of I-Au. The core peptide binding in this register, MBP5–16, would be favored relative to the MBPAc1–11 register, which places an unfavorable lysine (MBP4) in the p6 pocket. However, these earlier studies concluded that the naturally-processed peptides derived from whole MBP could not contain both MBPAc1–11 and MBP5–16 (10, 17). Our data demonstrating that MBP processing primarily generates NH2-terminal peptides containing both registers required us to reconsider this interpretation.
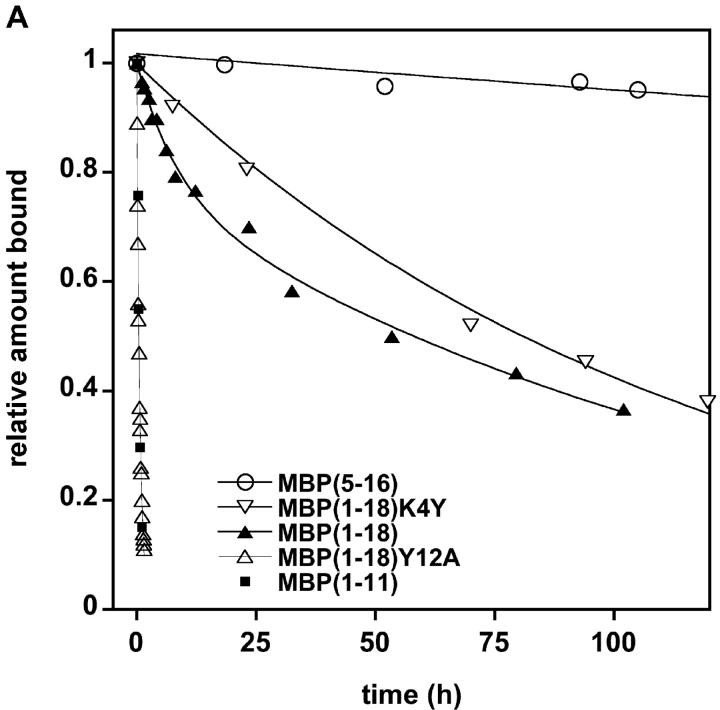
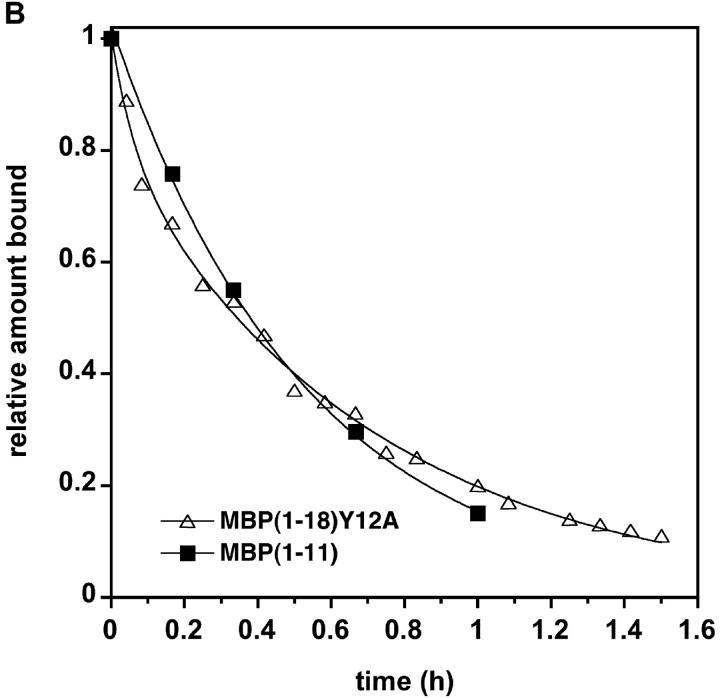
We investigated whether the NH2-terminal peptides could bind I-Au using both binding registers by analyzing the dissociation kinetics of MBPAc1–18 as well as smaller peptides representing the individual binding registers from solubilized I-Au. MBP5–16/I-Au dissociated so slowly that a half-life was difficult to measure (t1/2 > 1,000 h). The half-life of MBPAc1–11/I-Au was only 30 min. Interestingly, MBPAc1–18/I-Au dissociated with biphasic kinetics, suggesting that the peptide binds in a mixture of short-lived and long-lived complexes (Fig. 2 A). Under these binding conditions (incubation with MHC molecules for 1 h at 37°C), ∼38% and 62% of the MBPAc1–18 peptides formed short- and long-lived complexes, respectively. Increasing the incubation time from 1 to 16 h increased the amount of MBPAc1–18 peptide bound in the long phase to 77% (unpublished data). Dissociation of analogue peptides with mutations in the p6 anchor residues for each register was also analyzed. The MBPAc1–18Y12A substitution, which abrogates binding in the MBP5–16 register, exhibits single phase, rapid dissociation essentially identical to the dissociation of MBPAc1–11 (Fig. 2, A and B). Conversely, the MBPAc1–18K4Y substitution, which replaces the unfavorable lysine with a tyrosine in the anchor position of the MBPAc1–11 register, exhibits single phase, slow-dissociation kinetics with a half-life very similar to the slow-dissociation phase of native MBPAc1–18 (Fig. 2 A). These data confirm the utilization of two binding registers within MBPAc1–18 and suggest that most, but not all, of MBPAc1–18 is bound to I-Au in the MBP5–16 register.
Figure 2.
MBPAc1–18 demonstrates biphasic dissociation. (A) Dissociation of fluorescein-labeled MBP peptides from soluble I-Au. MBP peptides were incubated with soluble I-Au for 1 h at 37°C. For MBPAc1–18, ∼38% of the original peptide bound was bound in the faster phase: t1/2fast = 3.7 h. About 62% of the initial peptide is bound in the slow phase: t1/2slow = 117 h. (B) Expanded time scale of panel A showing the monophasic fast dissociation of both MBPAc1–18Y12A and MBPAc1–11.
MBPAc1–11–specific T Cells Recognize I-Au/Ac1–18 MBP Complexes on Fixed APCs.
The data described above suggest that priming of MBPAc1–11–specific T cells by immunization with whole MBP occurs through recognition of the small amount of MBPAc1–17 and Ac1–18 bound in the MBPAc1–11 register. This interpretation predicts that MBPAc1–11–specific T cells should respond to MBPAc1–18 without additional processing. To test this prediction, we analyzed proliferation of T cells from two different MBPAc1–11–specific TCR Tg lines (9, 11) using MBPAc1–18 and fixed or live APCs. While only live APCs could present whole MBP to the T cells, both T cells responded well to MBPAc1–18 and MBPAc1–11 on fixed APCs (SIs of 1476 and 784 for MBPAc1–18; 791 and 604 for MBPAc1–11).
Dendritic Cells Constitutively Present MBP121–150 Epitopes In Vivo.
Our results indicated that peptides bound in the MBP5–16 and MBP125–135 registers form very stable complexes with I-Au. This suggested that MBP121–150-specific TCR Tg T cells might detect MBP125–135/I-Au complexes on APCs directly ex vivo and allow us to identify the type of APC presenting these epitopes. Mononuclear cells from the spleen and lymph nodes were tested for their ability to stimulate MBP121–150–specific T cells in vitro without addition of exogenous MBP. MBP121–150–specific T cells isolated from MBP−/− TCR Tg mice proliferated specifically in response to either irradiated splenocytes, pooled brachial, axial, and inguinal lymph node cells or superficial cervical lymph node cells isolated from wild-type MBP+/+ but not MBP−/− mice (Fig. 3) . Splenocytes were fractionated based on expression of B220, CD11c, and F4/80. Only the CD11c+ population triggered significant proliferation (SI greater than 3), indicating that dendritic cells are the major cell type presenting endogenously-derived MBP121–150 epitopes. Although the amount of specific T cell proliferation varied for different tissues, no correlation was observed with either the number of CD11c+ cells or the level of I-Au expression on CD11c+ cells in different tissues, suggesting that differences in proliferation may reflect variation in the amount of MBP present in a tissue (unpublished data).
Discussion
We identified the peptides generated from processing whole MBP in B10.PL mice and the APCs that normally present these peptides in vivo. The most abundant MBP peptides eluted from I-Au are MBPAc1–17 and MBPAc1–18. A second set of nested peptides containing the core MBP125–135 was found at much lower abundance. The observation that more NH2-terminal than MBP125–135 peptides are presented by APCs is precisely the opposite of what our T cell tolerance studies predicted. This paradox is explained by the demonstration that MBPAc1–18 binds to I-Au using two binding registers, MBPAc1–11 and MBP5–16. The biphasic dissociation kinetics of MBPAc1–18, as well as the dissociation rates of analogue peptides shown in Fig. 2, suggest that most of the MBPAc1–17 and MBPAc1–18 peptides are bound in the MBP5–16 register. Processed NH2-terminal peptides truncated at the NH2 terminus (Table I) must also bind in the MBP5–16 register because they lack important residues required for binding in the MBPAc1–11 register (18). MBPAc1–11–specific T cells do not recognize NH2-terminal peptides bound in the MBP5–16 register and therefore do not encounter sufficient ligand to undergo tolerance induction.
Our data resolve the question of which peptides are generated by processing the NH2-terminal region of MBP. This question has been debated for many years because MBPAc1–11–specific T cells respond to processed whole MBP in vivo, yet MBPAc1–11 binds very poorly to I-Au in vitro. Structural studies confirm that MBPAc1–11 binds I-Au in an energetically unfavorable register in which one third of the MHC cleft is empty (18). MBP peptides that extend beyond MBPAc1–11 exhibit stronger binding via a second register (7, 10), however, it was hypothesized that processed peptides could not contain both registers because the more stable register would prevent binding in the MBPAc1–11 register. This idea was supported by the lack of MBPAc1–11–specific T cell hybridoma responses to peptides longer than MBPAc1–13 on fixed APCs in vitro (10). In contrast to these data, we demonstrate that T cells expressing two different Tg MBPAc1–11–specific TCRs respond well to MBPAc1–18 on fixed APCs. The Tg T cells may be more sensitive compared with the hybridomas used in earlier experiments. Interestingly, we show that MBPAc1–11 register binding is enhanced in the longer MBPAc1–18 peptide relative to MBPAc1–11. The rapid-dissociation phase for MBPAc1–18 gives an apparent half-time of 3.7 h as compared with the 30 min half-life observed for MBPAc1–11/I-Au complexes. This is consistent with our finding that 10-fold less MBPAc1–18 compared with MBPAc1–11 is needed to stimulate MBPAc1–11–specific T cells (unpublished data). Thus, the presence of the MBP5–16 register enhances presentation of the MBPAc1–11 register rather than inhibiting it.
It was initially surprising that ∼20 times more MBPAc1–17 and Ac1–18 than MBP125–135 peptides were detected in the eluted peptides because all of these peptides exhibit similar affinities for I-Au. Several factors may account for enrichment of NH2-terminal peptides. The most abundant murine MBP isoform purified from myelin includes the NH2-terminal region but excludes MBP125–135 (19), resulting in a fivefold enrichment of NH2-terminal sequences. The ends of a protein may also be easier to denature and facilitate processing than sequences in the middle of the protein. Lastly, the N-acetyl group may inhibit exopeptidases and protect peptides from further degradation, consistent with the observation that nonacetylated NH2-terminal peptides are detected at comparable levels to the MBP125–135 peptides (Table I) while MBPAc1–17 and Ac1–18 are more abundant.
Competition for binding to I-Au between the MBPAc1–11 and MBP5–16 binding registers provides a mechanistic explanation for the lack of tolerance induction in MBPAc1–11–specific T cells. The bias toward the MBP5–16 register observed in vitro after a 1 h incubation of MBPAc1–18 with I-Au could be even stronger in vivo because increasing the incubation to 16 h increased the percentage of peptide bound in the MBP5–16 register. The 1 h incubation time may underestimate the time that I-Au is exposed to MBP peptides in vivo, considering that there is approximately a 12 h delay before peptide derived from endocytosed protein is processed and presented on the surface of activated dendritic cells (20). H-2M activity may also influence register utilization and is likely to favor the more energetically stable MBP5–16 register.
The observation that a tolerogenic MBP epitope is detected ex vivo only on dendritic cells supports the dominant role believed to be played by these APCs in maintaining tolerance to auto-antigens. The fact that minute levels of MBP125–135 peptides on dendritic cells induces tolerance suggests that very little MBPAc1–17 and Ac1–18 is normally bound in the MBPAc1–11 register because it is insufficient to induce tolerance in MBPAc1–11–specific T cells. However, peptide bound in the MBPAc1–11 register is detected by activated MBPAc1–11–specific T cells in vivo during disease. Detection of MBPAc1–11-bound Ac1–18 may be enhanced during inflammation by increased presentation of all MBP epitopes due to increased MBP degradation, increased expression of costimulatory molecules on APCs and/or increased sensitivity of activated T cells to antigenic stimulation. Thus, the dynamic competition between two binding registers allows most MBPAc1–11–specific T cells to escape tolerance induction by dendritic cells presenting self-antigen under noninflammatory conditions but allows MBPAc1–11–specific T cells to see sufficient amounts of their epitope under inflammatory conditions to initiate autoimmunity.
Acknowledgments
The authors would like to acknowledge Dr. Thea Brabb, Sarah Cabbage, Antoine Perchellet, and Ingunn Stromnes for critical reading of the manuscript.
Research was funded by National Institutes of Health grants R01-NS35126 to J. Goverman and RO1 AI33993 to D.F. Hunt and T32 HG00035 supporting A. Seamons.
References
- 1.Anderson, G., N.C. Moore, J.J. Owen, and E.J. Jenkinson. 1996. Cellular interactions in thymocyte development. Annu. Rev. Immunol. 14:73–99. [DOI] [PubMed] [Google Scholar]
- 2.van Parijs, L., and A.K. Abbas. 1998. Homeostasis and self-tolerance in the immune system: turning lymphocytes off. Science. 280:243–248. [DOI] [PubMed] [Google Scholar]
- 3.Zamvil, S.S., and L. Steinman. 1990. The T lymphocyte in experimental allergic encephalomyelitis. Annu. Rev. Immunol. 8:579–621. [DOI] [PubMed] [Google Scholar]
- 4.Zamvil, S.S., D.J. Mitchell, A.C. Moore, K. Kitamura, L. Steinman, and J.B. Rothbard. 1986. T-cell epitope of the autoantigen myelin basic protein that induces encephalomyelitis. Nature. 324:258–260. [DOI] [PubMed] [Google Scholar]
- 5.Harrington, C.J., A. Paez, T. Hunkapiller, V. Mannikko, T. Brabb, M. Ahearn, C. Beeson, and J. Goverman. 1998. Differential tolerance is induced in T cells recognizing distinct epitopes of myelin basic protein. Immunity. 8:571–580. [DOI] [PubMed] [Google Scholar]
- 6.Loftus, C., E. Huseby, P. Gopaul, C. Beeson, and J. Goverman. 1999. Highly cross-reactive T cell responses to myelin basic protein epitopes reveal a nonpredictable form of TCR degeneracy. J. Immunol. 162:6451–6457. [PubMed] [Google Scholar]
- 7.Fairchild, P.J., R. Wildgoose, E. Atherton, S. Webb, and D.C. Wraith. 1993. An autoantigenic T cell epitope forms unstable complexes with class II MHC: a novel route for escape from tolerance induction. Int. Immunol. 5:1151–1158. [DOI] [PubMed] [Google Scholar]
- 8.Huseby, E.S., B. Sather, P.G. Huseby, and J. Goverman. 2001. Age-dependent T cell tolerance and autoimmunity to myelin basic protein. Immunity. 14:471–481. [DOI] [PubMed] [Google Scholar]
- 9.Goverman, J., A. Woods, L. Larson, L.P. Weiner, L. Hood, and D.M. Zaller. 1993. Transgenic mice that express a myelin basic protein-specific T cell receptor develop spontaneous autoimmunity. Cell. 72:551–560. [DOI] [PubMed] [Google Scholar]
- 10.Fairchild, P.J., H. Pope, and D.C. Wraith. 1996. The nature of cryptic epitopes within the self-antigen myelin basic protein. Int. Immunol. 8:1035–1043. [DOI] [PubMed] [Google Scholar]
- 11.Lafaille, J.J., K. Nagashima, M. Katsuki, and S. Tonegawa. 1994. High incidence of spontaneous autoimmune encephalomyelitis in immunodeficient anti-myelin basic protein T cell receptor transgenic mice. Cell. 78:399–408. [DOI] [PubMed] [Google Scholar]
- 12.Martenson, R.E., G.E. Deibler, and M.W. Kies. 1969. Microheterogeneity of guinea pig myelin basic protein. J. Biol. Chem. 244:4261–4267. [PubMed] [Google Scholar]
- 13.Luckey, C.J., J.A. Marto, M. Partridge, E. Hall, F.M. White, J.D. Lippolis, J. Shabanowitz, D.F. Hunt, and V.H. Engelhard. 2001. Differences in the expression of human class I MHC alleles and their associated peptides in the presence of proteasome inhibitors. J. Immunol. 167:1212–1221. [DOI] [PubMed] [Google Scholar]
- 14.Martin, S.E., J. Shabanowitz, D.F. Hunt, and J.A. Marto. 2000. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 72:4266–4274. [DOI] [PubMed] [Google Scholar]
- 15.Unanue, E.R. 2002. Perspective on antigen processing and presentation. Immunol. Rev. 185:86–102. [DOI] [PubMed] [Google Scholar]
- 16.Deng, H., R. Apple, M. Clare-Salzler, S. Trembleau, D. Mathis, L. Adorini, and E. Sercarz. 1993. Determinant capture as a possible mechanism of protection afforded by major histocompatibility complex class II molecules in autoimmune disease. J. Exp. Med. 178:1675–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakai, K., S.S. Zamvil, D.J. Mitchell, S. Hodgkinson, J.B. Rothbard, and L. Steinman. 1989. Prevention of experimental encephalomyelitis with peptides that block interaction of T cells with major histocompatibility complex proteins. Proc. Natl. Acad. Sci. USA. 86:9470–9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, X.L., C. Radu, J. Sidney, A. Sette, E.S. Ward, and K.C. Garcia. 2002. Structural snapshot of aberrant antigen presentation linked to autoimmunity: the immunodominant epitope of MBP complexed with I-Au. Immunity. 17:83–94. [DOI] [PubMed] [Google Scholar]
- 19.Barbarese, E., J.H. Carson, and P.E. Braun. 1978. Accumulation of the four myelin basic proteins in mouse brain during development. J. Neurochem. 31:779–782. [DOI] [PubMed] [Google Scholar]
- 20.Turley, S.J., K. Inaba, W.S. Garrett, M. Ebersold, J. Unternaehrer, R.M. Steinman, and I. Mellman. 2000. Transport of peptide-MHC class II complexes in developing dendritic cells. Science. 288:522–527. [DOI] [PubMed] [Google Scholar]