Abstract
We evaluated the effects of ectopic granulocyte/macrophage colony-stimulating factor (GM-CSF) signals on hematopoietic commitment and differentiation. Lineage-restricted progenitors purified from mice with the ubiquitous transgenic human GM-CSF receptor (hGM-CSFR) were used for the analysis. In cultures with hGM-CSF alone, hGM-CSFR–expressing (hGM-CSFR+) granulocyte/monocyte progenitors (GMPs) and megakaryocyte/erythrocyte progenitors (MEPs) exclusively gave rise to granulocyte/monocyte (GM) and megakaryocyte/erythroid (MegE) colonies, respectively, providing formal proof that GM-CSF signals support the GM and MegE lineage differentiation without affecting the physiological myeloid fate. hGM-CSFR transgenic mice were crossed with mice deficient in interleukin (IL)-7, an essential cytokine for T and B cell development. Administration of hGM-CSF in these mice could not restore T or B lymphopoiesis, indicating that enforced GM-CSF signals cannot substitute for IL-7 to promote lymphopoiesis. Strikingly, >50% hGM-CSFR+ common lymphoid progenitors (CLPs) and >20% hGM-CSFR+ pro-T cells gave rise to granulocyte, monocyte, and/or myeloid dendritic cells, but not MegE lineage cells in the presence of hGM-CSF. Injection of hGM-CSF into mice transplanted with hGM-CSFR+ CLPs blocked their lymphoid differentiation, but induced development of GM cells in vivo. Thus, hGM-CSF transduces permissive signals for myeloerythroid differentiation, whereas it transmits potent instructive signals for the GM differentiation to CLPs and early T cell progenitors. These data suggest that a majority of CLPs and a fraction of pro-T cells possess plasticity for myelomonocytic differentiation that can be activated by ectopic GM-CSF signals, supporting the hypothesis that the down-regulation of GM-CSFR is a critical event in producing cells with a lymphoid-restricted lineage potential.
Keywords: commitment, lineage conversion, cytokine, plasticity
Introduction
Hematopoietic stem cells (HSCs)*give rise to committed progenitors and undergo terminal differentiation into mature blood cells. Cytokines are one of the most important exogenous factors capable of regulating hematopoietic development. All developing blood cells express a variety of cytokine receptors, which transduce signals supportive of cell differentiation and survival. However, it is still unclear whether cytokines can play a role in hematopoietic lineage determination (1, 2). Lineage-specific action of cytokines could be achieved by several mechanisms. A diverse distribution of cytokine receptors in the hematopoietic system could directly represent the range of cell types that are responsive to cognate cytokines. Furthermore, each cell type could possess different accessibility for lineage-affiliated genes by epigenetic programs (3). These preconditions, specific for each cell type, could be important for cell lineage potential in response to cytokine signals.
It has been suggested that cytokine signals do not play a role in myeloid lineage determination. Mice with targeted mutations in myeloid cytokines or their receptors do not display a complete loss of specific lineage cells (4–7), suggesting that myeloid cytokine signals are largely redundant. Transgenic receptors for myeloid cytokines such as human GM-CSF (hGM-CSF; 8), human G-CSF (9, 10), and murine IL-5 (11) have been ubiquitously expressed in murine hematopoietic cells. In these models, binding of transgenic receptors by their cognate cytokines alone can induce the differentiation of megakaryocyte/erythroid (MegE) as well as granulocyte/monocyte (GM) lineage cells. Constitutive expression of the activated form of erythropoietin receptors (EpoRs) in bone marrow cells also supports GM differentiation (12). Finally, mice with a targeted chimeric mutation in the G-CSFR, where the cytoplasmic domain is replaced by the EpoR, do not display any abnormality in their hematopoietic development (13). Thus, signals from these myeloerythroid cytokine receptors might be largely redundant, which might play a permissive role in myeloerythroid commitment. Therefore, the lineage-specific action of these myeloid cytokines might be dependent on the lineage-specific expression of their receptors.
In contrast, T and B lymphoid development is supported by nonredundant IL-7 signals. IL-7R is a high affinity receptor complex composed of the IL-7Rα chain and the common cytokine receptor γ chain (γc). Genetic ablation of either IL-7, IL-7Rα, or γc causes a significant loss of T and B cells, resulting in SCID (14–17). IL-7 exerts “trophic” effects on αβ T cell development, but is indispensable for the rearrangement of the TCRγ chain (18) and the V segments of IgH genes (19). We and others have shown that IL-7R signals maintain T cell survival (20), and that enforced expression of Bcl-2 restored αβ T, but not γδ T or B cell development in IL-7Rα−/− and γc−/− mice (20–22). Nonetheless, the earliest lymphoid progenitors (common lymphoid progenitors [CLPs]) are present in IL-7−/− mice (23), suggesting that the role of IL-7 signals in lymphoid commitment could also be permissive.
It is of interest that the cytokine requirement for myelopoiesis is largely redundant, whereas for lymphopoiesis, it is highly dependent on lymphoid-specific IL-7 signals. It is possible that this apparent difference in cytokine dependence is simply due to differences in the distribution of cytokine receptors in the lymphoid and myeloid pathways. In fact, the earliest myeloid progenitors (common myeloid progenitors [CMPs]; 24) express multiple myeloerythroid cytokine receptors but do not express IL-7R, whereas lymphoid progenitors do not express myeloid cytokine receptors (25). Thus, it is possible that enforced GM-CSFR, like IL-7R, transduces signals that are supportive of lymphoid development because transgenic hGM-CSFR could transduce survival-promoting signals (26). Our recent studies, however, suggest that myeloid cytokine signals could exert effects on lymphoid development that are distinct from IL-7 signals. The enforced expression of IL-2 signals can instruct a fraction of CLPs to differentiate into GM cells at the expense of lymphoid differentiation (27). The ectopic IL-2 signals at the CLP stage may overlap with the GM-CSF signals, because a few percent of CLPs with retrovirally transduced hGM-CSFRα can also form GM colonies (27).
These data led us to directly analyze the effect of ectopic GM-CSF signals on lineage commitment at the lymphoid and myeloid branchpoints. We have identified myeloid- and lymphoid-committed progenitor populations in murine hematopoiesis, including CMPs, granulocyte/monocyte progenitors (GMPs), megakaryocyte/erythrocyte progenitors (MEPs; 24), and CLPs (23). For this study, we purified these progenitors from mice ubiquitously expressing the hGM-CSFRα/βc transgene (8), and tested the effect of hGM-CSF on their lineage potentials. Our results indicate that ectopic GM-CSF signals play a permissive role in lineage commitment throughout the myeloid, but not the lymphoid pathway, and induce a significant fraction of CLPs and pro-T cells to convert into the GM lineage at the expense of lymphoid differentiation. Thus, GM-CSF signals can reactivate GM-specific differentiation programs in early lymphoid progenitors, suggesting that the down-regulation of myeloid cytokine receptors is a critical event in lymphoid commitment.
Materials and Methods
Mice.
C57B6 (Ly5.1 or Ly5.2) and C57B6-H-2Ld-hGM-CSFRα/βc double transgenic mice (8) were used in this study. Expression of hGM-CSFR in all hematopoietic cells has been shown in previous studies (8). IL-7Rα−/− (14) and IL-7−/− mice (15) were obtained from The Jackson Laboratory and provided by R. Murray (DNAX Research Institute of Cellular and Molecular Biology, Palo Alto, CA), respectively. Both IL-7Rα−/− and IL-7−/− mice were crossed with C57B6 mice for seven generations. Mice were bred and maintained in the Research Animal Facility at the Dana-Farber Cancer Institute in accordance with their guidelines.
Cell Staining and Sorting.
Sorting of myeloid progenitors was accomplished by staining bone marrow cells with rat anti–IL-7Rα chain monoclonal antibodies (A7R34; eBioscience) and purified or PE-Cy5 (Tricolor)–conjugated rat antibodies specific for the following lineage markers: CD3 (CT-CD3), CD4 (RM4-5), CD8 (5H10), B220 (6B2), Gr-1 (8C5), TER119, and CD19 (6D5; Caltag). IL-7Rα+ Lin+ cells were removed with sheep anti–rat IgG–conjugated magnetic beads (Dynabeads M-450; Dynal) and the remaining cells were stained with goat PE-Cy5–conjugated anti–rat IgG (Caltag). Cells were then stained with PE-conjugated anti-FcγRII/III (2.4G2), FITC-conjugated anti-CD34 (RAM34), APC-conjugated anti-c-Kit (2B8), and biotinylated anti–Sca-1 (E13-161-7) monoclonal antibodies (BD Biosciences), followed by avidin-APC-Cy7 (Caltag). Myeloid progenitors were sorted as Lin− Sca-1− c-Kit+ CD34+ FcγRII/IIIlo (CMPs), Lin− Sca-1− c-Kit+ CD34+ FcγRII/IIIhi (GMPs), and as Lin− Sca-1− c-Kit+ CD34− FcγRII/IIIlo (MEPs) as previously described (24). Sorting of HSCs and CLPs was performed by staining bone marrow cells with lineage markers as well as FITC-conjugated anti–Sca-1, biotinylated anti–IL-7Rα, and APC-conjugated anti–c-Kit antibodies, followed by avidin-PE (Caltag). HSCs and CLPs were sorted as IL-7Rα− Lin− Sca-1hi c-Kithi and IL-7Rα+ Lin− Sca-1lo c-Kitlo populations, respectively (23). In some experiments, HSCs were further divided into CD34− long-term and CD34+ short-term HSC populations (28, 29). Detailed methodologies for staining and sorting can be found at http://www.metazoa.com (UPL2030 and UPL2001). Pro-T and pro-B cells were sorted as CD3− CD4− CD8− NK1.1− c-Kit+ CD25+ CD44+ and CD43+ B220+ IgM− cells, respectively (30). FITC-conjugated anti–mouse common β chain (βc; 9D3) antibodies (Upstate Biotechnology), and PE-conjugated anti–GM-CSFRα (S-20) antibodies (Santa Cruz Biotechnology, Inc.) were used to analyze the expression of these cytokine receptors. Cells were sorted using a highly modified double laser (488/350 nm Enterprise II plus 647 nm Spectrum) high speed FACS® (Moflo-MLS; DakoCytomation). The five-color sort using both positive and negative gates in multiple channels usually gives rise to cells that are >96% pure, avoiding co-sorting cells stained in a nonspecific manner. The sample line was washed out by 75% ethanol followed by saline between each round of sorting to eliminate remaining cells (31). The sorted cells were subjected to another sorting round using the same gate to eliminate contaminating cells and doublets (32). This second round of sorting always yielded target populations with >99% purity.
In Vitro Assays to Determine the Differentiation Potential of Progenitors.
To test myeloerythroid potential, stem or progenitor cells were cultured in IMDM-based methylcellulose media (Methocult H4100; StemCell Technologies Inc.), supplemented with 30% FBS, 1% bovine serum albumin, 2 mM l-glutamine, and 50 μM 2-mercaptoethanol. Cytokines such as 20 ng/ml murine steel factor (Slf), 10 ng/ml murine IL-3, 10 ng/ml murine IL-11, 10 ng/ml murine GM-CSF, 10 ng/ml murine thrombopoietin (Tpo), 1 U/ml human erythropoietin, and 20 ng/ml hGM-CSF were added at the start of the culture. All cytokines were purchased from R&D Systems. Colonies were enumerated under an inverted microscope consecutively from days 5 to 8. The CFU-Mix such as CFU-GEMMeg, CFU-GEM, and CFU-GEMeg was determined by Giemsa staining of cells that were picked from individual colonies using fine drawn-out Pasteur pipettes. To evaluate myeloid differentiation of CLPs and pro-T cells by hGM-CSF signals, purified single cells were sorted into Terasaki plates with IMDM (Invitrogen) containing 20% FBS. To evaluate B cell differentiation potential, cells were sorted onto irradiated (3,000 rad) S17 stromal layers in 96-well plates with IMDM with 20% FBS. All cultures were incubated at 37°C in a humidified chamber under 5% CO2.
In Vivo Reconstitution Assays.
6,000 purified CLPs from hGM-CSFR transgenic mice (C57B6-Ly5.2) were intravenously transplanted into congenic mice (C57B6-Ly5.1) after a sublethal irradiation dose (450 rad). Mice were killed on days 14 and 28 to evaluate their progeny by using Ly5.2 as a donor marker. Intrathymic injection was performed as previously described (33). 3,000 hGM-CSFR+ CLPs were directly injected into the thymus, and the phenotype of their progeny was evaluated on days 7 and 14. hGM-CSF (500 ng × 2/day) or PBS was subcutaneously administered for 5 d.
Analysis of Gene Expression from Total RNA.
Total RNA was extracted from 2,000 double-sorted cells for each population and reverse transcribed to cDNA as previously described (24). An aliquot of cDNA was analyzed for specific genes (primers provided upon request). The HPRT gene was also amplified and used as a control. PCR amplification consisted of an initial denaturation step at 94°C for 2 min, followed by 28–32 cycles at 94°C for 30 s, annealing for 30 s, and extension at 72°C for 90 s in each cycle. PCR products were subjected to electrophoresis on an ethidium bromide–stained 1.8% agarose gel. PCR amplification for each experiment was repeated for two or more independently prepared cDNA samples. Relative gene expression quantitation was accomplished by comparing the level of any transcript in the target samples to that in control cDNA prepared from 2 × 105 whole bone marrow cells, as previously reported (24). The PCR cycles for each target gene were at a point when the reaction is in the exponential phase, to obtain a linear correlation between pixel density units of the PCR products and the amount of control cDNA applied (24). After PCR amplification, each product was visualized by the Gel Doc 1000 Video Gel Documentation System, and pixel density units of each product was read by Molecular Analyst Software (Bio-Rad Laboratories).
Evaluation of TCRβ Rearrangement.
PCR analysis was used for detecting the rearrangement of the TCRβ genes as previously described (34). In brief, 2,000 sorted cells were incubated at 94°C for 10 min in 0.5% Tween D in PCR buffer before the PCR reaction. The primer sequences used for the TCRβ gene in the first PCR reaction were 5′-TAGGCACCTGTGGGGAAGAAAC-3′ (Dβ2.1 ext) and 5′-TGAGAGCTGTCTCCTACTATC-3′ (Jβ2.7 ext). 1/5 aliquots were further amplified with an internal primer pair, 5′-GTATCACGATGTAACATTGTG-3′ (Dβ2.1 int) and 5′-GGAAGCGAGAGATGTGAATC-3′ (Jβ2.7 int). The samples were denatured (at 94°C for 30 s), annealed (at 58°C for 2 min), and extended (at 72°C for 3 min) for 35 cycles. Amplified DNA was subjected to electrophoresis in a 1.2% agarose gel.
Results
Distribution of Cytokine Receptors in Hematopoiesis.
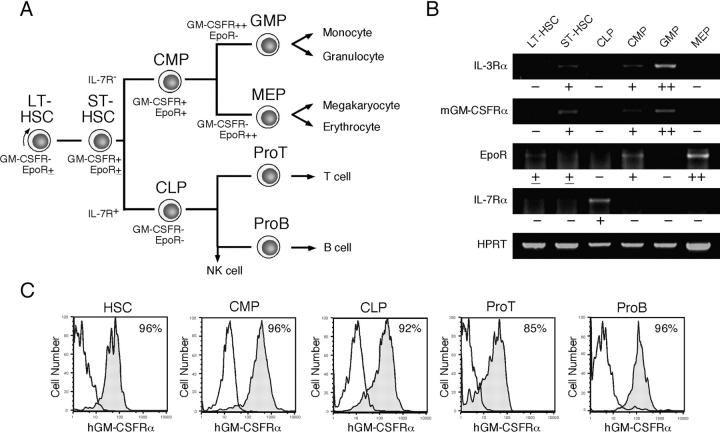
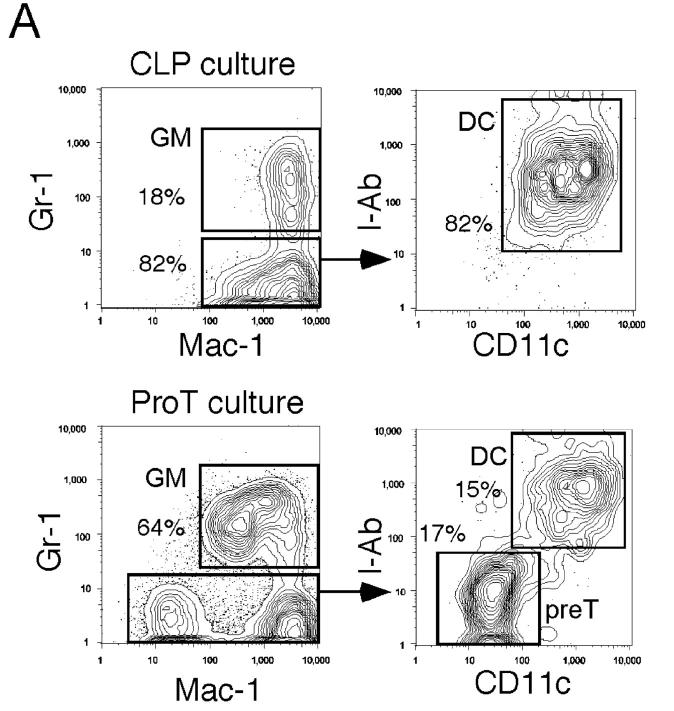
Fig. 1 A depicts the hematopoietic developmental tree and the progenitors used in this study. We analyzed the distribution of mGM-CSFRα, IL-3Rα, EpoR, and IL-7Rα by RT-PCR in murine stem and progenitor populations (23, 24). In this analysis, we separated Lin− Sca-1+ c-Kit+ HSC population (35) into CD34− Sca-1+ c-Kit+ HSCs that possess long-term reconstituting activity at the single cell level (28), and their descendants, CD34+ Sca-1+ c-Kit+ short-term HSCs (28, 29). As shown in Fig. 1 B, the expression of mGM-CSFRα and IL-3Rα is initiated at the short-term HSC stage, and gradually up-regulated along the myelomonocytic pathway including CMPs and GMPs, whereas they are down-regulated in CLPs and MEPs. FACS® analysis showed that murine βc is expressed in all hematopoietic cells (not depicted). Thus, the expression of βc-related cytokine receptors such as GM-CSFR and IL-3R are progressively up-regulated along the myelomonocytic differentiation pathway, but down-regulated if cells commit to the lymphoid or the MegE lineages. EpoR is up-regulated during the development from long-term HSCs to MEPs, but down-regulated in GMPs or CLPs. In contrast, lymphoid-related IL-7Rα is exclusively expressed in CLPs, but not in HSCs or myeloid progenitors. In H-2Ld-hGM-CSFRα/βc double transgenic mice (8), hGM-CSFRα/βc transgenes are driven by the MHC class I promoter, and most hematopoietic cells express hGM-CSFR. As shown in Fig. 1 C, 85–96% of cells in each progenitor population were stained with monoclonal anti–hGM-CSFRα antibodies. Because hGM-CSFR does not react with endogenous mGM-CSF, cells in H-2Ld-hGM-CSFRα/βc double transgenic mice never receive hGM-CSF signals in vivo in the absence of hGM-CSF (8). Thus, FACS® analysis showed that the frequency of each myeloid and lymphoid progenitor population in hGM-CSFR transgenic mice was identical to that of wild-type mice (unpublished data).
Figure 1.
Cytokine receptor expression in hematopoietic branch points. (A) Lineage relationships of prospectively purified lineage-restricted progenitors in steady-state hematopoiesis. LT-HSC, long-term HSCs (CD34− Lin− Sca-1+ c-Kit+); ST-HSC, short-term HSCs (CD34+ Lin− Sca-1+ c-Kit+). (B) Results of RT-PCR analyses of cytokine receptors targeted for 250 cells per progenitor species. The symbols under each lane depict relative amounts of mRNA in each population compared with control cDNA (2 × 105 cells, bands not shown) by the ratio pixel density units of target cDNA/pixel density units of control cDNA. −, <0.1; ±, 0.1–0.5; +, 0.5–1.5; ++, >1.5. (C) Expression of transgenic hGM-CSFRα in each purified population. Shaded and open lines show the staining of target populations by anti–hGM-CSFRα and control IgG antibodies, respectively. A majority of each stem and progenitor fraction express hGM-CSFRα.
hGM-CSF Signals Allow Differentiation throughout Myeloerythroid Development.
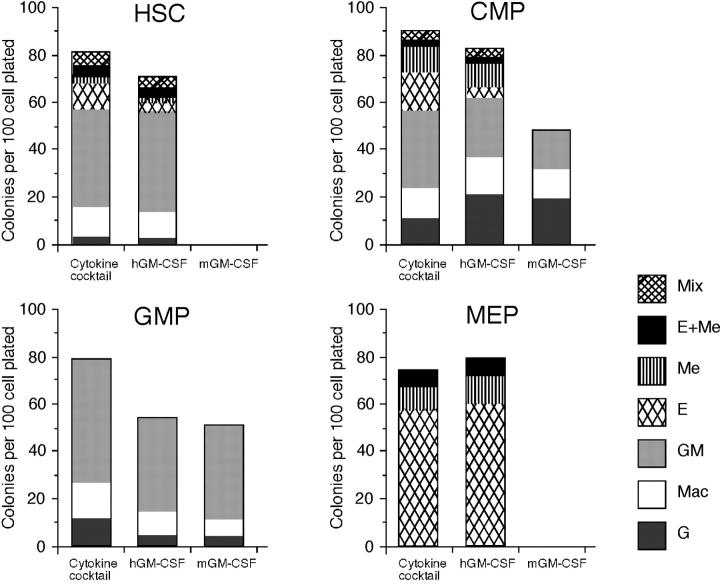
Previous studies have shown that enforced hGM-CSF signals can stimulate murine bone marrow cells to form a similar proportion of myeloerythroid colonies supported by a panel of cytokines, suggesting that hGM-CSF can stimulate maturation of cells without affecting lineage determination (8, 36). To test this hypothesis, we evaluated the effect of hGM-CSF on lineage readouts in HSCs (Lin− Sca-1+ c-Kit+) and myeloid progenitors purified from hGM-CSFR transgenic mice (Fig. 2) . Consistent with previous reports, hGM-CSFR+ HSCs and hGM-CSFR+ CMPs gave rise to a variety of myeloerythroid colonies in the presence of hGM-CSF alone. The efficiency and diversity of colony formation by hGM-CSF alone were almost identical to those produced by a cytokine cocktail containing Slf, IL-3, IL-11, mGM-CSF, Epo, and Tpo. Because HSCs cannot form colonies in the absence of Slf (37), enforced hGM-CSF signals could substitute for Slf signals in addition to myeloerythroid cytokine signals. Importantly, purified hGM-CSFR+ GMPs and hGM-CSFR+ MEPs exclusively generated GM and MegE colonies, respectively. These data formally prove that hGM-CSFR transduces permissive signals to myeloerythroid progenitors and does not affect their physiologically determined cell fate potential.
Figure 2.
In vitro colony-forming activity of purified stem and progenitors by ectopic hGM-CSF signaling. Clonogenic analysis of stem and progenitor cells purified from hGM-CSFR transgenic mice. Single cells were sorted into 96-well plates and cultured in methylcellulose for 8 d in the presence of cytokine cocktail (Slf, IL-3, IL-11, mGM-CSF, Epo, and Tpo), hGM-CSF alone, or mGM-CSF alone. 288 single cells were analyzed in each assay. Note that hGM-CSF alone, like the cytokine cocktail, can support formation of all myeloid lineage colonies.
hGM-CSF Signals Do Not Substitute for Lymphoid-specific IL-7.
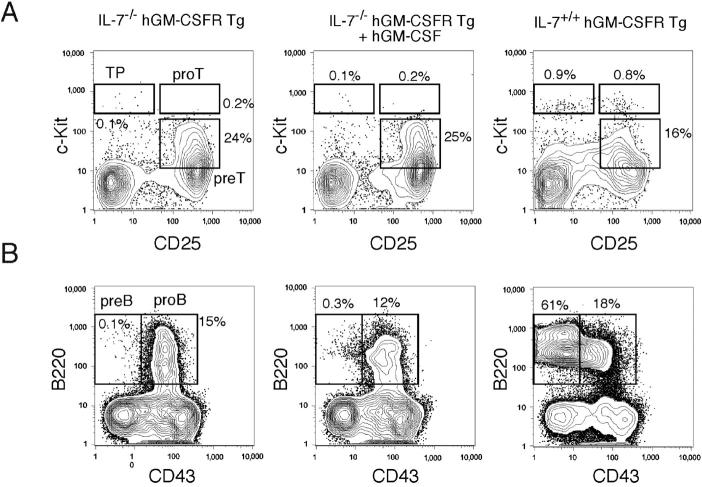
To test whether IL-7 signals in lymphoid development can be replaced by hGM-CSF, we crossed hGM-CSFR transgenic mice with IL-7−/− and IL-7Rα−/− mice. Both IL-7−/− hGM-CSFR and IL-7Rα−/− hGM-CSFR transgenic mice showed a significant loss of T and B cells, similar to IL-7−/− and IL-7Rα−/− mice (14, 15). First, we injected recombinant hGM-CSF into IL-7−/− hGM-CSFR transgenic mice. As shown in Fig. 3 , the impaired differentiation of T and B cell progenitors was not restored by hGM-CSF injection. IL-7−/− hGM-CSFR transgenic mice displayed a ∼50-fold reduction of thymocyte numbers as well as a decrease in the frequency of the earliest thymic precursors (38) and pro-T cells (Fig. 3 A), as observed in IL-7−/− mice (15). Subcutaneous injection of hGM-CSF (500 ng × 2/day for 8 d) resulted in >10-fold increases in the number of Gr-1+ Mac-1+ neutrophils and monocytes as well as Ter119+ erythroid cells in the spleen (not depicted). However, this treatment did not affect thymocyte numbers or progenitor frequencies in IL-7−/− hGM-CSF transgenic mice (Fig. 3 A). The number of CD4+ CD8− and CD4− CD8+ mature thymocytes was unchanged. Their mean numbers were ∼5 × 105 and ∼2 × 105 cells, respectively, which were ∼20-fold less than those in wild-type controls (not depicted). Thus, ectopic hGM-CSF signals cannot replace “trophic” IL-7 signals (20) in T lymphopoiesis.
Figure 3.
hGM-CSF signals cannot substitute for IL-7 in lymphoid development in vivo. (A) FACS® analysis of thymic progenitor subsets in IL-7–deficient hGM-CSFR transgenic mice after hGM-CSF injection. In these mice, absolute numbers of thymocytes were ∼50-fold less than in normal thymi (not depicted). Note that percentages of the earliest thymic progenitors (TP), pro-T, and pre-T cells did not change by hGM-CSF injection. (B) FACS® analysis of B cell progenitor subsets in IL-7–deficient hGM-CSFR transgenic mice after hGM-CSF injection. B cell differentiation is blocked at the pro-B stage, regardless of hGM-CSF injection. Representative data of four mice analyzed in each group.
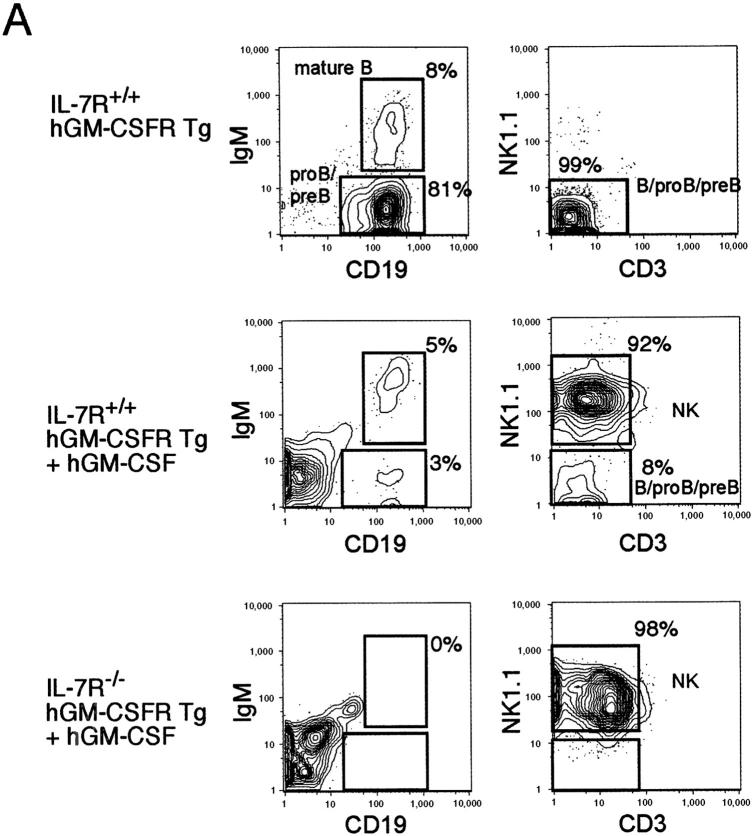
IL-7−/− mice displayed a differentiation block at the transition from the pro-B to pre-B stage. The impaired differentiation was not restored by the hGM-CSF injection (Fig. 3 B). Next, we purified IL-7Rα−/− hGM-CSFR+ and IL-7Rα+/+ hGM-CSFR+ B220+ CD43+ IgM− pro-B cells, and tested their differentiation on an S17 stromal layer (Fig. 4) . IL-7Rα+/+ hGM-CSFR+ pro-B cells differentiated into mature CD19+ IgM+ B cells in the absence of cytokines. The addition of 20 ng/ml hGM-CSF in the culture resulted in an unexpected expansion of CD19− B220+ CD3− NK1.1+ LGL (Fig. 4 B), which also expressed Mac-1 at a low level (not depicted). IL-7Rα−/− hGM-CSFR+ pro-B cells did not mature into B cells in vitro in the presence of hGM-CSF, indicating that GM-CSF signals cannot substitute for the nonredundant IL-7 in B lymphopoiesis. Instead, IL-7Rα−/− hGM-CSFR+ pro-B cells only gave rise to morphologically mature LGL cells in the presence of hGM-CSF. Thus, enforced hGM-CSF signals cannot substitute for IL-7 signals in T and B cell development but can stimulate proliferation of an NK-LGL subset. The NK-LGL population was phenotypically identical to the minor CD19− B220+ CD3-NK1.1+ population that is reportedly included in the B220+ CD43+ IgM− pro-B fraction (39). This is consistent with a report by Nishijima et al. (36), in which in vivo injection of a high dose of hGM-CSF into hGM-CSFR transgenic mice resulted in an expansion of Mac-1+ NK1.1+ NK-LGL cells.
Figure 4.
hGM-CSF signals cannot support pro-B cell differentiation but stimulate large granular lymphocytes with NK phenotype. (A) B220+ CD43+ IgM− pro-B cell fraction was cultured on an S17 stromal layer for 14 d, after which IL-7R+/+ hGM-CSFR+ pro-B cells gave rise to CD19+ IgM+ mature B cells in the absence of additional cytokines. IL-7R−/− hGM-CSFR+ pro-B fraction gave rise to CD19− B220+ NK1.1+ CD3− NK cells, but not mature B cells in the presence of hGM-CSF. (B) Morphology of CD19− B220+ NK1.1+ CD3− NK cells. These cells possess coarse cytoplasmic granules, which are specific to large granular lymphocytes.
Enforced hGM-CSF Signals Instruct Lineage Conversion into Myelomonocytic Cells In Vitro.
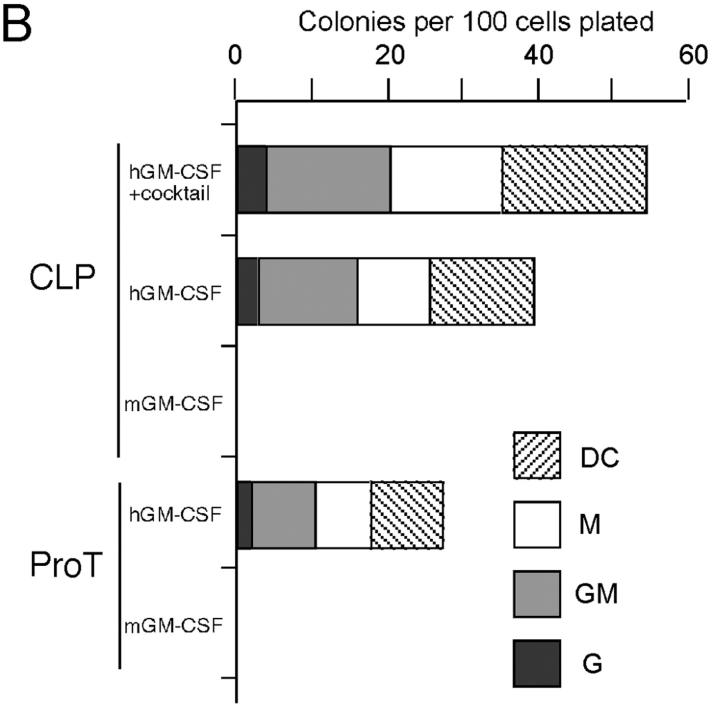
We tested the effect of ectopic hGM-CSF signals on myeloid development in lymphoid progenitors. hGM-CSFR+ CLPs, pro-T, pre-T, pro-B, and pre-B cells were cultured in liquid media containing only hGM-CSF (Fig. 5) . 6 d after the initiation of culture, CLPs gave rise only to Gr-1+ Mac-1+ GMs and Gr-1− Mac-1+ Class II (I-Ab)+ CD11c+ dendritic cells (Fig. 5 A). Because dendritic cells from normal GMPs, but not from CLPs or pro-T cells, express such high level of Mac-1 (40), the Mac-1+ I-Ab+ CD11c+ cells were categorized into “myeloid” dendritic cells. Pro-T cells also gave rise to all of these myeloid cells, whereas pre-T, pro-B, and pre-B cells did not generate any myeloid cells (not depicted). Clonogenic, single cell cultures with hGM-CSF alone revealed that >40% of CLPs and 20% of pro-T cells differentiate into myeloid cells (Fig. 5, B and C). The frequency of myeloid differentiation of hGM-CSFR+ CLPs increased up to 55% by the addition of other myeloid cytokines (mGM-CSF, Slf, IL-11, IL-3, Epo, and Tpo) to hGM-CSF (Fig. 5 B). Thus, a majority of CLPs can readout GM differentiation by ectopic hGM-CSF signals.
Figure 5.
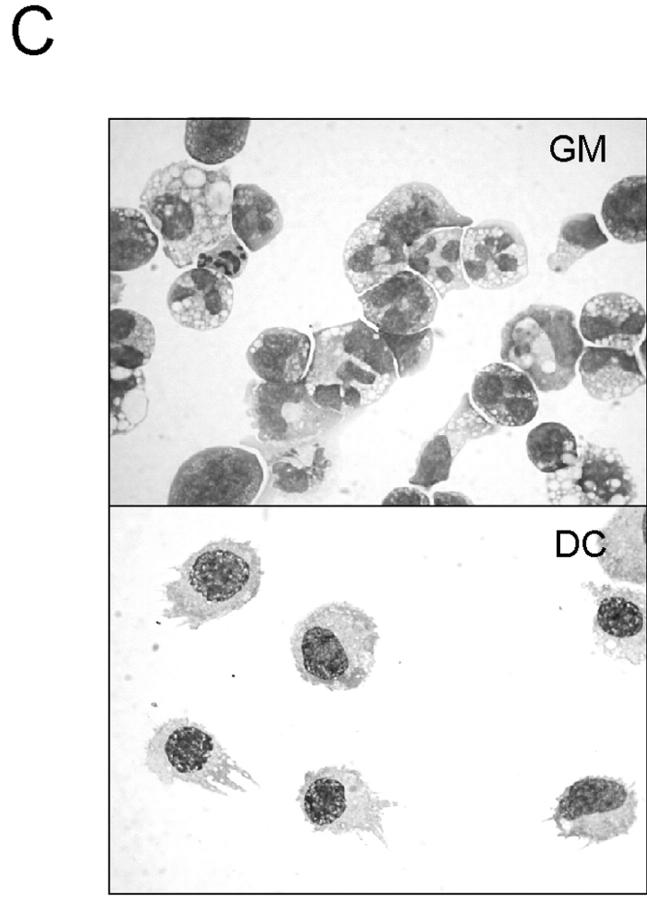
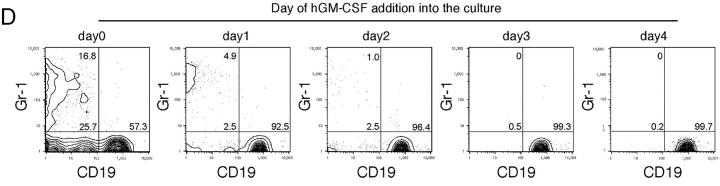
hGM-CSF signals can induce lineage switch from the lymphoid to the myeloid pathways in vitro. (A) FACS® analysis of 200 purified hGM-CSFR+ CLPs and pro-T cells cultured in the presence of hGM-CSF alone. Note that both CLPs and pro-T cells give rise to Gr-1+ Mac-1+ granulocytes/monocytes (GM) and Mac-1hi CD11c+ I-Ab+ myeloid dendritic cells (DC). (B) Clonogenic analysis of myeloid differentiation of CLPs and pro-T cells by ectopic hGM-CSF signals. Single cells were cultured in a Terasaki plate in the presence of cyto-kine cocktail (Slf, IL-3, IL-11, mGM-CSF, Epo, Tpo, and hGM-CSF), hGM-CSF alone, or mGM-CSF alone. hGM-CSFR+ CLPs and pro-T cells exhibit a significant differentiation potential into myeloid lineage cells including granulocytes, monocytes, and dendritic cells at high frequencies. 240 single cells were analyzed in each assay. (C) Morphology of a single pro-T cell derived GM colony (top) and a myeloid DC colony (bottom). Giemsa staining: ×1,000. (D) FACS® analysis of progeny of 40 hGM-CSFR+ CLPs on an S17 stromal layer in the presence of 10 ng/ml IL-7 for 10 d. 20 ng/ml hGM-CSF was added to the culture at the indicated time points. Myeloid progeny became undetectable if hGM-CSF was added >2 d after the initiation of culture.
We tested whether the myelomonocytic differentiation potential disappears as CLPs differentiate into the B cell lineage. We cultured hGM-CSFR+ CLPs on S17 stromal cells with IL-7 to promote B cell differentiation, and added hGM-CSF at different time points. The phenotype of their progeny was analyzed on day 10 (Fig. 5 D). If we added hGM-CSF together with IL-7 at the initiation of the culture, CLPs gave rise to 17% Gr-1+ GM cells, 26% Gr-1− CD19− (Mac-1+ CD11c+) dendritic cells, and 57% CD19+ B cells on day 10 under these culture conditions. Percentages of myeloid progeny gradually declined by the delayed addition of hGM-CSF. When we added hGM-CSF on day 3, CLPs only generated CD19+ B cells (Fig. 5 D). These data indicate that there is a 2-d window after which cultured CLPs cannot convert into the GM lineage. The disappearance of myeloid conversion occurs as cells differentiate into the B cell lineage. This result in turn suggests that the GM progeny, generated by ectopic hGM-CSF signals, was not derived from a minority of myeloid contaminants within the purified CLP fraction.
Myelomonocytic Lineage Conversion Can Occur Even after Cells Rearranged TCR Genes.
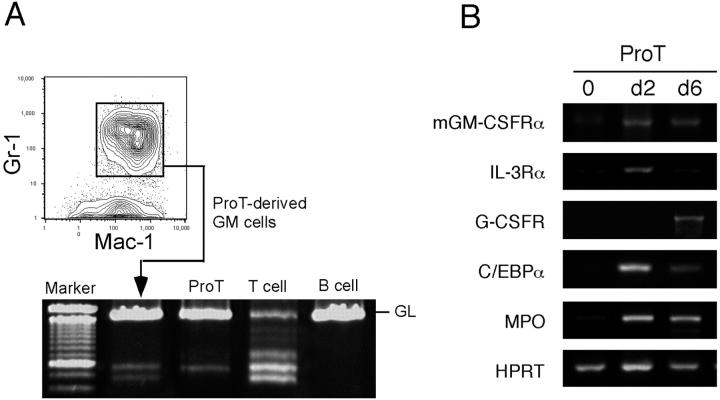
To confirm the T lymphoid origin of GM cells from hGM-CSFR+ pro-T cells, we purified the Gr-1+ Mac-1+ neutrophil/monocyte progeny of cultured hGM-CSFR+ pro-T cells and tested the rearrangement status of the TCR Dβ2-Jβ2. As shown in Fig. 6 A, a fraction of pro-T–derived Gr-1+ Mac-1+ cells rearranged Dβ2-Jβ2 genes, indicating that the conversion into the GM lineage can occur after the Dβ2-Jβ2 rearrangement.
Figure 6.
hGM-CSFR+ pro-T cells produce GMs with rearranged TCR genes and up-regulate GM-affiliated genes. (A) TCR rearrangement analysis of Gr-1+ Mac-1+ GMs derived from hGM-CSFR+ pro-T cells. 500 hGM-CSFR+ pro-T cells were cultured in the presence of hGM-CSF for 6 d and FACS®-sorted Gr-1+ Mac-1+ progeny was used for PCR analysis. (B) RT-PCR analysis of myeloid genes in hGM-CSFR+ pro-T cells cultured with hGM-CSF. Up-regulation of myeloid cytokine receptors and transcription factor is detected, indicating that hGM-CSF reactivates myeloid differentiation programs silenced in normal pro-T cells.
Activation of Transcription of Myeloid Genes in Lymphoid Progenitors During Myelomonocytic Lineage Conversion.
The hGM-CSFR+ pro-T cells stimulated by hGM-CSF rapidly up-regulated GM-affiliated cytokine receptors (mGM-CSFRα, IL-3Rα, and G-CSFR), transcription factor (C/EBPα), and myeloperoxidase (Fig. 6 B), but not MegE-affiliated EpoR or β globin (unpublished data). In contrast, hGM-CSFR+ pro-B cells did not up-regulate these myeloid genes by hGM-CSF stimulation (unpublished data), indicating that ectopic hGM-CSF signals can reactivate transcription of GM-affiliated genes only in early T, but not in B lymphoid progenitors.
Enforced hGM-CSF Signal-induced Myelomonocytic Lineage Conversion of CLPs Occurs at the Expense of Their Lymphoid Differentiation In Vivo.
We then tested the in vivo effect of ectopic hGM-CSF signals on differentiation of CLPs. hGM-CSFR+ CLPs were purified from hGM-CSFR transgenic mice (C57B6-Ly5.2), and transplanted into sublethally irradiated congenic mice (C57B6-Ly5.1) intravenously or intrathymically. Either hGM-CSF (500 ng × 2/day) or control PBS was administered in two groups of mice for 5 d after the transplantation.
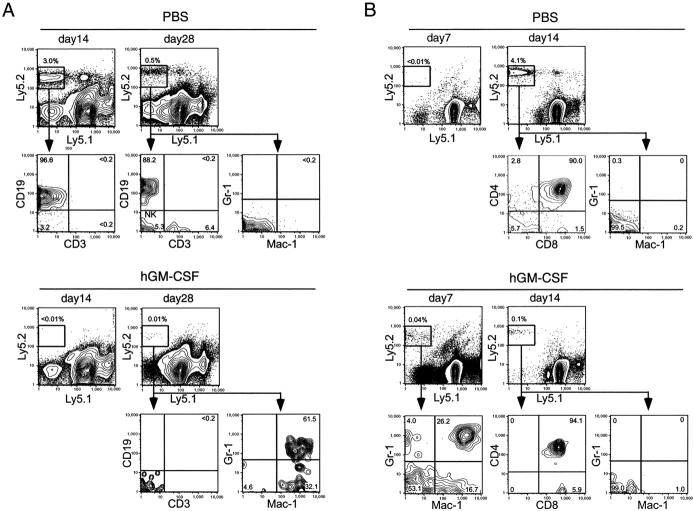
We have previously reported that normal CLPs generated B and T cells in the spleen peaking at days 14 and 28, respectively, after intravenous transplantation (23). In control mice treated with PBS, hGM-CSFR+ CLPs gave rise to B cells on day 14 and, T, B, and NK cells on day 28 in the spleen, but not to Gr-1+ Mac-1+ cells (Fig. 7 A, top). Developing thymocyte progeny was also found in the control thymi (not depicted). In contrast, in mice treated with hGM-CSF for the initial 5 d of transplantation, no detectable B or T cell progeny was observed on days 14 or 28 in the spleen (Fig. 7 A) or thymus (not depicted), whereas a small number of Mac-1+ Gr-1+ GM progeny was detectable on day 28 in the spleen (Fig. 7 A, bottom).
Figure 7.
Ectopic hGM-CSF signals inhibit lymphoid differentiation, but promote myelomonocytic lineage conversion of transplanted CLPs in vivo. (A) 6,000 hGM-CSFR+ CLPs (Ly5.2) were intravenously transplanted into congenic mice (Ly5.1). Control mice injected with PBS developed CD19+ B cells on day 14, and CD3+ T, CD19+ B, and CD3− CD19− (NK1.1+, not shown) NK cells on day 28 in the spleen. Lymphoid differentiation was completely inhibited in mice that received hGM-CSF, whereas a small number of Gr-1+ Mac-1+ cells were detectable within the day 28 spleen. (B) 3,000 hGM-CSFR+ CLPs (Ly5.2) were directly injected into the thymi of congenic mice (Ly5.1). On day 7, CLP-derived Gr-1+ Mac-1+ GM cells were found in the thymus of mice treated with hGM-CSF, whereas no progeny was detectable in control thymi at this time point. On day 14, hGM-CSFR+ CLPs gave rise to CD4+ CD8+ immature thymocytes and CD4+ or CD8+ mature thymocytes in control thymi, whereas numbers of CLP-derived thymocytes were significantly reduced (by >40-fold) in thymi of hGM-CSF–treated mice. In both experiments, two groups of mice (three mice per group) were subcutaneously injected with either hGM-CSF or control PBS for 5 d after transplantation. Representative data are shown.
CLPs need to home to the thymic environment for their T cell readouts (23). To evaluate the effect of ectopic hGM-CSF signals on thymic T cell development from CLPs, we injected hGM-CSFR+ CLPs directly into the thymus of congenic mice (Fig. 7 B). In control animals, progeny of hGM-CSFR+ CLPs was undetectable on day 7, whereas it became detectable on day 14, when hGM-CSFR+ CLPs differentiated into CD4+ CD8+ double positive cells as well as CD4+ CD8− or CD4− CD8+ single positive cells (Fig. 7 B, top). Gr-1+ Mac-1+ myelomonocytic cells were not found in the control thymi throughout the experiment. In mice treated with hGM-CSF, hGM-CSFR+ CLPs gave rise to Gr-1+ Mac-1+ GM progeny on day 7 in the thymus. These myelomonocytic cells disappeared on day 14, when only a small number of T cell progeny was detectable (Fig. 7 B, bottom). Importantly, the number of day 14 T cell progeny decreased by ∼40-fold in hGM-CSF–treated mice (Fig. 7 B). These data indicate that enforced hGM-CSF signals in vivo abrogate lymphoid potentials of CLPs, inducing their conversion into the GM lineage.
Discussion
Lineage potential of hematopoietic progenitors becomes progressively restricted as cells differentiate into mature cells. From a mechanistic point of view, it might be reasonable to separate the role of external signals in lineage commitment into two categories by their functions: (a) lineage permission and (b) lineage instruction. Uncommitted precursors could be directed to certain lineages, depending on either or both of these mechanisms. It has been suggested that the maintained accessibility for genes of multiple lineages represent the cells' immediate lineage potential (25, 41–43). In our hands, CMPs coexpress myeloerythroid genes, whereas CLPs coexpress T and B lymphoid genes at the single cell level (25), and these patterns of transcription might reflect their chromatin structures already open for multiple myeloid and lymphoid differentiation programs (3, 43). Thus, one of the simplest models for lineage commitment is that the permissive signals support to run differentiation programs that are being transcribed, whereas the instructive signals turn on silenced differentiation programs that are not transcribed before the stimulation. In this context, the “plasticity” of precursors for lineage commitment could be achieved if instructive signals can turn on differentiation programs outside of their physiological lineage potential.
Our data strongly suggest that GM-CSF signals allow the transcription of both GM and MegE differentiation programs in stem and myeloerythroid progenitor cells, but do not activate the GM program in MegE-committed cells. This is compatible with the previous data that ectopic myeloid cytokine signals can support both GM and MegE colony formation (8–11). Because commitment into GM or MegE lineages appears to occur in a cell-autonomous fashion at least in vitro (44, 45), the down-regulation of GM-CSFR as a result of commitment into the MegE lineage should cause MegE cells to become unresponsive to GM-CSF, thus enabling Epo to critically regulate erythropoiesis.
The “permissive” effect of hGM-CSF signals is myeloid specific. hGM-CSF signals cannot support lymphoid differentiation through maintaining or up-regulating lymphoid gene transcription at the CLP or its downstream lymphoid stages, and cannot replace IL-7 signals that are necessary for lymphoid development (Figs. 3 and 4; 20). Instead, ectopic hGM-CSF signals turn on the transcription of myeloid genes (Fig. 6 B), and induce the GM lineage conversion in a majority of CLPs and a considerable fraction of pro-T cells (Fig. 5). The GM lineage conversion can occur even in late pro-T cells with rearranged TCRβ genes (Fig. 6 A). Because lymphoid progenitors completely shut down myeloid gene transcription (3, 24, 25), this result strongly suggests that hGM-CSF signals can instruct lymphoid progenitors to become accessible to myeloid (GM but not MegE) differentiation programs, presumably through remodeling chromatin structures.
These data indicate that inactivation of genes for unselected lineages and activation of genes for committed lineages are equally critical in physiological lineage commitment and restriction. GM-CSFR is detectable in short-term HSCs and CMPs, but not CLPs or their downstream lymphoid progenitors. GM-CSF signals can induce GM but not MegE differentiation in CLPs and pro-T cells. Furthermore, in vivo reconstitution experiments showed that if CLPs receive ectopic GM-CSF signals after transplantation, they cannot undergo T, B, nor NK lymphoid differentiation, but instead convert into GMPs (Fig. 7). Thus, in physiological hematopoiesis, CLPs and pro-T cells might escape from GM commitment signals by losing GM-CSFR expression. We have recently found that enforced expression of GATA-1, a major MegE-affiliated transcription factor, can induce differentiation of CLPs into MegE lineage cells (46). Accordingly, CLPs possess plasticity for myeloerythroid differentiation. They can reactivate GM and MegE differentiation programs by ectopic signals of cytokines or transcription factors, which are silenced at the CLP stage in physiological hematopoiesis.
We have reported that ectopic GM-CSF signals suppressed T cell development in fetal thymic organ cultures especially at the transition from pro-T to pre-T cells (47), and that hGM-CSF injection induced the significant reduction in the number of thymocytes in adult hGM-CSFR transgenic mice (36). Here we show that the conversion into the GM lineage by ectopic GM-CSF signals occurs at the pro-T but not the pre-T stage in vitro, and that ectopic GM-CSF signals inhibit both T and B cell development from CLPs inducing their myeloid lineage conversion in vivo. Thus, our new findings strongly suggest that the inhibition of T lymphopoiesis in the previous report (36, 47) could result from the GM-CSF–induced myeloid conversion at the CLP and the pro-T stages.
As in the case of ectopic hGM-CSFR, ectopic IL-2R can mediate signals for GM differentiation in CLPs and pro-T cells, but not in pro-B cells (27). Interestingly, these cytokine signals cannot activate GM differentiation in pro-B cells. In our hands, single cell RT-PCR analysis showed that all 450 pro-B cells express Pax-5, a critical transcription factor for the B cell lineage (25). Pax-5–deficient pro-B cells are pluripotent (48, 49), and transduction of Pax-5 into myeloid progenitors can suppress proliferation through blunting responsiveness to myeloid cytokines (50). Therefore, it is possible that Pax-5 in pro-B cells may suppress their myeloid differentiation programs through inhibiting their responsiveness to ectopic myeloid cytokine signals. The hGM-CSF–induced lymphoid to GM lineage conversion progressively disappears as cells differentiate along the T cell pathway. hGM-CSF signals cannot reactivate GM differentiation programs at the further downstream pre-T stage. King et al. (51) reported that ectopic IL-2R–expressing pro-T cells with a TCR Dβ1-Jβ1 but not a Dβ2-Jβ2 rearrangement can differentiate into GM cells. The existence of the Dβ2-Jβ2 rearrangement in myeloid cells differentiated from hGM-CSFR+ pro-T cells shown here suggests that target lymphoid progenitors for lineage conversion might be slightly different between ectopic IL-2 and hGM-CSF signals.
We and others have reported that CLPs and pro-T cells but not pre-T or pro-B cells can differentiate into “lymphoid” dendritic cells in vitro (40, 52, 53). It is interesting that the natural loss of dendritic cell generative capacity and the revealed loss of myeloid conversion from the lymphoid pathway occur at similar differentiation stages. These data raise an interesting possibility that the development of “lymphoid” dendritic cells from CLPs and pro-T cells may represent the residue of their silenced myeloid potential.
The lineage conversion from the lymphoid to the myeloid pathway by ectopic hGM-CSF signals shown here may be linked to the developmental mechanism of leukemia with “mixed” phenotype (54). In a fraction of acute myelogenous leukemia, leukemic blasts possess lymphoid markers including monoclonal rearrangement of TCR genes (55). Conversely, acute lymphoblastic leukemia or even myeloma cells occasionally coexpress myeloid features (56, 57). Conversion from acute myelogenous leukemia to acute lymphoblastic leukemia or vice versa is also known to occur (58, 59). These intriguing features of leukemias have been speculated to represent lineage conversions induced by oncogenic events (lineage infidelity; 60, 61). The fact that the lineage conversion could be induced by ectopic cytokine signals provides a clue to understanding the pathogenesis of mixed lineage leukemia, because a number of tyrosine kinase fusions share their signaling pathways with cytokines. For example, BCR-ABL and TEL fusions directly activate Stat5 (62, 63), which is also a critical molecule for βc-mediated IL-3/IL-5/GM-CSF signaling. Thus, our data suggest that the mixed leukemia phenotype may represent plasticity or “lineage infidelity” of hematopoietic progenitors.
In summary, hGM-CSF signals alone can maintain the completion of MegE and GM but not lymphoid differentiation programs. Instead, ectopic hGM-CSF signals reactivate GM differentiation programs in CLPs and pro-T cells. These results demonstrate significant differences between cytokine signals required for myeloid and lymphoid development. Therefore, the organized down-regulation and up-regulation of cytokine receptors are equally important in the maintenance of myeloid and lymphoid hematopoiesis. Additional studies on target molecules activated by lineage-instructive cytokine signals will be crucial in elucidating the mechanism of lineage commitment in normal and malignant hematopoiesis.
Acknowledgments
We thank Maris A. Handley for technical assistance in FACS® operation, Shin-ichi Mizuno for technical advice, and D. Dalma-Weiszhausz for critically reviewing the manuscript.
This work was supported in part by grants from National Institutes of Health (2P01DK050654), Leukemia Research Foundation, Claudia Adams Barr Program, and Damon Runyon Cancer Research Program to K. Akashi, and Uehara Memorial Foundation to J. Iwasaki-Arai.
Footnotes
Abbreviations used in this paper: CLP, common lymphoid progenitor; CMP, common myeloid progenitor; EpoR, erythropoietin receptor; GM, granulocyte/monocyte; GMP, granulocyte/monocyte progenitor; hGM-CSF, human GM-CSF; HSC, hematopoietic stem cell; MegE, megakaryocyte/erythroid; MEP, megakaryocyte/erythrocyte progenitor; Slf, steel factor; Tpo, thrombopoietin.
References
- 1.Ogawa, M. 1993. Differentiation and proliferation of hematopoietic stem cells. Blood. 81:2844–2853. [PubMed] [Google Scholar]
- 2.Enver, T., C.M. Heyworth, and T.M. Dexter. 1998. Do stem cells play dice? Blood. 92:348–351. [PubMed] [Google Scholar]
- 3.Akashi, K., X. He, J. Chen, H. Iwasaki, C. Niu, B. Steenhard, J. Zhang, J. Haug, and L. Li. 2003. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 101:383–389. [DOI] [PubMed] [Google Scholar]
- 4.Lieschke, G.J., D. Grail, G. Hodgson, D. Metcalf, E. Stanley, C. Cheers, K.J. Fowler, S. Basu, Y.F. Zhan, and A.R. Dunn. 1994. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 84:1737–1746. [PubMed] [Google Scholar]
- 5.Stanley, E., G.J. Lieschke, D. Grail, D. Metcalf, G. Hodgson, J.A. Gall, D.W. Maher, J. Cebon, V. Sinickas, and A.R. Dunn. 1994. Granulocyte/macrophage colony-stimulating factor-deficient mice show no major perturbation of hematopoiesis but develop a characteristic pulmonary pathology. Proc. Natl. Acad. Sci. USA. 91:5592–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillessen, S., N. Mach, C. Small, M. Mihm, and G. Dranoff. 2001. Overlapping roles for granulocyte-macrophage colony-stimulating factor and interleukin-3 in eosinophil homeostasis and contact hypersensitivity. Blood. 97:922–928. [DOI] [PubMed] [Google Scholar]
- 7.Wu, H., X. Liu, R. Jaenisch, and H.F. Lodish. 1995. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 83:59–67. [DOI] [PubMed] [Google Scholar]
- 8.Nishijima, I., T. Nakahata, Y. Hirabayashi, T. Inoue, H. Kurata, A. Miyajima, N. Hayashi, Y. Iwakura, K. Arai, and T. Yokota. 1995. A human GM-CSF receptor expressed in transgenic mice stimulates proliferation and differentiation of hemopoietic progenitors to all lineages in response to human GM-CSF. Mol. Biol. Cell. 6:497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang, F.C., S. Watanabe, K. Tsuji, M.J. Xu, A. Kaneko, Y. Ebihara, and T. Nakahata. 1998. Human granulocyte colony-stimulating factor (G-CSF) stimulates the in vitro and in vivo development but not commitment of primitive multipotential progenitors from transgenic mice expressing the human G-CSF receptor. Blood. 92:4632–4640. [PubMed] [Google Scholar]
- 10.Yang, F.C., K. Tsuji, A. Oda, Y. Ebihara, M.J. Xu, A. Kaneko, S. Hanada, T. Mitsui, A. Kikuchi, A. Manabe, et al. 1999. Differential effects of human granulocyte colony-stimulating factor (hG- CSF) and thrombopoietin on megakaryopoiesis and platelet function in hG-CSF receptor-transgenic mice. Blood. 94:950–958. [PubMed] [Google Scholar]
- 11.Takagi, M., T. Hara, M. Ichihara, K. Takatsu, and A. Miyajima. 1995. Multi-colony stimulating activity of interleukin 5 (IL-5) on hematopoietic progenitors from transgenic mice that express IL-5 receptor alpha subunit constitutively. J. Exp. Med. 181:889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pharr, P.N., D. Hankins, A. Hofbauer, H.F. Lodish, and G.D. Longmore. 1993. Expression of a constitutively active erythropoietin receptor in primary hematopoietic progenitors abrogates erythropoietin dependence and enhances erythroid colony-forming unit, erythroid burst-forming unit, and granulocyte/macrophage progenitor growth. Proc. Natl. Acad. Sci. USA. 90:938–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semerad, C.L., J. Poursine-Laurent, F. Liu, and D.C. Link. 1999. A role for G-CSF receptor signaling in the regulation of hematopoietic cell function but not lineage commitment or differentiation. Immunity. 11:153–161. [DOI] [PubMed] [Google Scholar]
- 14.Peschon, J.J., P.J. Morrissey, K.H. Grabstein, F.J. Ramsdell, E. Maraskovsky, B.C. Gliniak, L.S. Park, S.F. Ziegler, D.E. Williams, C.B. Ware, et al. 1994. Early lymphocyte expansion is severely impaired in interleukin 7 receptor–deficient mice. J. Exp. Med. 180:1955–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Freeden-Jeffry, U., P. Vieira, L.A. Lucian, T. McNeil, S.E. Burdach, and R. Murray. 1995. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 181:1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao, X., E.W. Shores, J. Hu-Li, M.R. Anver, B.L. Kelsall, S.M. Russell, J. Drago, M. Noguchi, A. Grinberg, E.T. Bloom, et al. 1995. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 2:223–238. [DOI] [PubMed] [Google Scholar]
- 17.Ohbo, K., T. Suda, M. Hashiyama, A. Mantani, M. Ikebe, K. Miyakawa, M. Moriyama, M. Nakamura, M. Katsuki, K. Takahashi, et al. 1996. Modulation of hematopoiesis in mice with a truncated mutant of the interleukin-2 receptor gamma chain. Blood. 87:956–967. [PubMed] [Google Scholar]
- 18.Ye, S.K., K. Maki, T. Kitamura, S. Sunaga, K. Akashi, J. Domen, I.L. Weissman, T. Honjo, and K. Ikuta. 1999. Induction of germline transcription in the TCRgamma locus by Stat5: implications for accessibility control by the IL-7 receptor. Immunity. 11:213–223. [DOI] [PubMed] [Google Scholar]
- 19.Corcoran, A.E., A. Riddell, D. Krooshoop, and A.R. Venkitaraman. 1998. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 391:904–907. [DOI] [PubMed] [Google Scholar]
- 20.Akashi, K., M. Kondo, U. von Freeden-Jeffry, R. Murray, and I.L. Weissman. 1997. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 89:1033–1041. [DOI] [PubMed] [Google Scholar]
- 21.Maraskovsky, E., L.A. O'Reilly, M. Teepe, L.M. Corcoran, J.J. Peschon, and A. Strasser. 1997. Bcl-2 can rescue T lymphocyte development in interleukin-7 receptor-deficient mice but not in mutant rag-1−/− mice. Cell. 89:1011–1019. [DOI] [PubMed] [Google Scholar]
- 22.Kondo, M., K. Akashi, J. Domen, K. Sugamura, and I.L. Weissman. 1997. Bcl-2 rescues T lymphopoiesis, but not B or NK cell development, in common gamma chain-deficient mice. Immunity. 7:155–162. [DOI] [PubMed] [Google Scholar]
- 23.Kondo, M., I.L. Weissman, and K. Akashi. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672. [DOI] [PubMed] [Google Scholar]
- 24.Akashi, K., D. Traver, T. Miyamoto, and I.L. Weissman. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. [DOI] [PubMed] [Google Scholar]
- 25.Miyamoto, T., H. Iwasaki, B. Reizis, M. Ye, T. Graf, I.L. Weissman, and K. Akashi. 2002. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell. 3:137–147. [DOI] [PubMed] [Google Scholar]
- 26.Liu, R., C.B. Liu, M.G. Mohi, K. Arai, and S. Watanabe. 2000. Analysis of mechanisms involved in the prevention of gamma irradiation-induced apoptosis by hGM-CSF. Oncogene. 19:571–579. [DOI] [PubMed] [Google Scholar]
- 27.Kondo, M., D.C. Scherer, T. Miyamoto, A.G. King, K. Akashi, K. Sugamura, and I.L. Weissman. 2000. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 407:383–386. [DOI] [PubMed] [Google Scholar]
- 28.Osawa, M., K. Hanada, H. Hamada, and H. Nakauchi. 1996. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 273:242–245. [DOI] [PubMed] [Google Scholar]
- 29.Okuno, Y., H. Iwasaki, C.S. Huettner, H.S. Radomska, D.A. Gonzalez, D.G. Tenen, and K. Akashi. 2002. Differential regulation of the human and murine CD34 genes in hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 99:6246–6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akashi, K., L.I. Richie, T. Miyamoto, W.H. Carr, and I.L. Weissman. 2000. B lymphopoiesis in the thymus. J. Immunol. 164:5221–5226. [DOI] [PubMed] [Google Scholar]
- 31.Miyamoto, T., I.L. Weissman, and K. Akashi. 2000. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc. Natl. Acad. Sci. USA. 97:7521–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akashi, K., and I.L. Weissman. 1996. The c-Kit+ maturation pathway in mouse thymic T cell development: lineages and selection. Immunity. 5:147–161. [DOI] [PubMed] [Google Scholar]
- 33.Guidos, C.J., J.S. Danska, C.G. Fathman, and I.L. Weissman. 1990. T cell receptor–mediated negative selection of autoreactive T lymphocyte precursors occurs after commitment to the CD4 or CD8 lineages. J. Exp. Med. 172:835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mebius, R.E., P. Rennert, and I.L. Weissman. 1997. Developing lymph nodes collect CD4+ CD3− LTβ1 cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 7:493–504. [DOI] [PubMed] [Google Scholar]
- 35.Spangrude, G.J., S. Heimfeld, and I.L. Weissman. 1988. Purification and characterization of mouse hematopoietic stem cells. Science. 241:58–62. [DOI] [PubMed] [Google Scholar]
- 36.Nishijima, I., T. Nakahata, S. Watanabe, K. Tsuji, I. Tanaka, Y. Hirabayashi, T. Inoue, and K. Arai. 1997. Hematopoietic and lymphopoietic responses in human granulocyte-macrophage colony-stimulating factor (GM-CSF) receptor transgenic mice injected with human GM-CSF. Blood. 90:1031–1038. [PubMed] [Google Scholar]
- 37.Smith, L.G., I.L. Weissman, and S. Heimfeld. 1991. Clonal analysis of hematopoietic stem-cell differentiation in vivo. Proc. Natl. Acad. Sci. USA. 88:2788–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, L., R. Scollay, M. Egerton, M. Pearse, G.J. Spangrude, and K. Shortman. 1991. CD4 expressed on earliest T-lineage precursor cells in the adult murine thymus. Nature. 349:71–74. [DOI] [PubMed] [Google Scholar]
- 39.Rolink, A., E. ten Boekel, F. Melchers, D.T. Fearon, I. Krop, and J. Andersson. 1996. A subpopulation of B220+ cells in murine bone marrow does not express CD19 and contains natural killer cell progenitors. J. Exp. Med. 183:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manz, M.G., D. Traver, T. Miyamoto, I.L. Weissman, and K. Akashi. 2001. Dendritic cell potentials of early lymphoid and myeloid progenitors. Blood. 97:3333–3341. [DOI] [PubMed] [Google Scholar]
- 41.Cross, M.A., and T. Enver. 1997. The lineage commitment of haemopoietic progenitor cells. Curr. Opin. Genet. Dev. 7:609–613. [DOI] [PubMed] [Google Scholar]
- 42.Hu, M., D. Krause, M. Greaves, S. Sharkis, M. Dexter, C. Heyworth, and T. Enver. 1997. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11:774–785. [DOI] [PubMed] [Google Scholar]
- 43.Enver, T., and M. Greaves. 1998. Loops, lineage, and leukemia. Cell. 94:9–12. [DOI] [PubMed] [Google Scholar]
- 44.Nakahata, T., A.J. Gross, and M. Ogawa. 1982. A stochastic model of self-renewal and commitment to differentiation of the primitive hemopoietic stem cells in culture. J. Cell. Physiol. 113:455–458. [DOI] [PubMed] [Google Scholar]
- 45.Suda, T., J. Suda, and M. Ogawa. 1984. Disparate differentiation in mouse hemopoietic colonies derived from paired progenitors. Proc. Natl. Acad. Sci. USA. 81:2520–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwasaki, H., S.-I. Mizuno, R.A. Wells, and K. Akashi. 2002. GATA-1 instructs commitment and transdifferentiation into megakaryocyte and erythroid lineages, counteracting myelomonocytic differentiation programs. Blood. 100:60A. [Google Scholar]
- 47.Yasuda, Y., I. Nishijima, S. Watanabe, K. Arai, A. Zlotnik, and T.A. Moore. 1997. Human granulocyte-macrophage colony-stimulating factor (hGM-CSF) induces inhibition of intrathymic T-cell development in hGM-CSF receptor transgenic mice. Blood. 89:1349–1356. [PubMed] [Google Scholar]
- 48.Nutt, S.L., B. Heavey, A.G. Rolink, and M. Busslinger. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 401:556–562. [DOI] [PubMed] [Google Scholar]
- 49.Rolink, A.G., S.L. Nutt, F. Melchers, and M. Busslinger. 1999. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature. 401:603–606. [DOI] [PubMed] [Google Scholar]
- 50.Chiang, M.Y., and J.G. Monroe. 1999. BSAP/Pax5A expression blocks survival and expansion of early myeloid cells implicating its involvement in maintaining commitment to the B-lymphocyte lineage. Blood. 94:3621–3632. [PubMed] [Google Scholar]
- 51.King, A.G., M. Kondo, D.C. Scherer, and I.L. Weissman. 2002. Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling. Proc. Natl. Acad. Sci. USA. 99:4508–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, L., C.L. Li, and K. Shortman. 1996. Thymic dendritic cell precursors: relationship to the T lymphocyte lineage and phenotype of the dendritic cell progeny. J. Exp. Med. 184:903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Traver, D., K. Akashi, M. Manz, M. Merad, T. Miyamoto, E.G. Engleman, and I.L. Weissman. 2000. Development of CD8alpha-positive dendritic cells from a common myeloid progenitor. Science. 290:2152–2154. [DOI] [PubMed] [Google Scholar]
- 54.Hoffbrand, A.V., B.F. Leber, P.J. Browett, and J.D. Norton. 1988. Mixed acute leukaemias. Blood Rev. 2:9–15. [DOI] [PubMed] [Google Scholar]
- 55.Cheng, G.Y., M.D. Minden, B. Toyonaga, T.W. Mak, and E.A. McCulloch. 1986. T cell receptor and immunoglobulin gene rearrangements in acute myeloblastic leukemia. J. Exp. Med. 163:414–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grogan, T.M., B.G. Durie, C.M. Spier, L. Richter, and E. Vela. 1989. Myelomonocytic antigen positive multiple myeloma. Blood. 73:763–769. [PubMed] [Google Scholar]
- 57.Akashi, K., M. Harada, T. Shibuya, K. Fukagawa, N. Kimura, K. Sagawa, Y. Yoshikai, T. Teshima, M. Kikuchi, and Y. Niho. 1991. Simultaneous occurrence of myelomonocytic leukemia and multiple myeloma: involvement of common leukemic progenitors and their developmental abnormality of “lineage infidelity.” J. Cell. Physiol. 148:446–456. [DOI] [PubMed] [Google Scholar]
- 58.Stass, S., J. Mirro, S. Melvin, C.H. Pui, S.B. Murphy, and D. Williams. 1984. Lineage switch in acute leukemia. Blood. 64:701–706. [PubMed] [Google Scholar]
- 59.Murphy, S.B., S. Stass, D. Kalwinsky, and G. Rivera. 1983. Phenotypic conversion of acute leukaemia from T-lymphoblastic to myeloblastic induced by therapy with 2′-deoxycoformycin. Br. J. Haematol. 55:285–293. [DOI] [PubMed] [Google Scholar]
- 60.Smith, L.J., J.E. Curtis, H.A. Messner, J.S. Senn, H. Furthmayr, and E.A. McCulloch. 1983. Lineage infidelity in acute leukemia. Blood. 61:1138–1145. [PubMed] [Google Scholar]
- 61.McCulloch, E.A. 1987. Lineage infidelity or lineage promiscuity? Leukemia. 1:235. [PubMed] [Google Scholar]
- 62.Levy, D.E., and D.G. Gilliland. 2000. Divergent roles of STAT1 and STAT5 in malignancy as revealed by gene disruptions in mice. Oncogene. 19:2505–2510. [DOI] [PubMed] [Google Scholar]
- 63.Liu, Q., J. Schwaller, J. Kutok, D. Cain, J.C. Aster, I.R. Williams, and D.G. Gilliland. 2000. Signal transduction and transforming properties of the TEL-TRKC fusions associated with t(12;15)(p13;q25) in congenital fibrosarcoma and acute myelogenous leukemia. EMBO J. 19:1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]