Abstract
In developing lymphocytes, the recombination activating gene endonuclease cleaves DNA between V, D, or J coding and recombination signal (RS) sequences to form hairpin coding and blunt RS ends, which are fused to form coding and RS joins. Nonhomologous end joining (NHEJ) factors repair DNA double strand breaks including those induced during VDJ recombination. Human radiosensitive severe combined immunodeficiency results from lack of Artemis function, an NHEJ factor with in vitro endonuclease/exonuclease activities. We inactivated Artemis in murine embryonic stem (ES) cells by targeted mutation. Artemis deficiency results in impaired VDJ coding, but not RS, end joining. In addition, Artemis-deficient ES cells are sensitive to a radiomimetic drug, but less sensitive to ionizing radiation. VDJ coding joins from Artemis-deficient ES cells, which surprisingly are distinct from the highly deleted joins consistently obtained from DNA-dependent protein kinase catalytic subunit–deficient ES cells, frequently lack deletions and often display large junctional palindromes, consistent with a hairpin coding end opening defect. Strikingly, Artemis-deficient ES cells have increased chromosomal instability including telomeric fusions. Thus, Artemis appears to be required for a subset of NHEJ reactions that require end processing. Moreover, Artemis functions as a genomic caretaker, most notably in prevention of translocations and telomeric fusions. As Artemis deficiency is compatible with human life, Artemis may also suppress genomic instability in humans.
Keywords: Artemis, DNA repair, genomic instablility, telomere fusions, VDJ recombination
Introduction
DNA double strand breaks (DSBs)*are generated by damaging agents, such as ionizing radiation (IR), or as intermediates in normal DNA metabolism. If unrepaired or improperly repaired, DSBs can lead to cell death or chromosomal damage, including translocations (1). In mammalian cells, DSBs are predominantly repaired by one of two pathways (for review see reference 2), homologous recombination, or nonhomologous end joining (NHEJ). In addition, NHEJ is also used to repair DSBs that arise during early lymphocyte development in the context of VDJ recombination (for review see reference 3). The NHEJ pathway contains six known members including Ku70, Ku80, XRCC4, DNA ligase 4 (Lig4), DNA-dependent protein kinase catalytic subunit (DNA-PKcs), and Artemis (for review see reference 3). Previous gene targeted mutation studies have examined all NHEJ factors except Artemis. In addition to other defects (see below), NHEJ deficiency can lead to increased genomic instability (4–11). Moreover, NHEJ deficiencies, particularly in the context of cell cycle checkpoint deficiencies, cause increased tumorigenesis (1, 8, 10, 12–19). Ku70, Ku80, and DNA-PKcs are also required for the protection and maintenance of mammalian chromosome ends (for review see reference 20). Ku70, Ku80, and DNA-PKcs deficiencies result in increased levels of telomeric fusions (5, 6, 21–24), a phenotype indicative of telomere deprotection or defective telomere “capping.” In addition, Ku70 and Ku80 may function in regulating proper telomere length (7, 22–24). However, their precise roles in this process are not yet clear.
VDJ recombination is the process by which germline V, D, and J segments are assembled into complete Ig and TCR variable region genes (3, 25). RAG-1 and RAG-2 (RAG) initiate VDJ recombination by introducing DSBs between a pair of participating V, D, or J coding segments and flanking recombination signal (RS) sequences. RAG-mediated cleavage generates two types of end structures: blunt, 5′ phosphorylated RS ends and covalently closed hairpin coding ends. RAG-generated RS ends are precisely joined with no loss of nucleotides. In contrast, hairpin coding ends must be processed before ligation, with opening normally occurring at or within several nucleotides of the apex (25). Hairpin opening at points other than the apex results in overhanging flaps, which if incorporated into the join form nongermline-encoded palindromic (P) nucleotide additions that normally range from 1–2 bp in length (for review see references 25 and 26). In addition, “N-region” nucleotides might be added to joins by terminal deoxynucleotidyl transferase. Finally, a small number of nucleotides can be deleted from coding sequences, which along with P nucleotides and N-regions are a major source of somatic diversification of the Ig and TCR repertoire. NHEJ is required to seal both RS and coding ends (for review see reference 3).
Murine cells deficient in Ku70, Ku80, XRCC4, and Lig4, the four evolutionarily conserved NHEJ factors, exhibit premature senescence, sensitivity to agents that induce DSBs, and severely impaired VDJ recombination with respect to both coding and RS join formation (for review see reference 3). Inactivation of any of these four genes in mice leads to multiple defects including growth deficiency, SCID, and increased neuronal apoptosis. The precise functions of the various NHEJ components are not fully characterized. However, Ku70 and Ku80 are DNA end binding proteins, which upon binding to DSBs, can interact with and activate DNA-PKcs to generate the DNA-PK holoenzyme (for review see reference 2). DNA-PKcs deficiency also results in a SCID phenotype due to impaired VDJ recombination. However, DNA-PKcs–deficient cells do not exhibit growth defects, have variable IR sensitivity, and have severely impaired coding joining but relatively normal RS joining (27–29). The difference in severity of the Ku, XRCC4, and Lig4 deficiencies versus DNA-PKcs deficiencies suggests that the conserved NHEJ factors have functions independent of DNA-PKcs.
A precise role for DNA-PKcs has been difficult to elucidate as its in vivo kinase substrates are not known. However, DNA-PKcs has been proposed to function in end processing, potentially including hairpin opening as suggested by accumulation of covalently sealed hairpin ends in DNA-PKcs–deficient thymocytes (30, 31). Lack of such a function could indirectly lead to unusually large P nucleotide additions (30). Such elements are found in coding joins from DNA-PKcs–deficient cells (32–38). In this regard, several findings have suggested that putative DNA-PKcs hairpin opening functions may involve Artemis. Artemis was discovered as the mutated gene in human radiosensitive T− B− SCID (RS-SCID; reference 39), a disease characterized by lack of B and T lymphocytes and increased radiosensitivity of bone marrow cells and skin fibroblasts (40–42). VDJ recombination substrate assays of RS-SCID fibroblasts demonstrated that Artemis, like DNA-PKcs, is required primarily for coding versus RS joining in human cells (39, 42). Moreover, DNA-PKcs forms a complex with and phosphorylates Artemis in vitro, leading to the activation of an endonuclease activity that can cleave RAG-generated hairpins (43). The latter findings led to the suggestion that Artemis–DNA-PKcs complex may serve an end processing function required for a subset of NHEJ events. In this study, we have generated Artemis-deficient murine embryonic stem (ES) cells to further examine the role of Artemis in VDJ recombination, DNA repair, and genomic stability.
Materials and Methods
Generation of Artemis-deficient ES Cells.
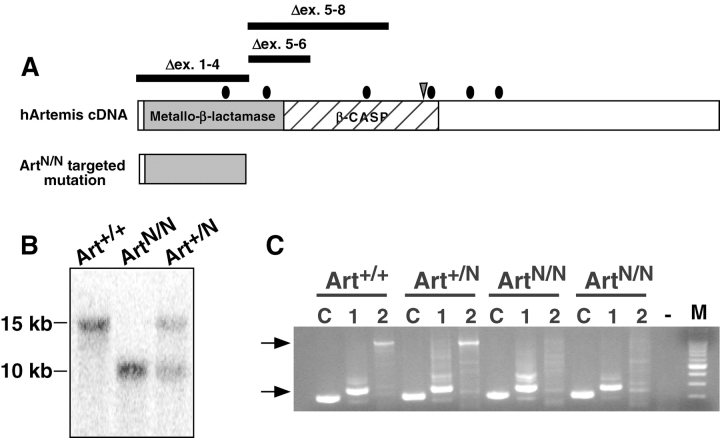
Two independent clones containing a single targeted Artemis allele (44) were selected in increasing concentrations of G418 as previously described (45). Clones were screened with a 3′ probe and double knockout cells were identified by their loss of the 15.1-kb germline band and retention of the 10-kb targeted band.
RT-PCR Analysis.
Total RNA was isolated from TC1, Art+/ N, and ArtN/N ES cells that had been passaged at least twice in the absence of feeder cells using Trizol (GIBCO BRL) and cDNA was made using Superscript II reverse transcriptase. The cDNA was then subjected to PCR as previously described (43).
DNA Damage Repair in Artemis-deficient ES cells.
ES cells of indicated genotypes were passaged at least twice in the absence of feeder cells. For IR sensitivity, cells were exposed to indicated doses of γ rays from a 137Cs source, plated at various dilutions, and then cultured for 7 d. For bleomycin sensitivity, ES cells were plated at various dilutions 16–24 h before exposure to indicated doses of bleomycin for 72 h. Cells were then allowed to recover in drug-free ES cell media for 4 d. For mitomycin C (MMC) sensitivity, ES cells were plated at various dilutions 16–24 h before exposure to indicated doses of MMC for 1 h, and allowed to recover in drug-free ES cell media for 7 d. Surviving colonies were stained with crystal violet and counted. Cells were plated in duplicate at three different dilutions and numbers of surviving colonies were averaged for each point. Percent survival was calculated as the number of treated colonies over untreated control. Data presented represents the average of three independent experiments.
Transient VDJ Recombination Assays.
Assays were performed as previously described (46). In brief, 8 × 106 ES cells were plated onto 10-cm gelatinized dishes and allowed to attach for 6–8 h. Full-length RAG-1 and RAG-2 expression constructs (36) and the pJH290 coding joining or pJH200 RS joining substrate plasmids (47) were transfected into the cells using the calcium phosphate method. The cells were harvested 48 h after transfection and the plasmids were isolated by alkaline lysis and then electroporated into Escherichia coli MC1061. VDJ recombination efficiencies were determined by calculating the ratio of 250 μg/ml amphenicol- and 20 μg/ml chloramphenicol-resistant colonies to amphenicol-resistant colonies. The fidelity of the RS joins was determined by PCR amplification of the recombining segment of pJH200 and subsequent digestion of the products with ApaLI. Coding join sequences were determined by isolating individual recombined pJH290 clones from the double selection plates and nucleotide sequencing.
Genomic Instability Assays.
ES cells were passaged at least two times in the absence of feeder cells. TC1, XRCC4−/−, and ArtN/N cells (3 × 106 cells) were plated on 10-cm gelatinized dishes and cultured for 24 h. The cells were exposed to 150 and 300 rads of γ radiation from a 137Cs source and then allowed to recover for 24 or 48 h as indicated. Unirradiated ES cells were cultured as described above without exposure to IR. Colcemid (KaryoMAX Colcemid solution; GIBCO BRL) was added to the cultures for 2 h and then the cells were harvested and fixed. Chromosomal aberrations were visualized either by staining with DAPI or by chromosome painting using spectral karyotyping (SKY)™ paints and analyzed using a Nikon Eclipse microscope equipped with an Applied Spectral Imaging interferometer and 40× and 63× objectives.
Fluorescence in Situ Hybridization.
Fluorescence in situ hybridization analysis of telomere sequences were performed as previously described (48) using a Cy3-labeled (CCCTAA)3 peptide nucleic acid probe (Applied Biosystems). Chromosomal fusions were analyzed using a Nikon Eclipse microscope.
Online Supplemental Material.
Fig. S1 shows sequences of coding joins recovered from ArtN/N and DNA-PKcsN/N ES cells complemented with human Artemis cDNA and murine DNA-PKcs cDNA expression constructs, respectively. Fig. S2 shows a representative ArtN/N metaphase containing a short-arm telomeric fusion. The online supplemental figures are available at http://www.jem.org/cgi/content/full/jem.20021891/DC1.
Results
Generation of Artemis-deficient ES Cell Lines.
Several different human Artemis mutations have been identified (39, 49) and all appeared to result in a null phenotype (Fig. 1 A). We designed a targeting strategy to mimic one such mutation in murine ES cells by replacing exons 5 and 6 of the Artemis gene with a neomycin resistance (NeoR) gene (Fig. 1 A; reference 44). Successfully targeted TC1 ES cell clones were subjected to selection in increased concentrations of G418 to obtain homozygous Artemis knockout ES cell lines (ArtN/N; reference 50). Clones were judged to be homozygous for the targeted mutation based on the loss of a 15-kb germline band and appearance of a 10-kb band by Southern blot analysis (Fig. 1 B). Two of the ArtN/N ES cell clones were subcloned and used in subsequent analyses. No obvious growth differences were observed between ArtN/N and TC1 ES cells (not depicted). RT-PCR analyses confirmed that the ArtN/N cells fail to make a transcript containing exons 1–12 (Fig. 1 C).
Figure 1.
Gene targeted mutation of Artemis. (A) Schematic representation of the targeting strategy. The human Artemis cDNA and the predicted truncated transcript from the ArtN/N targeted mutation are depicted. Conserved metallo-β lactamase and β CASP (reference 78) regions are denoted and mutations identified in human RS-SCID patients (reference 39) are indicated. •, point mutations; ▿, single base deletion; bars, genomic deletions of exons. (B) Representative Southern blot analysis of ES cells. Genomic DNA from the indicated cell lines was digested with BamHI and hybridized with the 3′ probe. The germline BamHI band is 15 kb and the targeted band is 10 kb. (C) RT-PCR analysis of transcripts. RT-PCR was performed on total RNA from ES cells using control ATM primers (C lanes) and Artemis-specific primers to detect transcripts containing exons 1–4 (1 lanes) or exons 1–12 (2 lanes).
Artemis- and DNA-PKcs–deficient ES Cells Exhibit Similar Sensitivities to DNA Damage Agents.
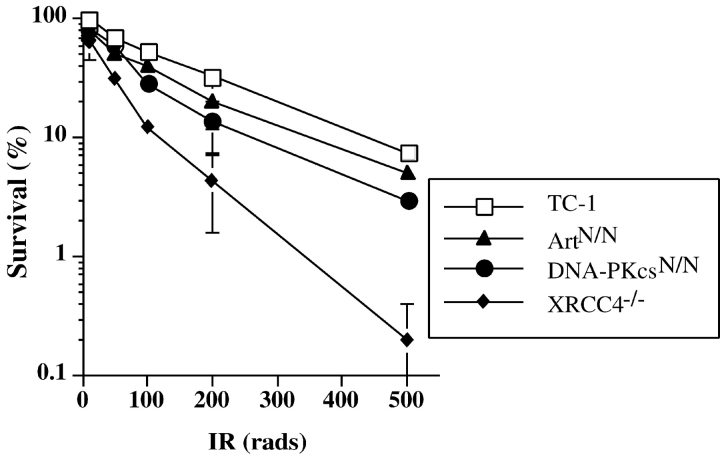
To determine the impact of Artemis deficiency on IR sensitivity, we compared the radiosensitivity of wild-type (TC1), ArtN/N, DNA-PKcsN/N, and XRCC4−/− ES cells using a colony survival assay (46, 51). At most, Artemis-deficient ES cells exhibited mildly increased IR sensitivity similar to that observed for DNA-PKcs–deficient ES cells whereas, as expected, XRCC4−/− ES cells were clearly hypersensitive (Fig. 2 and Table I). To assess sensitivity to the radiomimetic drug bleomycin, which induces DNA DSBs, ES cells were exposed to bleomycin for a period of 72 h and then allowed to recover for 4 d. In comparison to TC1 cells, XRCC4-, Artemis-, and DNA-PKcs–deficient ES cells all exhibited significantly increased bleomycin sensitivity (Table I). However, ArtN/N and DNA-PKcsN/N ES cells appeared somewhat less sensitive than XRCC4−/− cells (Table I). Together, our results suggest that Artemis, like DNA-PKcs, has a more restricted role than other NHEJ factors in repairing DSBs in ES cells (Fig. 2 and Table I; references 27, 46, and 51). In support of this notion, preliminary studies suggest that ArtN/N ES cells, as opposed to XRCC4−/− ES cells, did not exhibit significantly increased sensitivity when exposed to bleomycin for a short period of time (2 h; unpublished data).
Figure 2.
IR sensitivity of Artemis-deficient ES cells. Percent survival of TC1, ArtN/N, DNA-PKcsN/N, and XRCC4−/− ES cells is plotted as a function of IR dose (rads). The data represents the average of three independent experiments.
Table I.
Sensitivities of Artemis-deficient ES Cells to DNA Damaging Agents
| Cell line | IR 10–500 radsa |
Bleomycin 0.01–1 μg/ml |
MMC 0.02–1.2 μg/ml |
|---|---|---|---|
| TC1 (wild-type) | 140 ± 50b | 0.5 ± 0.03 | 0.5 ± 0.1 |
| ArtN/N | 70 ± 30 | 0.1 ± 0.06 | 0.6 ± 0.2 |
| DNA-PKcsN/N | 72 ± 13 | 0.06 ± 0.009 | 0.4 ± 0.2 |
| XRCC4−/− | 22 ± 10 | 0.03 ± 0.001 | ND |
| ERCC1−/− | ND | ND | 0.1 ± 0.07 |
All values are the mean IC50 ± SD of three independent experiments.
Range of DNA damaging agent used.
Concentration/level of DNA damaging agent that inhibited cell survival by 50% (IC50).
Artemis shares sequence similarity to members of the metallo-β lactamase superfamily, some of which are required for repair of interstrand cross-links (ICL; reference 39). Therefore, we measured sensitivity of ArtN/N ES cells to the ICL-inducing agent, MMC. We observed that neither Artemis- nor DNA-PKcs–deficient ES cells exhibited increased sensitivity to MMC (Table I). However, as previously reported, control ES cells deficient in the base excision repair factor, ERCC1, were markedly sensitive (Table I; references 52 and 53). Additional experiments failed to reveal increased sensitivity of ArtN/N ES cells to ICL agents cisplatin and melphalan (not depicted). Thus, we conclude that Artemis is not required for the resolution of ICL in ES cells.
Artemis Is Required for Efficient VDJ Coding, but Not RS, Joining in ES Cells.
To examine the impact of Artemis deficiency on coding versus RS joining in murine ES cells, we used a transient VDJ recombination assay (46, 47). Wild-type (TC1) and ArtN/N ES cells were transfected with full-length RAG-1 and RAG-2 expression constructs and plasmid recombination substrates designed to measure either coding or RS joining frequencies. Ku70−/−, XRCC4−/−, and DNA-PKcsN/N ES cells were assayed as controls. VDJ recombination on the substrate plasmids activates expression of the chloramphenicol acetyltransferase gene by deleting a transcriptional terminator that lies between the two coding/RS sequences. Relative recombination levels are determined by transforming plasmids recovered from the ES cells into bacteria and then assaying the ratio of ampicillin-resistant colonies (total plasmids recovered) versus ampicillin plus chloramphenicol-resistant colonies (recombined plasmids).
We observed that coding joining is substantially impaired in Artemis-deficient cells compared with wild-type TC1 ES cells (Table II). Defective coding joining in the Artemis-deficient ES cells appeared impaired to a roughly similar extent to that of DNA-PKcs– or XRCC4-deficient ES cells (Table II). In contrast to coding joins, levels of RS joins in Artemis-deficient ES cells were comparable to those observed in wild-type cells (Table II). Precise RS joins create the recognition site for the restriction endonuclease ApaLI. The fidelity of RS joins recovered from Artemis-deficient cells was assayed by ApaLI digestion of recovered plasmids, revealing that the majority of the RS joins analyzed were precise (31 of 32, 97%, of RS joins were digested by ApaLI). Thus, Artemis-deficient ES cells exhibit a severe impairment in coding joining but retain relatively normal ability to perform accurate RS joining. To confirm that the coding joining defect observed in ArtN/N ES cells was specifically due to the inactivation of Artemis, we cotransfected a cDNA construct expressing the human Artemis gene and observed full rescue of coding joining (Table II). Furthermore, we also fully complemented the DNA-PKcsN/N coding joining defect with the murine DNA-PKcs cDNA expression construct (Table II).
Table II.
Analysis of Coding and RS Joining in ArtN/N ES Cells
| Cell lines | (Ampr + Camr)/Ampr | Percent | Relative level |
|---|---|---|---|
| Experiment 1a | |||
| TC1 | 9,868/2,042,500 | 0.48 | 1.0 |
| ArtN/N (82.10.66) | 387/862,000 | 0.04 | 0.08 |
| ArtN/N (82.10.77) | 114/493,500 | 0.02 | 0.04 |
| XRCC4−/− | 6/52,133 | 0.01 | 0.02 |
| Experiment 2 | |||
| TC1 | 6,414/1,435,000 | 0.45 | 1.0 |
| ArtN/N (82.10.77) | 188/547,000 | 0.03 | 0.07 |
| DNA-PKcsN/N | 11/191,500 | 0.006 | 0.01 |
| XRCC4−/− | 21/246,500 | 0.009 | 0.02 |
| Experiment 3 | |||
| TC1 | 2,625/500,000 | 0.53 | 1.0 |
| ArtN/N (82.10.66) | 270/1,345,000 | 0.02 | 0.04 |
| ArtN/N (82.10.77) | 164/1,410,000 | 0.01 | 0.02 |
| DNA-PKcsN/N | 62/1,100,000 | 0.006 | 0.01 |
| Ku70−/− | 25/300,000 | 0.008 | 0.02 |
| Experiment 4 | |||
| TC1 | 33/3,199 | 1.0 | 1.0 |
| ArtN/N (82.10.66.2) | 50/126,000 | 0.05 | 0.05 |
| DNA-PKcsN/N | 12/90,000 | 0.01 | 0.01 |
| Experiment 5 | |||
| TC1 | 1,503/503,250 | 0.30 | 1.0 |
| ArtN/N (82.10.66.2) | 136/346,500 | 0.04 | 0.13 |
| DNA-PKcsN/N | 66/378,500 | 0.02 | 0.07 |
| Experiment 6 | |||
| TC1 | 2,499/844,500 | 0.30 | 1.0 |
| ArtN/N (82.10.66) | 80/107,933 | 0.07 | 0.2 |
| ArtN/N (82.10.77) | 10/74,133 | 0.01 | 0.04 |
| XRCC4−/− | 0/36,467 | <0.003 | <0.01 |
| Experiment 7 | |||
| TC1 | 188/17,667 | 1.1 | 1.0 |
| ArtN/N (82.10.66.2) | 36/19,000 | 0.19 | 0.17 |
| ArtN/N (82.10.77) | 48/59,867 | 0.08 | 0.07 |
| DNA-PKcsN/N (2-41-3) | 9/61,000 | 0.01 | 0.009 |
| DNA-PKcsN/N (2-35-2) | 4/54,400 | 0.007 | 0.006 |
| TC1 (no RAG) | 23/70,467 | 0.03 | 0.03 |
| + hArt cDNA | |||
| ArtN/N (82.10.66.2) | 248/17,333 | 1.4 | 1.3 |
| ArtN/N (82.10.77) | 410/69,133 | 0.59 | 0.55 |
| + mDNA-PKcs cDNA | |||
| DNA-PKcsN/N (2-41-3) | 181/27,667 | 0.65 | 0.59 |
| DNA-PKcsN/N (2-35-2) | 336/76,900 | 0.44 | 0.40 |
| Experiment 8 | |||
| TC1 | 3,850/640,000 | 0.6 | 1.0 |
| ArtN/N (82.10.66.2) | 107/147,000 | 0.07 | 0.12 |
| ArtN/N (82.10.77) | 70/292,000 | 0.02 | 0.04 |
| DNA-PKcsN/N (2-41-3) | 17/78,000 | 0.02 | 0.04 |
| DNA-PKcsN/N (2-35-2) | 12/65,300 | 0.02 | 0.04 |
| TC1 (no RAG) | 10/146,000 | 0.007 | 0.01 |
| + hArt cDNA | |||
| ArtN/N (82.10.66.2) | 118/6,940 | 1.7 | 2.8 |
| ArtN/N (82.10.77) | 1,037/148,000 | 0.7 | 1.2 |
| + mDNA-PKcs cDNA | |||
| DNA-PKcsN/N (2-41-3) | 2,389/115,800 | 2.1 | 3.4 |
| DNA-PKcsN/N (2-35-2) | 1,755/378,900 | 4.6 | 7.6 |
| Experiment 1b | |||
| TC1 | 552/107,266 | 0.51 | 1.0 |
| ArtN/N (82.10.66) | 450/57,533 | 0.78 | 1.5 |
| ArtN/N (82.10.77) | 551/70,400 | 0.78 | 1.5 |
| XRCC4−/− | 0/12,467 | <0.008 | <0.02 |
| Experiment 2 | |||
| TC1 | 5,643/852,500 | 0.66 | 1.0 |
| ArtN/N (82.10.77) | 4,319/394,500 | 1.09 | 1.7 |
| DNA-PKcsN/N | 526/141,000 | 0.37 | 0.6 |
| XRCC4−/− | 6/184,000 | 0.003 | 0.005 |
| Experiment 3 | |||
| TC1 | 2,011/414,000 | 0.49 | 1.0 |
| ArtN/N (82.10.77) | 3,540/615,000 | 0.58 | 1.2 |
| ArtN/N (82.10.66) | 5,240/765,000 | 0.68 | 1.4 |
| DNA-PKcsN/N | 1,016/659,500 | 0.15 | 0.3 |
| Ku70−/− | 25/473,500 | 0.005 | 0.01 |
| Experiment 4 | |||
| TC1 | 108/38,663 | 0.28 | 1.0 |
| ArtN/N (82.10.66.2) | 585/458,000 | 0.13 | 0.5 |
| DNA-PKcsN/N | 173/106,000 | 0.16 | 0.6 |
| Experiment 5 | |||
| TC1 | 902/478,500 | 0.19 | 1.0 |
| ArtN/N (82.10.66.2) | 1,128/451,400 | 0.25 | 1.3 |
| DNA-PKcsN/N | 1,559/255,500 | 0.61 | 3.2 |
pJH290 (coding joining) for Experiments 1–8.
pJH200 (signal joining) for Experiments 1–5.
Coding Joins Recovered from Artemis-deficient ES Cells Are Qualitatively Distinct.
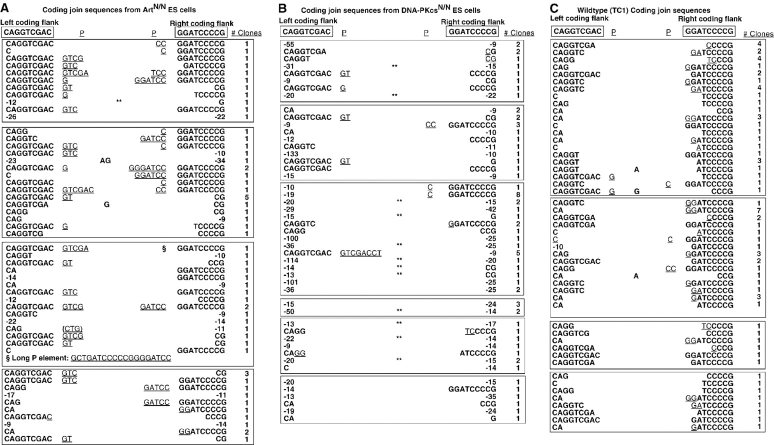
Those coding joins recovered from Artemis-deficient ES cells, as compared with TC1 and DNA-PKcsN/N ES cells were enriched for full-length coding flanks (45 vs. 20 and 11% for ArtN/N, TC1, and DNA-PKcsN/N ES cells, respectively; Fig. 3 and not depicted). Moreover, the majority of coding joins from Artemis-deficient ES cells contained P nucleotides (35 of 60 unique joins, 58%) in contrast to the relatively small percentage of joins with P nucleotides from wild-type (5 of 50 joins, 10%) and DNA-PKcsN/N ES cells (8 of 48 joins, 17%). In addition, P nucleotide additions observed in coding joins from ArtN/N ES cells typically ranged from one to seven nucleotides (mean = 3.6 ± 3.0 bases) whereas those from wild-type cells did not exceed two nucleotides. In DNA-PKcsN/N ES cells, the majority of P nucleotides were one or two nucleotides in length, although one long P nucleotide was observed (Fig. 3). Finally, coding joins containing deletions of greater than eight nucleotides from either end were observed at elevated levels in substrates recovered from Artemis-deficient versus wild-type ES cells (30 vs. 2%). Coding joins with larger deletions (>8 nucleotides) comprised the majority of sequences recovered from DNA-PKcsN/N ES cells (73%). Overall, coding join sequences isolated from DNA-PKcsN/N ES cells were qualitatively similar to the few that we have previously mentioned (27). However, the large number of sequences analyzed in this study has allowed us to make more quantitative comparisons (see Discussion). Introduction of the human Artemis and murine DNA-PKcs expression constructs into the respective mutant cell lines fully complemented the defects as joins recovered from complemented cells were similar to those of wild-type cells (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20021891/DC1).
Figure 3.
Sequence analysis of VDJ coding joins. Individual Camr/Ampr clones were isolated and sequenced. Top boxes, junctional coding sequences flanking the left and right RSs; underlined, Potential P elements; underlined/italicized, nucleotides that cannot be unequivocally assigned to either end; center column, random insertions; parentheses, potential P element possibly formed subsequent to open and shut join with nucleotide loss; **, homology-mediated joins; right column, number of clones with the same sequence. (A) ArtN/N coding join sequences. (B) DNA-PKcsN/N coding join sequences. (C) TC1 coding join sequences. Sequences were obtained from two independently derived ArtN/N and DNA-PKcsN/N ES clones. Independent experiments are indicated by boxes.
Artemis Deficiency Results in Increased Genomic Instability.
To assess the potential role of Artemis in maintenance of genomic stability, we examined chromosomes by DAPI staining and SKY. First, we measured the level of spontaneous chromosomal abnormalities in DAPI-stained metaphases from early passage ArtN/N and XRCC4−/− ES cells relative to those of TC1 cells (Table III). Two independently derived Artemis-deficient ES cell lines were analyzed and both exhibited significantly increased levels of chromosomal abnormalities, including chromosomal fragments, fusions, and detached centromeres (Table III). Thus, Artemis deficiency results in increased spontaneous genomic instability in ES cells, similar to that observed for XRCC4 deficiency (Table III, Spontaneous instability).
Table III.
Artemis-deficient ES Cells Exhibit Increased Genomic Instability
| TC1 WT | ArtN/N(82.10.66) | ArtN/N(82.10.77) | XRCC4−/− | |
|---|---|---|---|---|
| Spontaneous instability | ||||
| Metaphases | 56 | 30 | 21 | 22 |
| Fragmented chromosomes | 1 | 5 | 5 | 6 |
| Detached centromeres | 2 | 5 | 0 | 3 |
| Fusions | 0 | 4 | 3 | 3 |
| Unusual structures | 0 | 0 | 0 | 4 |
| Total anomalies (no. per metaphase) | 3 (0.05) | 14 (0.47) | 8 (0.38) | 16 (0.73) |
| Metaphases with abnormalities (percent) | 5 | 37 | 38 | 48 |
| Chromosomal aberrations 48 h after 150 rads IR | ||||
| Metaphases | 24 | 29 | 24 | 26 |
| Fragmented chromosomes | 4 | 9 | 5 | 12 |
| Detached centromeres | 0 | 5 | 2 | 3 |
| Fusions | 2 | 4 | 4 | 8 |
| Unusual structures | 0 | 1 | 0 | 0 |
| Total anomalies (no. per metaphase) | 6 (0.25) | 19 (0.66) | 11 (0.46) | 23 (0.88) |
| Metaphases with abnormalities (percent) | 25 | 48 | 42 | 54 |
| Chromosomal aberrations 48 h after 300 rads IR | ||||
| Metaphases | 24 | 29 | 31 | 26 |
| Fragmented chromosomes | 1 | 10 | 11 | 10 |
| Detached centromeres | 2 | 4 | 2 | 3 |
| Fusions | 3 | 12 | 13 | 18 |
| Unusual structures | 0 | 1 | 0 | 0 |
| Total anomalies (no. per metaphase) | 6 (0.25) | 27 (0.93) | 26 (0.84) | 31 (1.19) |
| Metaphases with abnormalities (percent) | 25 | 59 | 45 | 62 |
| SKY analysis of chromosomal aberrations 24 h after 300 rads IR | TC1 WT | ArtN/N | XRCC4−/− | |
| Metaphases | 22 | 20 | 22 | |
| Fragmented chromosomes | 22 | 12 | 24 | |
| Detached centromeres | 1 | 8 | 6 | |
| Fusions | 3 | 10 | 20 | |
| Translocations | 8 | 21 | 35 | |
| Metaphases with abnormalities (percent) | 42 | 75 | 82 | |
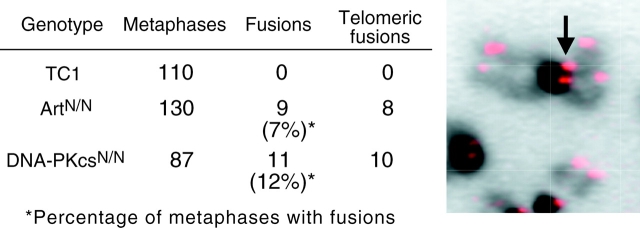
We also examined levels of chromosomal aberrations in early passage ES cells that had been exposed to 150 or 300 rads of IR and cultured for 48 h before analysis of recovered metaphases by DAPI staining. Upon exposure to IR, higher levels of chromosomal abnormalities, in particular chromosomal fragments and fusions, were observed in Artemis- and XRCC4-deficient cells in comparison to wild-type controls (Table III). Using SKY, a chromosomal painting technique that allows unambiguous identification of all murine chromosomes (54), we analyzed the types of chromosomal abnormalities present in metaphases from ArtN/N, XRCC4−/−, and wild-type ES cells that had been exposed to 300 rads of IR. We observed that Artemis deficiency, as well as XRCC4 deficiency, resulted in increased levels of chromosomal fusions and translocations in comparison to controls (Table III and Fig. 4) . Taken together, these results demonstrate that Artemis plays a significant role in maintaining genomic stability.
Figure 4.
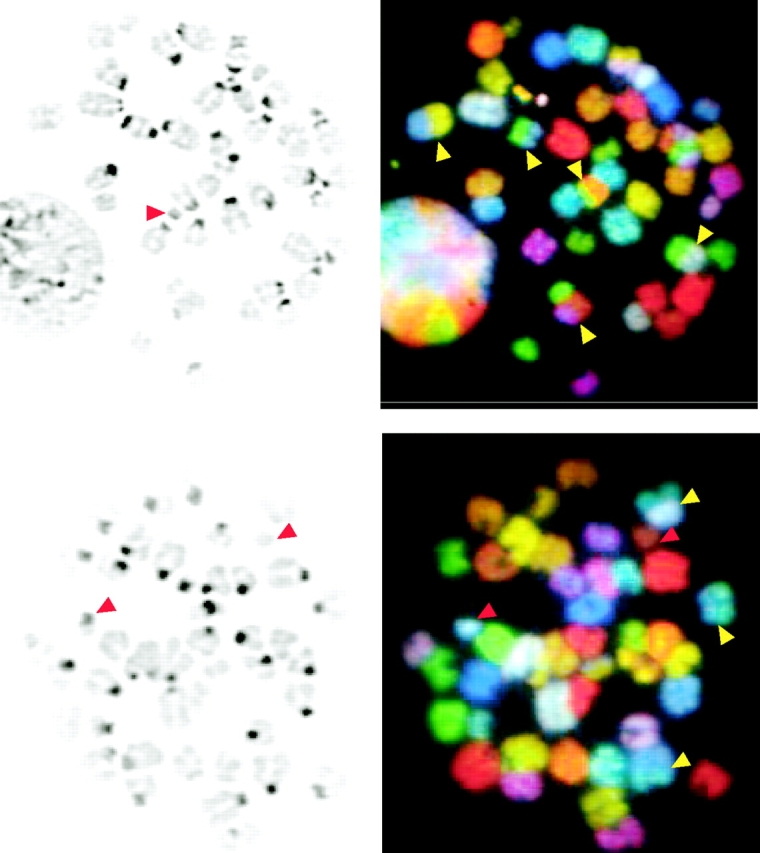
Chromosomal aberrations in metaphase spreads from irradiated ArtN/N ES cells. (A and B) Metaphase spreads of ArtN/N ES cells 24 h after 300 rads IR. On the left, DAPI-stained chromosomes are shown. On the right, SKY analysis is shown. Yellow arrows, chromosomal translocations; red arrow, detached centromeres and chromosomal fragments.
Telomeric Fusions in Artemis-deficient ES Cells.
Previously, Ku70, Ku80, and DNA-PKcs have been found localized at telomeric repeat sequences (7, 55) and Ku or DNA-PKcs deficiencies result in increased telomere fusions indicating that they may function in telomere capping (5, 6, 21–24). To determine whether Artemis plays a similar role, we further examined the nature of the chromosomal fusions observed in Artemis-deficient ES cells using a fluorescent peptide nucleic acid probe, which hybridizes to telomeric sequences (48). We observed increased numbers of fusions in Artemis-deficient cells in comparison to TC1 controls, and the majority contained telomeric sequence at the fusion junctions (Fig. 5 and Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20021891/DC1). We also found increased telomere fusions in DNA-PKcsN/N cells as has been previously reported (Fig. 5; references 5, 6, and 21).
Figure 5.
Telomere fusions arise in ArtN/N ES cells. (A) Telomeric sequence was identified using a Cy3-labeled peptide nucleic acid probe in metaphases from ArtN/N, DNA-PKcsN/N, and TC1 ES cells. (B) Representative short-arm telomeric fusion in an ArtN/N ES cell metaphase. Arrow represents two telomeric signals located at the point of fusion between cytogenetically invisible short arms. Adjacent, nonfused chromosomes would show four discrete signals.
Discussion
Artemis Plays a Role in a Subset of NHEJ Events.
We have generated Artemis-deficient murine ES cells by gene targeted mutation. In ES cells, Artemis is required for repair of bleomycin-induced DNA DSBs and for maintenance of chromosomal stability including telomeric fusions, indicating important genomic caretaker functions. As in RS-SCID (Artemis-deficient) human cells (39, 42, 56) and DNA-PKcs mutant murine cells (27, 36, 38, 57), VDJ recombination substrate coding joining is severely impaired in Artemis-deficient ES cells, but RS joining is relatively normal, both quantitatively and qualitatively. Moreover, VDJ substrate coding joins recovered from Artemis-deficient ES cells are qualitatively distinct from those obtained from wild-type cells, with a large proportion having P nucleotides. Overall, our observations support the suggestion that Artemis is a major factor involved in the processing of hairpin coding ends during VDJ recombination, but is not required for all NHEJ events (39, 43). The latter point is emphasized by the occurrence of normal RS joining and lack of severe IR sensitivity in ArtN/N ES cells, as opposed to the complete RS joining deficiency and severe IR sensitivity observed in XRCC4-deficient ES cells.
VDJ Coding Joins from Artemis-deficient ES Cells Have a Characteristic Structure.
Artemis-deficient ES cells are severely impaired in ability to generate coding joins recoverable via the standard transient VDJ recombination substrate assay. Yet, a large proportion of the coding joins recovered from these cells are relatively normal in that they do not have large deletions. In this regard, a significant proportion of attempted coding joins in NHEJ-deficient cells are not scored by the bacterial drug selection assay (36). Thus, it is likely that a significant proportion of coding joins in ArtN/N ES cells are also not rejoined or are joined with large deletions. In this regard, most unselected coding joins formed in the absence of DNA-PKcs also have extremely large deletions (58–64).
Coding joins that were recovered from Artemis-deficient ES cells have a high frequency of large (>2 bp) P nucleotides and a corresponding enrichment of coding flanks with no nucleotide loss (Fig. 3). These coding join structures are reminiscent of those recovered within reporter constructs transiently transfected into some DNA-PKcs mutant somatic cells (36–38). Lack of a hairpin opening function could indirectly lead to coding joins with either very large deletions or unusually long P elements (30). Thus, the structure of recovered Artemis-deficient coding joins is consistent with the notion that these joins result from a different route of hairpin opening and/or processing, supporting the proposal that Artemis acts directly to open hairpin coding ends (39, 43). Recently, we confirmed accumulation of hairpin coding ends in Artemis-deficient thymocytes (44) as previously observed in DNA-PKcs–deficient thymocytes (30, 31). Given that Artemis provides the main route of normal coding end hairpin opening, the low levels of “normal” coding joins in ArtN/N cells implies that hairpin coding ends can be opened by a much less efficient, alternative nicking activity. Proposed alternative nicking activities, based on in vitro studies, include the RAG-1/2 (RAG) or Mre11/RAD50/NBS1 complexes (65–68).
Differences between Recovered Coding Joins from Artemis- and DNA-PKcs–deficient ES Cells.
The majority of coding joins recovered from DNA-PKcsN/N ES cells contained large deletions with only a few exhibiting P nucleotides (Fig. 3). These coding join sequences are distinct from those of ArtN/N ES cells (Fig. 3) and also differ from joins recovered from certain DNA-PKcs mutant somatic cells in which a significant proportion contained unusually long P nucleotides (36–38). Joins recovered from ArtN/N somatic cells also showed some full-length coding flanks and P nucleotides, although not to the extent observed in ES cells (44). Although the basis for these differences is unclear, there are various, nonmutually exclusive possibilities. One is that there might be factors in ES cells as opposed to somatic cells or vice versa, which contribute to the altered spectrum of coding joints observed in different mutant cell types. In this context, ES cells do not have normal G1 checkpoints. Alternatively, apparent coding join differences between different DNA-PKcs mutants may reflect low level expression of catalytically inactive forms of DNA-PKcs resulting from certain DNA-PKcs mutations (69, 70), as opposed to the complete null generated by the DNA-PKcsN/N targeted mutation (27). Catalytically inactive forms of DNA-PKcs may retain residual activities including interaction with Artemis, potentially sequestering it in inactive complexes or altering its activities. Ultimately, the characterization of potential differences with respect to complete and partially inactivating DNA-PKcs mutations will require introduction of all mutations into the same cellular and genetic background.
There could be several different explanations for the striking qualitative differences between coding joins recovered from DNA-PKcsN/N and ArtN/N ES cells. DNA-PKcs likely has additional functions in the context of the DNA-PK holoenzyme, possibly in the activation and/or recruitment of other NHEJ factors (2). Thus, in the absence of Artemis, hairpin coding ends still might be captured within a DNA-PKcs–mediated synaptic complex (71) and become available to alternative, potentially DNA-PKcs–activated, hairpin opening activities. Likewise, in the absence of Artemis, ends opened in the context of a DNA-PKcs complex might be protected and less frequently processed, for example via an Artemis endonuclease or exonuclease activity (43). Additional studies will be needed to distinguish between these and other potential possibilities.
DSB Repair Defects in Artemis-deficient ES Cells.
ArtN/N and DNA-PKcsN/N ES cells exhibit markedly increased bleomycin sensitivity. However, at most they are mildly IR sensitive compared with XRCC4- or Ku-deficient ES cells (46, 51). Among the six known NHEJ factors, only Artemis and DNA-PKcs are not conserved in yeast. One potential explanation for the less profound radiosensitivity of Artemis- and DNA-PKcs–deficient ES cells, as opposed to those deficient in evolutionarily conserved NHEJ factors, is that Artemis and DNA-PKcs are used to process and repair a subset of DSBs, i.e., ends requiring endonucleolytic processing (27, 43). This notion is also consistent with the requirement for Ku and XRCC4 (46, 51), but not DNA-PKcs and Artemis, for RS joining in ES cells. In this context, decreased survival of DNA-PKcsN/N and ArtN/N ES cells observed upon sustained bleomycin exposure may result from accumulation of a class of DSBs requiring DNA-PKcs and Artemis for repair. Alternatively, prolonged bleomycin exposure may allow a threshold DSB level to be reached that can no longer be adequately repaired without Artemis or DNA-PKcs. Human RS-SCID fibroblasts, bone marrow cells (40–42), and ArtN/N MEFs (44) are markedly IR sensitive. Thus, as with DNA-PKcs deficiency (27), Artemis deficiency may exhibit cell type variation in the degree of its impact on IR sensitivity. Finally, Artemis-deficient ES cells do not display increased sensitivity to the ICL agent, MMC, despite sequence similarity to factors required for repair of ICL (39). Therefore, in murine ES cells, Artemis appears specifically required for repair of a subset of DNA DSBs but not for resolution of ICL
Artemis Is Required for Maintenance of Genomic Stability.
Artemis deficiency results in increased genomic instability in ES cells. The types of chromosomal aberrations observed include chromosomal fragmentation, detached centromeres, fusions, and translocations. Thus, Artemis plays an important role as a genomic caretaker. In addition, like Ku and DNA-PKcs (20), Artemis also may function in telomere capping based on the increased levels of telomere fusions observed in Artemis-deficient ES cells. In this regard, it will be of interest to determine potential roles of Artemis in maintenance of telomere length, as there are conflicting reports regarding the role of Ku in this context (7, 23, 24). Although the precise function of Artemis with respect to telomeres remains unclear, it is intriguing that Artemis possesses intrinsic exonuclease and endonuclease activities that are regulated by DNA-PKcs (43). Thus, the Artemis–DNA-PKcs complex may not only function in VDJ recombination and general DNA DSB repair, but also in telomere maintenance.
Genomic instability is a hallmark of tumorigenesis and mice deficient for Ku70 as well as some lines of DNA-PKcs–deficient mice are predisposed to T cell lymphomas (13, 14, 72). Lymphomagenesis caused by NHEJ deficiency is enhanced by inactivation of cell cycle checkpoints, as NHEJ/p53 double mutant mice succumb to aggressive pro–B cell lymphomas during young adulthood (1, 8, 10, 12, 15, 17, 19, 73). To date, inactivating mutations in XRCC4, Lig4, Ku70, Ku80, or DNA-PKcs have not been associated with human RS-SCID and several lines of evidence suggest that such mutations in humans may not support cell growth or survival (51, 74–77). Because Artemis deficiency is compatible with sustained human life, our finding that Artemis deficiency leads to genomic instability in murine cells suggests Artemis as a candidate tumor suppressor gene in humans.
Acknowledgments
We thank Dr. Craig Bassing for critical reading of the manuscript and Dr. Roland Kanaar for the gift of ERCC1-deficient ES cells.
This work was supported by National Institutes of Health (NIH) grants AI35714 and CA92625 to F.W. Alt and AI32524 to D.G. Schatz. F.W. Alt and D.G. Schatz are Investigators of the Howard Hughes Medical Institute. S. Fugmann is an Irvington postdoctoral fellow. J. Sekiguchi is a Special Fellow of the Leukemia and Lymphoma Society. D.O. Ferguson is supported by NIH grant K08 HL67580-02.
S. Rooney and J. Sekiguchi contributed equally to this work.
The online version of this article contains supplemental material.
Footnotes
Abbreviations used in this paper: DNA-PKcs, DNA-dependent protein kinase catalytic subunit; DSB, double strand break; ES, embryonic stem; ICL, interstrand cross-links; IR, ionizing radiation; Lig4, ligase 4; MMC, mitomycin C; NHEJ, nonhomologous end joining; P, palindromic; RS, recombination signal; RS-SCID, radiosensitive T− B− SCID; SKY, spectral karyotyping.
References
- 1.Ferguson, D.O., and F.W. Alt. 2001. DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene. 20:5572–5579. [DOI] [PubMed] [Google Scholar]
- 2.Jackson, S.P. 2002. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 23:687–696. [DOI] [PubMed] [Google Scholar]
- 3.Bassing, C.H., W. Swat, and F.W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 109:S45–S55. [DOI] [PubMed] [Google Scholar]
- 4.Gilley, D., H. Tanaka, M.P. Hande, A. Kurimasa, G.C. Li, M. Oshimura, and D.J. Chen. 2001. DNA-PKcs is critical for telomere capping. Proc. Natl. Acad. Sci. USA. 98:15084–15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey, S.M., M.N. Cornforth, A. Kurimasa, D.J. Chen, and E.H. Goodwin. 2001. Strand-specific postreplicative processing of mammalian telomeres. Science. 293:2462–2465. [DOI] [PubMed] [Google Scholar]
- 6.Goytisolo, F.A., E. Samper, S. Edmonson, G.E. Taccioli, and M.A. Blasco. 2001. The absence of the dna-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol. 21:3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.d'Adda di Fagagna, F., M.P. Hande, W.M. Tong, D. Roth, P.M. Lansdorp, Z.Q. Wang, and S.P. Jackson. 2001. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 11:1192–1196. [DOI] [PubMed] [Google Scholar]
- 8.Difilippantonio, M.J., J. Zhu, H.T. Chen, E. Meffre, M.C. Nussenzweig, E.E. Max, T. Ried, and A. Nussenzweig. 2000. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 404:510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson, D.O., J.M. Sekiguchi, S. Chang, K.M. Frank, Y. Gao, R.A. DePinho, and F.W. Alt. 2000. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. USA. 97:6630–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, Y., D.O. Ferguson, W. Xie, J. Manis, J. Sekiguchi, K.M. Frank, J. Chaudhuri, R.A. DePinho, and F.W. Alt. 2000. Interplay of p53 and DNA-repair protein XRCC4 in tumourigenesis, genomic stability and development. Nature. 404:897–900. [DOI] [PubMed] [Google Scholar]
- 11.Karanjawala, Z.E., U. Grawunder, C.L. Hsieh, and M.R. Lieber. 1999. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr. Biol. 9:1501–1504. [DOI] [PubMed] [Google Scholar]
- 12.Difilippantonio, M.J., S. Petersen, H.T. Chen, R. Johnson, M. Jasin, R. Kanaar, T. Ried, and A. Nussenzweig. 2002. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J. Exp. Med. 196:469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu, Y., K.J. Seidl, G.A. Rathbun, C. Zhu, J.P. Manis, N. van der Stoep, L. Davidson, H.L. Cheng, J.M. Sekiguchi, K. Frank, et al. 1997. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 7:653–665. [DOI] [PubMed] [Google Scholar]
- 14.Li, G.C., H. Ouyang, X. Li, H. Nagasawa, J.B. Little, D.J. Chen, C.C. Ling, Z. Fuks, and C. Cordon-Cardo. 1998. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol. Cell. 2:1–8. [DOI] [PubMed] [Google Scholar]
- 15.Lim, D.S., H. Vogel, D.M. Willerford, A.T. Sands, K.A. Platt, and P. Hasty. 2000. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol. Cell. Biol. 20:3772–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurley, K.E., K. Vo, and C.J. Kemp. 1998. DNA double-strand breaks, p53, and apoptosis during lymphomagenesis in scid/scid mice. Cancer Res. 58:3111–3115. [PubMed] [Google Scholar]
- 17.Nacht, M., A. Strasser, Y.R. Chan, A.W. Harris, M. Schlissel, R.T. Bronson, and T. Jacks. 1996. Mutations in the p53 and SCID genes cooperate in tumorigenesis. Genes Dev. 10:2055–2066. [DOI] [PubMed] [Google Scholar]
- 18.Frank, K.M., N.E. Sharpless, Y. Gao, J.M. Sekiguchi, D.O. Ferguson, C. Zhu, J.P. Manis, J. Horner, R.A. DePinho, and F.W. Alt. 2000. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell. 5:993–1002. [DOI] [PubMed] [Google Scholar]
- 19.Zhu, C., K.D. Mills, D.O. Ferguson, C. Lee, J. Manis, J. Fleming, Y. Gao, C.C. Morton, and F.W. Alt. 2002. Unrepaired DNA breaks in P53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 109:811–821. [DOI] [PubMed] [Google Scholar]
- 20.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene. 21:532–540. [DOI] [PubMed] [Google Scholar]
- 21.Bailey, S.M., J. Meyne, D.J. Chen, A. Kurimasa, G.C. Li, B.E. Lehnert, and E.H. Goodwin. 1999. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. USA. 96:14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu, H.L., D. Gilley, S.A. Galande, M.P. Hande, B. Allen, S.H. Kim, G.C. Li, J. Campisi, T. Kohwi-Shigematsu, and D.J. Chen. 2000. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 14:2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samper, E., F.A. Goytisolo, P. Slijepcevic, P.P. van Buul, and M.A. Blasco. 2000. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1:244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espejel, S., S. Franco, S. Rodriguez-Perales, S.D. Bouffler, J.C. Cigudosa, and M.A. Blasco. 2002. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 21:2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlissel, M.S. 2002. Does artemis end the hunt for the hairpin-opening activity in V(D)J recombination? Cell. 109:1–4. [DOI] [PubMed] [Google Scholar]
- 26.Lewis, S.M. 1994. P nucleotides, hairpin DNA and V(D)J joining: making the connection. Semin. Immunol. 6:131–141. [DOI] [PubMed] [Google Scholar]
- 27.Gao, Y., J. Chaudhuri, C. Zhu, L. Davidson, D.T. Weaver, and F.W. Alt. 1998. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 9:367–376. [DOI] [PubMed] [Google Scholar]
- 28.Taccioli, G.E., A.G. Amatucci, H.J. Beamish, D. Gell, X.H. Xiang, M.I. Torres Arzayus, A. Priestley, S.P. Jackson, A. Marshak Rothstein, P.A. Jeggo, et al. 1998. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 9:355–366. [DOI] [PubMed] [Google Scholar]
- 29.Kurimasa, A., H. Ouyang, L.J. Dong, S. Wang, X. Li, C. Cordon-Cardo, D.J. Chen, and G.C. Li. 1999. Catalytic subunit of DNA-dependent protein kinase: impact on lymphocyte development and tumorigenesis. Proc. Natl. Acad. Sci. USA. 96:1403–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth, D.B., J.P. Menetski, P.B. Nakajima, M.J. Bosma, and M. Gellert. 1992. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 70:983–991. [DOI] [PubMed] [Google Scholar]
- 31.Zhu, C., and D.B. Roth. 1995. Characterization of coding ends in thymocytes of scid mice: implications for the mechanism of V(D)J recombination. Immunity. 2:101–112. [DOI] [PubMed] [Google Scholar]
- 32.Carroll, A.M., and M.J. Bosma. 1991. T-lymphocyte development in scid mice is arrested shortly after the initiation of T-cell receptor delta gene recombination. Genes Dev. 5:1357–1366. [DOI] [PubMed] [Google Scholar]
- 33.Ferrier, P., L.R. Covey, S.C. Li, H. Suh, B.A. Malynn, T.K. Blackwell, M.A. Morrow, and F.W. Alt. 1990. Normal recombination substrate VH to DJH rearrangements in pre-B cell lines from scid mice. J. Exp. Med. 171:1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuler, W., N.R. Ruetsch, M. Amsler, and M.J. Bosma. 1991. Coding joint formation of endogenous T cell receptor genes in lymphoid cells from scid mice: unusual P-nucleotide additions in VJ-coding joints. Eur. J. Immunol. 21:589–596. [DOI] [PubMed] [Google Scholar]
- 35.Kienker, L.J., W.A. Kuziel, and P.W. Tucker. 1991. T cell receptor gamma and delta gene junctional sequences in SCID mice: excessive P nucleotide insertion. J. Exp. Med. 174:769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taccioli, G.E., G. Rathbun, E. Oltz, T. Stamato, P.A. Jeggo, and F.W. Alt. 1993. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 260:207–210. [DOI] [PubMed] [Google Scholar]
- 37.Taccioli, G.E., H.L. Cheng, A.J. Varghese, G. Whitmore, and F.W. Alt. 1994. A DNA repair defect in Chinese hamster ovary cells affects V(D)J recombination similarly to the murine scid mutation. J. Biol. Chem. 269:7439–7442. [PubMed] [Google Scholar]
- 38.Binnie, A., S. Olson, G.E. Wu, and S.M. Lewis. 1999. Gamma-irradiation directly affects the formation of coding joints in SCID cell lines. J. Immunol. 163:5418–5426. [PubMed] [Google Scholar]
- 39.Moshous, D., I. Callebaut, R. de Chasseval, B. Corneo, M. Cavazzana-Calvo, F. Le Deist, I. Tezcan, O. Sanal, Y. Bertrand, N. Philippe, et al. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 105:177–186. [DOI] [PubMed] [Google Scholar]
- 40.Cavazzana-Calvo, M., F. Le Deist, G. De Saint Basile, D. Papadopoulo, J.P. De Villartay, and A. Fischer. 1993. Increased radiosensitivity of granulocyte macrophage colony-forming units and skin fibroblasts in human autosomal recessive severe combined immunodeficiency. J. Clin. Invest. 91:1214–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolas, N., N.J. Finnie, M. Cavazzana-Calvo, D. Papadopoulo, F. Le Deist, A. Fischer, S.P. Jackson, and J.P. de Villartay. 1996. Lack of detectable defect in DNA double-strand break repair and DNA-dependent protein kinase activity in radiosensitive human severe combined immunodeficiency fibroblasts. Eur. J. Immunol. 26:1118–1122. [DOI] [PubMed] [Google Scholar]
- 42.Nicolas, N., D. Moshous, M. Cavazzana-Calvo, D. Papadopoulo, R. de Chasseval, F. Le Deist, A. Fischer, and J.P. de Villartay. 1998. A human severe combined immunodeficiency (SCID) condition with increased sensitivity to ionizing radiations and impaired V(D)J rearrangements defines a new DNA recombination/repair deficiency. J. Exp. Med. 188:627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma, Y., U. Pannicke, K. Schwarz, and M.R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 108:781–794. [DOI] [PubMed] [Google Scholar]
- 44.Rooney, S., J. Sekiguchi, C. Zhu, H.L. Cheng, J.P. Manis, S. Whitlow, J. DeVido, D. Foy, J. Chaudhuri, D. Lombard, et al. 2002. Leaky scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol. Cell. 10:1379–1390. [DOI] [PubMed] [Google Scholar]
- 45.Chen, J., R. Lansford, V. Stewart, F. Young, and F.W. Alt. 1993. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc. Natl. Acad. Sci. USA. 90:4528–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu, Y., S. Jin, Y. Gao, D.T. Weaver, and F.W. Alt. 1997. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl. Acad. Sci. USA. 94:8076–8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hesse, J.E., M.R. Lieber, M. Gellert, and K. Mizuuchi. 1987. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 49:775–783. [DOI] [PubMed] [Google Scholar]
- 48.Greenberg, R.A., L. Chin, A. Femino, K.H. Lee, G.J. Gottlieb, R.H. Singer, C.W. Greider, and R.A. DePinho. 1999. Short dysfunctional telomeres impair tumorigenesis in the INK4a(delta2/3) cancer-prone mouse. Cell. 97:515–525. [DOI] [PubMed] [Google Scholar]
- 49.Li, L., D. Moshous, Y. Zhou, J. Wang, G. Xie, E. Salido, D. Hu, J.P. de Villartay, and M.J. Cowan. 2002. A founder mutation in Artemis, an SNM1-like protein, causes SCID in Athabascan-speaking Native Americans. J. Immunol. 168:6323–6329. [DOI] [PubMed] [Google Scholar]
- 50.Chen, J., M. Trounstine, C. Kurahara, F. Young, C.C. Kuo, Y. Xu, J.F. Loring, F.W. Alt, and D. Huszar. 1993. B cell development in mice that lack one or both immunoglobulin kappa light chain genes. EMBO J. 12:821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao, Y., Y. Sun, K.M. Frank, P. Dikkes, Y. Fujiwara, K.J. Seidl, J.M. Sekiguchi, G.A. Rathbun, W. Swat, J. Wang, et al. 1998. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 95:891–902. [DOI] [PubMed] [Google Scholar]
- 52.Weeda, G., I. Donker, J. de Wit, H. Morreau, R. Janssens, C.J. Vissers, A. Nigg, H. van Steeg, D. Bootsma, and J.H. Hoeijmakers. 1997. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol. 7:427–439. [DOI] [PubMed] [Google Scholar]
- 53.Dronkert, M.L., J. de Wit, M. Boeve, M.L. Vasconcelos, H. van Steeg, T.L. Tan, J.H. Hoeijmakers, and R. Kanaar. 2000. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol. Cell. Biol. 20:4553–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liyanage, M., A. Coleman, S. du Manoir, T. Veldman, S. McCormack, R.B. Dickson, C. Barlow, A. Wynshaw-Boris, S. Janz, J. Wienberg, et al. 1996. Multicolour spectral karyotyping of mouse chromosomes. Nat. Genet. 14:312–315. [DOI] [PubMed] [Google Scholar]
- 55.Hsu, H.L., D. Gilley, E.H. Blackburn, and D.J. Chen. 1999. Ku is associated with the telomere in mammals. Proc. Natl. Acad. Sci. USA. 96:12454–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moshous, D., L. Li, R. Chasseval, N. Philippe, N. Jabado, M.J. Cowan, A. Fischer, and J.P. de Villartay. 2000. A new gene involved in DNA double-strand break repair and V(D)J recombination is located on human chromosome 10p. Hum. Mol. Genet. 9:583–588. [DOI] [PubMed] [Google Scholar]
- 57.Lieber, M.R., J.E. Hesse, S. Lewis, G.C. Bosma, N. Rosenberg, K. Mizuuchi, M.J. Bosma, and M. Gellert. 1988. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell. 55:7–16. [DOI] [PubMed] [Google Scholar]
- 58.Schuler, W., I.J. Weiler, A. Schuler, R.A. Phillips, N. Rosenberg, T.W. Mak, J.F. Kearney, R.P. Perry, and M.J. Bosma. 1986. Rearrangement of antigen receptor genes is defective in mice with severe combined immune deficiency. Cell. 46:963–972. [DOI] [PubMed] [Google Scholar]
- 59.Hendrickson, E.A., M.S. Schlissel, and D.T. Weaver. 1990. Wild-type V(D)J recombination in scid pre-B cells. Mol. Cell. Biol. 10:5397–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hendrickson, E.A., D.G. Schatz, and D.T. Weaver. 1988. The scid gene encodes a trans-acting factor that mediates the rejoining event of Ig gene rearrangement. Genes Dev. 2:817–829. [DOI] [PubMed] [Google Scholar]
- 61.Kim, M.G., W. Schuler, M.J. Bosma, and K.B. Marcu. 1988. Abnormal recombination of Igh D and J gene segments in transformed pre-B cells of scid mice. J. Immunol. 141:1341–1347. [PubMed] [Google Scholar]
- 62.Malynn, B.A., T.K. Blackwell, G.M. Fulop, G.A. Rathbun, A.J. Furley, P. Ferrier, L.B. Heinke, R.A. Phillips, G.D. Yancopoulos, and F.W. Alt. 1988. The scid defect affects the final step of the immunoglobulin VDJ recombinase mechanism. Cell. 54:453–460. [DOI] [PubMed] [Google Scholar]
- 63.Blackwell, T.K., B.A. Malynn, R.R. Pollock, P. Ferrier, L.R. Covey, G.M. Fulop, R.A. Phillips, G.D. Yancopoulos, and F.W. Alt. 1989. Isolation of scid pre-B cells that rearrange kappa light chain genes: formation of normal signal and abnormal coding joins. EMBO J. 8:735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Okazaki, K., S. Nishikawa, and H. Sakano. 1988. Aberrant immunoglobulin gene rearrangement in scid mouse bone marrow cells. J. Immunol. 141:1348–1352. [PubMed] [Google Scholar]
- 65.Paull, T.T., and M. Gellert. 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1:969–979. [DOI] [PubMed] [Google Scholar]
- 66.Paull, T.T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13:1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shockett, P.E., and D.G. Schatz. 1999. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol. Cell. Biol. 19:4159–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Besmer, E., J. Mansilla-Soto, S. Cassard, D.J. Sawchuk, G. Brown, M. Sadofsky, S.M. Lewis, M.C. Nussenzweig, and P. Cortes. 1998. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol. Cell. 2:817–828. [DOI] [PubMed] [Google Scholar]
- 69.Blunt, T., D. Gell, M. Fox, G.E. Taccioli, A.R. Lehmann, S.P. Jackson, and P.A. Jeggo. 1996. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl. Acad. Sci. USA. 93:10285–10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Araki, R., A. Fujimori, K. Hamatani, K. Mita, T. Saito, M. Mori, R. Fukumura, M. Morimyo, M. Muto, M. Itoh, et al. 1997. Nonsense mutation at Tyr-4046 in the DNA-dependent protein kinase catalytic subunit of severe combined immune deficiency mice. Proc. Natl. Acad. Sci. USA. 94:2438–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DeFazio, L.G., R.M. Stansel, J.D. Griffith, and G. Chu. 2002. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 21:3192–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jhappan, C., H.C. Morse, III, R.D. Fleischmann, M.M. Gottesman, and G. Merlino. 1997. DNA-PKcs: a T-cell tumour suppressor encoded at the mouse scid locus. Nat. Genet. 17:483–486. [DOI] [PubMed] [Google Scholar]
- 73.Vanasse, G.J., J. Halbrook, S. Thomas, A. Burgess, M.F. Hoekstra, C.M. Disteche, and D.M. Willerford. 1999. Genetic pathway to recurrent chromosome translocations in murine lymphoma involves V(D)J recombinase. J. Clin. Invest. 103:1669–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frank, K.M., J.M. Sekiguchi, K.J. Seidl, W. Swat, G.A. Rathbun, H.L. Cheng, L. Davidson, L. Kangaloo, and F.W. Alt. 1998. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 396:173–177. [DOI] [PubMed] [Google Scholar]
- 75.Barnes, D.E., G. Stamp, I. Rosewell, A. Denzel, and T. Lindahl. 1998. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 8:1395–1398. [DOI] [PubMed] [Google Scholar]
- 76.Li, G., C. Nelsen, and E.A. Hendrickson. 2002. Ku86 is essential in human somatic cells. Proc. Natl. Acad. Sci. USA. 99:832–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meek, K., L. Kienker, C. Dallas, W. Wang, M.J. Dark, P.J. Venta, M.L. Huie, R. Hirschhorn, and T. Bell. 2001. SCID in Jack Russell terriers: a new animal model of DNA-PKcs deficiency. J. Immunol. 167:2142–2150. [DOI] [PubMed] [Google Scholar]
- 78.Callebaut, I., D. Moshous, J.P. Mornon, and J.P. de Villartay. 2002. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 30:3592–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]