Abstract
The basal ganglia, a brain structure critical for sensorimotor and motivational aspects of behavior, contain very high levels of CB1 cannabinoid receptors. These receptors are activated by endogenous lipophilic ligands, and they are thought to mediate behavioral effects of cannabinoid drugs. To evaluate the role of the endogenous cannabinoid system in the regulation of basal ganglia pathways, we have investigated the effects of targeted deletion of CB1 receptors on gene expression of various neuropeptides and transmitter-related enzymes in basal ganglia neurons. Mice without CB1 receptors are extremely hypoactive in a test for exploratory behavior (open-field test), showing markedly reduced locomotion and rearing. These CB1 mutants display significantly increased levels of substance P, dynorphin, enkephalin, and GAD 67 mRNAs in neurons of the two output pathways of the striatum that project to the substantia nigra and the globus pallidus. Our findings demonstrate that elimination of CB1 receptors results in behavioral abnormalities and functional reorganization of the basal ganglia.
Cannabinoid drugs have various cognitive and behavioral effects, including euphoria, memory impairment, ataxia, and sedation, but they also seem to be useful as therapeutic agents, for example, in the treatment of nausea, glaucoma, chronic pain, and epilepsy (1). These pharmacological effects are thought to be mediated by the CB1 cannabinoid receptor subtype (2–4). However, the normal role of the CB1 receptor and its endogenous ligands (5–8) in brain function and behavior remains largely unknown. We have used targeted elimination of the CB1 receptor (34) to investigate the function of the endogenous cannabinoid system. The basal ganglia are among the brain structures with the highest levels of CB1 receptors (9–13). In the present study, we examined effects of CB1 receptor deletion on basal ganglia circuits and spontaneous motor behavior.
In the basal ganglia, CB1 receptors are expressed predominantly in neurons of the striatum that give rise to the two output pathways, one to the substantia nigra and the other to the globus pallidus (11–13). Both pathways use γ-aminobutyric acid (GABA) as their principal neurotransmitter (14), but they differ in neuropeptide cotransmitters. Striatonigral neurons, the so-called “direct” pathway, mostly express the neuropeptides substance P and dynorphin, whereas striatopallidal neurons, the initial segment of the “indirect” pathway, generally contain enkephalin (15). To assess whether striatal projection neurons are affected by disruption of the CB1 receptor, we investigated the expression of these neuropeptides and the GABA synthetic enzyme GAD 67 with quantitative in situ hybridization histochemistry.
Dopamine neurons in the substantia nigra and ventral tegmental area provide important afferents to the dorsal and ventral striatum and, in turn, receive inputs from these striatal regions. Although dopamine neurons do not appear to express CB1 receptors (11–13), recent results showed that cannabinoid agonists increase the firing rate of dopamine neurons (16, 17). Moreover, perinatal cannabinoid treatment was reported to affect the expression of the dopamine synthetic enzyme tyrosine hydroxylase in dopamine neurons (18). We thus also examined tyrosine hydroxylase expression in the substantia nigra/ventral tegmental area.
MATERIALS AND METHODS
Generation of CB1 Receptor-Deficient Mice.
CB1 mutants were generated as described (34). Briefly, the coding region of the CB1 gene was replaced between amino acids 32 and 448 with PGK-neo in embryonic stem cells. Chimeric mice derived from these cells were bred with C57BL/6J animals. Homozygous mutants (CB1−/−) and wild-type (CB1+/+) mice were produced with heterozygous intermatings. In these experiments, 12- to 20-week-old male and female mice were used. Animals were housed in groups under standard laboratory conditions (12-hr-light/12-hr-dark cycle) with food and water available ad libitum.
Open-Field Test.
An automated open-field system was used (box size, 27 × 27 cm; ENV-510; MED Associates, St. Albans, VT). Mice (CB1+/+, n = 11; CB1−/−, n = 8) were placed individually into the central area of the open field, and behaviors were recorded and analyzed from minutes 2 to 42. Ambulatory activity, local movements, and rearing events were assessed by using the activity 3.01 software.
Rotarod Test.
The rotarod test was performed by using an accelerating rotarod (4–40 rpm; Ugo Basile, Varese, Italy). Animals (CB1+/+, n = 9; CB1−/−, n = 11) were tested in three trials with 30-min intervals. Cut-off time was 5 min.
In Situ Hybridization Histochemistry.
Mice (n = 7, each genotype) were killed with CO2. The brains were removed rapidly and processed as described (19). Coronal sections (12 μm) were hybridized with 35S-labeled oligonucleotide probes (for substance P, complementary to bases 20–67, GenBank accession no. M68909; enkephalin, bases 304–351, M13227; dynorphin, bases 807–854, AF026537; GAD 67, bases 465–512, Y12257; tyrosine hydroxylase, bases 1435–1482, M69200; CB1, bases 1081–1128, U22948). The slide-mounted sections were apposed to x-ray film (Biomax MR; Kodak) for 3–11 days.
Measurement of Gene Expression.
Hybridization signals on film autoradiograms were measured with densitometry by using the public domain nih image program (developed by Wayne Rasband at the National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image). CB1, neuropeptide, and GAD 67 expression were analyzed in the following 11 areas: nucleus accumbens core and shell, at approximately 1.5 mm rostral to bregma (20); medial, central, and lateral striatal regions, at a rostral (1.2 mm), a middle (0.1 mm), and a caudal striatal level (−0.3 mm). For the nucleus accumbens, hybridization signals were measured in sample areas that covered the medial limb of the shell or most of the core. These sample areas were placed with the help of autoradiograms showing substance P expression, which is considerably higher in the shell than in the core (see Table 1). The sample areas in the striatum were selected based on regional variations in the levels of CB1 receptor expression in wild-type (CB1+/+) mice to facilitate a correlation analysis between changes in gene expression and normal levels of CB1 receptors in striatal regions. Thus, medial sample areas were always in the dorsomedial striatum, which shows minimal CB1 receptor expression (see Table 1). Following the distribution of maximal CB1 mRNA levels in wild-type mice, the lateral sample areas were in the dorsolateral striatum at the rostral level and in the ventrolateral striatum at middle and caudal levels. Central areas were in between. Examples of striatal sample areas are shown in Fig. 2. Tyrosine hydroxylase expression was measured on seven coronal sections spaced regularly throughout the rostrocaudal extent of substantia nigra/ventral tegmental area. All mean density values presented are background-corrected (minus mean density over corpus callosum). Genotype effects were determined with one-factor ANOVA. Illustrations of film autoradiograms depicted in Figs. 2 and 4 were generated from captured images (nih image).
Table 1.
CB1 receptor, neuropeptide, and GAD 67 mRNA expression in striatum and nucleus accumbens of CB1+/+ and CB1 −/− mice
| CB1
|
Dynorphin
|
Substance P
|
Enkephalin
|
GAD 67
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| +/+ | +/+ | −/− | +/+ | −/− | +/+ | −/− | +/+ | −/− | ||
| NAC | Shell | 1.9 ± 0.6 | 61.8 ± 2.7 | 64.9 ± 2.6 | 104.8 ± 2.1 | 94.6 ± 1.0*** | 64.9 ± 2.2 | 57.3 ± 3.2 | 69.6 ± 1.2 | 71.1 ± 1.1 |
| Core | 2.7 ± 0.4 | 53.7 ± 1.4 | 54.3 ± 2.9 | 49.7 ± 3.0 | 42.7 ± 2.0 | 70.2 ± 4.4 | 63.4 ± 3.8 | 51.1 ± 1.7 | 53.2 ± 1.2 | |
| S, rostral | Medial | 7.0 ± 0.4 | 22.9 ± 1.4 | 25.7 ± 1.4 | 53.7 ± 1.1 | 55.0 ± 1.3 | 58.1 ± 1.7 | 63.6 ± 1.8* | 48.8 ± 1.2 | 54.6 ± 0.8** |
| Central | 23.1 ± 1.7 | 10.9 ± 1.0 | 18.0 ± 1.7** | 50.5 ± 1.6 | 53.8 ± 1.0 | 64.3 ± 1.9 | 65.6 ± 1.2 | 55.1 ± 1.3 | 57.6 ± 0.5 | |
| Lateral | 50.4 ± 2.3 | 12.6 ± 1.3 | 18.8 ± 1.3** | 57.3 ± 2.6 | 62.3 ± 2.0 | 69.9 ± 1.4 | 72.7 ± 1.5 | 59.8 ± 1.2 | 64.4 ± 0.7** | |
| S, middle | Medial | 12.0 ± 0.4 | 21.5 ± 1.8 | 23.8 ± 1.8 | 43.5 ± 1.3 | 48.1 ± 1.7 | 58.6 ± 1.5 | 65.6 ± 1.0** | 44.3 ± 0.9 | 50.0 ± 0.8*** |
| Central | 40.5 ± 1.4 | 4.5 ± 0.3 | 8.3 ± 0.7*** | 37.1 ± 0.8 | 44.9 ± 1.0*** | 64.4 ± 2.4 | 71.4 ± 1.3* | 48.8 ± 1.1 | 53.9 ± 0.9** | |
| Lateral | 74.1 ± 1.2 | 5.9 ± 0.3 | 13.4 ± 1.3*** | 50.7 ± 1.4 | 61.3 ± 1.9*** | 68.7 ± 2.3 | 76.4 ± 0.8** | 61.5 ± 1.2 | 71.7 ± 1.6*** | |
| S, caudal | Medial | 4.7 ± 1.1 | 29.4 ± 2.5 | 32.8 ± 1.1 | 42.9 ± 1.4 | 43.9 ± 1.6 | 54.2 ± 1.4 | 58.8 ± 2.0 | 41.6 ± 1.2 | 45.4 ± 1.6 |
| Central | 26.6 ± 3.7 | 8.0 ± 1.6 | 15.3 ± 1.2** | 38.0 ± 1.8 | 42.9 ± 1.4 | 57.8 ± 2.6 | 61.7 ± 1.3 | 41.8 ± 1.4 | 48.6 ± 1.2** | |
| Lateral | 49.2 ± 3.7 | 11.2 ± 1.7 | 18.7 ± 1.2** | 46.7 ± 1.4 | 52.9 ± 1.3** | 67.5 ± 2.0 | 74.8 ± 1.4* | 44.7 ± 0.8 | 52.4 ± 1.7** | |
Data presented are mean density values (mean ± SEM) measured in CB1+/+ (n = 7) and CB1−/− mice (n = 7). NAC, nucleus accumbens; S, striatum.
, P < 0.001;
, P < 0.01;
, P < 0.05.
Figure 2.
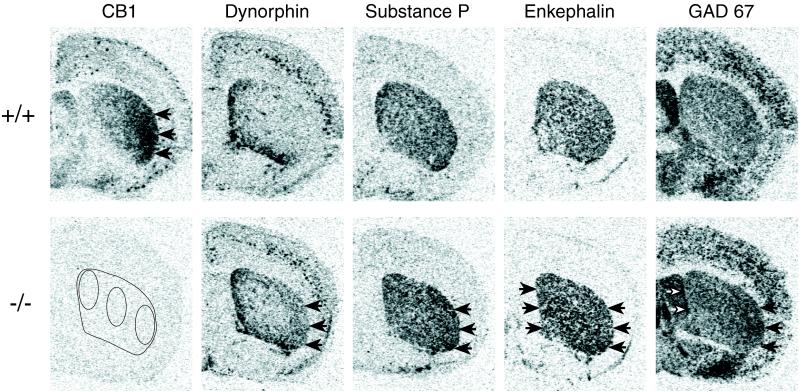
Gene expression in the striatum in CB1−/− mutants. Film autoradiograms depict CB1, dynorphin, substance P, enkephalin, and GAD 67 mRNA expression in coronal sections from the midstriatal level in CB1+/+ (Upper) and CB1−/− mice (Lower). Maximal hybridization signal is black. In wild-type (+/+) animals, CB1 receptor expression is maximal in the lateral striatum (arrows) and minimal in the medial striatum. In CB1−/− mice, increases in dynorphin and substance P mRNA expression (both in striatonigral neurons) are also maximal in the lateral striatum (arrows) and minimal in the medial striatum (for quantitative results, see Table 1). In contrast, increases in enkephalin mRNA expression (in striatopallidal neurons) are similar or even greater in some medial than lateral regions in CB1−/− mutants. GAD 67 mRNA expression (in both types of projection neurons) subsumes the changes seen for the neuropeptides, with an increase in most striatal regions that is somewhat stronger laterally. (Lower Left) The absence of CB1 mRNA in CB1−/− mutants and the sample areas in the middle striatum in which gene expression was measured are shown.
Figure 4.
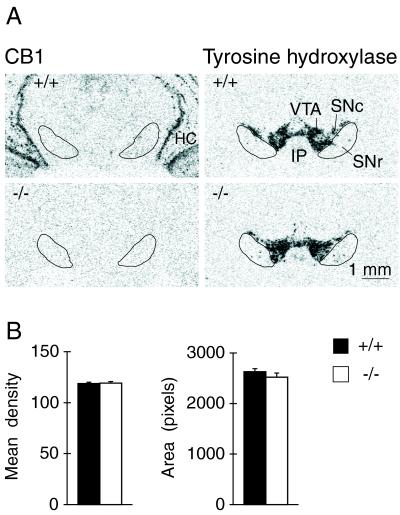
Expression of tyrosine hydroxylase mRNA in the ventral midbrain of CB1+/+ and CB1−/− animals. (A) Film autoradiograms depict CB1 (Left) and tyrosine hydroxylase expression (Right) in coronal brain sections containing the substantia nigra and ventral tegmental area for the two genotypes. Note the absence of CB1 mRNA expression in the area of tyrosine hydroxylase labeling (dopamine neurons) in wild-type (+/+) animals. The substantia nigra pars reticulata (SNr) was outlined by using Nissl-stained and GAD 67-labeled adjacent sections. HC, hippocampus; IP, interpeduncular nucleus; SNc, substantia nigra pars compacta; VTA, ventral tegmental area. (B) Mean density (mean ± SEM) (Left) and area (number of pixels with density values above background) (Right) for tyrosine hydroxylase expression in the substantia nigra/ventral tegmental area are given for CB1+/+ (n = 7) and CB1−/− mice (n = 7). Gene expression was measured in sections from seven rostrocaudal levels, and average values are presented.
RESULTS
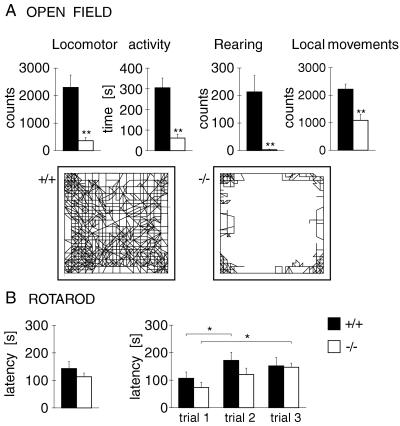
Open-field behavior of CB1 receptor-deficient mice was altered dramatically (Fig. 1). Compared with wild-type (CB1+/+) controls, CB1−/− mice displayed a large reduction in measures of locomotor activity (ambulation counts, ambulation time). Furthermore, although these mutants could rear, the rearing rate was reduced drastically (Fig. 1). In addition, CB1−/− mice were significantly less active in a ring catalepsy test (34). Despite these pronounced effects on locomotion and rearing, CB1−/− mice did not display gross deficits in motor coordination, posture, or gait. Also, local movements in the open field (e.g., grooming or shifting) were less affected (Fig. 1).
Figure 1.
Behavioral effects of CB1 receptor mutation. (A) Open-field behavior in CB1−/− mice. Animals (CB1+/+, n = 11; CB1−/−, n = 8) were tested for 40 min in an automated open field. Ambulation counts (mean ± SEM), ambulation time, vertical counts (rearing), and local counts (e.g., grooming, shifting) are presented (Upper). Illustrations of the path of ambulation during such a test are shown for a CB1+/+ (Lower Left) and a CB1−/− (Lower Right) animal. (B) Rotarod performance in CB1−/− mice. The average latency to fall (mean ± SEM) for three 5-min trials (Left) and the fall latencies for each of these three trials (Right) are shown for CB1+/+ (n = 9) and CB1−/− mice (n = 11). An accelerating-beam procedure (4–40 rpm over 5 min) was used. ∗∗, P < 0.01; ∗, P < 0.05.
To further evaluate motor coordination, we tested these mice in the rotarod apparatus, in which they have to balance on a rotating beam. We found no significant difference between the two genotypes in the latency to fall, even under relatively demanding test conditions, in which the speed of the beam was accelerated from 4 to 40 rpm over 5 min (Fig. 1). Importantly, both genotypes improved in their performance during subsequent trials (motor learning).
In the striatum, neuropeptide gene expression was increased significantly in CB1−/− mice (Table 1). However, neuropeptides in striatonigral and striatopallidal neurons were differentially affected, and changes in gene expression showed distinct regional variations. Increases in dynorphin and substance P mRNA levels (in striatonigral neurons) were maximal in the lateral striatum and minimal in the medial striatum (Table 1 and Fig. 2). In contrast, enkephalin expression (in striatopallidal neurons) was increased in lateral striatal regions and, in the rostral and middle striatum, also in medial regions (Table 1 and Fig. 2). For all neuropeptides, increases in gene expression were strongest at the midstriatal level and weaker at rostral and caudal levels (Table 1). With the exception of a decrease in substance P mRNA levels in the shell subdivision, no significant changes in neuropeptide expression were found in the nucleus accumbens (Table 1), which displays minimal expression of CB1 receptors (10–13, 21) (Table 1).
We also analyzed the expression of GAD 67 mRNA that is contained in both types of striatal projection neurons. These results confirmed the regionally distinct changes in gene regulation observed for the neuropeptides (Table 1 and Fig. 2). Thus, CB1−/− mice also showed significantly increased GAD 67 mRNA levels, and the regional patterns for these changes subsumed those seen for the neuropeptides.
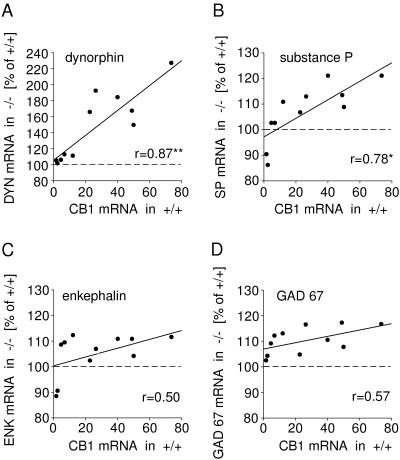
CB1 receptors display a distinct medial-to-lateral and rostral-to-caudal distribution in the rodent striatum (10–13, 21), with maximal levels in the lateral part of the middle striatum and decreasing levels toward the medial striatum and also in rostral and caudal directions (Table 1 and Fig. 2). This unique distribution allowed us to examine whether the magnitudes of changes in gene regulation were directly related to the normal levels of CB1 receptor expression, and, thus, to the degree of CB1 deficiency in CB1−/− mutants, in different regions of the striatum. Thus, we correlated the changes in gene expression in the 11 striatal regions with wild-type CB1 mRNA levels in these regions. The results of this analysis confirmed differential effects for gene regulation in striatonigral versus striatopallidal neurons (Fig. 3). Changes in dynorphin (r = 0.87, P < 0.01) and substance P mRNA expression (r = 0.78, P < 0.05) (striatonigral neurons) showed a significant positive correlation with CB1 mRNA levels in normal mice. In contrast, changes in enkephalin mRNA expression (r = 0.50) (striatopallidal neurons) and those for GAD 67 (r = 0.57) (both neuronal types) were uncorrelated with CB1 receptor expression.
Figure 3.
Relationship between increases in neuropeptide mRNA expression in the striatum of CB1−/− mutants and CB1 receptor expression in wild-type mice (CB1+/+). Dynorphin (A), substance P (B), enkephalin (C), and GAD 67 mRNA levels (D) were measured in 11 striatal regions of CB1−/− mice (n = 7) and expressed as percentage of levels in CB1+/+ mice (n = 7). These data were correlated with CB1 mRNA levels in CB1+/+ animals. Significant positive correlations were found for increases in dynorphin and substance P expression (both in striatonigral neurons), but not for enkephalin (in striatopallidal neurons) or GAD 67 expression (in both neuronal types). ∗∗, P < 0.01; ∗, P < 0.05.
Fig. 4 depicts tyrosine hydroxylase expression in the two genotypes. Gene expression was determined in sections from seven rostrocaudal levels across the substantia nigra/ventral tegmental area. In contrast to the effects on gene regulation in the striatum, we did not detect changes in tyrosine hydroxylase expression, neither for mRNA levels nor distribution, in CB1−/− mice (Fig. 4).
DISCUSSION
Projection neurons of the striatum contain CB1 receptors. To investigate the effects of CB1 receptor deletion on these neurons, we used neuropeptide markers that are expressed relatively selectively by the two subtypes of projection neurons. Our results demonstrate that the expression of neuropeptide genes in the striatum is increased in CB1 mutants. However, the two subtypes of projection neurons are differentially affected. Thus, increases in gene expression in striatonigral neurons (dynorphin, substance P) are directly related to the normal levels of CB1 receptors in different striatal regions, whereas increases in striatopallidal neurons (enkephalin) are not. These changes in gene regulation in CB1−/− mutants suggest that CB1 receptors tonically inhibit neurons of both striatal output pathways, yet in a region- and pathway-specific manner. Altered gene expression in striatal output pathways in CB1 receptor-deficient mice was accompanied by severely reduced locomotor activity and rearing behavior in an open-field test. In contrast, motor coordination (rotarod performance) hardly was impaired. Together, these findings indicate that initiation rather than coordination and maintenance of movement is affected by the CB1 disruption.
Effects of CB1 receptor stimulation include inhibition of adenylate cyclase, inhibition of voltage-dependent calcium channels, and activation of inwardly rectifying potassium channels (3, 4, 22). CB1 disruption thus may result in “disinhibition” of neurons that normally express CB1 receptors. Our findings, showing that increases in substance P and dynorphin gene expression in CB1−/− mice are positively correlated with the amount of CB1 receptors lost in different striatal regions, are consistent with a local CB1 inhibition for striatonigral neurons. However, striatopallidal neurons seem to be differentially regulated by CB1 receptors, at least in medial striatal regions. These regions contain low amounts of CB1 receptors in normal rodents (refs. 10–13, 21; Table 1), and increases in enkephalin (and GAD 67) mRNA expression were unrelated to these levels. Thus, these changes in gene regulation in striatopallidal neurons are likely the result of more complex alterations in these mutants. It is possible that they reflect network-level alterations in CB1−/− mice. For example, CB1 receptors also are found in the cerebral cortex (2, 10, 12), especially in the medial frontal cortex, which projects mostly to medial striatal regions. Input from the cortex also affects gene expression in striatal neurons (23), and a recent study indicates that cortical activation may preferentially increase gene expression in striatopallidal neurons (24). Therefore, it is conceivable that altered inputs from the cortex, or other brain areas, contribute to changes in gene expression in the striatum in CB1−/− mutants.
Behavioral effects of the CB1 deletion were investigated in a novel open field. The open-field test assesses various aspects of motivated behavior and is a sensitive assay for basal ganglia function. For example, changes in spontaneous open-field behavior have been reported for several dopamine receptor knockouts in which basal ganglia circuits are affected (25–28). CB1 mutants showed dramatically reduced ambulation and rearing during the open-field test, behaviors that reflect mostly exploratory activities in such a situation. In contrast, nonexploratory behaviors such as grooming and shifting were less reduced. These behavioral effects suggest that CB1 receptors are involved in mechanisms that regulate initiation of goal-directed behavior.
Recently, Ledent et al. (29) also generated a mouse strain with a deletion in the CB1 gene. They also found reduced rearing, but ambulatory activity was increased in these mutants. It remains to be determined whether this difference is due to factors such as experimental procedures or genetic background. However, our most recent tests, after further backcrossing (F3) into the C57BL/6J genetic background, indicate that the behavioral deficits in our mouse strain are stable, including the reduction in locomotor activity (data not shown).
In light of pharmacological results, reduced open-field behavior in CB1−/− mutants may seem paradoxical, because acute treatments with cannabinoid agonists, including the endogenous ligand, anandamide, produce behavioral inhibition such as immobility and catalepsy (30–32, 34). Interestingly, we also observed paradoxical hypoalgesia in CB1 knockout mice (34). It is possible that these “paradoxical” effects in CB1−/− mice do not reflect direct effects of CB1 receptor elimination, but rather are related to adaptive neuronal alterations in these mutant mice. For example, there is evidence that increased expression of neuropeptides in neurons of striatal output pathways represents such neuroadaptations. Previous work indicates that increased enkephalin and dynorphin functions in these striatal projection neurons are neuroadaptations that act to dampen activation of these pathways (33), an effect that likely alters the dynamics of basal ganglia circuits. Such neuronal changes can be expected to contribute to the behavioral changes seen in CB1−/− mutants.
In summary, our results show severe behavioral abnormalities and changes in gene expression in striatal output pathways in mice without CB1 cannabinoid receptors. Thus, these findings demonstrate that the endogenous cannabinoid system is critical for normal basal ganglia function and behavior.
Acknowledgments
We thank Sebastian Zimmer for help with behavioral experiments and Jennifer Hall for assistance. This work was supported in part by National Institute of Mental Health and U.S. Public Health Service Grants DA11261 (H.S.), NS20702, and NS26473 (S.T.K).
Footnotes
A Commentary on this article begins on page 5338.
References
- 1.Grinspoon L, Bakalar J B. Marihuana, the Forbidden Medicine. New Haven, CT: Yale Univ. Press; 1997. [Google Scholar]
- 2.Matsuda L A, Lolait S J, Brownstein M J, Young A C, Bonner T I. Nature (London) 1990;346:561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- 3.Abood M E, Martin B R. Int Rev Neurobiol. 1996;39:197–221. doi: 10.1016/s0074-7742(08)60667-4. [DOI] [PubMed] [Google Scholar]
- 4.Axelrod J, Felder C C. Neurochem Res. 1998;23:575–581. doi: 10.1023/a:1022418217479. [DOI] [PubMed] [Google Scholar]
- 5.Devane W A, Hanus L, Breuer A, Pertwee R G, Stevenson L A, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 6.Devane W A, Axelrod J. Proc Natl Acad Sci USA. 1994;91:6698–6701. doi: 10.1073/pnas.91.14.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stella N, Schweitzer P, Piomelli D. Nature (London) 1997;388:773–778. doi: 10.1038/42015. [DOI] [PubMed] [Google Scholar]
- 8.Mechoulam R, Fride E, Di Marzo V. Eur J Pharmacol. 1998;359:1–18. doi: 10.1016/s0014-2999(98)00649-9. [DOI] [PubMed] [Google Scholar]
- 9.Herkenham M, Lynn A B, Little M D, Johnson M R, Melvin L S, de Costa B R, Rice K C. Proc Natl Acad Sci USA. 1990;87:1932–1936. doi: 10.1073/pnas.87.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herkenham M, Lynn A B, Johnson M R, Melvin L S, de Costa B R, Rice K C. J Neurosci. 1991;11:563–583. doi: 10.1523/JNEUROSCI.11-02-00563.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herkenham M, Lynn A B, de Costa B R, Richfield E K. Brain Res. 1991;547:267–274. doi: 10.1016/0006-8993(91)90970-7. [DOI] [PubMed] [Google Scholar]
- 12.Mailleux P, Vanderhaeghen J J. Neuroscience. 1992;48:655–668. doi: 10.1016/0306-4522(92)90409-u. [DOI] [PubMed] [Google Scholar]
- 13.Matsuda L A, Bonner T I, Lolait S J. J Comp Neurol. 1993;327:535–550. doi: 10.1002/cne.903270406. [DOI] [PubMed] [Google Scholar]
- 14.Kita H, Kitai S T. Brain Res. 1988;447:346–352. doi: 10.1016/0006-8993(88)91138-9. [DOI] [PubMed] [Google Scholar]
- 15.Gerfen C R. Trends Neurosci. 1992;15:133–139. doi: 10.1016/0166-2236(92)90355-c. [DOI] [PubMed] [Google Scholar]
- 16.French E D, Dillon K, Wu X. NeuroReport. 1997;8:649–652. doi: 10.1097/00001756-199702100-00014. [DOI] [PubMed] [Google Scholar]
- 17.Gessa G L, Melis M, Muntoni A L, Diana M. Eur J Pharmacol. 1998;341:39–44. doi: 10.1016/s0014-2999(97)01442-8. [DOI] [PubMed] [Google Scholar]
- 18.Bonnin A, de Miguel R, Rodriguez-Manzaneque J C, Fernandez-Ruiz J J, Santos A, Ramos J A. Dev Brain Res. 1994;81:147–150. doi: 10.1016/0165-3806(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 19.Steiner H, Gerfen C R. J Neurosci. 1993;13:5066–5081. doi: 10.1523/JNEUROSCI.13-12-05066.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franklin K B J, Paxinos G. The Mouse Brain in Stereotaxic Coordinates. New York: Academic; 1997. [Google Scholar]
- 21.Tsou K, Brown S, Sanudo-Pena M C, Mackie K, Walker J M. Neuroscience. 1998;83:393–411. doi: 10.1016/s0306-4522(97)00436-3. [DOI] [PubMed] [Google Scholar]
- 22.Rhee M H, Vogel Z, Barg J, Bayewitch M, Levy R, Hanus L, Breuer A, Mechoulam R. J Med Chem. 1997;40:3228–3233. doi: 10.1021/jm970126f. [DOI] [PubMed] [Google Scholar]
- 23.Wang J Q, McGinty J F. In: Pharmacological Regulation of Gene Expression in the CNS. Merchant K M, editor. Boca Raton, FL: CRC; 1996. pp. 81–113. [Google Scholar]
- 24.Parthasarathy H B, Graybiel A M. J Neurosci. 1997;17:2477–2491. doi: 10.1523/JNEUROSCI.17-07-02477.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drago J, Gerfen C R, Lachowicz J E, Steiner H, Hollon T R, Love P E, Ooi G T, Grinberg A, Lee E J, Huang S P, et al. Proc Natl Acad Sci USA. 1994;91:12564–12568. doi: 10.1073/pnas.91.26.12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu M, Moratalla R, Gold L H, Hiroi N, Koob G F, Graybiel A M, Tonegawa S. Cell. 1994;79:729–742. doi: 10.1016/0092-8674(94)90557-6. [DOI] [PubMed] [Google Scholar]
- 27.Baik J-H, Picetti R, Saiardi A, Thiriet G, Dierich A, Depaulis A, Le Meur M, Borrelli E. Nature (London) 1995;377:424–428. doi: 10.1038/377424a0. [DOI] [PubMed] [Google Scholar]
- 28.Accili D, Fishburn C S, Drago J, Steiner H, Lachowicz J E, Park B-H, Gauda E B, Lee E J, Cool M H, Sibley D R, et al. Proc Natl Acad Sci USA. 1996;93:1945–1949. doi: 10.1073/pnas.93.5.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ledent C, Valverde O, Cossu G, Petitet F, Aubert J F, Beslot F, Bohme G A, Imperato A, Pedrazzini T, Roques B P, et al. Science. 1999;283:401–404. doi: 10.1126/science.283.5400.401. [DOI] [PubMed] [Google Scholar]
- 30.Crawley J N, Corwin R L, Robinson J K, Felder C C, Devane W A, Axelrod J. Pharmacol Biochem Behav. 1993;46:967–972. doi: 10.1016/0091-3057(93)90230-q. [DOI] [PubMed] [Google Scholar]
- 31.Romero J, de Miguel R, Garcia-Palomero E, Fernandez-Ruiz J J, Ramos J A. Brain Res. 1995;694:223–232. doi: 10.1016/0006-8993(95)00835-e. [DOI] [PubMed] [Google Scholar]
- 32.Fride E, Mechoulam R. Dev Brain Res. 1996;95:131–134. doi: 10.1016/0165-3806(96)00087-9. [DOI] [PubMed] [Google Scholar]
- 33.Steiner H, Gerfen C R. Exp Brain Res. 1998;123:60–76. doi: 10.1007/s002210050545. [DOI] [PubMed] [Google Scholar]
- 34.Zimmer A, Zimmer A M, Hohmann A G, Herkenham M, Bonner T I. Proc Natl Acad Sci USA. 1999;96:5780–5785. doi: 10.1073/pnas.96.10.5780. [DOI] [PMC free article] [PubMed] [Google Scholar]