Abstract
We have previously shown that DNA circles containing telomeric repeats and a marker gene can promote the recombinational elongation of telomeres in Kluyveromyces lactis by a mechanism proposed to involve rolling-circle DNA synthesis. Wild-type cells acquire a long tandem array at a single telomere, while telomerase deletion (ter1-Δ) cells, acquire an array and also spread it to multiple telomeres. In this study, we further examine the factors that affect the formation and spread of telomeric tandem arrays. We show that a telomerase+ strain with short telomeres and high levels of subtelomeric gene conversion can efficiently form and spread arrays, while a telomere fusion mutant is not efficient at either process. This indicates that an elevated level of gene conversion near telomeres is required for spreading but that growth senescence and a tendency to elongate telomeres in the absence of exogenously added circles are not. Surprisingly, telomeric repeats are frequently deleted from a transforming URA3-telomere circle at or prior to the time of array formation by a mechanism dependent upon the presence of subtelomeric DNA in the circle. We further show that in a ter1-Δ strain, long tandem arrays can arise from telomeres initially containing a single-copy insert of the URA3-telomere sequence. However, the reduced rate of array formation in such strains suggests that single-copy inserts are not typical intermediates in arrays formed from URA3-telomere circles. Using heteroduplex circles, we have demonstrated that either strand of a URA3-telomere circle can be utilized to form telomeric tandem arrays. Consistent with this, we demonstrate that 100-nucleotide single-stranded telomeric circles of either strand can promote recombinational telomere elongation.
Telomeres are DNA-protein structures that protect the ends of linear chromosomes (for reviews, see references 1, 5, and 22). In the great majority of eukaryotic cells, telomeres are maintained by the reverse-transcriptase enzyme telomerase. However, work with both yeast and mammalian cells has shown that cells lacking telomerase can sometimes maintain telomeres through mechanisms independent of telomerase (3, 24). Research on such mechanisms has gained increasing significance since it has been found that ∼5 to 10% of cancers maintain their telomeres in the absence of telomerase (2). Telomerase-independent telomere maintenance in mammalian cells has been called “alternate lengthening of telomeres” (ALT). Cells displaying ALT have telomeres of heterogeneous sizes but often much longer than those of telomerase-containing cells. There has been recent evidence that ALT in mammalian cells is dependent on homologous recombination (2, 7, 9).
Studies of yeast have shown that when telomerase is deleted, most of the cells senesce and eventually die as telomeres gradually shrink to critically short lengths. Cell death is preceded by induction of a damage response that resembles, but appears distinct from, the DNA damage checkpoint (8, 14). The small minority of cells that live beyond this point have been termed postsenescence survivors. Two distinct types of postsenescence survivors have been described in Saccharomyces cerevisiae, both of which depend upon recombination, as indicated by their dependence upon RAD52 function (6, 16, 29, 30). Type I survivors have subtelomeric amplifications and maintain short telomeric arrays, while type II survivors have elongated telomeric arrays and lack the subtelomeric amplifications. In addition to their differing telomeric structures, type I and type II survivors also have different genetic requirements for their formation. Type I survivors require Rad51, Rad54, and Rad57, while type II survivors require Rad50, Rad59, and Sgs1 (6, 13, 29).
Postsenescence survivors of Kluyveromyces lactis cells lacking telomerase resemble the type II survivors of S. cerevisiae in having telomeric expansions that depend upon RAD52 (18). Recombination in the vicinity of K. lactis telomeres has been shown to be enormously elevated when telomeres are very short (20). This is likely due to the telomeres losing their protective capping function and being subject to recombinational repair in a manner similar to that which would occur with a broken DNA end. The high rate of telomeric recombination in senescing cells was proposed to be crucial to the formation of postsenescence survivors (20).
Recently, it has been proposed that the elongated telomeres of K. lactis postsenescence survivors arise through two types of recombinational processes (25). In one, a very small circle of telomeric DNA is used as a template for the extension of at least one short telomere by a rolling-circle-type mechanism. Once a single long telomere is formed, that sequence can be used as a template by other telomeres in the cell. This “roll-and-spread” model arose initially from the observation that recombinational telomere elongation in K. lactis cells containing two types of telomeric repeats generated telomeres with repeating patterns that were common to most or all telomeres within a given survivor (25). Direct evidence for the extension of telomeres by circles came from the finding that circles of DNA containing URA3 and K. lactis telomeric repeats led to the formation of long tandem arrays at telomeres when transformed into K. lactis cells. These tandem arrays were normally found at a single telomere when transformed into wild-type cells but were found at many or most telomeres in transformants of telomerase deletion mutants. Mixing experiments using two slightly different URA3-telomere circles showed that all copies of the integrated sequence in a particular clone were derived from a single transforming circular DNA molecule. In this study, we further examine recombinational telomere elongation promoted by URA3-telomere circles, as well as show that circles as small as 100 nucleotides (nt) can promote telomere elongation in vivo.
MATERIALS AND METHODS
Strains.
The ter1-Taq, ter1-AccSna, msh2-Δ, and ter1-Δ msh2-Δ strains are derivatives of haploid K. lactis 7B520 (Ura− His− Trp−) (35). The ter1-Taq, ter1-Δ, and ter1-AccSna strains have been described previously (19, 20). The TER1/ter1-Δ msh2-Δ heteroallele strain was obtained from the laboratory of V. Lundblad (28). The ter1-Δ msh2-Δ and TER1 msh2-Δ strains were constructed by the loop-out of TER1 or ter1-Δ, respectively, from the TER/ter1-Δ msh2-Δ heteroallele strain as described before (19). ter1-16T was generated by the loop-in-loop-out procedure. The ter1 allele used for the loop-in had a point mutation in the Rap1 binding site of the template region (D. Underwood, C. Carroll, and M. McEachern, submitted for publication).
IUT-WT1 was generated when a single copy of circle S was incorporated at a telomere in a TER1 strain that was transformed with circle S (25). To generate IUT-Taq, we mated ter1-Taq to IUT-WT1. However, the TER1 and ter1-Taq haploid spores that were produced from this mating had single circle S inserts at two or three telomeres (data not shown), apparently because the insert had spread to other telomeres. This spreading occurred prior to the formation of ascospores, as the URA3 gene was detectable in 50 to 100% of spores instead of the expected 50%. The initial presence of short telomeres in the diploid that came from the ter1-Taq parent may have led to increased recombination events at the telomeres. We then used an IUT-WT haploid strain that had single-copy circle S inserts at two telomeres to mate with a ter1-Δ strain to generate IUT-Del. Tetrad dissection of TER1/ter1-Δ diploids produced haploid spores of ter1-Δ and TER1 that had single-copy circle S inserts at one, two, or three telomeres (data not shown). Examples of these strains were then used to test whether the 1.6-kb circle S inserts could expand into long tandem arrays. As a control, the single-copy circle S insert in the TER1 strain was introduced into another TER1 strain by mating to generate additional IUT-WT strains with the single circle S insert at one telomere. Serial restreaks of the strains generated was done by passaging a mixture of colonies on yeast-peptone-dextrose (YPD) agar.
Mating and sporulation.
Matings were performed on malt extract agar for 2 days, and diploids were selected on synthetic defined plates lacking uracil and histidine. The diploids were allowed to sporulate on minimal sporulation medium (10% potassium acetate) for 2 days. Individual spores from tetrad dissection were allowed to grow on YPD for 3 days before further analysis.
Southern hybridization and quantitation of URA3 copy number.
Hybridizations to telomeric, subtelomeric, and URA3 probes and quantitation of URA3 copy numbers were done as described previously (25). A second subtelomeric fragment of 113 bp was used to determine if there were any subtelomeric deletions of the tandem arrays formed in circle N transformants. This 113-bp fragment was obtained as a BsrBI-HindIII fragment from the plasmid Stumaker II, which is a derivative of pBluescript containing telomeric and subtelomeric sequences from a chromosome end of K. lactis (20). The 113-bp subtelomeric fragment was hybridized at 50°C. Hybridization washes were done in 200 mM Na2HPO4 and 2% sodium dodecyl sulfate for a total of 15 min. URA3-RAD52 hybridizations were done using a 2.2-kb DraI-BglII fragment from the plasmid p52ΔC:UB, a derivative of pBluescript containing the K. lactis RAD52 gene disrupted by URA3 from S. cerevisiae. Hybridization was done at 65°C. Hybridization washes were done in 100 mM Na2HPO4 and 2% sodium dodecyl sulfate for a total of 45 min. URA3 copy numbers were determined using a phosphorimager.
Construction and isolation of the circle with no subtelomeric region (circle N), heteroduplex circles (circle H), and the 100-nt circle.
A circle with no subtelomeric sequence, circle N, was isolated as described previously for circle S and circle P (25) by ligation of a gel-purified BamHI-BglII fragment. The plasmid construct used for generating circle N was pMH3-Tel-NoST, which lacked subtelomeric sequence. pMH3-Tel-NoST was created by introducing an XhoI-BsrBI telomeric fragment from pMH3-Tel (25) into pMH3 (XhoI-NruI site), which resulted in a telomeric insert present in the orientation opposite to that originally present in pMH3-Tel. All plasmids were maintained in Escherichia coli XL1.
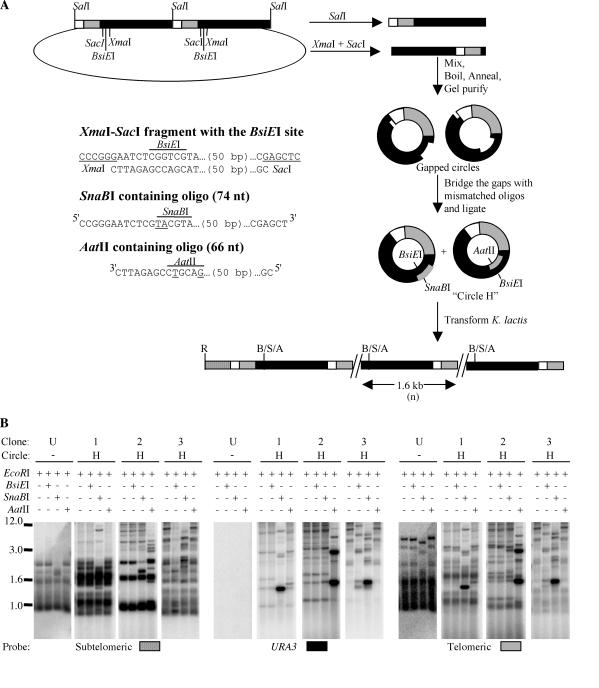
A heteroduplex circle, circle H, was made using a construct derived from pMH3-2UraTel that had two tandem copies of the BamHI-BglII 1.6-kb URA3-telomere sequence (see Fig. 5A). This was constructed by introducing a second copy of the BamHI-BglII fragment containing the URA3-telomere sequence into the BglII site of pMH3-Tel. Two fragments were gel purified from this plasmid; a 1.6-kb SalI fragment that had the entire length of the URA3-telomere sequence and an XmaI-SacI fragment that contained all of the 1.6-kb URA3-telomere sequence except for a 70-bp region at the end of the URA3 gene that contained a BsiEI site. Two oligomers that spanned the 70-bp region were made as complementary strands that could fill the gap in the XmaI-SacI fragment. One of the two oligomers, which corresponded to the strand that contained the G-rich telomeric sequence, had two mismatched bases that created a SnaBI site and eliminated the BsiEI site. The other oligomer, which corresponded to the strand that contained the C-rich telomeric sequence, had two mismatched bases that created an AatII site and destroyed the BsiEI site. The SalI and XmaI-SacI fragments were mixed in equal proportions, boiled for 5 min, and allowed to anneal by slow cooling (∼1 h) to 25°C in the presence of the SnaBI- and AatII-containing oligomers to form two linear structures (the original SalI and XmaI-SacI fragments) and two nicked circular structures (one containing a SalI-derived Watson strand and an XmaI-SacI-derived Crick strand bridged by one oligomer and the other containing a SalI-derived Crick strand and an XmaI-SacI-derived Watson strand bridged by the other oligomer). The mixture was run on 1% agarose without ethidium bromide at 6 V/cm for 4 h to separate the nicked circular forms from the linear DNA. The nicked circles were gel purified and ligated in the presence of an excess of the two oligonucleotides that bridged the gap created by the XmaI-SacI fragment to generate circle H. Thus, the circle H preparation contained two types of 1.6-kb circles with two-base-mismatched heteroduplexes each, one that had a BsiEI-SnaBI mismatch (BsiEI on the telomeric G strand and SnaBI on the C strand) and another that had a BsiEI-AatII mismatch (BsiEI on the telomeric C strand and AatII on the G strand).
FIG. 5.
Absence of strand bias in utilization of a URA3-telomere circle during formation of telomeric long tandem arrays. (A) Process by which heteroduplex circles (circle H) were generated and expected structure of a tandem array at a telomere derived by transforming a circle H mixture. Plasmid pMH3-2UraTel was digested with SalI and XmaI-SacI to generate two linear fragments, which were boiled together, annealed, and gel purified as gapped circles. The gaps were filled with oligonucleotides that contained mismatches that created SnaBI and AatII restriction sites on different strands of the circle, as shown. R, EcoRI; B, BsiEI; S, SnaBI; A, AatII. The sequence of the XmaI-SacI fragment that was absent in the gapped circles and the sequences of the oligonucleotides used to bridge the gaps are also shown. (B) Southern blots of three transformants obtained in a ter1-Δ msh2-Δ double mutant. Every transformant is represented by four digests (indicated above each lane; +, present; −, absent) and three hybridizations (indicated below the blots). A digested untransformed control (U) is also shown with each hybridization. For all four digests of each transformant, the primary digest is EcoRI, which generates telomeric fragments, and the secondary digest is either BsiEI, SnaBI, or AatII. The EcoRI-SnaBI digest, which is in the third lane of each set of digests, cleaves some of the telomeric bands to a slightly smaller size. Positions of size markers (in kilobases) are indicated at left.
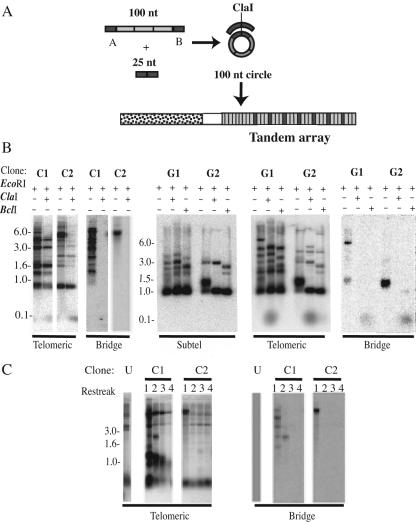
A 100-nt oligomer composed of three 25-bp telomeric repeats flanked on either side by nontelomeric sequence (A on the 5′ side and B on the 3′ side) was synthesized. This oligomer was circularized by bringing the ends together with another 25-nt oligomer composed of sequence complementary to the nontelomeric sequence of the 100-nt oligomer (B′-A′) in the presence of T4 DNA ligase (see Fig. 6A). One hundred-nucleotide circles composed of either the C- or the G-rich telomeric strand were made in the same way. The G-strand 100-nt telomeric circle transformants were derived from circles composed of telomeric repeats that had a BclI restriction site; 250 to 300 ng of the 100-nt circle, along with 0.5 to 1 μg of p1B3, an autonomously replicating sequence (ARS)-containing plasmid (35), was used for each transformation.
FIG. 6.
Sequence from a 100-nt telomeric circle can form tandem arrays at telomeres. (A) In vitro generation of a 100-nt circle and expected structure of a tandem array formed by the integration of its sequence at a telomere. The partially single-stranded circle was generated by ligating the ends of a 100-nt oligonucleotide brought together by annealing it to a bridging oligonucleotide that is complementary to a 25-nt nontelomeric region of the 100-nt oligonucleotide. The shaded boxes represent telomeric repeats, and the solid boxes represent nontelomeric sequence that forms the bridge between the telomeric repeats in the circle. The stippled box represents the region of subtelomeric DNA used as a probe. (B) Southern blots of two C-strand 100-nt circle ter1-Taq transformants (C1 and C2) and two G-strand 100-nt circle ter1-Taq transformants (G1 and G2) that have a telomeric array formed with sequence derived from the circles. The probes used are indicated beneath the blots. Each C-strand circle transformant is shown as two digests (EcoRI and EcoRI plus ClaI). EcoRI generates the telomeric fragments, and ClaI cuts once within each unit of the array, leading to the formation of a 100-nt band. The G-strand circle transformants are shown as thee digests: EcoRI, EcoRI plus ClaI, and EcoRI plus BclI. BclI cleaves telomeric repeats derived from the G-strand circle. +, present; −, absent; Subtel, subtelomeric. (C) Southern blots of serial restreaks of the same two C-strand 100-nt circle ter1-Taq transformants (C1 and C2) shown in panel B digested with XbaI. XbaI generates smaller telomeric fragments for 9 of the 12 telomeres in TER1 K. lactis, making it easier to detect the telomere(s) that has been elongated by formation of an array(s) derived from the 100-nt circle. Hybridizations to telomeric and bridge probes are shown.
RESULTS AND DISCUSSION
Formation and spread of long tandem arrays in a telomerase+ strain transformed with a URA3-telomere circle.
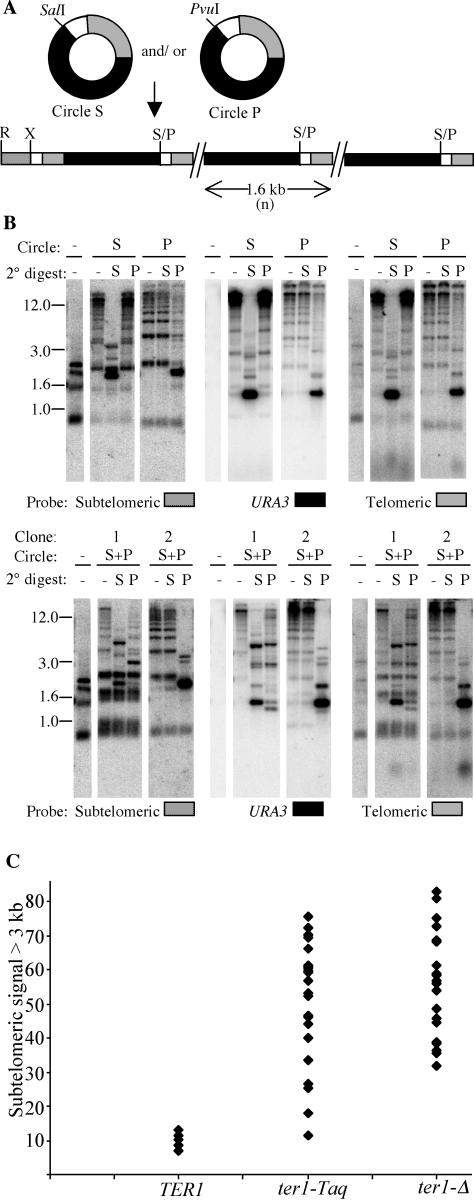
It was previously reported that single molecules of a DNA circle containing URA3 and telomeric repeats could promote the formation of tandem arrays at telomeres in both TER1 and ter1-Δ strains (25). Transformants of the TER1 strain typically had a tandem array at only a single telomere, whereas transformants of the ter1-Δ strain invariably had tandem arrays at multiple telomeres. These results suggested that while both strains could form a tandem array from a URA3-telomere circle, only the ter1-Δ strain could spread the sequence from the initial array to other telomeres. We wanted to test if the URA3-telomere DNA circle could lead to both the formation and spread of tandem arrays in a telomerase+ strain that is not prone to recombinational telomere elongation. To do this, we transformed circular DNA molecules containing telomeric repeats and URA3 into ter1-Taq, a strain which has been shown to have stably short telomeres and high rates of subtelomeric recombination but no detectable tendency to elongate telomeres via recombination (20). This mutant contains a single-base change in one of the terminal 5-bp repeats of the telomerase RNA template. Its phenotype of having fewer telomeric repeats per chromosome end appears to be due to an aberrant translocation step that both interferes with proper telomerase function and leads to telomerase synthesizing abnormally long 31-bp telomeric repeats (Underwood and McEachern, submitted). We introduced two types of 1.6-kb URA3-telomere circles (Fig. 1A) that differed at one restriction site (circle S, which had a SalI site, and circle P, which had a PvuI site) either singly or as a mixture. Transformants that grew on medium lacking uracil were analyzed by Southern blotting. The top blot of Fig. 1B shows one each of ter1-Taq transformants derived from circle S and circle P. The use of a subtelomeric probe that hybridizes to 11 of the 12 telomeric EcoRI fragments showed that some telomeres in 32 of 32 transformants examined had become greatly elongated. When cleaved with either SalI or PvuI (2° digest), these extended telomeres were largely cleaved down to units near 1.6 kb in size that hybridized intensely to URA3 and telomeric probes. These data indicated that sequence from the URA3-telomere circles integrated at telomeres in ter1-Taq cells as long tandem arrays. When ter1-Taq was transformed with a mixture of circle S and circle P, almost all of the resulting transformants had arrays that were composed entirely of sequence derived from a single type of circle: 63 out of 64 ter1-Taq transformants had arrays that could be cleaved by either SalI or PvuI, but not both (Fig. 1B, bottom, and data not shown). The single transformant that had arrays that could be cleaved by both SalI and PvuI is shown in the bottom blot of Fig. 1B (clone 1). Aside from this exceptional transformant, these data are consistent with our past results and indicate that all copies of the integrated 1.6-kb sequence present in a transformant are ultimately derived from a single circular URA3-telomere molecule.
FIG. 1.
Long tandem arrays form at multiple telomeres in ter1-Taq when transformed with circle S or circle P. (A) Structures of circle S and circle P and expected structure of a tandem array formed by circle S and/or circle P at a telomere. The solid boxes are the URA3 gene, the shaded boxes represent telomeric repeats, and the small open boxes represent subtelomeric sequence present in circles S and P. The stip-pled box represents internal subtelomeric sequence not present in the circles. Restriction sites: S, SalI; P, PvuI; R, EcoRI; X, XbaI. (B) Southern hybridizations of one circle S and one circle P transformant of ter1-Taq are shown in the top row. Southern hybridizations of two circle S plus P transformants (S+P) are shown in the bottom row. An untransformed control (−) is shown in each case for comparison. Every transformant is represented by three lanes for three types of restriction digests: EcoRI, EcoRI plus SalI, and EcoRI plus PvuI. The primary digest (−) is always EcoRI, which cuts at a subtelomeric position that can vary among telomeres. The secondary digests are either SalI (S) or PvuI (P). The type of circle (S or P) used for the transformation is represented above the blots for each transformant. Size markers in kilobases are also indicated. The same membrane was probed sequentially with three probes: XbaI-EcoRI subtelomeric fragment, URA3, and telomeric probes, as indicated at the bottom of each blot. (C) The percentage of elongated telomeres was estimated as the amount of subtelomeric signal found above 3 kb relative to the total subtelomeric signal in a lane of EcoRI-digested DNA for URA3-telomere circle transformants of TER1 (9 transformants), ter1-Taq (24 transformants), and ter1-Δ (24 transformants) strains. Eleven of the 12 telomeres hybridize to the subtelomeric probe.
Overall, the URA3-telomere circle transformants of ter1-Taq bore more resemblance to circle transformants of ter1-Δ than to those of TER1 cells. Most notably, ter1-Taq and ter1-Δ transformants had multiple telomeres that had acquired arrays. We estimated the number of telomeres that had elongated to form tandem arrays in a given transformant by quantitation of the percentage of subtelomeric hybridization signal above 3 kb in EcoRI-digested genomic DNA (Fig. 1C). Three kilobases is larger than the size of the largest EcoRI telomeric fragment detected by the subtelomeric probe in DNA from untransformed cells, and any subtelomeric signal above this size must be due to incorporation of sequence from the URA3-telomere circle. In TER1 transformants, only 1, or at most 2, of the 12 telomeres had tandem arrays, whereas in ter1-Taq and ter1-Δ transformants, typically, many of the 12 telomeres had arrays. The total number of integrated copies of the 1.6-kb sequence derived from the circle was obtained by determining the hybridization intensities of URA3 relative to a single-copy gene control, as described previously (25). Among the 17 ter1-Taq transformants that were analyzed, we estimated that the range of URA3 copy numbers varied between 16 and 145. This is similar to the 30 to 180 copies seen earlier in ter1-Δ transformants. Another similarity of ter1-Taq circle transformants to those of ter1-Δ is the presence of extrachromosomal ladders of bands that may correspond to linear and circular forms of the URA3-telomere sequence that have been excised out of the array (reference 23 and data not shown). Such extrachromosomal species were not detected in TER1 transformants (25).
Thus, we have shown that transformation with URA3-telomere circles can lead to both formation of long tandem arrays and the spreading of the arrays to multiple telomeres in ter1-Taq, a telomerase+ strain with stably short telomeres. We conclude that the efficient spread of tandem arrays to multiple telomeres requires neither senescence nor the tendency to elongate telomeres by using recombination in the absence of exogenously added telomeric circles. However, spreading does appear to require elevated gene conversion rates near telomeres.
The URA3-telomere circle frequently deletes telomeric repeats prior to formation of tandem arrays.
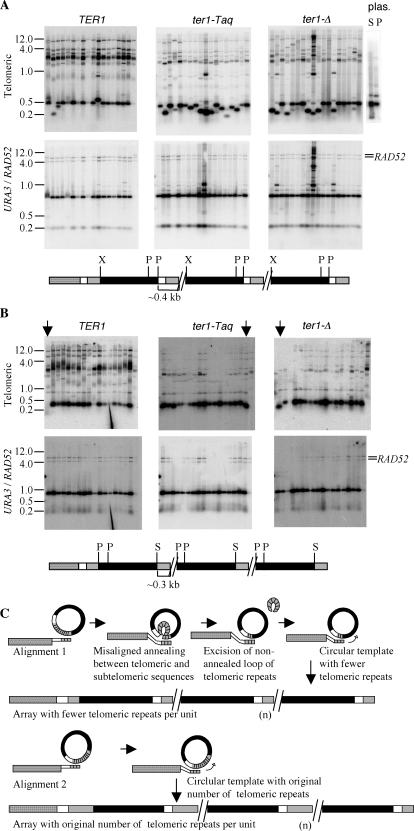
When either circle S or circle P was transformed into K. lactis cells (TER1, ter1-Taq, or ter1-Δ), the resulting tandem arrays were often seen to be composed of unit copies that were slightly smaller than 1.6 kb when digested with SalI or PvuI, respectively, and probed with URA3 (data not shown). We analyzed circle S and circle P transformants further to determine the location of the deleted sequence. To do this, DNA from randomly selected circle S and circle P transformants was digested with XhoI and PstI, which cleaved the URA3-telomere unit into three fragments: two URA3 fragments (of the expected sizes of ∼1.0 and 0.2 kb) and one telomeric fragment (of the expected size, ∼0.4 kb). The top row of Fig. 2A shows hybridization to a telomeric probe of XhoI-PstI-digested circle S and circle P transformants of TER1, ter1-Taq, and ter1-Δ strains and a control digestion of the plasmids from which circles S and P were derived. The bottom row of Fig. 2A shows hybridizations to a URA3 probe of the same filters as in the top row. The sizes of the two URA3 fragments are constant (∼1.0 and 0.2 kb) in all examined transformants from each of the three strains, indicating that there are no deletions in the URA3 regions of the arrays. In contrast, the telomeric fragment (∼0.4 kb) was observed to be variable in size between transformants, particularly in ter1-Taq and ter1-Δ strains. This showed that the deletions in the URA3-telomere units were in the region of the circle derived from the cloned K. lactis telomere. This telomeric region contained 113 bp of subtelomeric DNA in addition to the tract of telomeric repeats. The restriction enzyme BsrBI recognizes a site just 3 bp from the border of subtelomeric and telomeric sequences, and separation of the two regions by BsrBI cleavage showed that all deletions mapped within the telomeric repeats (data not shown).
FIG.2.
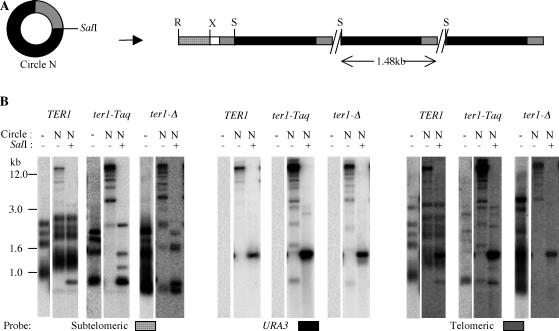
The presence of subtelomeric sequence in a URA3-telomere circle promotes deletion of telomeric repeats. (A) Southern blots of circle S transformants obtained in TER1, ter1-Taq, and ter1-Δ strains. DNAs from the transformants were digested with XhoI and PstI and probed with a telomeric probe (top) and a URA3/RAD52 probe (bottom). The RAD52 signal was used as a single-copy control. The telomeric and URA3 fragments that are formed are detected with the two probes. The XhoI-PstI fragment containing the telomeric repeats is apparent in the top panel at ∼0.2 to 0.4 kb. The plasmids (plas.) from which circles S and P were derived were also digested with XhoI-PstI-BglII to release the equivalent telomeric fragment and probed with the telomeric probe (top) to show that telomeric deletions are not present in the original transforming molecule (bright band at 0.4 kb; the slightly bigger and much fainter band is signal from vector sequences). The expected map of a telomeric array formed by circle S is shown below the Southern blots. Solid boxes, URA3 gene; shaded box, telomeric repeats; open and stippled boxes, subtelomeric sequence not present in circle S or circle P. (B) Southern blots of circle N (no subtelomeric region) transformants obtained in TER1, ter1-Taq, and ter1-Δ strains are shown. The transformants were digested with SmaI and PstI and probed with a telomeric probe (top) and a URA3/RAD52 probe (bottom). The expected map of a telomeric array formed by circle N is shown below the Southern blots. Fragments containing telomeric repeats are visible near ∼0.3 kb (top). P, PstI; S, SmaI. The three lanes that show the three clones with a deletion in the telomeric part of the array are marked with arrows. (C) Model for how subtelomeric DNA present in a transforming circle may lead to deletions within the block of telomeric repeats. The thinner boxes represent single-stranded DNA. In short telomere strains, different alignments of the subtelomeric and telomeric regions of the circle and a strand-invading chromosome end could occur (e.g., alignments 1 and 2 as shown). Alignment 1 can lead to a single-stranded loop between two regions of paired duplex DNA if base pairing extends into the subtelomeric sequence present on the circle (open box). Excision of this loop followed by ligation of the ends would then lead to deletion of some telomeric repeats from the circle (which originally has 11.5 telomeric repeats). Copying the resultant circle would lead to arrays composed of units with fewer repeats.
The presence of subtelomeric sequence in circle S and circle P might influence strand invasion by a native telomeric sequence into either circle. We therefore determined if this short subtelomeric sequence was required for the deletions to occur. This was done by first constructing a circle with no subtelomeric homology (circle N). To first confirm that circle N could promote the formation of tandem arrays, it was introduced into TER1, ter1-Taq, and ter1-Δ strains, and DNA from the transformants was examined. As shown in Fig. 3, the lack of subtelomeric sequences did not hamper the formation of tandem arrays in any strain. The arrays that formed had largely the same features as the ones transformed from circle S and circle P in TER1, ter1-Δ, and ter1-Taq strains (Fig. 1B) (25). Intense 1.6-kb bands were observed in URA3 and telomeric hybridizations when the arrays were cut with a restriction enzyme that cuts once in each unit of the array. Additionally, as judged by the relative amounts of subtelomeric signal above 3 kb in size, multiple telomeres had arrays in ter1-Taq and ter1-Δ transformants, while TER1 transformants had arrays at only one or two telomeres.
FIG. 3.
Long tandem arrays form after transformation of a URA3-telomere circle lacking subtelomeric sequences. (A) Structure of a circle with no subtelomeric sequence (circle N) and expected structure of a tandem array templated by circle N. The solid box is the URA3 gene, the shaded box represents telomeric repeats, and the open and stippled boxes represent subtelomeric sequences. X, XbaI; R, EcoRI; S, SalI. (B) Southern hybridizations of one transformant each of TER1, ter1-Taq, and ter1-Δ circle N, as well as untransformed controls (−). Every transformant is represented by two lanes for two types of restriction digests: EcoRI and EcoRI plus SalI. The primary digest is always EcoRI, which cuts at a subtelomeric position as shown in panel A. The secondary digest is SalI as indicated at the top of each lane (+, present; −, absent). Size markers are also indicated. The same membrane was probed sequentially with three probes: subtelomeric, URA3, and telomeric probes as indicated at the bottom of each blot. The subtelomeric “stump” released can be seen with the subtelomeric probe at a position below 1 kb in the TER1 transformant. Hybridization to the telomeric probe does not reveal the unelongated telomeres well, especially in the lanes representing the ter1-Δ transformant.
In circle N transformants, deletions within the ∼1.6-kb band were greatly reduced. Among seventeen clones each of TER1, ter1-Taq, and ter1-Δ, Southern analysis showed that only one clone of each had a deletion in the telomeric part of the array (SmaI-PstI digests and hybridizations are shown in Fig. 2B). These results suggest that the presence of subtelomeric sequence in circle S or P actively promotes telomeric-repeat deletions within the circle prior to or at the time of formation of a tandem array at a telomere. Although we cannot rule out the possibility that the subtelomeric sequence is a hot spot for recombination, we suggest that it acts by complicating the possibilities for strand invasion by a native K. lactis telomere. Since the telomeres are composed of perfect tandem repeats, strand invasion of a short telomere at a native chromosome end could occur in multiple different registers into a DNA circle containing multiple telomeric repeats (Fig. 2C). If the DNA circle also contained adjacent subtelomeric sequence, continued strand annealing extended into that sequence could in principle result in a single-stranded loop of telomeric repeats being extruded from the middle of the annealed DNA. If this loop was excised and the break resealed by ligation, the resulting DNA circle would contain fewer telomeric repeats. Incorporation of the sequence from the deleted circle by a rolling-circle mechanism would then result in a tandem array with all units containing the deletion.
The formation of long tandem arrays is greatly reduced in a mutant with fused telomeres.
We predicted that the absence of free telomeric ends in a ter1 mutant that had circular chromosomes due to telomere fusions might prevent URA3-telomere circles from forming tandem arrays. The ter1-AccSna mutant has two substitutions in the template region of the telomerase RNA gene that result in telomeric repeats with a disrupted Rap1 binding site. Early passages of this mutant have short telomeres, but within ∼20 restreaks (400 to 500 cell generations), most or all of the telomeres have undergone fusions, resulting in the apparent circularization of the six chromosomes (21). These fusions retain telomeric repeats that could serve as targets for homologous recombination with a URA3-telomere circle. Circle S and an ARS plasmid control were introduced into ter1-AccSna cells by transformation. By normalizing transformation frequencies using the ARS plasmid, we found that transformation of circle S into ter1-AccSna was reduced by ∼20-fold relative to a TER1 strain (Table 1). It can be concluded that the fused telomeres of the ter1-AccSna mutant cannot efficiently integrate sequence from the URA3-telomere circle.
TABLE 1.
The number of circle S transformants is greatly reduced in a strain with fused telomeresa
| Strain | No. of transformants
|
Relative circle S transformation frequency | |
|---|---|---|---|
| Circle S | ARS plasmid | ||
| TER1 | 267 | 16,000 | 1.00 |
| ter1-AccSna | 4 | 5,000 | 0.05 |
The number of circle S transformants obtained in TER1 and the fused telomere ter1-AccSna strain is shown compared to the number of ARS plasmid transformants obtained.
Southern analysis of ter1-AccSna transformants that did occur showed that 4 of 10 had patterns consistent with telomeric tandem arrays. In each of these four transformants, we found one of the six original chromosomal fusion bands to be shifted up to limit mobility in an EcoRI digest. This band was cleaved down to a band of near 1.6 kb and a second small, diffuse band (the probable telomeric end of the array) by digestion with EcoRI and SalI (data not shown). This indicated that the arrays were limited to one or two telomeric ends and probably had structures very similar to those formed in TER1 and ter1-Δ cells.
How tandem arrays can sometimes form in ter1-AccSna cells is not clear. Some free telomeric ends may be present in populations of ter1-AccSna cells. Although the telomere fusions in ter1-AccSna cells are fairly stable, they may be subject to occasional resolution to free telomeres, as is thought to occur in other ter1 template mutants that lead to fusions (21). The formation of a tandem array may therefore occur due to the occasional presence of these free telomeric ends. Alternatively, the process of integration of circle S at the telomeric repeats within a fusion may have led to DNA strand breaks that resolved the fusion and thereby permitted array formation. Even if tandem arrays initially formed at a nonfused telomere, the presence of the Ter1-AccSna telomerase would lead to dysfunctional telomeric repeats being added onto the ends, which in turn would be expected to lead to a new telomere fusion. We have not determined whether URA3-telomere arrays in ter1-AccSna transformants are present in fusions, though the presence of a small diffuse band after SalI digestion (data not shown) suggested they are not.
Testing the ability of a single 1.6-kb URA3-telomere insert to expand into a long tandem array.
A prediction of the rolling-circle replication model is that a long tandem array can arise in a single step. In principle, however, long tandem arrays could arise from multiple unequal recombination events (Fig. 4B) starting from just a single-copy insert of the URA3-telomere sequence. We therefore wanted to test if a single-copy URA3-telomere insert present at one or more telomeres could lead to the formation of tandem arrays in host cells carrying different ter1 alleles. Among the circle S TER1 transformants that we initially obtained (25), we had one transformant that had a single copy of circle S integrated at a telomere. The existence of this clone indicated that expansion of a single insert into a long tandem array was not an automatic event in a TER1 strain.
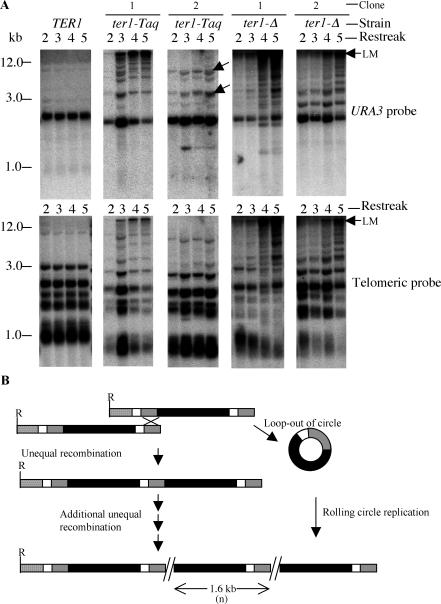
FIG. 4.
A single copy of circle S integrated at a telomere can expand to form tandem arrays in short telomere mutants but not in a wild-type strain. (A) Southern hybridizations of serial restreaks of one TER1, two ter1-Taq, and two ter1-Δ clones that initially had one or more telomeres containing a single copy of the URA3-telomere insert. The top row shows hybridization of EcoRI-digested genomic DNA to URA3, and the bottom row shows the same filters hybridized to a telomeric probe. The arrows point to bands that are dimers or trimers of the 1.6-kb insert. LM, limit mobility. (B) Models for expansion of a single-copy URA3-telomere insert into tandem arrays at many telomeres. Multiple nonreciprocal unequal recombination events combined with nonreciprocal translocations could generate and spread long tandem arrays. Alternatively, excision of a circle followed by rolling-circle gene conversion might generate arrays. The solid box represents the URA3 gene, the shaded box represents telomeric repeats, and the open and stippled boxes represent subtelomeric sequences.
To more thoroughly examine the potential ability of the single-copy URA3-telomere insert to expand into a long tandem array, we generated haploid TER1, ter1-Taq, and ter1-Δ strains (called IUT-WT, IUT-Taq, and IUT-Del, respectively), carrying one or more telomeres with the single insert and followed them over multiple streaks of growth (see Materials and Methods for details of strain construction). Results from following the IUT-WT, IUT-Taq, and IUT-Del strains over five consecutive streaks (100 to 125 cell divisions) are shown in Fig. 4A. The top row shows hybridization to a URA3 probe of two clones each of IUT-Taq and IUT-Del, as well as an IUT-WT control. The bottom row shows hybridization of the same filters to a telomeric probe. As seen previously, the IUT-WT strain maintained the fragment with the single-copy circle S insert at its original size (2.6 kb) with no sign of hybridization signal at higher positions in the gel. Moreover, the intensity of the 2.6-kb URA3 band did not change over serial restreaks, which indicated that there was no obvious duplication of the URA3 band through spreading to other telomeres. In contrast, the IUT-Taq and IUT-Del strains were able to expand the single circle S inserts into long tandem arrays (visible as URA3 and telomeric signal running at limit mobility [Fig. 4A]). In 14 out of 19 IUT-Taq clones and each of 6 IUT-Del clones that we analyzed, tandem arrays derived from URA3-telomere sequence formed by the fourth restreak. In the second and third restreaks of the strains shown, although long arrays are often detectable, most of the URA3 signal is at ∼2.6 kb, the position of the single-copy band. This indicates that the URA3-telomere signal remained at its original size in most cells in the population examined. Only at approximately the fourth streak did arrays appear to be present in the bulk of the cells of the population. In many IUT-Taq clones, the additional URA3-hybridizing bands acquired after passaging the clones were of sizes that were consistent with their being dimers or trimers of the 1.6-kb insert (Fig. 4A, ter1-Taq clone 2). This suggests that unequal crossing over, as diagrammed in Fig. 4B, may be occurring. In some IUT-Taq and most IUT-Del clones, many additional URA3-hybridizing bands migrating at high molecular weights were acquired after serial passaging. These probably represent telomeres with multiple URA3-telomere inserts and excised ladders of extrachromosomal URA3-telomere sequence. Such species have been shown to be abundant in ter1-Δ cells containing long tandem URA3-telomere arrays (25).
Our results indicate that a single URA3-telomere insert at a telomere is not capable of expanding into a long tandem array in TER1 cells. This rules out unequal crossover (Fig. 4B) as a possible mechanism for the formation of long tandem arrays by URA3-telomere circles in a strain with normal telomeres. Although long tandem arrays can form from single URA3-telomere inserts in ter1-Taq and ter1-Δ cells, the kinetics of their formation appear inadequate to account for the formation and spread of long tandem arrays in cells of the same mutant strains that have been transformed with a URA3-telomere circle. In the latter situation, formation and spreading of arrays reliably occurs within no more than ∼30 cell divisions. We conclude that single URA3-telomere inserts are unlikely to be common intermediates of long tandem array formation in URA3-telomere circle transformants of ter1-Taq and ter1-Δ cells.
Multiple unequal crossover events could be one way by which telomeres could expand a single-copy insert of circle S to tandem copies. Alternatively, the expansion at telomeres could also be triggered by circular URA3-telomere DNA molecules that loop out from the single-insert molecule (Fig. 4B). We suggest that a combination of unequal crossover and rolling-circle gene conversion might contribute to the formation of tandem arrays in strains with highly recombinogenic telomeres that initially have only single-copy URA3-telomere inserts.
Either strand of a URA3-telomere circle can be used for the formation of tandem arrays.
An important question about the long tandem array formation promoted by the URA3-telomere circle is whether there is a strand bias in the utilization of a circular template to form tandem arrays at telomeres. Both native telomeric ends and DNA double-strand breaks are processed to produce 3′ single-stranded overhangs (11, 17, 32-34). This suggests that the G-rich telomeric strand would be expected to initiate a strand invasion event and that the C-rich strand of a circular telomeric template might be used exclusively to template the formation of tandem arrays. In order to differentiate between the use of the C-rich and the G-rich telomeric strands of a URA3-telomere circle, we assembled two heteroduplex circles (circles H1 and H2, collectively referred to as circle H), as shown in Fig. 5A. The heteroduplex circles had the same sequence as circle S, except for two-base mismatches that eliminate a BsiEI site and create unique restriction enzyme sites (SnaBI on the telomeric C strand of circle H1 and AatII on the telomeric G-strand of circle H2). The preparation of a mixture of these two heteroduplex circles was introduced by transformation into ter1-Δ msh2-Δ cells and TER1 msh2-Δ cells. Strains deficient in mismatch repair were used so that the mismatches in the heteroduplex circles would not be repaired.
Heteroduplex DNA could in principle produce sectored transformants, with different sectors having sequence information derived from different strands. Therefore, care was taken to maximize the chances of detecting sectored transformants by using entire colonies from the transformation plates to inoculate cultures for DNA preparations. As before, genomic DNAs from transformants were examined by Southern blotting. Digestion with EcoRI was used to visualize telomeric fragments containing intact tandem arrays. Secondary digests that included BsiEI, SnaBI, or AatII were used to see which enzyme could cut the telomeric arrays down to units of 1.6 kb. Southern analysis of DNAs from circle H transformants showed that, as expected, the great majority produced hybridization patterns consistent with the formation of tandem arrays. Three examples of ter1-Δ msh2-Δ transformants are shown in Fig. 5B.
Table 2 shows the distribution of the restriction sites present in the arrays of the ter1-Δ msh2-Δ and TER1 msh2-Δ transformants that were analyzed. More than half of all transformants in both strains (38 of 67 ter1-Δ msh2-Δ and 25 of 48 TER1 msh2-Δ transformants) were found to have arrays containing only a single type of the three restriction sites. This indicates that utilization of sequence from only one strand of circle H was the most common means of generating the telomeric tandem arrays. Many transformants, however, had arrays that contained more than one of the three types of restriction sites (one example is shown in Fig. 5B). In some cases SnaBI and AatII sites were present together (17 of 67 ter1-Δ msh2-Δ and 3 of 48 TER1 msh2-Δ transformants). This indicated that arrays had been derived from at least two transforming molecules. In other cases, when BsiEI sites were present together with one other type of site, arrays may have been derived either from two different molecules or from two strands of the same molecule. Multiple subclones of five ter1-Δ msh2-Δ transformants of this type contained the same restriction site profiles as the parental clone. This suggests that the original transformants were not mixtures of two kinds of cells that each contained a single type of restriction site (as might have occurred from segregation of the URA3-telomere heteroduplex). Arrays in a few ter1-Δ msh2-Δ transformants were not cleaved by any of the three enzymes. These may have resulted from errors in the oligonucleotides that were used in the construction of circle H.
TABLE 2.
Restriction sites derived from individual strands of a heteroduplex circle mix in telomeric tandem arrays of transformantsa
| Restriction site(s) present in arrays | No. of transformants
|
|
|---|---|---|
| ter1-Δ msh2-Δ | TER1 msh2-Δ | |
| SnaBI only | 2 | 3 |
| AatII only | 7 | 5 |
| BsiEI only | 29 | 17 |
| BsiEI and SnaBI | 5 | 7 |
| BsiEI and AatII | 8 | 4 |
| SnaBI and AatII | 4 | 1 |
| BsiEI, SnaBI, and AatII | 9 | 2 |
| Arrays not cleaved by BsiEI, SnaBI, or AatII | 3 | 0 |
| Single-copy 1.6-kb insert at one telomere | 0 | 7b |
| Aberrant structure | 0 | 2 |
| Total no. of transformants analyzed | 67 | 48 |
The number of transformants that had tandem arrays with BsiEI, SnaBI, and/or AatII are shown. Numbers of transformants are shown for two strains.
Of these seven, four had a BsiEI site, two had a SnaBI site, and one had an AatII site.
The most striking result from our data was that SnaBI sites and AatII sites were present in arrays with similar frequencies. This was true in both ter1-Δ and TER1 strains. Combining results from both recipient strains, 35 of 115 transformants had arrays with at least some SnaBI sites and 41 of 115 had arrays with at least some AatII sites. If the C-rich telomeric strand of URA3-telomere circles had been the only strand used as the template to generate the arrays, AatII sites would not be present at all in the transformants. We conclude that array formation can arise with roughly equal frequency from copying either the C-rich or the G-rich strand of a transforming URA3-telomere circle. With that assumption, it would be expected that transformants containing arrays with BsiEI, SnaBI, and AatII sites would occur in a 2:1:1 ratio, respectively. However, transformants containing arrays with only BsiEI sites were distinctly more abundant than those containing only one of the other enzyme sites. This may have arisen from lingering discontinuities in the strands of circle H that included annealed oligonucleotides.
The relatively high percentage of transformants with arrays containing more than one type of restriction site was somewhat surprising. We suspect that this was largely or entirely due to cells that took up more than one heteroduplex molecule. Although experiments with circle S and circle P (this work and reference 25) indicated that incorporation of sequences from two molecules during array formation was rare, the circle H transformations were carried out using a greater amount of DNA. A higher concentration of transforming DNA would be expected to increase the likelihood of cells taking up more than one URA3-telomere circle.
How the 1.6-kb URA3-telomere circle led to the generation of tandem arrays at telomeres is unclear. The fact that introduction of the URA3-telomere circle into TER1 cells typically produces a tandem array at only a single telomere suggests that array formation occurs in a single step. Conceivably, an additional step might be used to integrate the array at a telomere. Array formation has been proposed to be due to some type of rolling-circle replication (25), a possibility that raises many interesting questions. One such question is, what primes the replication? Our data suggest that a mechanism exists for priming DNA synthesis on either strand of circle H. Strand invasion of a telomeric 3′ end into a URA3-telomere circle is expected to be able to prime replication of the telomeric C strand. Processing of recombination intermediates could also routinely generate means of priming replication of the telomeric G strand. Alternatively, the displaced telomeric sequence of the G strand of the URA3-telomere circle might be able to recruit a DNA polymerase. G-strand telomeric DNA provides binding sites for the telomere binding protein Cdc13, which is known to be able to bind to DNA polymerase α (26). Another question is whether the DNA synthesis that generates tandem arrays resembles break-induced replication. In break-induced replication events, a replication fork is established after invasion of a 3′ end, leading to replication that can proceed all the way to a telomere (10, 12). Tandem array formation by a rolling-circle synthesis by its nature involves displacement DNA synthesis. This would seem to preclude a typical replication fork.
One hundred-nucleotide telomeric circles of either strand can generate tandem arrays at telomeres.
Sequencing data suggested that the elongated telomeric arrays in ter1-Δ survivors containing two kinds of telomeric repeats can be composed of repeating units of 100 bp (25). We therefore wanted to test if circles of that size and composed mainly of telomeric repeats could promote the formation of tandem arrays at telomeres. We first created single-stranded 100-nt circles composed of three telomeric repeats (75 nt of C-rich strand) and 25 nt of nontelomeric sequence (including a ClaI restriction site) by annealing, in the presence of DNA ligase, the two ends of a linear 100-nt oligonucleotide with a 25-nt bridge oligonucleotide that was complementary to the nontelomeric sequence in the presence of DNA ligase (see Materials and Methods) (Fig. 6A). Formation of circles was confirmed by showing that denatured ligation reactions could be hybridized to a labeled bridge oligonucleotide in a Southern blot while identical samples that had not been ligated could not. We introduced the 100-nt circle into ter1-Taq cells by cotransforming it with p1B3, an ARS-containing plasmid that had a URA3 marker gene. The ter1-Taq strain was chosen because it had short recombinogenic telomeres but did not display the growth senescence and unstable telomere lengths of ter1-Δ.
We initially screened pools of ∼10 transformants each for the presence of an ∼100-bp ClaI fragment that hybridized to a telomeric probe. A fragment of this size is expected if the 100-nt circle integrates in the form of tandem arrays (Fig. 6A). By screening 30 such transformant pools, we found 4 that released a small fragment when cut with ClaI. We then attempted to isolate the individual transformants containing the 100-bp telomeric ClaI fragment from all four pools by restreaking individual colonies from each pool, isolating DNA from each, and testing for the presence of bands that hybridized to a probe made with the bridge oligonucleotide. Two transformants that had bands that hybridized to the bridge oligonucleotide were successfully isolated. One of these (C1) had multiple hybridizing bands, while the other (C2) had only a prominent pair of closely spaced bands (Fig. 6B, left). The DNA fragments that hybridized to the bridge oligonucleotide also appeared to hybridize to telomeric and subtelomeric probes (Fig. 6B and data not shown). As observed with the pooled transformants, cleavage of C1 and C2 with ClaI also produced telomere-hybridizing bands of 100 bp. We conclude from these data that sequence from the 100-nt circle had become incorporated as tandem arrays at one or more telomeres.
We next tested whether 100-nt circles composed of the G-rich telomeric strand could also lead to the formation of tandem arrays. As before, 100-nt “G-strand” circles were generated in vitro using a 100-nt oligonucleotide containing three telomeric repeats and a 25-nt bridge oligonucleotide. The telomeric repeats in the G-strand circle were each constructed to have a single-base-pair change that makes a BclI restriction site. The Bcl mutant repeats are functionally normal but serve as tags that can be readily identified by digestion with BclI (23). As a control, 100-nt “C-strand” circles were generated again. Each type of circle was then transformed into ter1-Taq cells along with p1B3. From each cotransformation, DNAs from 35 pools (each composed of ∼10 Ura+ transformants) were then isolated and examined by Southern blotting. Consistent with our initial results, at least six pools derived from the C-strand circle were found to exhibit EcoRI fragments that hybridized to a bridge oligonucleotide probe, as well as 100-bp ClaI fragments that hybridized to a telomeric probe. Results with the G-strand circle were similar. At least 9 of the 35 pools exhibited EcoRI fragments that hybridized to a bridge oligonucleotide, as well as 100-bp ClaI fragments that hybridized to a telomeric probe. Four clones positive for hybridization to the bridge oligonucleotide were isolated from G-strand circle transformant pools. DNAs from two of these clones (G1 and G2) are shown in Fig. 6B hybridized with subtelomeric, telomeric, and bridge probes. These clones again displayed the expected characteristics for having telomeres that had been extended by tandem copies of the transforming circular sequence. Each produced ClaI fragments of ∼100 bp that hybridized to a telomeric probe and BclI fragments of still smaller size that hybridized to a bridge oligonucleotide. The G2 clone was particularly notable. As judged from the subtelomeric hybridization, the majority of the 12 telomeres in these cells had acquired arrays derived from the G-strand circle. Judging by the sizes of restriction fragments in a gel run for a longer distance (not shown), we estimate that the extended telomeres in the G2 clone were shortened by an average of ∼430 bp from BclI cleavage and ∼380 bp from ClaI cleavage. These studies show that 100-nt circles of either strand can lead to recombinational telomere elongation through the formation of telomeric tandem arrays.
To examine the stability of the integrated telomere bridge tandem arrays, the two original C-strand circle ter1-Taq transformants were grown for several serial restreaks on YPD plates. Shown in Fig. 6C is a Southern blot of XbaI-cleaved DNA from these cells after hybridization to telomeric and bridge oligonucleotide probes. The results from this analysis showed that the integrated arrays were highly unstable. The C1 transformant lost all but one band that hybridized to the bridge oligonucleotide by the second streak and had lost all bands by the third streak. The C2 transformant completely lost its bands that hybridized to the bridge oligonucleotide by the second streak. Telomeric tandem arrays from other clones, including the G1 and G2 clones shown in Fig. 2B, seemed less unstable (data not shown). Because the ter1-Taq telomerase normally maintains telomeres at a short length, gradual loss of the elongated telomeres was to be expected. However, the reason for the high rate of instability in some clones is not clear. Conceivably, the presence of nontelomeric DNA (the bridge sequence) at multiple positions throughout an elongated telomere can destabilize it. In S. cerevisiae, it has been shown that even normal telomeric sequences can be subject to rapid large deletions (4).
The simplest model for recombinational telomere elongation using a circular DNA template would be for the 3′ single-stranded end of a telomere to strand invade a telomeric circle (annealing to the C-rich strand) and act directly as the primer for DNA synthesis around the circle. Our results with both 100-nt circles and the 1.6-kb heteroduplex circles argue strongly against this model being the only mechanism for a telomere to acquire the sequence from copying a circle. Instead, there must be efficient mechanisms for circles of either strand to serve as templates for generating elongated arrays of telomeric repeats. One possibility is that rolling-circle replication often occurs extrachromosomally. Once a telomeric array is generated, it could readily be incorporated at a chromosome end. How extrachromosomal rolling-circle replication would be primed is not clear. Conceivably, small single-stranded telomeric pieces that can anneal to the circle are generated. Another possibility for the priming of G-strand telomeric circles might be to use a mechanism related to that which primes telomeric Okazaki fragment synthesis. The yeast single-strand protein-binding complex, which includes Cdc13, is thought to help recruit DNA polymerase α to a telomeric end and to bring about synthesis of the second strand (26). It is conceivable that a single-stranded G-strand telomeric circle could recruit a DNA polymerase in the same manner.
The recombinational telomere elongation that occurs in ter1 K. lactis mutants in the absence of exogenously added DNA circles results in telomeres that have been extended by only moderate amounts, typically hundreds to low thousands of base pairs. Based on this, we had postulated that if rolling-circle replication of ∼100-nt or -bp telomeric circles was responsible for the generation of elongated telomeres in ter1 postsenescence survivors, then the extent of telomere elongation produced by 100-nt circles should be equally moderate. Our data here are consistent with that prediction. While transformation of ter1-Taq with the 1.6-kb URA3-telomere circle leads to telomeric bands that routinely extend to limit mobility in agarose gels, transformation of the same strain with 100-nt circles typically produces much less extensive telomere elongation.
Our data presented here provide further support for the roll-and-spread model (25) by showing that exogenously added 100-nt circles can promote recombinational telomere elongation. However, for a rolling-circle mechanism to account for the telomere elongation that occurs in postsenescence survivors, ter1 cells would have to be able to occasionally generate similarly small telomeric circles. Circular DNA composed of telomeric repeats has been found in some human cells (27). DNA circles as small as ∼110 bp have been found that are derived from the unusual mitochrondrial telomeres of some yeasts, including Candida salmanticensis (31). Additionally, in vitro evidence has shown that DNA circles composed of telomeric repeats can act as catalytic templates for DNA polymerases to synthesize long telomeric tracts in vitro (15). Although we have not been able to find small telomeric circles in ter1 deletion mutants (data not shown), we have observed small extrachromosomal telomeric DNA from a ter1 mutant with highly elongated telomeres (unpublished data). Some of this material hybridized to a telomeric probe from one strand but not the other and was resistant to exonuclease, suggesting that it was composed of single-stranded telomeric circles. This suggests that at least some K. lactis cells are capable of generating small telomeric circles.
Acknowledgments
We thank Sidney Kushner, Anna Karls, Marcus Fechheimer, and Claiborne Glover for critical reading of the manuscript. We also thank Neil Odum for assistance with experiments.
This work was supported by grants from the American Cancer Society (RPG GMC-99746) and from the National Institutes of Health (GM61645-01).
REFERENCES
- 1.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 2.Bryan, T. M., A. Englezou, L. Dalla-Pozza, M. A. Dunham, and R. R. Reddel. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines Nat. Med. 3:1271-1274. [DOI] [PubMed] [Google Scholar]
- 3.Bryan, T. M., A. Englezou, J. Gupta, S. Bacchetti, and R. R. Reddel. 1995. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14:4240-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucholc, M., Y. Park, and A. J. Lustig. 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cervantes, R. B., and V. Lundblad. 2002. Mechanisms of chromosome-end protection. Curr. Opin. Cell Biol. 14:351-356. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunham, M. A., A. A. Neumann, C. L. Fasching, and R. R. Reddel. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26:447-450. [DOI] [PubMed] [Google Scholar]
- 8.Enomoto, S., L. Glowczewski, and J. Berman. 2002. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2626-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ford, L. P., Y. Zou, K. Pongracz, S. M. Gryaznov, J. W. Shay, and W. E. Wright. 2001. Telomerase can inhibit the recombination-based pathway of telomere maintenance in human cells. J. Biol. Chem. 276:32198-32203. [DOI] [PubMed] [Google Scholar]
- 10.Haber, J. E. 1999. DNA recombination: the replication connection. Trends Biochem. Sci. 24:271-275. [DOI] [PubMed] [Google Scholar]
- 11.Henderson, E. R., and E. H. Blackburn. 1989. An overhanging 3′ terminus is a conserved feature of telomeres. Mol. Cell. Biol. 9:345-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes, A. M., and J. E. Haber. 1999. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96:415-424. [DOI] [PubMed] [Google Scholar]
- 13.Huang, P., F. E. Pryde, D. Lester, R. L. Maddison, R. H. Borts, I. D. Hickson, and E. J. Louis. 2001. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11:125-129. [DOI] [PubMed] [Google Scholar]
- 14.Ijpma, A. S., and C. W. Greider. 2003. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14:987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindstrom, U. M., R. A. Chandrasekaran, L. Orbai, S. A. Helquist, G. P. Miller, E. Oroudjev, H. G. Hansma, and E. T. Kool. 2002. Artificial human telomeres from DNA nanocircle templates. Proc. Natl. Acad. Sci. USA 99:15953-15958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 17.Makarov, V. L., Y. Hirose, and J. P. Langmore. 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88:657-666. [DOI] [PubMed] [Google Scholar]
- 18.McEachern, M. J., and E. H. Blackburn. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10:1822-1834. [DOI] [PubMed] [Google Scholar]
- 19.McEachern, M. J., and E. H. Blackburn. 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376:403-409. [DOI] [PubMed] [Google Scholar]
- 20.McEachern, M. J., and S. Iyer. 2001. Short telomeres in yeast are highly recombinogenic. Mol. Cell 7:695-704. [DOI] [PubMed] [Google Scholar]
- 21.McEachern, M. J., S. Iyer, T. B. Fulton, and E. H. Blackburn. 2000. Telomere fusions caused by mutating the terminal region of telomeric DNA. Proc. Natl. Acad. Sci. USA 97:11409-11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331-358. [DOI] [PubMed] [Google Scholar]
- 23.McEachern, M. J., D. H. Underwood, and E. H. Blackburn. 2002. Dynamics of telomeric DNA turnover in yeast. Genetics 160:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murnane, J. P., L. Sabatier, B. A. Marder, and W. F. Morgan. 1994. Telomere dynamics in an immortal human cell line. EMBO J. 13:4953-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natarajan, S., and M. J. McEachern. 2002. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 22:4512-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi, H., and V. A. Zakian. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 14:1777-1788. [PMC free article] [PubMed] [Google Scholar]
- 27.Regev, A., S. Cohen, E. Cohen, I. Bar-Am, and S. Lavi. 1998. Telomeric repeats on small polydisperse circular DNA (spcDNA) and genomic instability. Oncogene 17:3455-3461. [DOI] [PubMed] [Google Scholar]
- 28.Rizki, A., and V. Lundblad. 2001. Defects in mismatch repair promote telomerase-independent proliferation. Nature 411:713-716. [DOI] [PubMed] [Google Scholar]
- 29.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 30.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomaska, L., J. Nosek, A. M. Makhov, A. Pastorakova, and J. D. Griffith. 2000. Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res. 28:4479-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wellinger, R. J., K. Ethier, P. Labrecque, and V. A. Zakian. 1996. Evidence for a new step in telomere maintenance. Cell 85:423-433. [DOI] [PubMed] [Google Scholar]
- 33.Wellinger, R. J., A. J. Wolf, and V. A. Zakian. 1993. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72:51-60. [DOI] [PubMed] [Google Scholar]
- 34.White, C. I., and J. E. Haber. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wray, L. V., M. M. Witte, R. C. Dickson, and M. I. Riley. 1987. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1111-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]