Abstract
Natural killer (NK) cells play a critical role in the innate immune response against cytomegalovirus (CMV) infections. Although CMV encodes several gene products committed to evasion of adaptive immunity, viral modulation of NK cell activity is only beginning to be appreciated. A previous study demonstrated that the mouse CMV m152-encoded gp40 glycoprotein diminished expression of ligands for the activating NK cell receptor NKG2D on the surface of virus-infected cells. Here we have defined the precise ligands that are affected and have directly implicated NKG2D in immune responses to CMV infection in vitro and in vivo. Murine CMV (MCMV) infection potently induced transcription of all five known retinoic acid early inducible 1 (RAE-1) genes (RAE-1α, RAE-1β, RAE-1δ, RAE-1ɛ, and RAE-1γ), but not H-60. gp40 specifically down-regulated the cell surface expression of all RAE-1 proteins, but not H-60, and diminished NK cell interferon γ production against CMV-infected cells. Consistent with previous findings, a m152 deletion mutant virus (Δm152) was less virulent in vivo than the wild-type Smith strain of MCMV. Treatment of BALB/c mice with a neutralizing anti-NKG2D antibody before infection increased titers of Δm152 virus in the spleen and liver to levels seen with wild-type virus. These experiments demonstrate that gp40 impairs NK cell recognition of virus-infected cells through disrupting the RAE-1–NKG2D interaction.
Keywords: NKG2D, RAE-1, cytomegalovirus, NK cell, gp40
Introduction
CMVs are characterized by an impressive array of immunomodulatory mechanisms. Both human CMV (HCMV)*and murine CMV (MCMV) encode several glycoproteins that are involved in down-regulation of MHC class I molecules on the surface of infected cells (1, 2). These proteins function by a diverse set of mechanisms at distinct stages in MHC class I biosynthesis. MCMV has three gene products, encoded by m04, m06, and m152, which inhibit expression of MHC class I on infected cells (1, 2). Interestingly, the products of the m04, m06, and m152 genes are not homologous to the HCMV class I modulators. In addition, they down-regulate MHC class I expression by distinctly different mechanisms, suggesting that they evolved independently. gp34, encoded by m04, complexes with class I in the ER, is transported to the cell surface, and can inhibit CTL activity (3, 4). gp48, the product of the m06 gene, binds to class I molecules and redirects them to the lysosome for degradation (5). m152 encodes gp40, which is involved in retention of class I complexes in ER to Golgi intermediate compartments (6), and is a potent inhibitor of virus-specific CTL activity (7). Although MHC class I down-regulation can interfere with CD8+ T cell–mediated lysis of virus-infected cells, these cells might be more susceptible to NK cell cytotoxicity because NK cells preferentially kill cells with low MHC class I expression (“missing self hypothesis”; 8).
The critical role of NK cells in controlling infection by HCMV and MCMV has long been appreciated (9, 10). NK cells respond to target cells by integrating signals from activating and inhibitory receptors expressed on the cell surface. NKG2D, an activating receptor expressed on NK cells, CD8+ T cells, and γδ T cells in humans and mice, associates with the adaptor protein DAP10, which contains a YXXM motif for binding of the p85 subunit of PI-3 kinase (11). In mice, an isoform of NKG2D on activated NK cells can also associate with the immunoreceptor tyrosine-based activation motif–bearing DAP12 adaptor protein (12). Engagement of NKG2D by its ligands can override inhibitory signals delivered by self–MHC class I (13, 14), thereby reinforcing NKG2D as a potent receptor mediating NK cell activity.
Ligands for human and mouse NKG2D all share some homology with MHC class I proteins, although they do not associate with β2-microglobulin or bind peptides. Two families of ligands for human NKG2D are known: the polymorphic MHC class I chain–related molecules, MICA and MICB (15), and the UL16-binding proteins, ULBP-1, ULBP-2, and ULBP-3 (16). Interestingly, the ULBPs were identified by expression cloning for molecules that bind to a HCMV glycoprotein, UL16. Ligands for murine NKG2D include a family of retinoic acid early inducible 1 gene (RAE-1) products (RAE-1α, RAE-1β, RAE-1γ, RAE-1δ, and RAE-1ɛ), H-60 (a minor histocompatibility antigen; 17, 18), and a murine UL16-binding protein-like transcript-1 glycoprotein (19). RAE-1 is abundantly expressed in mouse embryonic tissue and in many mouse tumor cell lines, but is rare in adult tissue.
Recent studies by Krmpotic et al. (20) demonstrated that MCMV m152 affects NK cell immunity against MCMV by modulation of ligands for NKG2D. We sought to precisely identify the NKG2D ligands that are regulated by MCMV infection. In this study, we have examined viral modulation of RAE-1 and the functional consequences of RAE-1–NKG2D interactions in NK cell responses to MCMV in vitro and in vivo.
Materials and Methods
Mice.
BALB/c and C57BL/6 mice were purchased from The Jackson Laboratory and Charles River, respectively. For in vivo experiments, 6–10-wk-old female mice were used.
Cell Lines and Transfectants.
BALB/c 3T3 cells were obtained from the American Type Culture Collection. 293T cells and TpnT fibroblasts were provided by T. Kitamura (University of Tokyo, Tokyo, Japan) and A. Hill (Oregon Health Sciences University, Portland, OR), respectively.
Transient transfection of 293T cells was performed using Lipofectamine 2000 reagent (Invitrogen). pMX-pie vector encoding RAE-1α, RAE-1β, RAE-1γ, RAE-1δ, RAE-1ɛ, or Flag-H-60 was mixed with an equal amount of either pMX-neo alone or pMX-neo vector encoding m152. Lipofectamine 2000 was then added to plasmid DNA and this mixture was incubated with 5 × 105 293T cells at 37°C. 48 h after transfection, cells were analyzed by flow cytometry.
Viruses.
Wild-type MCMV Smith and a well-characterized recombinant m152 deletion mutant strain (96.24), which has been previously described (21), were provided by A. Hill.
Immunofluorescence and Flow Cytometry.
BALB/c 3T3 cells were infected with Smith virus or Δm152 at a multiplicity of infection (MOI) of 1. 72 h after infection, cells were trypsinized and 2 × 105 cells were stained on ice for 30 min. The following staining reagents were used to detect NKG2D ligands: mouse NKG2D-Ig fusion protein, consisting of the extracellular domain of mouse NKG2D fused to the Fc domain of human IgG (17); biotinylated CX1, a rat mAb that recognizes mouse RAE-1α, RAE-1β, and RAE-1γ (22); and 186107, a rat mAb that recognizes RAE-1α, RAE-1β, RAE-1γ, RAE-1δ, and RAE-1ɛ. CX1 reacts strongly with RAE-1γ and weakly with RAE-1α and RAE-1β, but does not bind to RAE-1δ, RAE-1ɛ, or H-60. mAb 186107 was generated by immunizing LOU/MWS1 rats with a soluble recombinant protein consisting of the extracellular domain of RAE-1δ. This antibody recognizes all RAE-1 proteins, but not H-60. The Flag epitope on Flag-H-60 was detected using the mouse anti-Flag M2 mAb (Sigma-Aldrich). We previously showed that the Flag epitope tag does not interfere with the binding of mouse NKG2D-Ig to H-60 (23). Human MHC class I molecules were detected by staining with DX17, a mouse anti–human pan HLA class I mAb. Mouse β1-integrin (CD29) was detected by staining with HMβ1-1 mAb (BD Biosciences), followed by biotinylated goat anti–hamster Ig and streptavidin-PE. PE-conjugated secondary antibodies (Jackson Immunoresearch Laboratories) were used for detection of human Ig fusion protein and mouse and rat mAbs. Flow cytometry was performed by using a FACSCalibur® with CellQuest™ software (Becton Dickinson) or a Guava Personal Cytometer with Guava ViaCount and Guava Express Software (Guava Technologies).
TaqMan Quantitative RT-PCR.
Quantitative TaqMan PCR was performed on RNA extracted from BALB/c and C57BL/6 resident peritoneal macrophages collected from 6-wk old female mice. After harvest, macrophages were plate-adhered for 24 h, B cells were removed by washing cells with PBS, and macrophages were infected with Smith or Δm152 at an MOI of 1. Total RNA was extracted from macrophages 48 h after MCMV infection, RNA was DNase treated, and cDNA was synthesized using standard methods. To control for the possibility of amplifying genomic DNA by TaqMan PCR, 25% of total RNA was used in a cDNA synthesis reaction without reverse transcriptase. Samples without reverse transcriptase were used as templates in parallel with samples treated with reverse transcriptase. DNA amplified from samples without reverse transcriptase accounted for <10% of the total signal generated. Primer and probe sets specific for RAE-1α, RAE-1β, RAE-1δ, RAE-1γ, RAE-1ɛ, and H-60, previously described (22), were used with macrophage cDNA for TaqMan PCR. All samples were normalized to the signal generated from a housekeeping gene, HPRT. TaqMan PCR was performed using the ABI PRISM® 7700 Sequence Detection System.
IFN-γ Production.
The levels of IFN-γ produced by NK cells cocultured with MCMV-infected cells were determined by a standard ELISA using a BD Biosciences OptEIA™ Set. TpnT fibroblasts were infected with Smith or Δm152 virus at an MOI of 1 and incubated at 37°C. 48 h after infection, cells were trypsinized, fixed with fresh 4% paraformaldehyde, and washed three times with DMEM. Effector NK cells were prepared from C57BL/6 splenocytes and cultured for 7 d in RPMI 1640 medium with 10% heat-inactivated FBS, 2-mercaptoethanol, and 4,000 U/ml recombinant human IL-2 provided by the National Cancer Institute Biological Resources Branch Pre-clinical Repository. Before coculture with target cells, NK cells were either left untreated or treated for 30 min with control Ig, anti-NKG2D mAb (CX6; reference 22), or anti-Ly49H mAb (1F8), provided by M. Bennett (University of Texas Southwestern Medical Center, Dallas, TX). Infected target cells were incubated with NK cells at a 1:1 ratio for 24 h at 37°C. Supernatants were harvested and used in ELISA for detection of IFN-γ. Quantitation of IFN-γ production was determined by averaging triplicate wells and using linear regression from a standard curve.
In Vivo Infection and Determination of Viral Titers.
Five BALB/c mice were used in each of four experimental groups. 2 d before infection, BALB/c mice were injected intraperitoneally with either 100 μg rat anti–mouse NKG2D mAb CX5 (22) or 100 μg control rat IgG (eBioscience). On the day of MCMV infection (48 h after antibody treatment), 106 PFU of Smith or Δm152 virus was resuspended in DMEM without serum and used to intraperitoneally infect mice treated with anti-NKG2D antibody or control Ig. Spleens and livers were harvested from mice 3 d after infection and transferred to DMEM supplemented with 10% FBS. Liver weights were determined and organs were homogenized. Tissue homogenates were centrifuged at 3,400 rpm to pellet cell debris. Serial dilutions were made of the supernatants and a standard plaque assay was performed on BALB/c 3T3 cells to determine viral titers in each organ. Viral titers for spleen are expressed as PFU per organ whereas liver viral titers were calculated as PFU per gram tissue.
The rat anti–mouse NKG2D mAb CX5 blocks binding of NKG2D to the RAE-1 and H-60 ligands and inhibits NKG2D-dependent NK cell–mediated cytotoxicity against NKG2D ligand-bearing tumors in vitro and in vivo (22). When injected in vivo, anti-NKG2D mAb CX5 either blocked or modulated NKG2D, but did not deplete NK cells because the frequency of DX5+ CD3− NK cells in BALB/c mice and CD3− NK1.1+ NK cells in C57BL/6 mice was not reduced compared with normal, untreated mice (22). As a control to confirm modulation of NKG2D on NK cells from anti-NKG2D–treated mice, spleens from control Ig– and anti-NKG2D–treated mice were harvested 48 h after antibody injection, and expression of NKG2D on NK cells was analyzed. In brief, total splenocytes were stained with H-60-Ig fusion protein (consisting of the extracellular domain of H-60 fused to human IgG1 Fc), followed by biotinylated donkey anti–human Ig (Jackson ImmunoResearch Laboratories). Cells were then stained with streptavidin-PE, FITC-conjugated anti-pan NK cell mAb DX5 (VLA-2), and CyChrome-conjugated anti-CD3ɛ mAb (all from BD Biosciences). Samples were analyzed by flow cytometry.
Statistics.
For in vivo MCMV experiments, the Student's two-tailed t test with unequal variance was used to test the difference between arithmetic mean values of each set of PFU.
Results
Transcriptional Induction of RAE-1 Genes by MCMV Infection.
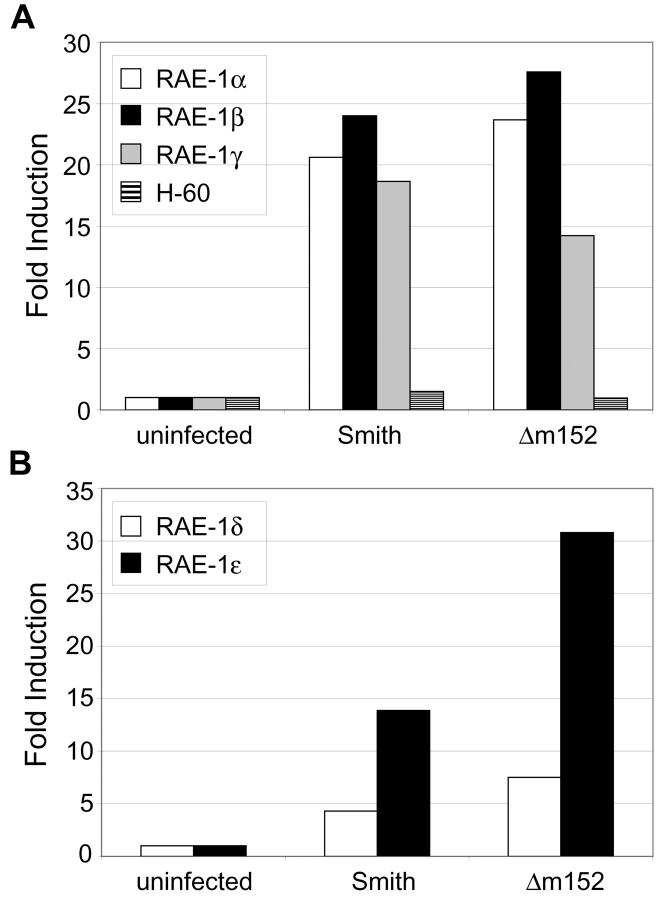
The products of at least six different genes have been identified as ligands for mouse NKG2D: RAE-1α, RAE-1β, RAE-1γ, RAE-1δ, RAE-1ɛ, and H-60 (17, 18). RAE-1α, RAE-1β, RAE-1γ, and H-60 are expressed in BALB/c mice (but not C57BL/6 mice) whereas RAE-1δ and RAE-1ɛ are expressed in C57BL/6 mice (but not BALB/c mice; unpublished data). To examine the effect of MCMV infection on transcription of RAE-1 genes, we analyzed RAE-1 and H-60 transcripts from MCMV-infected peritoneal macrophages. Resident peritoneal macrophages from BALB/c and C57BL/6 mice were isolated and infected with the wild-type Smith strain or an m152 deletion mutant (Δm152) at an MOI of 1. Total RNA was extracted from infected macrophages 48 h after infection and cDNA was synthesized. cDNA was then used in TaqMan quantitative PCR to analyze RAE-1 and H-60 transcripts. TaqMan primer and probe sets were designed to recognize H-60 and distinguish each of the five RAE-1 gene products. MCMV infection transcriptionally induced RAE-1α, RAE-1β, and RAE-1γ in peritoneal macrophages from BALB/c mice (Fig. 1 A) and RAE-1δ and RAE-1ɛ in infected C57BL/6 macrophages (Fig. 1 B). Both the wild-type and mutant virus induced similar levels of RAE-1 transcripts in infected cells. However, neither virus induced H-60 transcripts.
Figure 1.
RAE-1 transcripts are induced in MCMV-infected peritoneal macrophages. BALB/c and C57BL/6 peritoneal macrophages were left untreated or were infected with Smith or Δm152 (MOI = 1). RNA was extracted from cells 48 h after infection and cDNA was synthesized. TaqMan quantitative PCR was performed on BALB/c cDNA using primer/probe sets specific for RAE-1α, RAE-1β, RAE-1γ, or H-60 (A), and on C57BL/6 cDNA with primer/probe sets specific for RAE-1δ and RAE-1ɛ (B). Signals generated from each sample were normalized to HPRT signals from each set of cDNA. Data are expressed as the fold induction of transcription in infected cells compared with uninfected cells. Standard deviations of triplicate samples were <1%. Results shown are representative of three independent experiments.
Down-regulation of NKG2D Ligands by MCMV.
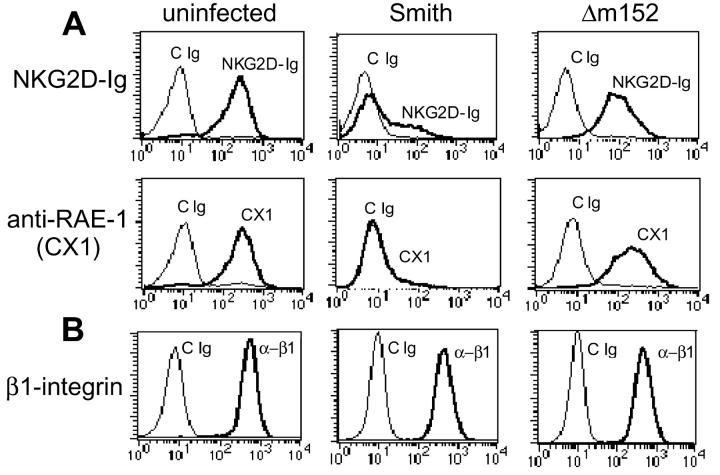
Given this potent induction of RAE-1 transcription, we examined RAE-1 protein levels on MCMV-infected cells. We infected BALB/c 3T3 cells, a permissive cell line from BALB/c mice that constitutively expresses RAE-1 on the cell surface, with wild-type virus at an MOI of 1. 72 h after infection, cells were stained for surface expression of NKG2D ligands using two different staining reagents: a mouse NKG2D-Ig fusion protein (17) and CX1, a mAb that recognizes RAE-1α, RAE-1β, and RAE-1γ (22). MCMV infection resulted in dramatic down-regulation of NKG2D ligands compared with uninfected cells when analyzed by flow cytometry using NKG2D-Ig or the anti–RAE-1–specific mAb CX1 (Fig. 2 A). Thus, RAE-1 appeared to be a prominent target for modulation by virus infection. To assess the specificity of down-regulation of NKG2D ligands, infected cells were also stained with anti–β1-integrin, which was unaffected by MCMV infection (Fig. 2 B). These experiments demonstrate that infection with MCMV results in specific down-regulation of RAE-1.
Figure 2.
MCMV infection results in down-regulation of NKG2D ligands. BALB/c 3T3 cells were infected with wild-type MCMV (Smith) or Δm152 (MOI = 1). 72 h after infection, infected cells were stained with control Ig (cIg), NKG2D-Ig fusion protein, or anti–RAE-1 mAb (CX1; A). As a control, cells were stained with anti–β1-integrin (B). Representative results are shown based on similar findings in three independent experiments.
One viral gene product, gp40, encoded by the MCMV m152 gene, has been shown to down-regulate MHC class I (6) as well as an undefined NKG2D ligand (20). We infected BALB/c 3T3 cells with wild-type or Δm152 mutant virus and stained infected cells with the mouse NKG2D-Ig fusion protein or anti–RAE-1 mAb. Interestingly, cells infected with Δm152 down-regulated expression of NKG2D ligands much less efficiently than wild-type virus (Fig. 2 A). This finding is consistent with that of Krmpotic et al. (20) who showed, using an NKG2D tetramer, that Δm152-infected cells maintained expression of NKG2D ligands compared with strong down-regulation observed in cells infected with wild-type virus. These experiments demonstrate that gp40 modulates expression of ligands for NKG2D in addition to down-regulating MHC class I proteins on the surface of MCMV-infected cells.
gp40 Down-regulates RAE-1α, RAE-1β, RAE-1γ, RAE-1δ, and RAE-1ɛ, but not H-60.
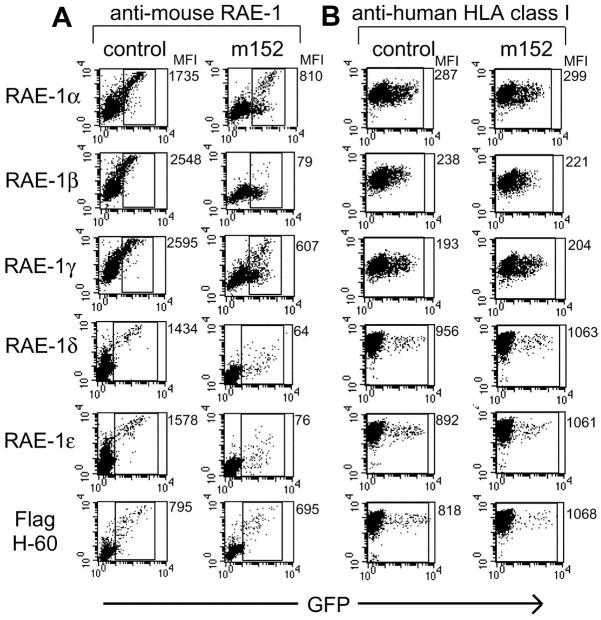
Studies of MHC class I down-regulation by gp40 have revealed an allele-specific preference, with certain MHC class I haplotypes affected more than others (24). To determine whether there are differential effects of gp40 on the five different RAE-1 proteins, we transiently transfected either m152 cDNA or a control empty vector, along with RAE-1α, RAE-1β, RAE-1γ, RAE-1δ, or RAE-1ɛ cDNA into human 293T cells. Mouse RAE-1 cDNA were encoded by a vector that also expressed green fluorescent protein (GFP [internal ribosome entry site (IRES)-GFP]) whereas m152 cDNA was encoded by a non-GFP vector. 48 h after transfection of RAE-1 cDNA with or without m152 cDNA, cells were examined for mouse RAE-1 expression by staining with anti–RAE-1 mAbs (CX1 mAb for detection of RAE-1α, RAE-1β, and RAE-1γ, or 186107 mAb for detection of RAE-1δ and RAE-1ɛ). Gating on the GFP+ population allowed us to analyze only RAE-1–expressing cells. Transfection of RAE-1 cDNA with a control empty vector resulted in high mouse RAE-1 expression as determined by anti–RAE-1 mAb staining (Fig. 3 A). In contrast, cotransfection of m152 along with any of the RAE-1 cDNAs (α, β, γ, δ, or ɛ) resulted in a substantial down-regulation. All of the RAE-1 proteins were found to be susceptible to the impact of gp40, in particular RAE-1β. As a control, the transfected human cells were also stained with an anti-HLA class I mAb. Anti–human HLA staining of human 293T cells was unaffected by m152 transfection (Fig. 3 B), suggesting that m152 down-regulation of RAE-1 proteins was specific and did not affect human MHC class I expression.
Figure 3.
gp40 selectively down-regulates RAE-1 proteins but not H-60. RAE-1 cDNAs or H-60 cDNA (with a 5′ Flag epitope tag) were cotransfected with a control empty vector, or with a vector encoding the m152 gene into human 293T cells. RAE-1 and H-60 cDNAs were encoded on vectors carrying an IRES-GFP whereas a non-GFP vector was used for m152 cDNA and for the control vector. 48 h after transfection, cells were stained with control Ig (cIg), biotinylated anti–RAE-1 mAb CX1 (recognizing RAE-1α, RAE-1β, and RAE-1γ), rat anti–RAE-1 mAb 186107 (recognizing RAE-1δ and RAE-1ɛ), or anti-Flag (A). As a control, cells were stained with anti–human HLA class I mAb DX17 (B). Mean fluorescence intensities (MFI) are shown to the right of each dot plot. MFI in A reflect RAE-1 or H-60 expression, calculated based on gated, GFP+ cells. MFI of human MHC class I expression (B) were determined based on the total cell population. Results are representative of similar findings in three independent experiments.
In a similar 293T transfection system, the effect of gp40 on H-60 expression was analyzed. Due to the lack of an antibody against H-60, a Flag epitope tag was inserted onto the 5′ terminus of the H-60 cDNA and expressed by using an IRES-GFP vector. Flag-H-60 cDNA was cotransfected with m152 cDNA or with a control vector into 293T cells. Cells were examined for H-60 expression by anti-Flag mAb staining 48 h after transfection. m152 expression had little or no effect on levels of Flag-H-60 (Fig. 3 A). Cotransfection of m152 and native H-60 without the Flag epitope tag and staining with mouse NKG2D-Ig fusion protein also indicated that m152 did not effect expression of H-60 (unpublished data). Thus, although gp40 potently down-regulates RAE-1α, RAE-1β, RAE-1γ, RAE-1δ, and RAE-1ɛ, gp40 does not appear to influence H-60 expression.
MCMV Infection Induces NKG2D-mediated IFN-γ Production by NK Cells.
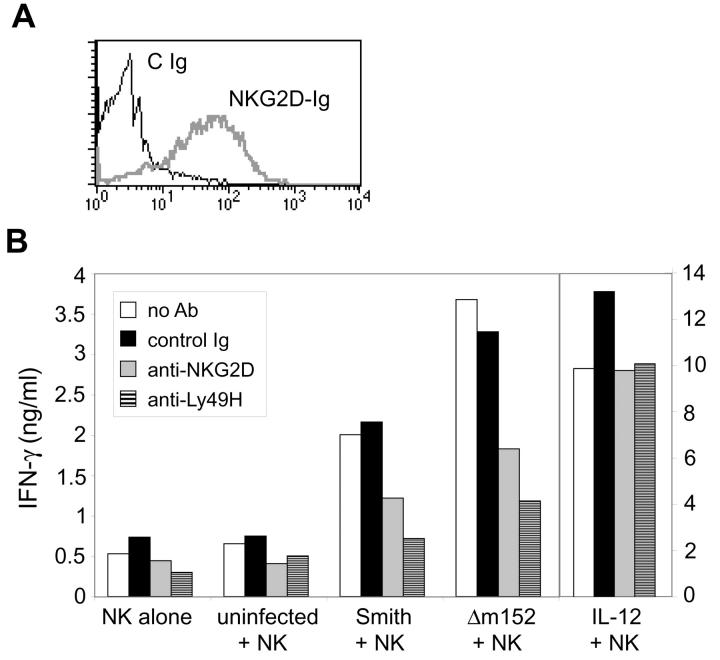
To evaluate the functional consequences of RAE-1 down-regulation by gp40, we analyzed IFN-γ production by NK cells cocultured with MCMV-infected cells. For this experiment, TpnT cells (mouse embryonic fibroblasts derived from C57BL/6 mice) were used for infection. These cells stained positively with the NKG2D-Ig fusion protein (Fig. 4 A), indicating that they constitutively expressed relatively high levels of RAE-1 proteins. TpnT fibroblasts were infected with wild-type or Δm152 mutant virus at an MOI of 1, and 72 h after infection cells were trypsinized and fixed with 4% paraformaldehyde. Infected cells and uninfected control cells were then cocultured with IL-2–activated C57BL/6 NK cells at a 1:1 ratio for 24 h in the presence or absence of blocking antibodies against mouse NKG2D, mouse Ly49H, or a control mAb. As additional controls, NK cells were cultured alone to determine background production of IFN-γ, or cultured with IL-12 to stimulate maximal IFN-γ production. Supernatants were collected from the cocultured cells and analyzed by ELISA for IFN-γ. Coculture of NK cells with uninfected cells generated low amounts of IFN-γ comparable to levels of IFN-γ produced by NK cells alone (Fig. 4 B). In contrast, coculture with virus-infected cells, particularly Δm152-infected cells, resulted in high IFN-γ production, which was partially blocked by culturing NK cells with anti-NKG2D mAb. Consistent with previous findings (25), anti-Ly49H mAb also substantially blocked IFN-γ production. Blocking antibodies did not effect IFN-γ production by NK cells cultured with IL-12. These data demonstrate that modulation of RAE-1 molecules by gp40 has functional consequences for NK cell function: the presence of RAE-1 molecules on Δm152-infected cells allows for NKG2D-dependent activation of NK cell IFN-γ production. The ability to block this effect by addition of anti-NKG2D antibody highlights the potential significance of RAE-1–NKG2D interactions for NK cell activation during virus infection.
Figure 4.
MCMV infection costimulates NK cell IFN-γ production through NKG2D. (A) TpnT cells were stained with a control antibody or with the NKG2D-Ig fusion protein. (B) TpnT cells were infected with Smith or Δm152 virus (MOI = 1) and cocultured with IL-2–activated C57BL/6 NK cells. Before coculture, NK cells were left untreated or treated with control Ig, anti-NKG2D (CX6), or anti-Ly49H (IF8), as indicated. As controls, NK cells were also cultured alone or in the presence of IL-12. Standard deviations of triplicate samples were <1%. This experiment was performed several times with comparable results.
m152 Contributes to the Virulence of MCMV by Disrupting NKG2D Functions.
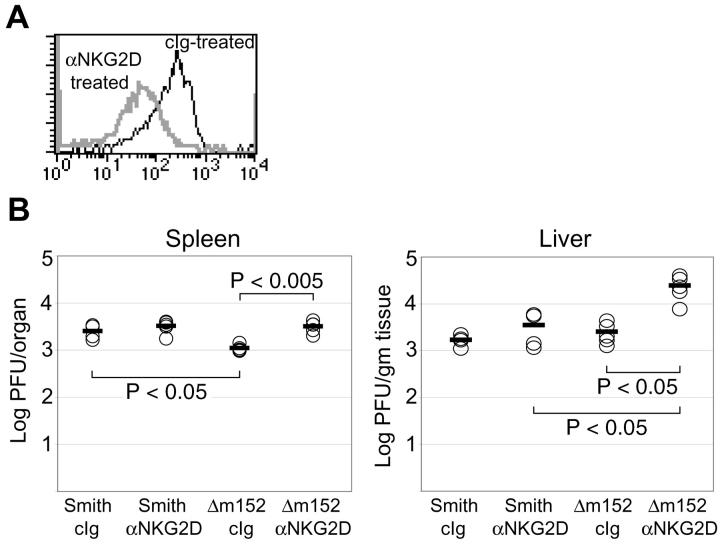
Based on the in vitro experiments described above, gp40-mediated down-regulation of RAE-1 molecules during MCMV infection seemed to impair the ability of NK cells to respond to MCMV-infected cells. To examine the physiological relevance of these findings, we evaluated the role of RAE-1–NKG2D interactions during viral infection in MCMV-susceptible BALB/c mice. In BALB/c mice, which lack the Ly49H receptor, the importance of NKG2D during MCMV infection could be investigated in the absence of the dominant effects of Ly49H in C57BL/6 mice (26), which may obscure the effects of gp40 (20). 2 d before infection, BALB/c mice were treated intraperitoneally with a blocking anti-NKG2D mAb (CX5) or an isotype-matched control antibody. Treatment of mice with anti-NKG2D blocked or modulated expression of the NKG2D receptor, but did not deplete NK cells, as determined by detection of normal frequencies of DX5+ CD3− NK cells in the anti-NKG2D mAb-treated mice (22). On the day of infection, NKG2D blocking was verified by staining splenocytes from anti-NKG2D– or control Ig–treated mice with an H-60-Ig fusion protein. As shown in Fig. 5 A, treatment of mice with anti-NKG2D resulted in decreased NKG2D expression on DX5+ CD3− splenocytes. For MCMV infection, 106 PFU of Smith or Δm152 virus were injected intraperitoneally into anti-NKG2D– or control Ig–treated mice. 3 d after infection, spleens and livers were harvested from infected mice and homogenized. Virus was extracted from organ homogenates and used in a plaque assay to determine viral titers in each organ.
Figure 5.
Virulence of Δm152 virus is restored by blocking NKG2D in vivo. (A) BALB/c mice were injected intraperitoneally with control Ig or anti-NKG2D mAb (CX5). As a control to ensure NKG2D modulation, spleens were harvested from control Ig– and anti-NKG2D–treated animals 48 h after antibody treatment. Total splenocytes were stained with H-60-Ig fusion protein (to detect NKG2D), biotinylated goat anti–human Ig, streptavidin-PE, FITC-conjugated anti-pan NK cell mAb DX5 (VLA-2), and CyChrome-conjugated anti-CD3ɛ mAb. Overlaid histograms represent H-60-Ig staining of DX5+ CD3− cells from control Ig– or anti-NKG2D–treated mice. Although equivalent frequencies of DX5+ CD3− NK cells were present in control Ig– and anti-NKG2D mAb–treated mice, NKG2D was modulated on NK cells of mice treated with anti-NKG2D mAb in vivo. (B) 48 h after antibody treatment, mice were infected with 106 PFU of Smith or Δm152. 3 d after infection, mice were killed and spleens and livers were harvested. Plaque assays were performed on organ homogenates to determine viral titers in spleen and liver. Five mice were used per group. Two independent experiments were performed with comparable results; a representative experiment is shown. Statistical analysis was performed using the Student's two-tailed t test with unequal variance.
As depicted in Fig. 5 B, in spleens of control Ig–treated mice, Δm152 infection resulted in significantly lower viral titers than Smith infection (P < 0.05). It is likely that this is due to the expression of RAE-1 on the surface of Δm152-infected cells, which facilitates NK cell–mediated clearance. Consistent with this interpretation, in the anti-NKG2D–treated mice infected with Δm152, viral titers in the spleen and liver were comparable to titers in mice infected with wild-type virus, which down-regulates RAE-1 on infected cells. During infection with wild-type virus, anti-NKG2D mAb treatment consistently resulted in a slight, but not significant, increase in viral titers, as compared with control Ig–treated mice. This is not unexpected because cells infected with wild-type virus may have very little RAE-1 on the cell surface, due to its down-regulation by gp40. Collectively, these experiments suggest that gp40, encoded by m152, may provide a means of MCMV escape from NK cell recognition at early times after infection.
Discussion
In this study, we have precisely defined another target of the MCMV immunomodulatory gene product gp40. In addition to MHC class I down-regulation (6), gp40 selectively prevents cell surface expression of RAE-1 and impairs NK cell recognition of virus-infected cells. MCMV gp40 does not reduce transcription of the RAE-1 genes, but appears to prevent expression of RAE-1 glycoproteins on the cell surface by a posttranscriptional mechanism. The ability of gp40 to affect expression of both conventional H-2 class I and the RAE-1 proteins is remarkable, considering that RAE-1 shares such limited amino acid homology with MHC class I (<20%; 17, 18). RAE-1 glycoproteins lack an α3 domain, do not bind peptides, do not associate with β2-microglobulin, and are glycosylphosphatidylinositol-anchored proteins. Like MHC class I, though, RAE-1 glycoproteins have an extracellular domain containing α1 and α2 helices, which are believed to be the sites of gp40 interaction with MHC class I (27). Additional studies will reveal whether gp40 binds to RAE-1 and H-2 in a similar fashion.
Although gp40 impacts all five known RAE-1 molecules, RAE-1α, RAE-1β, RAE-1γ, RAE-1δ, and RAE-1ɛ, interestingly it does not seem to affect surface expression of H-60, another cell surface MHC class I–like glycoprotein with α1 and α2 domains that binds to NKG2D with high affinity (28). Although the RAE-1 proteins share a high degree of similarity (94–98% identical), H-60 has only ∼25% homology to the RAE-1 family (17, 18). It is tempting to speculate that MCMV may have evolved gp40 to bind the RAE-1 proteins, but not H-60, because of selective pressure exerted by the immune system. The abundance of RAE-1 RNA in infected cells may have driven the evolution of a viral protein that can specifically prevent cell surface expression of RAE-1 glycoproteins in addition to conventional H-2 class I. Because H-60 transcription does not appear to be induced by MCMV infection of macrophages, gp40 binding to H-60 proteins may confer little selective advantage to the virus.
A recent paper by Krmpotic et al. (20) also demonstrated down-regulation of NKG2D ligands by gp40. In an effort to address whether the susceptibility of BALB/c mice to MCMV is caused by virus-induced subversion of NK cell–mediated immunity, these investigators examined the ability of two MCMV-encoded MHC class I modulators, gp40 and gp48, to regulate NK cell activation. They found that the presence of gp40 during viral infection resulted in diminished NK cell control of MCMV in the lungs of BALB/c, but not C57BL/6 mice. Thus, this study convincingly demonstrated the importance of gp40 in viral pathogenesis in vivo in BALB/c mice using an m152 deletion mutant virus (20). Based on previous evidence that BALB/c, but not C57BL/6 mice can express H-60 (29), these investigators proposed that gp40 might preferentially affect expression of H-60. However, no evidence was provided to verify that H-60 is present in MCMV-infected cells or that gp40 can directly modulate H-60 expression. In addition to differences in H-60 expression, C57BL/6 and BALB/c mice differentially express RAE-1 genes: RAE-1α, RAE-1β, and RAE-1γ are expressed in BALB/c but not in C57BL/6 mice whereas RAE-1δ and RAE-1ɛ are expressed in C57BL/6 but not in BALB/c mice. Therefore, the different immune responses to MCMV observed in BALB/c compared with C57BL/6 mice must take into account the expression of different RAE-1 genes and different activating Ly49 receptors, as well as other background differences, in these two mouse strains.
Immunity to MCMV requires both the action of NK cells and cytotoxic T cells (CTL), both of which express NKG2D. In this regard, it is interesting that gp40 has evolved to block cell surface expression of conventional MHC class I and RAE-1, a mechanism that might be particularly effective in evading detection by CTL. Our present studies have focused on the early NK cell–mediated events after viral infection. We have shown that transcription of RAE-1 genes was induced by MCMV infection of primary cells, but that viral gp40 efficiently prevented cell surface expression of the RAE-1 glycoproteins. The physiological importance of this was revealed by comparing the virulence of wild-type and Δm152 virus in MCMV-susceptible BALB/c mice. Consistent with previous findings (20), the m152-deficient virus showed reduced virulence during in vivo infection of BALB/c mice, possibly due to the elimination of RAE-1–bearing virus-infected cells by an NKG2D-dependent mechanism. This hypothesis was tested by treating BALB/c mice with anti-NKG2D mAb before infection. Treatment of mice with anti-NKG2D mAb restored viral titers of Δm152-infected mice to levels comparable to wild-type virus-infected mice. These findings support an important role for m152 in viral evasion of NKG2D-dependent immune surveillance. Blockade of NKG2D in mice infected with wild-type virus showed only modest effects on viral titers early after infection, a result predicted if gp40 efficiently suppresses RAE-1 expression in infected cells.
The ability of gp40 to down-regulate MHC class I has been clearly implicated in evasion of CTL-mediated immunity late after viral infection. However, during the early stages of infection dominated by NK cell immunity, loss of inhibitory MHC class I should render virus-infected cells more susceptible to NK cell attack. In mice infected with m152-deficient virus, which does not efficiently block H-2 expression, however, viral titers in the spleen were in fact lower than in mice infected with wild-type virus at early times after infection. As previously noted, blockade of NKG2D restored virulence of the m152-deficient virus, suggesting that NKG2D may permit NK cells to recognize and eliminate virus-infected cells, even if MHC class I is expressed on the infected cells. Similarly, recent studies have shown that NK cells are capable of rejecting tumors bearing MHC class I if they express NKG2D ligands (13, 14).
Based on experiments with gene-deficient mice, it was found that a perforin-dependent NK cell–mediated cytotoxicity is critical for control of MCMV in the spleen, but in the liver, NK cell–derived IFN-γ is a predominant mechanism of viral clearance (30). We have shown that C57BL/6 NK cells cultured with wild-type or Δm152-infected cells produced IFN-γ by a mechanism that is at least in part dependent on NKG2D. We have also shown that IFN-γ produced by C57BL/6 NK cells cocultured with virus-infected cells is largely due to Ly49H-dependent activation because this effect is blocked by anti-Ly49H antibody. Ly49H, expressed in C57BL/6 but not BALB/c mice, recognizes m157, a MCMV-encoded MHC class I–like glycoprotein that is expressed on the surface of virus-infected cells (25, 31). Engagement of Ly49H by m157 results in NK cell activation and contributes to NK cell protection against MCMV in C57BL/6 mice (26, 32, 33). Our in vitro results imply that NKG2D may costimulate Ly49H-dependent NK cell activation, thereby providing more efficient immunity in MCMV-resistant C57BL/6 mice. However, it appears that Ly49H is the dominant player in this response in vivo because previous studies failed to demonstrate a substantial difference in viral titers in C57BL/6 mice infected with wild-type or m152 deletion mutant viruses (20). Interestingly, we were unable to observe IFN-γ production by BALB/c NK cells in vitro in response to MCMV-infected BALB/c-derived fibroblasts (unpublished data). These data suggest that Ly49H might be necessary for robust IFN-γ production in response to MCMV infection.
In summary, by down-regulating expression of RAE-1 molecules, thereby impairing NK cell recognition of virus-infected cells, gp40 may effectively undermine the early NK cell response to MCMV infection. This mechanism of immune evasion likely contributes to the complex interplay of viral persistence and NK cell protection inherent to herpesvirus infections.
Acknowledgments
We thank Dr. Ann Hill for generously providing viruses, Dr. Lenore Pereira and Dr. Jill Bechtel for helpful discussion, and Mr. Chad Borchert for expert assistance in the production of monoclonal antibodies.
This work is supported by National Institutes of Health grants CA89189, CA95137, and AI30363. K. Ogasawara is supported by Human Frontier Science Program Long-Term Fellowship, J.A. Hamerman is supported by an Irvington Foundation Fellowship, H. Arase is supported by a Japan Society for the Promotion of Science Postdoctoral Fellowship, and L.L. Lanier is an American Cancer Society Research Professor.
H. Arase's present address is Division of Molecular Genetics, Chiba University Graduate School of Medicine, 1-8-1 Inohana, Chuou-ku, Chiba 260-8670, Japan.
Footnotes
Abbreviations used in this paper: GFP, green fluorescent protein; HCMV, human CMV; IRES, internal ribosome entry site; MCMV, murine CMV; MOI, multiplicity of infection; RAE-1, retinoic acid early inducible 1 gene.
References
- 1.Tortorella, D., B.E. Gewurz, M.H. Furman, D.J. Schust, and H.L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861–926. [DOI] [PubMed] [Google Scholar]
- 2.Mocarski, E.S., Jr. 2002. Immunomodulation by cytomegaloviruses: manipulative strategies beyond evasion. Trends Microbiol. 10:332–339. [DOI] [PubMed] [Google Scholar]
- 3.Kleijnen, M.F., J.B. Huppa, P. Lucin, S. Mukherjee, H. Farrell, A.E. Campbell, U.H. Koszinowski, A.B. Hill, and H.L. Ploegh. 1997. A mouse cytomegalovirus glycoprotein, gp34, forms a complex with folded class I MHC molecules in the ER which is not retained but is transported to the cell surface. EMBO J. 16:685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kavanagh, D.G., U.H. Koszinowski, and A.B. Hill. 2001. The murine cytomegalovirus immune evasion protein m4/gp34 forms biochemically distinct complexes with class I MHC at the cell surface and in a pre-Golgi compartment. J. Immunol. 167:3894–3902. [DOI] [PubMed] [Google Scholar]
- 5.Reusch, U., W. Muranyi, P. Lucin, H.-G. Burgert, H. Hengel, and U.H. Koszinowski. 1999. A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J. 18:1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziegler, H., R. Thale, P. Lucin, W. Muranyi, T. Flohr, H. Hengel, H. Farrell, W. Rawlinson, and U.H. Koszinowski. 1997. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity. 6:57–66. [DOI] [PubMed] [Google Scholar]
- 7.Kavanagh, D.G., M.C. Gold, M. Wagner, U.H. Koszinowski, and A.B. Hill. 2001. The multiple immune-evasion genes of murine cytomegalovirus are not redundant. M4 and m152 inhibit antigen presentation in a complementary and cooperative fashion. J. Exp. Med. 194:967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ljunggren, H.-G., and K. Karre. 1985. Host resistance directed selectively against H-2–deficient lymphoma variants: analysis of the mechanism. J. Exp. Med. 162:1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukowski, J.F., J.F. Warner, G. Dennert, and R.M. Welsh. 1985. Adoptive transfer studies demonstrating the antiviral effect of natural killer cells in vivo. J. Exp. Med. 161:40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biron, C.A., K.S. Byron, and J.L. Sullivan. 1989. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 320:1731–1735. [DOI] [PubMed] [Google Scholar]
- 11.Wu, J., Y. Song, A.B.H. Bakker, S. Bauer, V. Groh, T. Spies, L.L. Lanier, and J.H. Phillips. 1999. An activating receptor complex on natural killer and T cells formed by NKG2D and DAP10. Science. 285:730–732. [DOI] [PubMed] [Google Scholar]
- 12.Diefenbach, A., E. Tomasello, M. Lucas, A.M. Jamieson, J.K. Hsia, E. Vivier, and D.H. Raulet. 2002. Selective associations with signaling proteins determine stimulatory versus costimulatory activity of NKG2D. Nat. Immunol. 3:1142–1149. [DOI] [PubMed] [Google Scholar]
- 13.Cerwenka, A., J.L. Baron, and L.L. Lanier. 2001. Ectopic expression of retinoic acid early inducible-1 gene (RAE-1) permits natural killer cell-mediated rejection of a MHC class I-bearing tumor in vivo. Proc. Natl. Acad. Sci. USA. 98:11521–11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diefenbach, A., E.R. Jensen, A.M. Jamieson, and D.H. Raulet. 2001. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 413:165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer, S., V. Groh, J. Wu, A. Steinle, J.H. Phillips, L.L. Lanier, and T. Spies. 1999. Activation of natural killer cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 285:727–730. [DOI] [PubMed] [Google Scholar]
- 16.Cosman, D., J. Mullberg, C.L. Sutherland, W. Chin, R. Armitage, W. Fanslow, M. Kubin, and N.J. Chalupny. 2001. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 14:123–133. [DOI] [PubMed] [Google Scholar]
- 17.Cerwenka, A., A.B. Bakker, T. McClanahan, J. Wagner, J. Wu, J.H. Phillips, and L.L. Lanier. 2000. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 12:721–727. [DOI] [PubMed] [Google Scholar]
- 18.Diefenbach, A., A.M. Jamieson, S.D. Liu, N. Shastri, and D.H. Raulet. 2000. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat. Immunol. 1:119–126. [DOI] [PubMed] [Google Scholar]
- 19.Carayannopoulos, L.N., O.V. Naidenko, D.H. Fremont, and W.M. Yokoyama. 2002. Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J. Immunol. 169:4079–4083. [DOI] [PubMed] [Google Scholar]
- 20.Krmpotic, A., D.H. Busch, I. Bubic, F. Gebhardt, H. Hengel, M. Hasan, A.A. Scalzo, U.H. Koszinowski, and S. Jonjic. 2002. MCMV glycoprotein gp40 confers virus resistance to CD8+ T cells and NK cells in vivo. Nat. Immunol. 3:529–535. [DOI] [PubMed] [Google Scholar]
- 21.Gold, M.C., M.W. Munks, M. Wagner, U.H. Koszinowski, A.B. Hill, and S.P. Fling. 2002. The murine cytomegalovirus immunomodulatory gene m152 prevents recognition of infected cells by M45-specific CTL but does not alter the immunodominance of the M45-specific CD8 T cell response in vivo. J. Immunol. 169:359–365. [DOI] [PubMed] [Google Scholar]
- 22.Ogasawara, K., J.A. Hamerman, H. Hsin, S. Chikuma, H. Bour-Jordan, T. Chen, T. Pertel, C. Carnaud, J.A. Bluestone, and L.L. Lanier. 2003. Impairment of NK cell function by NKG2D modulation in NOD mice. Immunity. 18:41–51. [DOI] [PubMed] [Google Scholar]
- 23.Cerwenka, A., C.A. O'Callaghan, J.A. Hamerman, R. Yadav, W. Ajayi, D.C. Roopenian, S. Joyce, and L.L. Lanier. 2002. Cutting edge: the minor histocompatibility antigen H60 peptide interacts with both H-2K(b) and NKG2D. J. Immunol. 168:3131–3134. [DOI] [PubMed] [Google Scholar]
- 24.Wagner, M., A. Gutermann, J. Podlech, M.J. Reddehase, and U.H. Koszinowski. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arase, H., E.S. Mocarski, A.E. Campbell, A.B. Hill, and L.L. Lanier. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 296:1323–1326. [DOI] [PubMed] [Google Scholar]
- 26.Brown, M.G., A.O. Dokun, J.W. Heusel, H.R. Smith, D.L. Beckman, E.A. Blattenberger, C.E. Dubbelde, L.R. Stone, A.A. Scalzo, and W.M. Yokoyama. 2001. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 292:934–937. [DOI] [PubMed] [Google Scholar]
- 27.Gutermann, A., A. Bubeck, M. Wagner, U. Reusch, C. Menard, and U.H. Koszinowski. 2002. Strategies for the identification and analysis of viral immune-evasive genes–cytomegalovirus as an example. Curr. Top. Microbiol. Immunol. 269:1–22. [DOI] [PubMed] [Google Scholar]
- 28.O'Callaghan, C.A., A. Cerwenka, B.E. Willcox, L.L. Lanier, and P.J. Bjorkman. 2001. Molecular competition for NKG2D. H60 and RAE1 compete unequally for NKG2D with dominance of H60. Immunity. 15:201–211. [DOI] [PubMed] [Google Scholar]
- 29.Malarkannan, S., P.P. Shih, P.A. Eden, T. Horng, A.R. Zuberi, G. Christianson, D. Roopenian, and N. Shastri. 1998. The molecular and functional characterization of a dominant minor H antigen, H60. J. Immunol. 161:3501–3509. [PubMed] [Google Scholar]
- 30.Tay, C.H., and R.M. Welsh. 1997. Distinct organ-dependent mechanisms for the control of murine cytomegalovirus infection by natural killer cells. J. Virol. 71:267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, H.R., J.W. Heusel, I.K. Mehta, S. Kim, B.G. Dorner, O.V. Naidenko, K. Iizuka, H. Furukawa, D.L. Beckman, J.T. Pingel, et al. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. USA. 99:8826–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daniels, K.A., G. Devora, W.C. Lai, C.L. O'Donnell, M. Bennett, and R.M. Welsh. 2001. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J. Exp. Med. 194:29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, H.-S., S. Gerard, D. Macina, M. Busa, A. Zafer, A. Belouchi, P. Gros, and S.M. Vidal. 2001. Susceptibility to mouse cytomegalovirus is associated with depletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat. Genet. 28:42–45. [DOI] [PubMed] [Google Scholar]