Abstract
It is widely assumed that the ability of Candida albicans to switch between different morphologies is required for pathogenesis. However, most virulence studies have used mutants that are permanently locked into either the yeast or filamentous forms which are avirulent but unsuitable for discerning the role of morphogenetic conversions at the various stages of the infectious process. We have constructed a strain in which this developmental transition can be externally modulated both in vitro and in vivo. This was achieved by placing one copy of the NRG1 gene (a negative regulator of filamentation) under the control of a tetracycline-regulatable promoter. This modified strain was then tested in an animal model of hematogenously disseminated candidiasis. Mice injected with this strain under conditions permitting hyphal development succumbed to the infection, whereas all of the animals injected under conditions that inhibited this transition survived. Importantly, fungal burdens were almost identical in both sets of animals, indicating that, whereas filament formation appears to be required for the mortality resulting from a deep-seated infection, yeast cells play an important role early in the infectious process by extravasating and disseminating to the target organs. Moreover, these infecting Candida yeast cells still retained their pathogenic potential, as demonstrated by allowing this developmental transition to occur at various time points postinfection. We demonstrate here the importance of morphogenetic conversions in C. albicans pathogenesis. This engineered strain should provide a useful tool in unraveling the individual contributions of the yeast and filamentous forms at various stages of the infectious process.
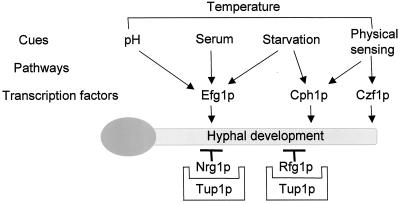
Candida albicans, a commensal organism normally found in mammals, is the most common causative agent recovered from immunocompromised patients succumbing to a fungal infection. Even with aggressive drug therapy the prognosis is not good, with mortality rates ranging from 30 to 50% (36, 37); moreover, there is a significant economic burden associated with treatment (29, 38). The organism can exist in several different forms, principally as yeast cells, pseudohyphae, or true hyphae, depending on the environmental conditions in which it is growing. Although some of the details remain to be elucidated, a much clearer picture of the pathways and mechanisms involved in these morphogenetic changes has begun to emerge over the last few years (1, 6, 7, 10, 21). There appears to be several signal transduction pathways that can be activated to begin this developmental transition with some, such as the mitogen-activated protein kinase and cyclic AMP/protein kinase A pathways, being better characterized than others. Of course, the final outcome of any such signal, irrespective of the environmental stimulus originally received, is a change in expression of a particular subset of genes, which thereby facilitates the change in the mode of growth from one morphology to another. This change in gene expression is achieved through the combined action of both positive and negative regulators. As shown in Fig. 1, key transcription factors that can be activated in response to the various stimuli include Efg1p, Cph1p, and Czf1p (8, 22, 23, 35), whereas negative regulation, which prevents expression of hypha-specific genes during growth in the yeast form, is provided by the action of Tup1p (3) and the more recently discovered Nrg1p and Rfg1p proteins (5, 16, 25). Despite an early critique warning of the pitfalls of interpretation of data obtained by using such mutants (18), the acceptance of the critical role played by this ability of C. albicans to change morphology in the disease process has come, almost exclusively, from studies with strains defective in one or more of these proteins.
FIG. 1.
Schematic representation of pathways leading to hyphal development in C. albicans. In contrast to the transcription factors Efg1p and Cph1p, whose activation is required for hyphal development, Nrg1p functions as a negative regulator, preventing the transcription of hyphal specific genes in yeast form cells. Nrg1p is a DNA-binding protein that associates with elements upstream of hyphal specific genes and forms a complex with the general repressor protein Tup1p. In contrast to TUP1, which is constitutively transcribed, NRG1 expression is downregulated after appropriate stimuli, thereby facilitating hypha-specific gene transcription (adapted from reference 7).
At first, it was thought that the hyphal form may represent the pathogenic state of C. albicans since mutants that are locked into the yeast form are avirulent (23, 35). The subsequent construction and characterization of the Δtup1 mutant, which is constitutively filamentous but also avirulent (2, 3), has led to the present widely held belief that it is the ability to switch between the yeast and hyphal forms, rather than the individual morphologies per se, that is the principal determinant in the development of the disease. However, despite this widespread acceptance, as stated in a recent review, “there are currently no molecular data which unambiguously establish a role for yeast-to-hypha morphogenesis as a virulence factor in C. albicans” (14).
The principal reasons for this is the fact that all of the mutant strains previously used to study this phenomenon are locked into either the yeast or filamentous form (and therefore require the use of a wild-type strain alongside the mutant for comparative purposes) and the means by which all of these strains were constructed. Since C. albicans has no haploid stage in its life cycle, targeted mutants are constructed by using the URA-blaster technique (11). This method involves the sequential disruption of both alleles of the gene of interest through recycling of the URA3 auxotrophic marker gene, resulting in a mutant strain containing a single copy of the URA3 gene integrated at the modified locus. Generally, this mutant strain is then compared to a wild-type strain, heterozygous at the URA3 locus, in the process under study. Unfortunately, it has recently become apparent that the chromosomal location of the URA3 gene can have profound effects on its level of transcription (20, 34), raising great doubts regarding the validity of such comparisons: this is particularly important since C. albicans Δura3 mutants are unable to survive in an animal host and therefore appear to be avirulent (17). In addition, it has recently been suggested that exposure of C. albicans to 5-fluoroorotic acid (the compound used in the counterselection step between the two allelic disruptions) can cause chromosomal rearrangements (M. Wellington and E. Rustchenko, Abstr. 6th Am. Soc. Microbiol. Conf. Candida Candidiasis, abstr. 2, 2002), casting further doubt over the genetic homogeneity and thus on any comparisons made between the two strains under study.
To overcome these concerns and more directly assess the role of the morphogenetic transition and the individual impact that the yeast and hyphal forms have at the various stages in the infectious process, we have constructed a C. albicans strain in which this important developmental response can be tightly controlled through external manipulation. This was achieved by placing one copy of the recently characterized NRG1 gene, a negative regulator of filament formation (5, 25), under the control of a tetracycline-regulatable promoter. As indicated in Fig. 1, Nrg1p functions as a transcriptional repressor of hypha-specific genes and of genes required for the switch in C. albicans. The NRG1 gene is transcribed maximally in yeast cells but is rapidly downregulated upon receipt of an appropriate stimulus to remove this repression and facilitate hyphal development (5, 24, 25). As such, we believed NRG1 represented an ideal candidate for manipulation since, we argued, placing only one copy of the gene under the control of a strong regulatable promoter should drive sufficient NRG1 expression to inhibit the yeast-to-hypha transition and thereby allow external manipulation of this developmental process. A further advantage of using this construction strategy was that it removed the necessity of exposing the cells to the potentially genotoxic effects of the 5-fluoroorotic acid counterselection agent used in recycling the URA3 marker (as mentioned above).
MATERIALS AND METHODS
Strains and media.
The yeast strains used in the present study were the wild-type strain CAF2-1 (11), the TR transactivator gene-containing strain THE1 (26) and the tet-NRG1 strains SSY50-B, SSY50-F, and SSY50-H constructed herein. All of these strains were routinely maintained and grown on yeast extract-peptone-dextrose (YPD)-rich medium, whereas selections for uracil prototrophy were performed on minimal SD plates lacking uridine (30). All plasmid manipulations were performed with the Escherichia coli strain DH5α.
Strain construction and in vitro analysis.
The strain SSY50-B was constructed as follows: first, two regions spanning positions −566 to −98 (NRGA) and positions −33 to +446 (NRGB) relative to the ATG start codon of the NRG1 open reading frame (GenBank accession no. AF321521) were PCR amplified by using the primer pairs NRGA.FOR 5′-CCAAACGGTACCAAGACATG-3′ with NRGA.REV 5′-CAGATTCTCGAGGATACTTGAAC-3′ and NRGB.FOR 5′-CCCTACTAGTTTCATTAAG-3′ with NRGB.REV 5′-GGGCCGCGGATAAGGAGGAGCAGCATACTG-3′. These amplification products were digested with KpnI/XhoI and SpeI/SacII at the sites engineered into the primers (underlined) and ligated sequentially between these sites in the proximal and distal cloning regions of the p97CAU1 plasmid (26). The entire 2.8-kb promoter-replacing construct was then liberated from the new plasmid as a KpnI-SacII fragment and transformed into the TR transactivator gene-containing C. albicans strain THE1 (26) by using a modified polyethylene glycol-lithium acetate transformation method. Genomic DNA was prepared from several of the Ura+ transformants obtained by using a commercial kit (Masterpure, Epicentre Technologies, Madison, Wis.), digested with StyI or XbaI, transferred to a nylon membrane (Nytran; Schleicher & Schuell, Keene, N.H.) and subjected to Southern blot analysis by an established method (9): this identified the strain SSY50-B (and strains SSY50-F and SSY50-H), which was used throughout the present study.
Total RNA was isolated from the wild-type CAF2-1 and modified SSY50-B strains after 5 h of growth under a variety of conditions by using a previously published bead beater protocol (28) and separated through a formaldehyde-containing agarose gel. Transfer to a nylon membrane and hybridization followed the method described above for the Southern blot. The probe used was the NRGB PCR product described in the cloning procedure outlined above.
To determine the effect of doxycycline on the C. albicans response to hypha-inducing conditions in strain SSY50-B, samples from an overnight culture grown in YPD at 25°C were diluted 1:20 (i) into YPD containing 20 μg of doxycycline/ml and incubated at either 25, 30, or 37°C or (ii) into YPD alone at 25 and 37°C or YPD plus 10% fetal calf serum at 37°C. Samples were taken from these cultures at various time points, and their morphology was evaluated microscopically.
Murine virulence assay.
Cultures of strain SSY50-B for injection were grown overnight at 25°C in YPD without doxycycline. Cells were harvested by centrifugation and washed three times in sterile pyrogen-free saline. After cells were counted with a hemocytometer, appropriate dilutions were made, and the required dosage of cells was injected in a final volume of 200 μl into the lateral tail veins of 6- to 8-week-old female BALB/c mice that had been placed on either 5% sucrose (−DOX) or 5% sucrose containing 2 mg of doxycycline/ml (+DOX); this treatment was started 3 days prior to infection. Confirmation of the number and viability of cells present in the infecting inocula was performed by plate count. We used groups of six to eight mice for each condition (+DOX or −DOX) at every dosage tested. For the delayed switch experiments, animals were injected as before while on 5% sucrose and switched onto 5% sucrose containing doxycycline (2 mg/ml) at 3, 8, or 14 days postinfection. On the days on which the mice died were recorded, moribund animals were euthanized and recorded as dying the following day. To determine the fungal burden, mice on either sucrose alone or sucrose containing doxycycline were sacrificed either 6 h after infection with 5 × 106 cells or 3 days after infection with 5 × 105 cells. In all experiments, one kidney was processed for histopathology, whereas the other kidney, the brain, and the spleen were homogenized, and fungal loads were determined by plating dilutions onto Sabouraud agar plates. All experiments were performed in accordance with institutional regulations in an Association for Assessment and Accreditation of Laboratory Animal Care facility at the University of Texas Health Sciences Center at San Antonio. Mice were allowed a one week acclimatization period before experiments were started.
Histopathology.
Kidneys excised from deceased or sacrificed mice were fixed in 10% buffered formalin and stored at 4°C until required. After kidneys were embedded in paraffin, thin tissue slices were removed and stained with Grocott-Gomori methenamine-silver (15) prior to microscopic evaluation.
Statistical analysis.
Survival data and differences between groups were analyzed by using the Kaplan-Meier and log-rank tests. Organ fungal burden was monitored by determining the total CFU per gram of organ in the kidneys, brain, and spleen. Thereafter, logarithmic values for the different groups were obtained, and results are expressed as geometric means and standard deviations. The Mann-Whitney test was used to determine statistical significance for CFU data. Analyses were performed by using InStat and Prism by GraphPad Software, Inc. (San Diego, Calif.).
RESULTS
Regulatable strain construction.
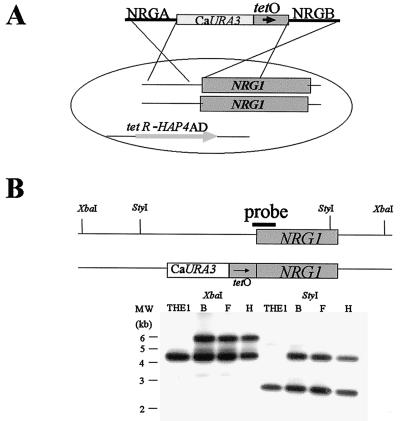
The modified strains were constructed through the integration of a DNA fragment in which a small region (position −98 to −33 relative to the ATG start codon) upstream of the C. albicans NRG1 gene had been replaced with the bacterially derived tetO sequence into the genome of the strain THE1 that contains the transactivator gene required for TR expression (Fig. 2A). This transactivator gene is composed of the bacterial tetR sequence fused to that encoding the activation domain of the highly active Saccharomyces cerevisiae Hap4p transcription factor previously codon corrected to function in C. albicans (26). This chimeric protein dimerizes to drive high expression from the tetO sequence wherever it may be introduced in this strain: addition of doxycycline to the cells leads to dissociation of the two subunits, thereby inactivating the transcription factor and effectively switching off expression from the tetO promoter (13). Correct integration of this promoter-modifying fragment at the NRG1 locus was verified for three independent transformants (designated SSY50-B, SSY50-F, and SSY50-H) via Southern blot analysis (Fig. 2B). Preliminary analysis of these three independently isolated strains determined that they were phenotypically indistinguishable; therefore, all of the subsequent experiments were performed with only the SSY50-B strain.
FIG. 2.
Construction of strain SSY50-B. (A) Diagram depicting the construction of the NRG1 promoter replacing construct and its introduction into strain THE1. This yeast strain contains the TR transactivator gene required to drive expression from the tetO promoter sequence introduced upstream of NRG1. (B) Southern blot analysis confirming integration of the promoter replacing fragment at the NRG1 locus in strains SSY50-B, -F, and -H. Genomic DNA prepared from the three Ura+ transformants and the parental THE1 was digested with StyI or XbaI and probed with the NRGB PCR product described in the text. Note that the DNA prepared from the Ura+ strains has an additional larger hybridizing fragment as a consequence of the integration of the URA3 and tetO sequences into one copy of the NRG1 gene (both alleles are the same size in the THE1 parental strain).
NRG1 gene expression is modulated by doxycycline in the modified strain.
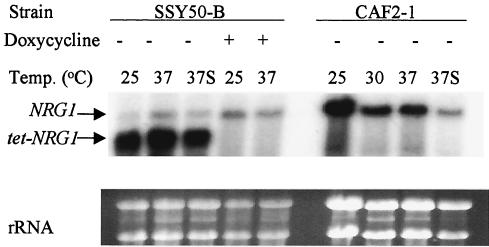
To determine whether NRG1 transcription could actually be modulated by doxycycline in these modified strains, RNA was extracted from both the modified SSY50-B and wild-type CAF2-1 strains grown under a variety of conditions and subjected to Northern blot analysis. The results obtained with the RNA prepared from the SSY50-B strain grown in YPD with or without serum in the presence or absence of doxycycline compared to those obtained with the CAF2-1 are shown (Fig. 3). Interestingly, this analysis revealed that the modified tet-NRG1 allele produced a smaller transcript than the wild-type copy (presumably as a consequence of a shorter 5′ untranslated leader sequence since the tetO sequence was integrated very close to the ATG start codon). This fortuitous size difference allowed the analysis of the effects of environmental changes on the transcription of both the native and modified NRG1 alleles in the same experiment. As predicted, transcription from the modified allele was completely dependent on the presence or absence of doxycycline, irrespective of the growth temperature or addition of serum to the medium, whereas that produced from the unaltered wild-type gene was unaffected by the antibiotic but was regulated as in the wild-type strain CAF2-1 (Fig. 3). This analysis also demonstrated that the level of NRG1 transcription produced from the modified, tetracycline-regulated allele was significantly higher than that produced from the unaltered NRG1 gene copy and provided further reassurance that modification of only one allele would be sufficient to facilitate external manipulation of the morphology of the modified strain.
FIG. 3.
Northern blot analysis displaying the effect of doxycycline on the wild-type and modified NRG1 alleles in strain SSY50-B. Total RNA was prepared from the CAF2-1 (wild-type) and SSY50-B strains after growth for 5 h under various conditions and then hybridized with an NRG1 probe. The conditions under which the cells were grown are indicated above the blot; lanes labeled 37S are those supplemented with 10% fetal calf serum. Note that the modified strain contains two hybridizing bands: one identical in size and regulated like the band seen in the CAF2-1 wild-type strain and a second, smaller band whose expression is completely dependent on the presence or absence of doxycycline in the medium. Also note the elevated expression, with respect to the message seen in the wild-type strain, of the smaller band in the absence of doxycycline.
Cellular response to serum is modulated by doxycycline in the modified strain.
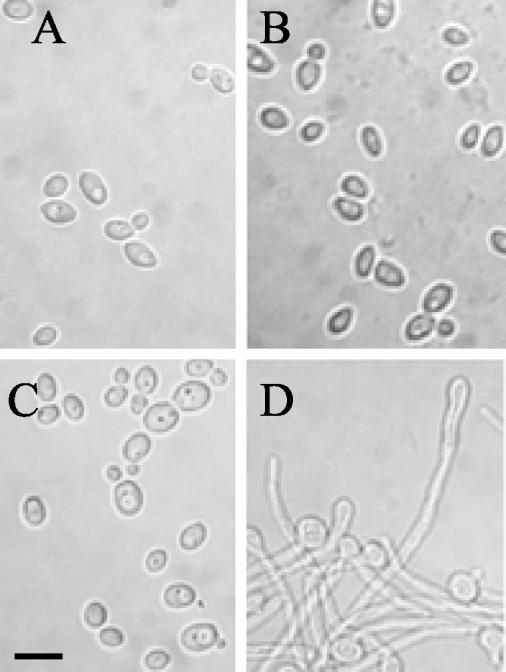
Having established that NRG1 expression from the modified allele could be tightly controlled by the presence or absence of doxycycline in the SSY50-B strain, it was now necessary to determine whether the elevated expression levels driven from the heterologous tetO promoter would be sufficient to affect the normal environmental cues leading to filament formation. This was assessed through microscopic examination of the modified strain grown under the same conditions as for the Northern blot analysis described above. This confirmed that the normal physiological response to serum at 37°C (filament formation) is completely blocked in the absence of doxycycline (i.e., when NRG1 is constitutively overproduced) in strain SSY50-B (Fig. 4). This NRG1-driven inhibition of hyphal development was not limited to YPD medium containing serum but was also observed under every condition tested (S. P. Saville and J. L. Lopez-Ribot, unpublished data). More interesting, perhaps, was the observation that the presence of the antibiotic in the medium removed the requirement of serum for this developmental response at elevated (37°C) but not lower (25 and 30°C) temperatures (compare Fig. 4C to D). The growth rate of the strain was otherwise unaffected by the presence of doxycycline at 25, 30, and 37°C in a variety of media (data not shown).
FIG. 4.
Microscopic evaluation of the effect of doxycycline on the morphogenetic transition in the SSY50-B modified strain. Samples were taken from cultures grown under various yeast- and hypha-inducing conditions for 5 h in the presence or absence of doxycycline, and their morphology was examined microscopically. (A) YPD without doxycycline at 37°C; (B) YPD plus serum without doxycycline at 37°C (cells fail to form filaments under inducing conditions); (C) YPD plus doxycycline at 25°C; (D) YPD plus doxycycline at 37°C (cells form filaments without the need for serum). Scale bar, 10 μm.
Doxycycline affects the pathogenic potential of the modified strain in vivo.
One of the principal reasons for selecting the tetracycline-regulated promoter system to construct our engineered strain was that it can be manipulated within an animal host simply by the addition or omission of doxycycline from the drinking water (26). This allowed us to directly examine the effect or role of the yeast-to-hypha transition in the widely used murine model of hematogenously disseminated candidiasis. After we determined earlier (data not shown) that the presence of doxycycline had no effect on the virulence of the wild-type parental strain CAF2-1 within the mouse host, the pathogenic potential of the modified strain was tested at various doses in matched groups of mice with or without doxycycline in their drinking water. At all infective inocula tested, mice on doxycycline succumbed to candidiasis and died at rates similar to those seen in studies with wild-type strains (Fig. 5). In contrast, every single mouse not exposed to the antibiotic survived for the duration of the experiment even at doses as high as 5 × 106 organisms per animal (Fig. 5). demonstrating the necessity for NRG1 downregulation in C. albicans virulence.
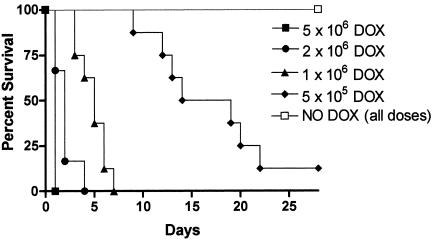
FIG. 5.
Control of NRG1 expression by doxycycline affects the outcome of haematogenously disseminated candidiasis caused by strain SSY50-B. Groups of mice either on 5% sucrose alone (NO DOX) or on sucrose containing doxycycline (DOX) were injected, as indicated, with different doses of the modified strain SSY50-B grown overnight in the absence of doxycycline, and their survival was monitored over a period of 28 days. While mice on doxycycline succumbed to the infection at rates similar to those seen with wild-type strains, every mouse not exposed to the antibiotic survived even at the highest dose tested (5 × 106 CFU). Statistically significant differences were observed between each two groups of mice injected with the same yeast inoculum in the presence or absence of doxycycline in their drinking water (P < 0.01 for all comparisons).
Fungal burdens in the infected mice are not affected by the doxycycline.
To assess whether the reduced virulence seen in animals not treated with doxycycline reflected an inability of our modified strain to leave the bloodstream and enter the tissues, we determined the fungal burden in organs recovered from mice which were either inoculated with a high dose of cells and sacrificed after 6 h or given a lower infective dose and euthanized after 3 days. Surprisingly, the organ loads were almost identical in both doxycycline-treated and untreated mice in the early hours after inoculation (Fig. 6A) and, more strikingly perhaps, were present in slightly greater numbers in animals not exposed to the antibiotic and that usually survived the infection (Fig. 6B). This demonstrated that the survival of the mice not treated with doxycyline did not reflect an inability of the modified strain to reach the target organs in sufficient numbers but some other NRG1-regulated process.
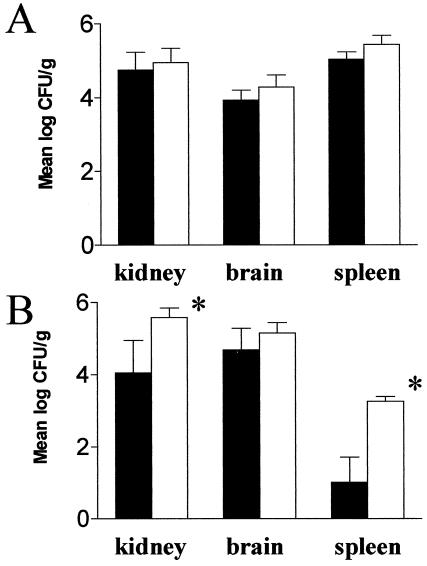
FIG. 6.
Fungal burden in the kidney, brain, and spleen of mice 6 h after infection with 5 × 106 cells (A) and 3 days after infection with 5 × 105 cells (B) of strain SSY50-B. Solid bars represent results obtained with mice on 5% sucrose containing 2 mg of doxycycline/ml; open bars represent results obtained with mice on sucrose only. Note the remarkable result that the fungal burdens are similar, irrespective of the animal's exposure to doxycycline, implying that the modified strain is able to efficiently extravasate while prevented from making the yeast-to-hypha transition. ✽, Statistically significant differences (P < 0.01).
Candida cell morphology within the kidneys is affected by doxycycline.
Since it was still possible that the modified strain had formed hyphae within the animal host through an NRG1-independent mechanism, we performed a histological analysis of kidneys recovered from both mice that had succumbed to the infection and kidneys that had been sacrificed at timed intervals. This examination revealed that the C. albicans cells in the organs of doxycycline-treated mice displayed the characteristic elongated hyphal morphology and associated tissue lesions normally seen in mice succumbing to a candidal infection (Fig. 7C and D). In stark contrast, examination of the kidneys recovered from animals not exposed to the antibiotic revealed no significant damage, with all of the invading fungal cells, whether isolated or in aggregates, present only in the yeast form (Fig. 7A and B).
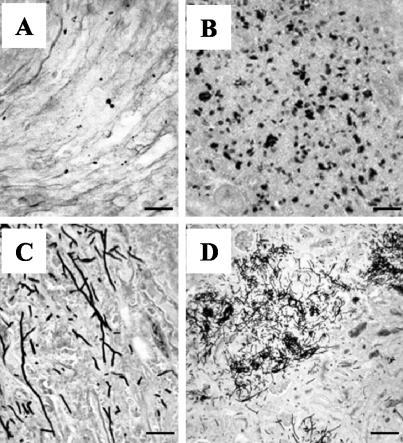
FIG. 7.
Histopathological analysis of kidneys retrieved from mice infected with strain SSY50-B in the presence or absence of doxycycline. (A and B) Scattered yeast cells (A) and yeast microabscess (B) in the kidneys of antibiotic-free mice sacrificed 3 days after infection. (C and D) Hyphal elements (C) and extensive mycelial lesions (D) in kidneys recovered from doxycycline-treated mice succumbing to infection. Scale bars: 20 μm (A, B, and C) and 80 μm (D).
Yeast form cells in the organs retain their pathogenic potential.
All of the in vivo virulence studies described above were performed with mice that were not treated with doxycycline or were placed on doxycycline 3 days prior to infection. To further demonstrate that NRG1 gene expression could be regulated in vivo and support our belief that the modified strain would be ideally suited out to study the impact of the morphogenetic transition at various stages of the infectious process, we performed a trial where mice without doxycycline in their water were switched to antibiotic treatment at various time points after injection. As before, animals not exposed to doxycycline survived for the duration of the experiment and those already on the antibiotic prior to infection rapidly died (Fig. 8): the mice that were switched to antibiotic treatment postinoculation, however, succumbed to the infection to various degrees depending on the length of time elapsed between injection and the switch. This demonstrates that the invading yeast form cells present within the organs are still capable of displaying their full pathogenic potential once NRG1 overexpression is relieved.
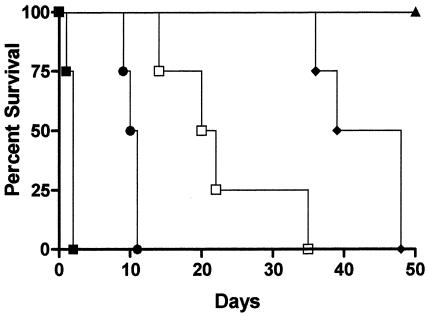
FIG. 8.
Delayed switch experiments to demonstrate that the yeast cells within the animal host are still capable of undergoing the morphogenetic transition. Groups of mice (n = 4) that were either pretreated (▪), not treated (▴), or switched to doxycycline at 3 (•), 8 (□), or 14 (♦) days after injection with 2 × 106 cells of strain SSY50-B were monitored for their ability to survive the infection. All comparisons between survival curves for each two groups resulted in statistically significant differences (P ≤ 0.01).
DISCUSSION
Over the last few years a lot of circumstantial evidence has accumulated that indicates that the yeast-to-hypha transition is required for the dissemination and virulence of C. albicans (4, 10, 21-23, 35). Practically all of the previous studies have made use of strains locked into either the yeast (Δefg1/Δefg1 and Δefg1/Δefg1 Δcph1/Δcph1) or filamentous (Δtup1/Δtup1) lifestyles, necessitating the use of a wild-type strain for comparative purposes and, more importantly, preventing an analysis of the role of the morphogenetic transition at the various stages of the infectious process. The strain we constructed and describe here, however, retains its capacity to undergo this morphogenetic transition but is either prevented or allowed to do so by the absence or presence of doxycycline in the strain's environment. Analysis of this modified strain both in vitro and in vivo reveals some novel, hitherto unseen, aspects of the morphogenetic transition in C. albicans pathogenicity and the role that NRG1 plays in them.
The first surprise was the observation that the presence of doxycycline in the medium was sufficient to drive filament formation in the absence of serum in the modified strain. This was particularly unexpected since Northern blot analysis had already shown that, as we predicted, transcription from the modified allele of NRG1 is completely dependent on the presence or absence of doxycycline and that from theunaltered gene copy responds normally to its usual environmental cues. (This ability to differentiate between the expression level derived from each of the two alleles was made possible by the fortuitous difference in the size of their respective transcripts.) While it is easy to envisage how the greatly elevated expression level driven from the modified NRG1 allele in the absence of doxycycline would be sufficient to block the normal developmental response to serum at 37°C, the observation that switching off expression from only one gene copy of NRG1 is sufficient to allow filament formation is, at first sight, more puzzling. Neither of the previous publications describing the identification and characterization of the C. albicans NRG1 gene describes a phenotypic consequence of haploinsufficiency (5, 25). It should be noted, however, that NRG1 expression was found to be still in excess of 50% of that seen in yeast form cells 90 min after the switch to inducing conditions (when filaments would certainly have started to appear) in one of those studies (5). An alternative explanation could be that the environmental cue inducing this developmental transition has two components—a “trigger” and a “propagation” signal—both of which are required since cultures of the modified strain incubated at 25 and 30°C in the presence of doxycycline (when the expression of NRG1 is clearly reduced enough to allow filament formation) grow in the yeast form, and only when the cells are exposed to an elevated temperature (the trigger) do they respond. In contrast, cells grown at 37°C in the absence of doxycycline do not form filaments even in the presence of serum because, although they have been triggered, constitutive elevated NRG1 expression blocks the propagation signal facilitating hyphal development.
The second surprising result was the observation that the fungal burdens in the organs of mice not exposed to doxycycline (and which normally survive the infection) were very similar to those with the antibiotic in their drinking water as early as 6 h postinfection. Although we appreciate that hyphal elements may have a lower plating efficiency than yeast cells and therefore may be underestimated, these striking results still suggest that the widely accepted paradigm that filament formation is required for C. albicans cells to leave the bloodstream and disseminate to the organs may not, in fact, be the case. An explanation for the discrepancy seen between our results and those presented previously is likely to come from differences between the C. albicans strains used for the respective studies. Almost all of the previous studies on the role of filament formation have made use of the Δefg1/Δefg1 single or Δefg1/Δefg1 Δcph1/Δcph1 double mutant strains. Examination of the organs of animals infected with these strains show very few infecting cells, and those that are present, while not hyphal, have an elongated morphology (23). It has been assumed that these few infecting cells had somehow managed to form filaments in vivo through an Efg1p/Cph1p-independent mechanism, possibly through activation of the transcription factor Czf1p (8, 12), which had enabled them to extravasate and disseminate to the organs, with supporting evidence for this proposal coming from in vitro studies showing that these strains are unable to penetrate and injure cultured endothelial cell lines (27). However, as outlined above, the Efg1p protein is a transcription factor that has been shown to be involved in a whole plethora of functions in C. albicans, some of which are not involved in the yeast-to-hypha transition (19, 31-33). It is therefore possible that the apparent inability of the Δefg1/Δefg1 or Δefg1/Δefg1 Δcph1/Δcph1 strains to extravasate is not a consequence of a deficiency in filament formation but rather is due to another cellular attribute important in the infectious process, such as adhesion to the blood vessel wall. This could also explain the differences between our data and the data obtained with the constitutively filamentous Δtup1/Δtup1 mutant. While the latter strain is sometimes grown in glycerol-containing medium to diminish filament formation and facilitate injection into the animal host, it is possible that the cells are expressing hypha-specific (rather than yeast form-specific) cell surface proteins, due to the absence of the Tup1p protein, and are therefore also unable to adhere to the blood vessel wall. Alternatively, the Δtup1/Δtup1 may form filaments prematurely once injected into the animal host, thereby preventing extravasation in sufficient numbers to cause death. There also remains the formal possibility that our modified strain becomes filamentous through an NRG1-independent mechanism that facilitates its ability to leave the bloodstream: this process would have to be both transient and rapid, however, since we have only ever observed yeast form cells in the organs of these infected mice. Ongoing studies with our engineered strain should further elucidate the mechanism behind this surprising observation.
Another issue to be addressed is whether it is the block of the morphological transition or some other NRG1-dependent process that leads to the survival of the mice infected with our engineered strain in the absence of doxycycline. While it is true that overproduction of Nrg1p will lead to the repression of many genes in the modified strain, including potential virulence determinants such as SAP5 and iron assimilation genes among others (24), that may be required for the full pathogenic potential of the organism to be realized, it is clear that these played no part in its dissemination to the organs. Furthermore, it could reasonably be argued that the expression of these potential virulence factor encoding genes is inherent to, and therefore a characteristic of, the filamentous lifestyle. The individual contribution that each of these genes makes to the virulence associated with a deep-seated Candida infection is not yet known and may well be cumulative. What is certain is that the repression of this subset of C. albicans genes through constitutive NRG1 overexpression completely inhibits the pathogenicity of this organism even when present in high numbers within the organs of the infected host. The fact that the infecting cells, if held in the yeast form, are eventually cleared from the organs naturally by the host (Saville and Lopez-Ribot, unpublished) may open up new strategies for the treatment of systemically derived Candida infections. Finally, the manipulability of our modified strain represents an important development that should prove invaluable in assessing how morphological changes impact at each stage of the infectious process.
In summary, the in vivo data presented here provide the most unequivocal evidence to date in support of the role of the morphogenetic transition in C. albicans virulence. Significantly, the results obtained indicate that the yeast form may, in fact, represent the disseminating form of the organism (from the bloodstream at least) and thus plays a critical role in the early stages of the C. albicans disease process.
Acknowledgments
We thank H. Nakayama and M. Arisawa (Nippon Roche) for providing the materials required for the C. albicans TR system.
The work presented here was funded in part by by Public Health Service grant R03 A1054447 (J.L.L.-R.). J.L.L.-R. is the recipient of a New Investigator Award in Molecular Pathogenic Mycology from the Burroughs Wellcome Fund.
REFERENCES
- 1.Berman, J., and P. E. Sudbery. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918-930. [DOI] [PubMed] [Google Scholar]
- 2.Braun, B. R., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor, TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 4.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20:4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, A. J., and N. A. Gow. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 7:333-338. [DOI] [PubMed] [Google Scholar]
- 7.Brown, A. J. P. 2002. Morphogenetic signaling pathways in Candida albicans, p. 95-106. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 8.Brown, D. H., Jr., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651-662. [DOI] [PubMed] [Google Scholar]
- 9.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst, J. F. 2000. Transcription factors in Candida albicans: environmental control of morphogenesis. Microbiology 146:1763-1774. [DOI] [PubMed] [Google Scholar]
- 11.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giusani, A. D., M. Vinces, and C. A. Kumamoto. 2002. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160:1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gossen, M., A. L. Bonin, and H. Bujard. 1993. Control of gene activity in higher eukaryotic cells by prokaryotic regulatory elements. Trends Biochem. Sci. 18:471-475. [DOI] [PubMed] [Google Scholar]
- 14.Gow, N. A., A. J. Brown, and F. C. Odds. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366-371. [DOI] [PubMed] [Google Scholar]
- 15.Grocott, R. G. 1955. A stain for fungi in tissue sections and smears. Using Gomori's methenamine-silver nitrate technique. Am. J. Clin. Pathol. 25:975-979. [DOI] [PubMed] [Google Scholar]
- 16.Kadosh, D., and A. D. Johnson. 2001. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21:2496-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirsch, D. R., and R. R. Whitney. 1991. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect. Immun. 59:3297-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, S. D., and J. E. Cutler. 1998. Candida albicans hyphal formation and virulence: is there a clearly defined role? Trends Microbiol. 6:92-94. [DOI] [PubMed] [Google Scholar]
- 19.Lachke, S. A., T. Srikantha, and D. R. Soll. 2003. The regulation of EFG1 in white-opaque switching in Candida albicans involves overlapping promoters. Mol. Microbiol. 48:523-536. [DOI] [PubMed] [Google Scholar]
- 20.Lay, J., L. K. Henry, J. Clifford, Y. Koltin, C. E. Bulawa, and J. M. Becker. 1998. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect. Immun. 66:5301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4:728-735. [DOI] [PubMed] [Google Scholar]
- 22.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 23.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 24.Murad, A. M., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1, and CaNrg1. Mol. Microbiol. 42:981-993. [DOI] [PubMed] [Google Scholar]
- 25.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakayama, H., T. Mio, S. Nagahashi, M. Kokado, M. Arisawa, and Y. Aoki. 2000. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect. Immun. 68:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phan, Q. T., P. H. Belanger, and S. G. Filler. 2000. Role of hyphal formation in interactions of Candida albicans with endothelial cells. Infect. Immun. 68:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramage, G., S. P. Saville, B. L. Wickes, and J. L. Lopez-Ribot. 2002. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 68:5459-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rentz, A. M., M. T. Halpern, and R. Bowden. 1998. The impact of candidemia on length of hospital stay, outcome, and overall cost of illness. Clin. Infect. Dis. 27:781-788. [DOI] [PubMed] [Google Scholar]
- 30.Sherman, F., G. R. Fink, and J. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Sohn, K., C. Urban, H. Brunner, and S. Rupp. 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47:89-102. [DOI] [PubMed] [Google Scholar]
- 32.Sonneborn, A., D. P. Bockmuhl, and J. F. Ernst. 1999. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 67:5514-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonneborn, A., B. Tebarth, and J. F. Ernst. 1999. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect. Immun. 67:4655-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staab, J. F., and P. Sundstrom. 2003. URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol. 11:69-73. [DOI] [PubMed] [Google Scholar]
- 35.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albi-cans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viudes, A., J. Peman, E. Canton, P. Ubeda, J. L. Lopez-Ribot, and M. Gobernado. 2002. Candidemia at a tertiary-care hospital: epidemiology, treatment, clinical outcome and risk factors for death. Eur. J. Clin. Microbiol. Infect. Dis. 21:767-774. [DOI] [PubMed] [Google Scholar]
- 37.Wey, S. B., M. Mori, M. A. Pfaller, R. F. Woolson, and R. P. Wenzel. 1988. Hospital-acquired candidemia: the attributable mortality and excess length of stay. Arch. Intern. Med. 148:2642-2645. [DOI] [PubMed] [Google Scholar]
- 38.Wilson, L. S., C. M. Reyes, M. Stolpman, J. Speckman, K. Allen, and J. Beney. 2002. The direct cost and incidence of systemic fungal infections. Value Health 5:26-34. [DOI] [PubMed] [Google Scholar]