Abstract
Due to ordered, stage-specific T cell receptor (TCR)-β and -α locus gene rearrangements and cell division during T cell development, a given, ancestral TCR-β locus VDJ rearrangement might be selected into the mature T cell repertoire as a small cohort of “half-sibling” progeny expressing identical TCR-β chains paired with different TCR-α chains. The low frequency of such a cohort relative to the total αβ TCR repertoire precludes their direct identification and characterization in normal mice. We considered it possible that positive selection constraints might limit the diversity of TCR-α chains selected to pair with β chains encoded by an ancestral VDJ-β rearrangement. If so, half-sibling T cells expressing structurally similar, but different TCR-α chains might recognize the same foreign antigen. By single cell polymerase chain reaction analysis of antigen-specific TCRs selected during a model anti-tumor response, we were able to identify clusters of T cells sharing identical VDJ-β rearrangements but expressing different TCR-α chains. The amplification of residual DJ-β rearrangements as clonal markers allowed us to track T cells expressing different TCR-α chains back to a common ancestral VDJ-β rearrangement. Thus, the diversity of TCR-α's selected as partners for a given VDJ-β rearrangement into the mature TCR repertoire may indeed be very limited.
Keywords: CD8+ T lymphocytes, antigen, receptors, sequence analysis, cell lineage
Introduction
Specific recognition of foreign Ags by T lymphocytes is a function of the highly diverse, clonally distributed αβ TCRs (for review see reference 1). The diversity of the TCRs derives from the largely random and imprecise somatic recombination of variable (V), diversity (D), and junctional (J) genes, the addition of one to a few nucleotides at the junctions, and the pairing of the α and β chains encoded by the rearranged genes. General principles for Ag recognition (for review see references 2 and 3) have begun to emerge from the first analyses of crystal structures that show the interaction of αβ TCRs with their MHC class I peptide (pMHC)*ligands (4–10). All display a similar diagonal orientation of the αβ TCR with respect to its pMHC ligand. The receptor–ligand interaction occurs mainly via loops formed by the six CDRs (CDR1, CDR2, and CDR3) of the α and β chains. The CDR1 and CDR2 loops are directly encoded by the various TCR Vβ and Vα genes whereas the highly diverse CDR3 loops are encoded by the VDJ (for TCR-β) and VJ (for TCR-α) junctions. As predicted previously (1), αβ TCRs make extensive contacts with the MHC-bound peptide via the highly diverse CDR3 loops.
The size of the available αβ TCR repertoire is difficult to determine, but Casrouge et al. (11) estimate that there are nearly 2 million different αβ TCRs in mouse spleen. Interestingly, the diversity of TCR-α chains appears to be several fold greater than that of TCR-β chains, implying that a given β chain could pair with more than one α chain (in different T cells; reference 11). As the authors pointed out, this would be compatible with current concepts of T cell development in the thymus. Rearrangements occur first at the TCR-β locus where D to J recombinations are initiated, followed by V to DJ. Thymocytes making a successful in-frame VDJ-β rearrangement undergo an estimated six to seven rounds of cell division (12–14). These progeny undergo independent rearrangements at the TCR-α locus during a later developmental stage. Those expressing αβ TCRs at the cell surface undergo extensive positive and negative selection pressures that mold a mature TCR repertoire toward a general recognition of foreign, but not self-, peptides in the context of self-MHC molecules (for review see reference 15). Consequently, only a few descendants (an estimated three to seven cells) from a given VDJ-β recombination are expected to be recruited as mature T cells (11, 14).
Intriguing studies with TCR-β transgenic (Tg) mice suggest that for a given VDJ-β rearrangement the potential diversity of paired TCR-α chains that can be selected into the mature T cell repertoire might be considerably more constrained than that of normal mice, possibly reflecting a limited diversity of positively selecting pMHC ligands (16–19). The overwhelming diversity of TCR VDJ-β rearrangements in normal mice makes the identification of small cohorts of such “half-sibling” αβ T cells problematic. As an indirect approach, we considered the possibility that if some of the progeny issuing from a given VDJ-β rearrangement express structurally similar (but different) TCR-α chains, then some of them might recognize similar foreign pMHC ligands. If that were the case, then it might be possible to detect the expansion of such cohorts by a detailed analysis of Ag-specific αβ TCR repertoires selected by individual mice during an immune response.
We have previously shown that DBA/2 mice immunized with P815 cells transfected with the human MHC class I gene HLA-CW3 (P815-CW3 cells) display a high magnitude CD8 T cell response (hereafter called the CW3 response) focused mainly on a single epitope defined by peptide 170–179 of HLA-CW3 (20–22). TCRs expressed by CW3-specific CTL clones display structural similarities that facilitate direct and quantitative repertoire analysis by single cell PCR (23–26). In this study, we ask whether different CW3-specific αβ TCRs can arise from the same ancestral TCR VDJ-β rearrangement. By using an efficient single cell RT-PCR analysis to characterize extensively the αβ TCR repertoires of individual mice, we identify clusters of T cell clones that all express TCR-β chains encoded by the same nucleotide sequence but express different TCR-α chains. Because some of these might represent unrelated clones that by chance express identical VDJ-β rearrangement sequences, we sought a direct way to assess clonal relatedness. At the TCR-β loci of both chromosomes, a mature T cell may have up to three different DJ-β rearrangements in addition to its functional VDJ-β rearrangement. Because of their sequence diversity, we decided to use DJ-β rearrangements as clonal markers. We set up a sensitive single cell PCR protocol to amplify and sequence the TCR VDJ-β and VJ-α rearrangements, together with potential DJ-β rearrangements. With this approach we now demonstrate that a given VDJ-β rearrangement may indeed give rise to different half-sibling CW3-specific T cell clones in which the common, ancestral TCR-β chain is paired with distinct but structurally similar TCR-α chains.
Materials and Methods
Construction of H-2Kd-CW3 Peptide Multimers.
H-2Kd peptide monomers were generated as previously described (27, 28). In brief, recombinant β2-microglobulin and H-2Kd heavy chain containing the BirA recognition sequence in-frame at its C terminus (provided by S. Nathenson, Albert Einstein College of Medicine, Bronx, NY, and J.-P. Abastado, Centre de Recherches Biomedicales des Cordeliers, Paris, France, respectively) were produced as inclusion bodies in BL21(DE3)pLysS bacteria, dissolved in 8 M urea, and refolded by dilution with 10 μM CW3 peptide 170–179 (RYLKNGKETL). After biotinylation using BirA (Avidity), the monomeric pMHC complexes were purified by anion exchange chromatography. Multimerization was performed by reaction with PE-labeled extravidin (Sigma-Aldrich) at a 4:1 molar ratio.
Immunization, Staining, and Cell Sorting.
8–10-wk-old female DBA/2 mice (Iffa Credo) were injected intraperitoneally with 20 million viable P815-CW3 transfectant cells maintained as ascites in Swiss nu/nu mice (Iffa Credo) as previously described (22). 2 wk later mice (M) were killed and lymphocytes from blood (M-2, M-3, and M-33) or spleen (M-41, M-42, and M-43) were isolated as previously described (22). Cells from M-2 and M-3 were stained with antibodies specific for Vβ10, CD62L, and CD8 as previously described (24). Cells from M-33, M-41, M-42, and M-43 were first incubated with pCW3Kd multimers for 30 min at room temperature and then washed twice. Staining was continued on ice by a 10-min preincubation with anti-CD32/16 (anti-Fc receptor, clone 2.4G2) followed by incubation with anti–TCRβV10-FITC (B21.5; BD Biosciences) and anti–CD8α-biotin (53-6.7) prepared in our laboratory. After washing, cells were incubated with streptavidin CyChrome (BD Biosciences) to reveal the biotinylated antibody. Cells gated as either Vβ10+CD62L−CD8+ or pCW3Kd+Vβ10+CD8+ were sorted as single cells using the automatic cell deposition unit of a FACStarPlus™ (for M-2, M-3, and M-33) or FACSVantage-SE (for M-41, M-42, and M-43; Becton Dickinson).
Primers.
Reverse primers 3′ of the AJ35 gene segment were designed using the DNA sequence of the productive TCR-α locus rearrangement of CTL clone CW3/1.1 (these sequence data are available from GenBank/EMBL/DDBJ under accession no. X67432; reference 23). Because the TCR-α and -β loci for the DBA/2 strain of mice have not been completely sequenced, DNA sequences for known members of the Vα3, Vα4, and Vα8 families (29) and for the BD1 (D1) to BJ2 (J2) region of the TCR-β locus (these sequence data are available from GenBank/EMBL/DDBJ under accession no. AE000665) were used. Primers were designed using the Primer3 program, available at http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi (30). Potential primer problems and incompatibilities were assessed with the Amplify 1.2 program, available at http://engels.genetics.wisc.edu/amplify/. Oligonucleotides used as primers for PCR and sequencing reactions (Fig. 1) were purchased from Eurogentec.
Figure 1.

Sequences of oligonucleotide primers. Forward (A) and reverse (B) primers are named for gene segments with a laboratory code in parentheses. Primers used for the first or second rounds of RT-PCR or DNA-PCR or for sequencing (Seq) are indicated. Optional (opt) primers were used to obtain shorter PCR products for sequencing or for confirmation.
Single Cell RT-PCR Protocol.
Our RT-PCR protocol was adapted from that of Correia-Neves et al. (19). Vβ10+ CD62L−CD8+ or pCW3Kd+Vβ10+CD8+ cells were sorted as single cells into 10 μl cDNA reaction mixture in individual 0.2-ml PCR microtubes (Dominique Dutscher). The cDNA reaction mixture contained 90 U M-MLV reverse transcriptase (GIBCO BRL) with recommended 1× RT buffer, 2% Triton X-100, 1 μg BSA (GIBCO BRL), 500 μM dNTP mix (Roche), 50 ng Oligo pd (T)12–18 (Amersham Biosciences), and 8 U RNasin (Promega). cDNA synthesis was performed by over-night incubation at 37°C, after which the microtubes were stored at −20°C until further use. The entire 10-μl cDNA reaction was used for the first PCR reaction in a final volume of 50 μl containing 1 U Taq polymerase in the manufacturer's 1× reaction buffer (Roche), 2.85 mM MgCl2 (Roche), 200 μM of each dNTP (Promega), and 100 nM of each of the primers specific for Vβ10, Cβ, Vα3, Vα4, Vα8, or Cα (Fig. 1). The first PCR program begins at 95°C for 2 min, continues with 35 cycles of 10 s at 95°C, 45 s at 59°C, and 45 s at 72°C, and then ends with 5 min at 72°C. A 0.5-μl aliquot of the first PCR reaction was used for each second PCR in a final volume of 50 μl containing 0.5 U Taq polymerase with the recommended 1× reaction buffer (Roche), 1.75 mM MgCl2 (Roche), 200 μM of each dNTP (Promega), and 100 nM of each primer. Separate second PCR reactions were performed with Vβ10 and Cβ primers or with pairs of Vα3, Vα4, or Vα8 and Cα primers (Fig. 1). The second PCR program begins at 95°C for 2 min then 72°C for 5 s followed by 35 cycles of 10 s at 95°C, 60 s at 61°C, and 30 s at 72°C, and then ends with 5 min at 72°C.
Single Cell DNA-PCR Protocol.
Single cell DNA-PCR was performed as previously described (24, 26), using the primers shown in Fig. 1 and with the following modifications. Cells gated as pCW3Kd+Vβ10+CD8+ were sorted as single cells into tubes containing 20 μl 1× PCR buffer (Roche) and 4 μg/ml 16S rRNA (Roche). Tubes were frozen immediately at −80°C and then stored at −20°C. After proteinase K digestion (24), a first PCR reaction that amplifies rearrangements of Vβ10, D1, or D2 gene segments to any J1 or J2 segment, or those of Vα3, Vα4, or Vα8 genes to Jα35 was performed. Two separate second PCR reactions using nested primers were performed on all samples to amplify either Vβ10 to Jβ rearrangements or Vα3, Vα4, or Vα8 to Jα35 rearrangements. For selected cells of interest we performed three additional, separate second PCR reactions with D1-J1, D1-J2, or D2-J2 combinations of nested primers. In some cases, additional second PCR reactions were performed using optional primers (Fig. 1, opt) to obtain shorter PCR products for sequencing. All putative unrearranged, germline (GL) D2-J2 PCR products were confirmed by performing an additional second PCR using the DB2(432) primer and a nested primer, JB2(923), located in the intron between the D2 and BJ2S1 (Jβ2.1) gene segments.
Precautions Against PCR Contamination.
For at least every eight tubes amplified in the first step PCR, we amplified an additional control tube without sorted cells but otherwise prepared and treated identically to the sorted samples. These “blanks” were reamplified in the second PCR step. The same number of additional “negative” control tubes, containing only the second PCR reaction mixture, were amplified during the second step PCR. None of these (732 total) control tubes led to a PCR product, however, one series of amplified cells was eliminated due to obvious contamination in that experiment.
Identification and Sequencing of PCR Products.
Cells with successful amplifications were identified by migration of 7 μl of the second PCR reaction on 2% agarose gel (Eurobio). The RT-PCR products were purified using PCR purification columns (QIAGEN) according to the manufacturer's instructions. Sequencing of the purified RT-PCR products was performed in 20 μl reaction mixture of 10 μl purified DNA, 0.8 μM specific primer, and using 6 μl ready reaction of dye terminator cycle DNA sequencing kit (Applied Biosystems). Sequences were analyzed on an ABI 373 A DNA sequencer (PerkinElmer). The DNA-PCR products were sequenced directly in 20 μl reaction mixture of 1 μl second PCR product diluted to one fifth in sterile water (Aguettant), 8 μM specific primer, and 4 μl BigDye™ Terminator v2.0 cycle sequencing kit (Applied Biosystems). Sequences were analyzed on an ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems).
For convenience, each unique TCR nucleotide sequence is assigned a three-part code. The TCR Vβ10 nucleotide sequence codes are as previously defined (26) and indicate the Jβ segment used followed by a number and letter code to identify the amino acid (aa; number) and nucleotide sequence. Due to the location of the primers, the sequence of the Vα segment was only partially determined. Correspondence with TCR AV3 (Vα3), -4, or -8 family members in the nomenclature of Arden et al. (29), was verified using the ImMunoGeneTics website (available at http://imgt.cines.fr). The letter P is used in the code to designate the preliminary assignment of a subfamily member. As an example, for the TCR-α sequence code 3P5-2a, “3P5” represents at least partial identity to the AV3S5 subfamily and “2a” identifies the aa (2 = SAKGFASAL) and nucleotide (2a = AGC GCG AAG GGC TTT GCA AGT GCG CTG) sequence of the CDR3 region. Sequence data are available from GenBank/EMBL/DDBJ under accession numbers AY177428, AY177429, AY177430, AY177431, AY177432, AY177433, AY177434, AY177435, AY177436, AY177437, AY177438, AY177439, AY177440, AY177441, AY177442, AY177443, AY177444, AY177445, AY177446, AY177447, AY177448, AY177449, AY177450, AY177451, AY177452, AY177453, AY177454, AY177455, AY177456, AY177457, AY177458, AY177459, AY177460, AY177461, AY177462, AY177463, AY177464, AY177465, AY177466, AY177467, AY177468, AY177469, AY177470, AY177471, AY177472, AY177473, AY177474, AY177475, AY177476, AY177477, AY177478, AY177479, AY177480, AY177481, AY177482, AY177483, AY177484, AY177485, AY177486, AY177487, AY177488, AY177489, AY177490, AY177491, AY177492, AY177493, AY177494, AY177495, AY177496, AY177497, AY177498, AY177499, AY177500, AY177501, AY177502, AY177503, AY177504, AY177505, AY177506, AY177507, AY177508, AY177509, AY177510, AY177511 and AY177587, AY177588, AY177589, AY177590, AY177591, AY177592, AY177593, AY177594, AY177595, AY177596, AY177597, AY177598, or from the authors.
Online Supplemental Material.
Supplemental Figs. S1, S2, and S3 display complete data from the single cell RT-PCR analysis of M-2, M-3, and M-33, respectively, and are available at http://www.jem.org/cgi/content/full/jem.20021945/DC1.
Results
Clusters of CW3-specific αβ TCR Clones Expressing β Chains Identical at the Nucleotide Level Paired with Different α Chains.
To characterize directly and relatively easily a large number of CW3-specific αβ TCRs from individual mice, we set up a highly efficient single cell RT-PCR protocol. Because all CW3-specific CTL clones and most CD8 T cells stained with pCW3Kd multimers (23, 31, 32, and unpublished data) express Vβ10 TCRs, and the TCR-α chains of most CW3-specific CTL clones use Vα3, -4, or -8 gene segments (23, 31), we used primers specific for these V gene segments. The amplification efficiency for Vβ10 sequences from sorted Vβ10+CD8+ single cells was >90% when only primers for Vβ10 and Cβ were included in the first PCR (unpublished data). When primers were mixed to amplify both TCR-β and TCR-α sequences, the amplification efficiency ranged from 63.2 to 79.9% for Vβ10 and from 40.3 to 63.2% for TCR-α for the three mice analyzed (Table I). Paired CW3-like (defined below) αβ TCRs could be amplified in approximately one third to one half of the sorted cells (Table I).
Table I.
Efficient Single Cell RT-PCR and DNA-PCR Amplification of CW3-specific αβ TCRs from Individual Mice a
|
|
M-2RT-PCR
|
M-3RT-PCR
|
M-33RT-PCR
|
M-41DNA-PCR
|
M-42DNA-PCR
|
M-43DNA-PCR
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | n | % | |
| Amplification:b | ||||||||||||
| TCR-β+ cells | 91 | 63.2 | 114 | 79.2 | 115 | 79.9 | 70 | 54.7 | 62 | 48.4 | 79 | 41.1 |
| TCR-α1 cells | 76 | 52.8 | 91 | 63.2 | 58 | 40.3 | 85 | 66.4 | 70 | 54.7 | 135 | 70.3 |
| Total cells amplified | 144 | 144 | 144 | 128 | 128 | 192 | ||||||
| Cells with seq's for:c | ||||||||||||
| CW3-TCR-β | 78 | 54.2 | 114 | 79.2 | 115 | 79.9 | nae | na | na | na | na | na |
| CW3-TCR-α | 71 | 49.3 | 90 | 62.5 | 57 | 39.6 | na | na | na | na | na | na |
| CW3-αβ TCR | 48 | 33.3 | 79 | 54.9 | 45 | 31.2 | 46 | 35.9 | 37 | 28.9 | 57 | 29.7 |
| No. of different CW3-TCRs:d | ||||||||||||
| αβ TCRs | 15 | 18 | 10 | 15 | 11 | 16 | ||||||
| Additional TCR-β's | 1 | 3 | 5 | na | na | na | ||||||
| Additional TCR-α's | 3 | 0 | 0 | na | na | na | ||||||
2 wk after immunization with P815-CW3 transfectants, Vβ10+CD8 T cells gated as CD62L− (M-2 and M-3) or pCW3Kd+ (M-33, M-41, M-42, and M-43) were sorted as single cells and TCR sequences were amplified by RT-PCR or DNA-PCR.
The number and percent of cells for which a TCR-β or TCR-α sequence was amplified.
The number and percent of cells for which a CW3-like TCR-β, TCR-α, or paired αβ TCR was amplified.
The number of different CW3-like αβ TCRs and the number of additional CW3-like TCR-β or TCR-α sequences found without a partner.
Not applicable because we only determined sequences for cells from which both a TCR-α and TCR-β PCR product was detected.
We had previously shown that Vβ10+CD62L−CD8+ T cells from DBA/2 mice immunized with P815-CW3 transfectants were highly enriched for CW3-specific T cells (22, 24, 25, 33). Most of the TCR-β and TCR-α sequences (Fig. 2 and Figs. S1–S3, available at http://www.jem.org/cgi/content/full/jem.20021945/DC1) that we identified by single cell RT-PCR from cells sorted either as Vβ10+ CD62L−CD8+ (M-2 and M-3) or as pCW3Kd+Vβ10+ CD8+ (M-33) displayed canonical “CW3-like” features previously identified for the CW3-specific TCRs expressed by CTL clones (23). For the 175 cells from which we amplified both a Vβ10 and an in-frame TCR-α sequence, nearly all (172 or 98.3%) express CW3-like αβ TCRs. In such paired αβ TCRs, most (166 out of 172) TCR-β chains display a 6aa length CDR3 region with an SXGXXX motif, although exceptions in sequence (SFSSDY) or length (7aa) presumably corresponding to rare CW3-specific TCRs were also found. Most (171 out of 173) in-frame TCR-α sequences that were identified in cells with CW3-like TCR-β sequences had a CDR3 length of 9aa ending with the motif “GFASAL” encoded by the Jα35 segment, as had those found in CW3-specific CTL clones. One exception was a sequence (nucleotide sequence = 8P29–009a: SDRGLASAL) with an apparent single base substitution in the Jα region that transformed phenylalanine (F) into leucine (L). It is likely that the other noncanonical in-frame TCR-α sequence, as well as six out of frame sequences (Figs. S1–S3, available at http://www.jem.org/cgi/content/full/jem.20021945/DC1), represent secondary TCR-α rearrangements because these are frequently found in T cells (34).
Figure 2.
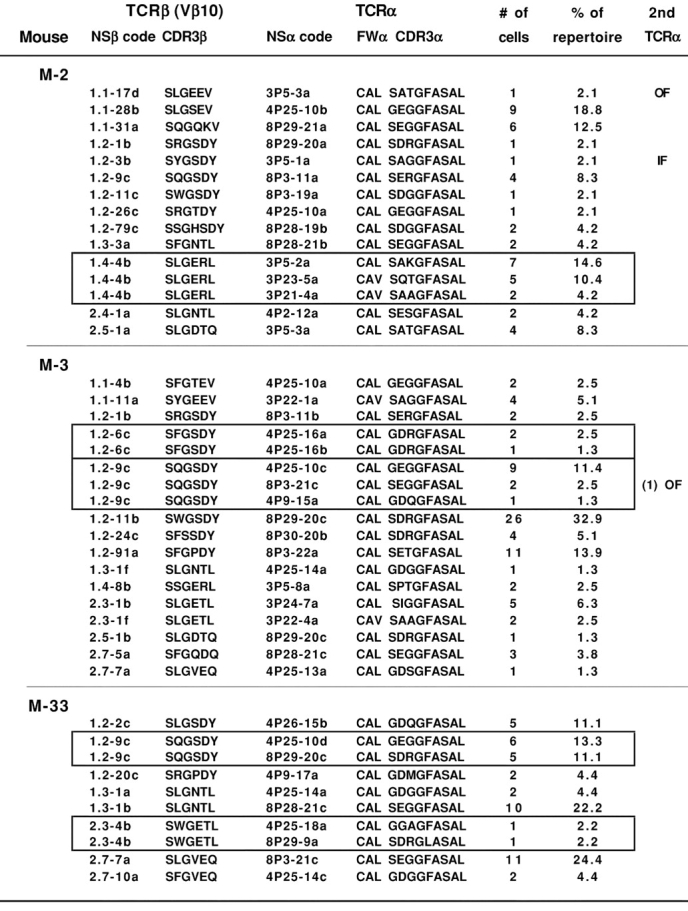
Single cell RT-PCR analysis reveals clusters of CW3-specific clones expressing β chains encoded by identical VDJ-β nucleotide sequences paired with different TCR-α chains. The cDNA from tubes containing single CW3-specific CD8 T cells sorted from PBL of M-2 and M-3 or M-33 was subjected to RT-PCR, and PCR products were sequenced and assigned a TCR sequence code (NS). The complete analysis is shown in Figs. S1, S2, and S3, available at http://www.jem.org/cgi/content/full/jem.20021945/DC1. Represented here are those cells for which both a CW3-like Vβ10 sequence and a CW3-like Vα3, Vα4, or Vα8 sequence were amplified. The deduced aa sequences of the TCR junctions are shown. All TCR-α sequences incorporate the Jα35 sequence. Also shown are the number (#) of cells found for each αβ TCR clone and its corresponding percentage (%) within the αβ TCR repertoire defined here for each mouse. Each cluster of αβ TCR clones sharing an identical Vβ10 TCR nucleotide sequence is framed. Cells for which an in-frame (IF) or out of frame (OF) second TCR-α rearrangement was also amplified are indicated.
A total of 43 different CW3-specific αβ TCRs were identified, distributed as 10–18 clones per mouse (Table I). Clonal frequencies varied widely, with as few as three clones accounting for more than half of the CW3-specific CD8 T cells of each mouse (Fig. 2 and Figs. S1–S3, available at http://www.jem.org/cgi/content/full/jem.20021945/DC1), confirming our previous conclusion based on a partial analysis of the TCR-β repertoire (25). The number of clones detected is likely to be an underestimate of the total number of CW3-specific clones because many αβ TCRs were found in only one or two cells. Several additional clones probably use the “orphan” CW3-like TCR-α or TCR-β sequences that were not found together with a CW3-like partner.
Within individual animals, we could identify one or two clusters of clones that shared a common TCR VDJ-β nucleotide sequence but expressed different CW3-like TCR-α chains (Fig. 2). Among the 12 clones defining these clusters, 8 were found as multiple (2–9) cell copies. The TCR-α and TCR-β sequences of these clones were confirmed by a repeat second PCR amplification and sequencing. None of the TCR-α sequences found in a cluster of clones was detected in other clones from the same animal, making it unlikely that they represent contaminants. It is noteworthy that we found up to three CW3-specific TCR-α sequences as well as an out of frame TCR-α sequence per cluster. Because T cells have only two TCR-α loci, these patterns cannot be explained by the PCR amplification of only one or the other of two different CW3-specific TCR-α sequences from each cell.
DJ-β Rearrangements as Clonal Markers for αβ T Cells.
Our strategy for demonstrating a common origin for CW3-specific clones sharing only the TCR-β sequence was to search for identical residual DJ-β rearrangements as clonal markers. This required setting up a new single cell DNA-PCR protocol for the efficient, simultaneous amplification of multiple rearrangements including (a) the Vβ10 gene to a BJ1 or BJ2 gene, (b) the Vα3, -4, or -8 genes to the AJ35 gene, (c) the D1 gene to a J1 or J2 gene, and (d) the D2 gene to a J2 gene as well as the amplification of sequences corresponding to unrearranged (GL) D-J loci. For maximum specificity and sensitivity, we used a two-step PCR with nested primers for each of the separate second PCR amplifications. New primers were designed for the Vα3, -4, and -8 genes using published sequences as were reverse primers located 3′ of the AJ35 gene segment, forward primers located 5′ of the D1 and D2 genes, and reverse primers 3′ of the BJ1S7 and BJ2S7 genes. In control experiments using the complete mixture of primers, we succeeded in amplifying six or seven D1 to J1 or D2 to J2 GL sequences, respectively, from eight tubes of sorted single P815 cells (unpublished data). Because these GL sequences represent the longest targets to be amplified in the first PCR, we believe our PCR efficiency must be fairly high.
The Vβ10+pCW3Kd+ CD8 T cells from three mice immunized 2 wk previously with P815-CW3 transfectants were sorted for single cell PCR. The efficiency of amplification for the Vβ10 TCR ranged from 41.1 to 54.7% for the three mice, slightly lower than we achieved by single cell RT-PCR (Table I), but not unexpected because only one target DNA sequence of each TCR rearrangement is available per cell during the first amplification. The amplification of TCR-α sequences, however, was as good or slightly better than RT-PCR, perhaps due to the use of nested Vα primers.
Because our goal was to search for new clusters of clones sharing a VDJ-β sequence but expressing different CW3-specific VJ-α rearrangements, we mixed primers to amplify all rearrangements in the first PCR, but initially performed second PCR reactions only to amplify VDJ-β and VJ-α rearrangements. We then sequenced only those PCR products amplified from cells in which both (a TCR-α and a TCR-β) rearrangements were amplified to establish αβ TCR repertoires for the three mice. This identified two clusters of clones, one sharing the Vβ10-Jβ1.2–2i sequence in M-42 and the other sharing Vβ10-Jβ1.4–4a in M-43 (Fig. 3) . The first PCR reaction products from the cells forming these clusters were then subjected to three separate second PCR reactions to amplify potential D1-J1, D1-J2, or D2-J2 rearrangements or unrearranged D-J GL sequences.
Figure 3.
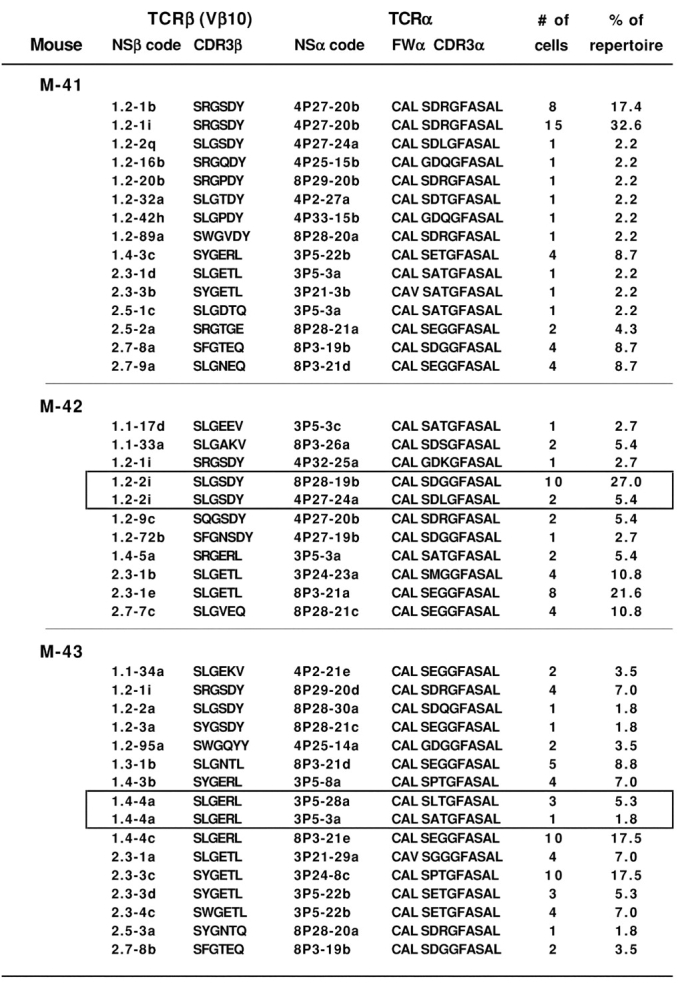
Repertoire analysis by single cell DNA-PCR for coamplification of CW3-specific TCR VDJ-β and VJ-α sequences together with DJ-β rearrangements as potential clonal markers. For each of three mice (M-41, M-42, and M-43), single pCW3Kd+Vβ10+CD8+ splenocytes were sorted 2 wk after immunization with P815-CW3 tumor cells. The rearranged TCR-α and TCR-β nucleotide sequences were amplified by single cell DNA-PCR. PCR products from cells with successful amplifications for TCR-α and TCR-β rearrangements were sequenced to identify paired αβ TCRs. Other details are as described for Fig. 2.
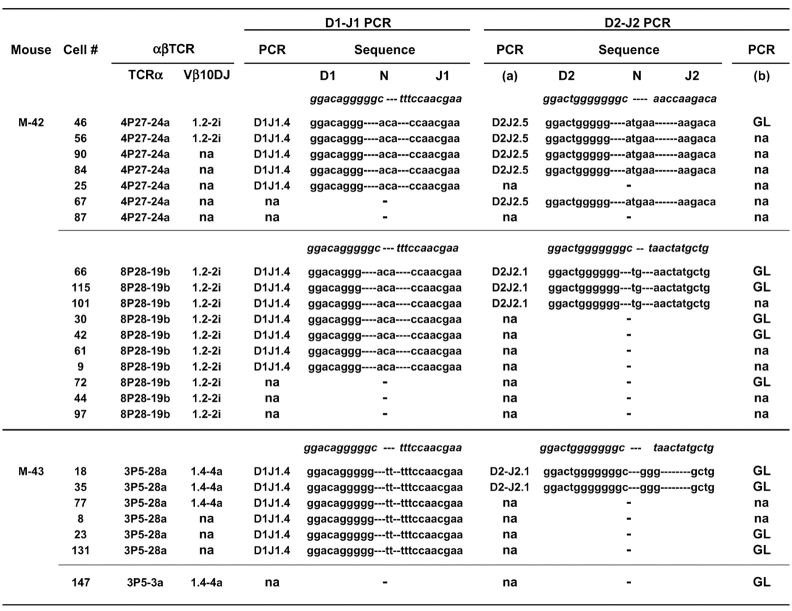
The two clones from the cluster expressing the Vβ10-Jβ1.2–2i TCR-β sequence expressed either a Vα4 (4P27-24a) or Vα8 (8P28-19b) CW3-specific TCR-α (Fig. 3). As shown in Fig. 4 , an identical DJ-β1.4 nucleotide sequence was amplified from cells of both clones. Because only two cells (nos. 46 and 56) expressing the Vα4 TCR had been identified for this cluster, we searched for other cells from the same mouse for which we had amplified an orphan Vα4 TCR without its Vβ10 partner, and for which we could amplify DJ PCR products. Three out of the five additional cells that expressed an identical Vα4 (4P27-24a) TCR were also found to express the identical DJ-β1.4 rearrangement indicating they do indeed belong to the same clone (Fig. 4). For this cluster, two different D2J2 rearrangements were identified, including a D2Jβ2.1 rearrangement found only in cells with the Vα8 TCR and a D2Jβ2.5 rearrangement found only in cells with the Vα4 TCR (Fig. 4). GL sequences corresponding to unrearranged D2-J2 loci were also amplified for cells from both clones in the cluster, thus accounting for all of the four Jβ loci in each of the two clones.
Figure 4.
Two clones expressing CW3-specific αβ TCRs encoded by an identical VDJ-β but different VαJα sequences share an identical Dβ1-Jβ1.4 rearrangement. Individual cells (identified by #) from M-42 or M-43 were amplified by DNA-PCR as described in Fig. 3 to identify their CW3-specific αβ TCRs and their potential DJ-β rearrangements. Shown for each cell are the codes for the TCR-α and TCR-β sequences, the amplified DJ rearrangements together with a code to indicate D and J gene segment usage, and the unrearranged D2-J2 GL sequences. Where indicated, there was no detectable PCR amplification (na) using the indicated primer combinations. No Dβ1 to Jβ2 rearrangements were found for any of these cells. Sequences in italics shown above each series represent the 3′ or 5′ ends of the relevant genomic D or J sequences, respectively. DJ nucleotide sequences are separated to indicate the D, N-nucleotide, and J portions.
For the cluster identified in M-43, two clones expressing the same Vβ10 sequence (1.4-4a) were found to express different Vα3 TCRs (Figs. 3 and 4). We searched for and found additional cells expressing one of these Vα3 (3P5-28a) TCRs in samples from which we were unable to amplify the partner TCR-β sequences. All six cells with this TCR-α sequence expressed an identical DJ-β1.4 rearrangement. We also amplified identical DJ-β2.1 rearrangements or D2-J2 GL sequences from two or four of the cells, respectively. The sequences of these DJ rearrangements were clearly different from those found previously in the M-42 clones (Fig. 4). For the cell that expressed the other Vα3 (3P5-3a) TCR, we were only able to amplify a D2-J2 GL sequence. In the absence of additional cells from this clone to analyze for DJ rearrangements, we are unable to determine whether the two clones in this cluster are related.
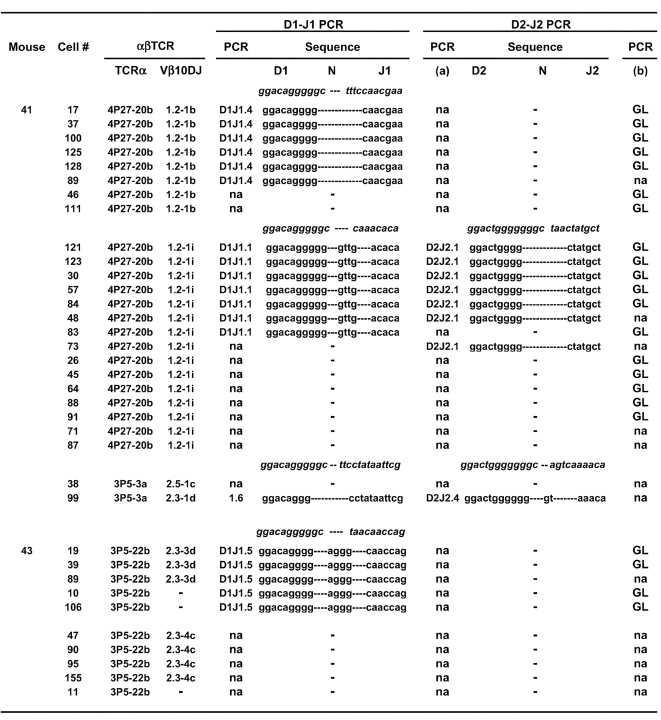
We occasionally found groups of clones that expressed different CW3-specific TCR-β rearrangements but appeared to share the same TCR-α rearrangement. However, because we only amplified and sequenced part of the Vα segment, we cannot be certain that the same subfamily members were used. Interestingly, for one of these groups that appeared to express identical CW3-specific TCR-α's (4P27-20b), the two TCR-β sequences differ only by one nucleotide in the VDJ junctional region. We amplified the DJ rearrangements from these cells to look for clonal markers that might support somatic mutation as their origin. Although no such evidence was found, the data demonstrate further the reproducibility of our single cell PCR protocol for finding identical DJ rearrangements among cells defined by the expression of identical αβ TCRs (Fig. 5) . In this case, for one clone (with Vβ10-1.2-1b), we amplified a D1Jβ1.4 rearrangement and a D2-J2 GL sequence and for the other (with Vβ10-1.2-1i) we amplified D1Jβ1.1 and D2Jβ2.1 rearrangements and a D2-J2 GL sequence. All DJ sequences were clearly different from those found previously in this study (Figs. 4 and 5), confirming their potential diversity and usefulness as markers. DJ rearrangements involving the Jβ1.6, 1.5, or 2.4 genes were found in clones from groups apparently sharing the TCR-α sequences 3P5-3a or 3P5-22b (Fig. 5) but unfortunately, we could not amplify any DJ rearrangements or GL sequences from the other clones of these groups (Fig. 5). Interestingly, however, for several cells from two of the clones, we could account for all 4 Jβ loci. In the first (cell no. 99 of M-41), Vβ10 is rearranged to Jβ2.3, meaning that the DJ-β1.6 and DJ-β2.4 rearrangements must be present on the other chromosome. Likewise, in the second (cell nos. 19 and 39 of M-43), the Vβ10 gene is rearranged to the Jβ2.3 gene, so its DJ-β1.5 rearrangement and D2-J2 GL sequence must be on the second chromosome.
Figure 5.
Analysis of DJ-β rearrangements for clones that appear to share CW3-specific TCR-α chains encoded by identical nucleotide sequences. Note that the V gene portions were only partially sequenced, so identity of the TCR-α rearrangements with the same nucleotide sequence code might be only partial. The analysis and presentation of the data is the same as that described for Fig. 4. No Dβ1 to Jβ2 rearrangements were found for any of these cells.
A Striking Correlation between Vα Family and Jβ Gene Segment Usage Among CW3-specific TCRs.
As might be expected from our earlier work (23, 31), a high proportion (42.4%) of the Vβ10 rearrangements identified herein used the Jβ1.2 gene segment (Table II). However, from this large collection of 85 different CW3-specific αβ TCRs, it is evident that Jβ1.2 usage is not uniform, but is highly skewed (35 out of 36) toward TCR-β's pairing with α chains of the Vα4 or Vα8 families (Table II). In contrast, 9 out of 10 TCRs with β chains using Jβ1.4 are paired with TCR-α chains using Vα3. Biases for Jβ2.3 and Jβ2.7 toward Vα3 and Vα8, respectively, are also apparent. The correlation between Jβ segment and Vα family usage is apparent not only when the 85 αβ TCRs are considered individually but also when the relative clonal frequency of each is taken into account (Table II).
Table II.
Correlation between Jβ and Vα Usage among CW3-specific TCRs
| Gene segment usage in theCW3-specific αβ TCR repertoirea | ||||
|---|---|---|---|---|
| Vα3 | Vα4 | Vα8 | All Vα's | |
| Jβ 1.1 | 1.6 (3) | 4.1 (3) | 3.0 (2) | 8.7 (8) |
| Jβ 1.2 | 0.3 (1) | 21.0 (19) | 21.1 (16) | 42.4 (36) |
| Jβ 1.3 | 0 | 1.0 (2) | 5.9 (3) | 6.9 (5) |
| Jβ 1.4 | 10.0 (9) | 0 | 3.0 (1) | 13.0 (10) |
| Jβ 2.3 | 10.1 (9) | 0.4 (1) | 4.0 (2) | 14.5 (12) |
| Jβ 2.4 | 0 | 0.7 (1) | 0 | 0.7 (1) |
| Jβ 2.5 | 1.8 (2) | 0 | 1.2 (3) | 3.0 (5) |
| Jβ 2.7 | 0 | 1.0 (2) | 10.0 (6) | 11.0 (8) |
| All Jβ's | 23.9 (24) | 28.1 (28) | 48.1 (33) | 100 (85) |
The repertoires of CW3-specific αβ TCR clones identified in the study were combined, with the repertoire of each of the six mice weighted equally. The values represent the percent of clones in the combined repertoire that used each combination of Jβ and Vα gene segments. The numbers in parentheses indicate the number of different TCR nucleotide sequences (out of a total of 85) that were identified for each Jβ and Vα gene combination.
Discussion
We demonstrate in this study that individual mice can select groups of Ag-specific T cells in which identical TCR-β chains encoded by the same VDJ-β nucleotide sequence are paired with one of several different TCR-α chains in different T cells. In principle, these may have arisen by independent VDJ-β rearrangement events that by chance generated the same nucleotide sequence. Alternatively, they may represent half-sibling descendants of the same, ancestral VDJ-β rearrangement that express different TCR-α chains due to independent TCR-α locus rearrangements. We were able to establish a common TCR rearrangement lineage for one of the cohorts by amplifying and sequencing DJ-β rearrangements as clonal markers. Systematic searches in individual responders for clusters of T cell clones sharing an ancestral TCR-β rearrangement but expressing different TCR-α chains have not been reported previously. However, an intriguing study of Epstein-Barr virus–specific CTL (35) showed two clones from the same individual that express identical VDJ-β, but different VJ-α nucleotide sequences, suggesting that such a process may also be involved in the generation of other Ag-specific repertoires.
Single cell PCR represents a powerful strategy for not only identifying the αβ TCR sequences but also the DJ rearrangement patterns expressed by large numbers of T cells. Negatives arising from PCR failure can be compensated for by the possibility to analyze the DJ rearrangement status of multiple cells from each αβ TCR–bearing clone, in particular for responses that involve Ag-driven clonal expansion. In this study, we were able to detect at least one DJ rearrangement in 7 out of the 10 αβ TCR clones analyzed, and for 6 of these we could account for all of the 4 BJ loci (Figs. 4 and 5). All of the amplified DJ rearrangements were sequenced and each distinct nucleotide sequence was associated with only one clone. In accordance with the high level of diversity expected, different rearrangements using the same BJ gene segment differed not only in sequence but also in the extent of D or J segment trimming and in the number and sequence of N-nucleotide additions.
As shown at the top of Fig. 6 , the development of αβ T cells in the mouse thymus involves the ordered expression of a series of cell surface markers and TCR gene segment rearrangements (for review see references 36–40). Several stages based on the relative levels of CD44 and CD25 expression can be defined for the most immature precursors that are negative for both CD4 and CD8 (DN1 to DN4). DJ-β rearrangements initiate at the DN2 to DN3 transition and occur at multiple loci before the V to DJ rearrangements that take place later in the DN3 stage (41–43). Cells with an in-frame TCR VDJ-β rearrangement that allows expression of a functional β chain together with an invariant, surrogate pre–TCR-α chain (44) as part of an immature pre-TCR complex undergo cell division at the DN3 to DN4 stages (for review see references 45 and 46). Further V to DJ rearrangements are inhibited or allelically excluded by a signal from the pre-TCR (47). TCR-α locus rearrangements occur subsequently, mainly during the CD4+CD8+ (double positive [DP]) stage.
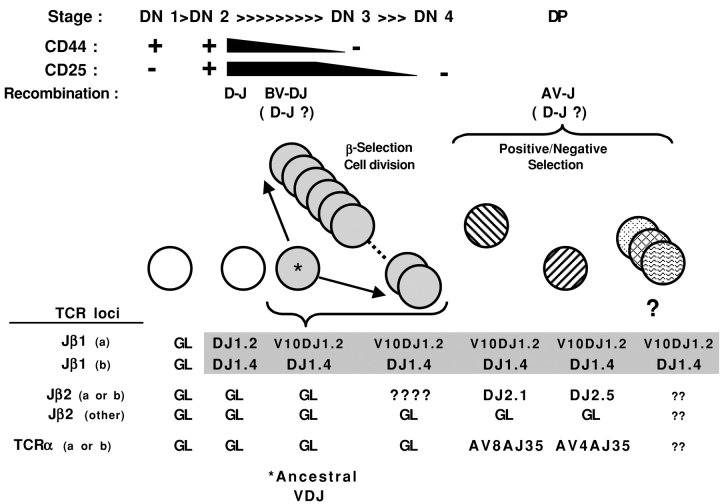
Figure 6.
A model linking two different CW3-specific αβ TCR clones to the same, ancestral VDJ-β precursor. At the top of the figure, T cell development in the thymus is schematized for the CD4−CD8− (DN) to CD4+CD8+ (DP) stages, showing the progressive changes in surface expression of CD44 and CD25 that define the DN1 to DN4 stages. The stages at which different TCR gene rearrangements occur are indicated, with potential stages for continued D to J recombination shown in parentheses. The lower part shows our model for the rearrangements at the different TCR loci on both chromosomes (arbitrarily designated a or b) at different stages of development for the two clones from M-42 that share the Vβ10Dβ1Jβ1.2-2i and Dβ1Jβ1.4 rearrangements shown in Fig 4. The cell making the ancestral VDJ-β rearrangement that is shared by all progeny is indicated with *. Potential (?) additional progeny expressing other TCR-α and/or Dβ2Jβ2 rearrangements are also shown.
The number of residual DJ-β rearrangements that a mature αβ T cell can express and their usefulness as clonal markers for tracing TCR rearrangement lineages depends on several factors. With the exception of the Vβ14 gene segment that is located 3′ of the D-J-C loci and rearranges by inversion, recombination of V gene segments to the J2 locus delete the J1 locus with the intervening DNA. This implies that most cells with VDJ1 or VDJ2 rearrangements can have a maximum of three or two DJ-β rearrangements, respectively. However, some loci may remain in GL configuration and in some cells the second chromosome may also have a VDJ rearrangement although this would appear to be rare (34, 47). Determining the profiles of DJ rearrangements in individual cells during T cell development presents a technical challenge. In a pioneering study using single cell PCR to analyze VDJ and DJ rearrangements in double negative thymocytes (47), Aifantis et al. mention that rearrangements to the BJ1 locus must be excluded from analysis because they might be present on DNA excision loops at this stage. At the population level, it seems that many BJ loci have already rearranged by the DN3 to DN4 stages (48), making it likely that most cells should have at least one additional DJ-β rearrangement already in place before β selection and cell division. Due to the organization of the TCR-β locus, new DJ rearrangements do not delete previous ones and allelic exclusion preventing further V to DJ rearrangement should preserve DJ rearrangements from loss at a later stage.
We propose the following TCR rearrangement lineage for the M-42 cluster of CW3-specific T cells sharing the Vβ10-1.2–2i sequence (Fig. 6). Two DJ rearrangements, DJ-β1.2 and DJ-β1.4, occurred before the recombination of the Vβ10 gene segment with the DJ-β1.2 sequence. Cell division at the DN3 to DN4 stages produced multiple copies of cells sharing both the ancestral Vβ10-1.2–2i rearrangement and the DJ-β1.4 sequence. At the DP stage, at least two of the progeny rearranged either a Vα4 or a Vα8 gene segment to a Jα35 gene segment and the cells were positively selected into the mature T cell repertoire. These cells, or their descendants, were later recruited into the CW3 response. How did the additional, different DJ-β2.1 or DJ-β2.5 rearrangements arise in the two CW3-specific clones? All of the TCR BJ loci can be accounted for because both clones have rearrangements at both BJ1 loci (Vβ10-1.2–2i and DJ-β1.4) and at one BJ2 locus (either DJ-β2.1 or DJ-β2.5), and the second BJ2 locus is in GL configuration (Fig. 4). This suggests that the DJ-β2.1 and DJ-β2.5 rearrangements occurred at a stage subsequent to the first cell division of a precursor carrying both the Vβ10-1.2–2i and DJ-β1.4 rearrangements. Further DJ rearrangement during this phase of cell division would presumably be deleterious due to chromosome breakage, unless the process occurs rapidly enough or at a narrow window between cycles (48). Alternatively, the additional DJ rearrangements may have occurred later in the DN4 or DP stages, coincident with Vα to Jα rearrangement. In support of the latter, Whitehurst et al. (49) reported that Dβ-Jβ signal ends and signal joints could be detected among DP cells, implying that allelic exclusion at the TCR-β locus applies mainly to V to DJ rather than D to J rearrangements. It seems probable that many of the other clusters identified in the first part of this study (Fig. 2) were also derived from ancestral VDJ-β rearrangements. However, their analysis by RT-PCR precludes the amplification of DJ-β rearrangements, and a more extensive study will be required to address the frequency of ancestral versus independent origins of such clusters.
In a number of responses against viral (50–52), tumor (53), or foreign protein (54) epitopes in mouse or man, identical VDJ-β nucleotide sequences can be found in TCR repertoires selected by different individuals. This implies that some sequences are selected more frequently than others not only at the protein level, but also due to a bias in the recombination or coding end processing during rearrangement. If so, they might also be expected to occur more frequently within an individual. In some cases, these frequently found sequences lack N-nucleotide additions, suggesting they might be preferentially established early in ontogeny in the absence of terminal deoxynucleotidyl transferase activity (55, 56). In this context, the Vβ10-1.2-9c sequence lacking N-nucleotides was found in four out of six mice in this study (Figs. 2 and 3). Moreover, this sequence identified clusters of clones that expressed different TCR-α chains in both M-3 and M-33. Because T cells of the latter mice were sorted for RT-PCR, we could not amplify their DJ-β sequences to determine whether they arose from independent or ancestral VDJ-β rearrangements. However, the two mechanisms are not mutually exclusive and the progeny from multiple independently derived, identical VDJ-β rearrangements might also select structurally similar TCR-α chains, further expanding the potential repertoire for a given foreign pMHC ligand.
Previous hints for processes leading to the selection of highly restricted Ag-specific TCR repertoires come from key experiments with TCR-β Tg mice, which compared with normal mice, display biased Vα gene segment usage or restricted combinatorial Vα/Jα usage and CDR3 region diversity (16–19). For TCR-β Tg mice in which the diversity of selecting pMHC ligands is artificially constrained, the TCR-α diversity of mature T cells is even further restricted (16, 57). The impact of ligand selection events on individual cohorts of immature T cells expressing the same VDJ-β rearrangement becomes apparent in a clever model of TCR-β Tg mice that carry inactivated TCR-α loci and a Vα-Jα minilocus to reduce TCR-α diversity to a manageable level (58). In this “limited mouse” model, the initial pool of immature T cells expresses more highly diverse α chains than do mature T cells, suggesting a major role for TCR–ligand interactions rather than gene recombination or αβ chain pairing constraints in limiting the TCR-α diversity of mature T cells expressing a given TCR-β chain.
The identity of peptides that function in vivo to positively select TCRs with a particular specificity for a foreign pMHC ligand is difficult to determine, but a recent study by Santori et al. (59) used two independent biologic or bioinformatic approaches to identify naturally occurring peptides that function in assays that mimic thymic selection in vitro. The peptides found were from proteins unrelated to the Ag (pMHC) recognized by TCR studied, but shared structural similarity in TCR-accessible residues. In addressing the relationship between positive selection and TCR bias in a recent review of crystal structures of TCR–pMHC interactions, Rudolph and Wilson (3) suggest that the αβ TCR CDR1 and CDR2 loops may interact mainly with the MHC helices and possibly with the peptide backbone structure. In interactions with self-MHC molecules, Vα displays a more conservative interaction with the pMHC complex than does Vβ, suggesting a potential docking role for Vα (2, 3, 10). Among the 85 distinct CW3-specific TCRs analyzed in this study, we identified an unexpected correlation between the usage of Vα and Jβ gene segments, not only for individual TCRs but also for those within clusters of clones sharing a Vβ10DJ rearrangement. Molecular modeling is currently underway to search for a structural basis for this correlation. It will also be interesting to look for similar correlations in other Ag-specific repertoires, but this may require the analysis of a large collection of paired αβ TCR sequences. TCR gene usage biases in Ag-specific responses are well documented, however, we are unaware of others characterized by such a clear correlation between V genes used in one chain and J element usage in the opposite chain. One interesting possibility is that immature T cells expressing a CW3-like Vβ10-Jβ1.2 TCR-β chain could positively select CW3-like TCR-α chains using either Vα3, -4, or -8, but most of those using Vα3 would be lost by negative selection, possibly via homologous peptides from the mouse MHC (H-2 Kd, Dd, or Ld) molecules as previously postulated by our group (60). Alternatively, a differential docking onto the Kd molecule by a CW3-like Vα3 TCR-α chain rather than one using Vα4 or Vα8 may preclude a paired CW3-like Vβ10-β chain using a Jβ1.2 element from interacting appropriately either with a positively selecting pMHC ligand in the thymus or with the CW3/Kd ligand during the CW3 response.
In this study, we have identified residual DJ-β rearrangements to demonstrate that Ag-selected T cells expressing the same TCR-β chains but different TCR-α chains can be traced back to the same ancestral VDJ-β rearrangement. In normal mice, it is estimated that fewer than 10 progeny of a VDJ-β rearrangement are selected into the mature T cell pool (11, 14). The existence of structurally similar TCR-α chains within such small ancestral VDJ cohorts is intriguing and may reflect a process proposed by others (61–65) involving selection by a common pMHC ligand during receptor editing at the TCR-α locus.
Acknowledgments
J.L. Maryanski is grateful to Drs. A. Wilson and H.T. Petrie for helpful discussions. We thank Dr. M. Correia-Neves for technical advice, and M. Rossi and C. Bella for help with cell sorting.
We gratefully acknowledge financial support from INSERM and La Ligue le Cancer (Rhône). A. Hamrouni was supported by a fellowship from the Ministère de l'Enseignement Supérieur of Tunisia.
The online version of this article contains supplemental material.
Footnotes
Abbreviations used in this paper: aa, amino acid; DP, double positive; GL, germline; pMHC, MHC class I peptide; Tg, transgenic.
References
- 1.Davis, M.M., and P.J. Bjorkman. 1988. T-cell antigen receptor genes and T-cell recognition. Nature. 334:395–402. [DOI] [PubMed] [Google Scholar]
- 2.Garcia, K.C., L. Teyton, and I.A. Wilson. 1999. Structural basis of T cell recognition. Annu. Rev. Immunol. 17:369–397. [DOI] [PubMed] [Google Scholar]
- 3.Rudolph, M.G., and I.A. Wilson. 2002. The specificity of TCR/pMHC interaction. Curr. Opin. Immunol. 14:52–65. [DOI] [PubMed] [Google Scholar]
- 4.Garboczi, D.N., P. Ghosh, U. Utz, Q.R. Fan, W.E. Biddison, and D.C. Wiley. 1996. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 384:134–141. [DOI] [PubMed] [Google Scholar]
- 5.Garcia, K.C., M. Degano, R.L. Stanfield, A. Brunmark, M.R. Jackson, P.A. Peterson, L. Teyton, and I.A. Wilson. 1996. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 274:209–219. [DOI] [PubMed] [Google Scholar]
- 6.Ding, Y.H., K.J. Smith, D.N. Garboczi, U. Utz, W.E. Biddison, and D.C. Wiley. 1998. Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity. 8:403–411. [DOI] [PubMed] [Google Scholar]
- 7.Garcia, K.C., M. Degano, L.R. Pease, M. Huang, P.A. Peterson, L. Teyton, and I.A. Wilson. 1998. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science. 279:1166–1172. [DOI] [PubMed] [Google Scholar]
- 8.Speir, J.A., K.C. Garcia, A. Brunmark, M. Degano, P.A. Peterson, L. Teyton, and I.A. Wilson. 1998. Structural basis of 2C TCR allorecognition of H-2Ld peptide complexes. Immunity. 8:553–562. [DOI] [PubMed] [Google Scholar]
- 9.Reiser, J.B., C. Darnault, A. Guimezanes, C. Gregoire, T. Mosser, A.M. Schmitt-Verhulst, J.C. Fontecilla-Camps, B. Malissen, D. Housset, and G. Mazza. 2000. Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat. Immunol. 1:291–297. [DOI] [PubMed] [Google Scholar]
- 10.Reiser, J.B., C. Gregoire, C. Darnault, T. Mosser, A. Guimezanes, A.M. Schmitt-Verhulst, J.C. Fontecilla-Camps, G. Mazza, B. Malissen, and D. Housset. 2002. A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex. Immunity. 16:345–354. [DOI] [PubMed] [Google Scholar]
- 11.Casrouge, A., E. Beaung, S. Dalle, C. Pannetier, J. Kanellopoulos, and P. Kourilsky. 2000. Size estimate of the alpha beta TCR repertoire of naive mouse splenocytes. J. Immunol. 164:5782–5787. [DOI] [PubMed] [Google Scholar]
- 12.Dudley, E.C., H.T. Petrie, L.M. Shah, M.J. Owen, and A.C. Hayday. 1994. T cell receptor beta chain gene rearrangement and selection during thymocyte development in adult mice. Immunity. 1:83–93. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman, E.S., L. Passoni, T. Crompton, T.M. Leu, D.G. Schatz, A. Koff, M.J. Owen, and A.C. Hayday. 1996. Productive T-cell receptor beta-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 10:948–962. [DOI] [PubMed] [Google Scholar]
- 14.Penit, C., B. Lucas, and F. Vasseur. 1995. Cell expansion and growth arrest phases during the transition from precursor (CD4−8−) to immature (CD4+8+) thymocytes in normal and genetically modified mice. J. Immunol. 154:5103–5113. [PubMed] [Google Scholar]
- 15.Sebzda, E., S. Mariathasan, T. Ohteki, R. Jones, M.F. Bachmann, and P.S. Ohashi. 1999. Selection of the T cell repertoire. Annu. Rev. Immunol. 17:829–874. [DOI] [PubMed] [Google Scholar]
- 16.Sant'Angelo, D.B., P.G. Waterbury, B.E. Cohen, W.D. Martin, L. Van Kaer, A.C. Hayday, and C.A. Janeway, Jr. 1997. The imprint of intrathymic self-peptides on the mature T cell receptor repertoire. Immunity. 7:517–524. [DOI] [PubMed] [Google Scholar]
- 17.Burns, R.P., Jr., K. Natarajan, N.J. LoCascio, D.P. O'Brien, J.A. Kobori, N. Shastri, and R.K. Barth. 1998. Molecular analysis of skewed Tcra-V gene use in T-cell receptor beta-chain transgenic mice. Immunogenetics. 47:107–114. [DOI] [PubMed] [Google Scholar]
- 18.Sant'Angelo, D.B., B. Lucas, P.G. Waterbury, B. Cohen, T. Brabb, J. Goverman, R.N. Germain, and C.A. Janeway, Jr. 1998. A molecular map of T cell development. Immunity. 9:179–186. [DOI] [PubMed] [Google Scholar]
- 19.Correia-Neves, M., C. Waltzinger, J.M. Wurtz, C. Benoist, and D. Mathis. 1999. Amino acids specifying MHC class preference in TCR V alpha 2 regions. J. Immunol. 163:5471–5477. [PubMed] [Google Scholar]
- 20.Maryanski, J.L., R.S. Accolla, and B. Jordan. 1986. H2-restricted recognition of cloned HLA class I gene products expressed in mouse cells. J. Immunol. 136:4340–4347. [PubMed] [Google Scholar]
- 21.MacDonald, H.R., J.L. Casanova, J.L. Maryanski, and J.C. Cerottini. 1993. Oligoclonal expansion of major histocompatibility complex class I-restricted cytolytic T lymphocytes during a primary immune response in vivo: direct monitoring by flow cytometry and polymerase chain reaction. J. Exp. Med. 177:1487–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker, P.R., T. Ohteki, J.A. Lopez, H.R. MacDonald, and J.L. Maryanski. 1995. Distinct phenotypes of antigen-selected CD8 T cells emerge at different stages of an in vivo immune response. J. Immunol. 155:3443–3452. [PubMed] [Google Scholar]
- 23.Casanova, J.L., J.C. Cerottini, M. Matthes, A. Necker, H. Gournier, C. Barra, C. Widmann, H.R. MacDonald, F. Lemonnier, B. Malissen, et al. 1992. H-2–restricted cytolytic T lymphocytes specific for HLA display T cell receptors of limited diversity. J. Exp. Med. 176:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maryanski, J.L., C.V. Jongeneel, P. Bucher, J.L. Casanova, and P.R. Walker. 1996. Single-cell PCR analysis of TCR repertoires selected by antigen in vivo: a high magnitude CD8 response is comprised of very few clones. Immunity. 4:47–55. [DOI] [PubMed] [Google Scholar]
- 25.Maryanski, J.L., V. Attuil, P. Bucher, and P.R. Walker. 1999. A quantitative, single-cell PCR analysis of an antigen-specific TCR repertoire selected during an in vivo CD8 response: direct evidence for a wide range of clone sizes with uniform tissue distribution. Mol. Immunol. 36:745–753. [DOI] [PubMed] [Google Scholar]
- 26.Attuil, V., P. Bucher, M. Rossi, M. Mutin, and J.L. Maryanski. 2000. Comparative T cell receptor repertoire selection by antigen after adoptive transfer: a glimpse at an antigen-specific preimmune repertoire. Proc. Natl. Acad. Sci. USA. 97:8473–8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altman, J.D., P.A. Moss, P.J. Goulder, D.H. Barouch, M.G. McHeyzer-Williams, J.I. Bell, A.J. McMichael, and M.M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science. 274:94–96. [DOI] [PubMed] [Google Scholar]
- 28.Garboczi, D.N., D.T. Hung, and D.C. Wiley. 1992. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. USA. 89:3429–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arden, B., S.P. Clark, D. Kabelitz, and T.W. Mak. 1995. Mouse T-cell receptor variable gene segment families. Immunogenetics. 42:501–530. [DOI] [PubMed] [Google Scholar]
- 30.Rozen, S., and H.J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Bioinformatics Methods and Protocols: Methods in Molecular Biology. S. Misener and S.A. Krawetz, editors. Humana Press, Totowa, NJ. 365–386. [DOI] [PubMed]
- 31.Casanova, J.L., F. Martinon, H. Gournier, C. Barra, C. Pannetier, A. Regnault, P. Kourilsky, J.C. Cerottini, and J.L. Maryanski. 1993. T cell receptor selection by and recognition of two class I major histocompatibility complex-restricted antigenic peptides that differ at a single position. J. Exp. Med. 177:811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bousso, P., J.P. Levraud, P. Kourilsky, and J.P. Abastado. 1999. The composition of a primary T cell response is largely determined by the timing of recruitment of individual T cell clones. J. Exp. Med. 189:1591–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker, P.R., A. Wilson, P. Bucher, and J.L. Maryanski. 1996. Memory TCR repertoires analyzed long-term reflect those selected during the primary response. Int. Immunol. 8:1131–1138. [DOI] [PubMed] [Google Scholar]
- 34.Casanova, J.L., P. Romero, C. Widmann, P. Kourilsky, and J.L. Maryanski. 1991. T cell receptor genes in a series of class I major histocompatibility complex–restricted cytotoxic T lymphocyte clones speqcific for a Plasmodium berghei nonapeptide: implications for T cell allelic exclusio and antigen-specific repertoire. J. Exp. Med. 174:1371–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burrows, S.R., S.L. Silins, D.J. Moss, R. Khanna, I.S. Misko, and V.P. Argaet. 1995. T cell receptor repertoire for a viral epitope in humans is diversified by tolerance to a background major histocompatibility complex antigen. J. Exp. Med. 182:1703–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Godfrey, D.I., and A. Zlotnik. 1993. Control points in early T-cell development. Immunol. Today. 14:547–553. [DOI] [PubMed] [Google Scholar]
- 37.Borst, J., H. Jacobs, and G. Brouns. 1996. Composition and function of T-cell receptor and B-cell receptor complexes on precursor lymphocytes. Curr. Opin. Immunol. 8:181–190. [DOI] [PubMed] [Google Scholar]
- 38.Fehling, H.J., and H. von Boehmer. 1997. Early alpha beta T cell development in the thymus of normal and genetically altered mice. Curr. Opin. Immunol. 9:263–275. [DOI] [PubMed] [Google Scholar]
- 39.Muljo, S.A., and M.S. Schlissel. 2000. Pre-B and pre-T-cell receptors: conservation of strategies in regulating early lymphocyte development. Immunol. Rev. 175:80–93. [PubMed] [Google Scholar]
- 40.Livak, F., and H. Petrie. 2002. Access roads for RAG-ged terrains: control of T cell receptor gene rearrangement at multiple levels. Semin. Immunol. 14:297–309. [DOI] [PubMed] [Google Scholar]
- 41.Capone, M., R.D. Hockett, Jr., and A. Zlotnik. 1998. Kinetics of T cell receptor beta, gamma, and delta rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44(+)CD25(+) pro-T thymocytes. Proc. Natl. Acad. Sci. USA. 95:12522–12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrie, H.T., F. Livak, D. Burtrum, and S. Mazel. 1995. T cell receptor gene recombination patterns and mechanisms: cell death, rescue, and T cell production. J. Exp. Med. 182:121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak, F., M. Tourigny, D.G. Schatz, and H.T. Petrie. 1999. Characterization of TCR gene rearrangements during adult murine T cell development. J. Immunol. 162:2575–2580. [PubMed] [Google Scholar]
- 44.Saint-Ruf, C., K. Ungewiss, M. Groettrup, L. Bruno, H.J. Fehling, and H. von Boehmer. 1994. Analysis and expression of a cloned pre-T cell receptor gene. Science. 266:1208–1212. [DOI] [PubMed] [Google Scholar]
- 45.von Boehmer, H., and H.J. Fehling. 1997. Structure and function of the pre-T cell receptor. Annu. Rev. Immunol. 15:433–452. [DOI] [PubMed] [Google Scholar]
- 46.von Boehmer, H., I. Aifantis, J. Feinberg, O. Lechner, C. Saint-Ruf, U. Walter, J. Buer, and O. Azogui. 1999. Pleiotropic changes controlled by the pre-T-cell receptor. Curr. Opin. Immunol. 11:135–142. [DOI] [PubMed] [Google Scholar]
- 47.Aifantis, I., J. Buer, H. von Boehmer, and O. Azogui. 1997. Essential role of the pre-T cell receptor in allelic exclusion of the T cell receptor beta locus. Immunity. 7:601–607. [DOI] [PubMed] [Google Scholar]
- 48.Tourigny, M.R., S. Mazel, D.B. Burtrum, and H.T. Petrie. 1997. T cell receptor (TCR)-beta gene recombination: dissociation from cell cycle regulation and developmental progression during T cell ontogeny. J. Exp. Med. 185:1549–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitehurst, C.E., S. Chattopadhyay, and J. Chen. 1999. Control of V(D)J recombinational accessibility of the D beta 1 gene segment at the TCR beta locus by a germline promoter. Immunity. 10:313–322. [DOI] [PubMed] [Google Scholar]
- 50.Argaet, V.P., C.W. Schmidt, S.R. Burrows, S.L. Silins, M.G. Kurilla, D.L. Doolan, A. Suhrbier, D.J. Moss, E. Kieff, and T.B. Suclley. 1994. Dominant selection of an invariant T cell antigen receptor in response to persistent infection by Epstein-Barr virus. J. Exp. Med. 180:2335–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lehner, P.J., E.C. Wang, P.A. Moss, S. Williams, K. Platt, S.M. Friedman, J.I. Bell, and L.K. Borysiewicz. 1995. Human HLA-A0201–restricted cytotoxic T lymphocyte recognition of influenza A is dominated by T cells bearing the V beta 17 gene segment. J. Exp. Med. 181:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim, A., L. Trautmann, M.A. Peyrat, C. Couedel, F. Davodeau, F. Romagne, P. Kourilsky, and M. Bonneville. 2000. Frequent contribution of T cell clonotypes with public TCR features to the chronic response against a dominant EBV-derived epitope: application to direct detection of their molecular imprint on the human peripheral T cell repertoire. J. Immunol. 165:2001–2011. [DOI] [PubMed] [Google Scholar]
- 53.Levraud, J.P., C. Pannetier, P. Langlade-Demoyen, V. Brichard, and P. Kourilsky. 1996. Recurrent T cell receptor rearrangements in the cytotoxic T lymphocyte response in vivo against the p815 murine tumor. J. Exp. Med. 183:439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cibotti, R., J.P. Cabaniols, C. Pannetier, C. Delarbre, I. Vergnon, J.M. Kanellopoulos, and P. Kourilsky. 1994. Public and private V beta T cell receptor repertoires against hen egg white lysozyme (HEL) in nontransgenic versus HEL transgenic mice. J. Exp. Med. 180:861–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feeney, A.J. 1991. Junctional sequences of fetal T cell receptor beta chains have few N regions. J. Exp. Med. 174:115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bogue, M., S. Candeias, C. Benoist, and D. Mathis. 1991. A special repertoire of alpha:beta T cells in neonatal mice. EMBO J. 10:3647–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fukui, Y., O. Hashimoto, A. Inayoshi, T. Gyotoku, T. Sano, T. Koga, T. Gushima, and T. Sasazuki. 1998. Highly restricted T cell repertoire shaped by a single major histocompatibility complex–peptide ligand in the presence of a single rearranged T cell receptor beta chain. J. Exp. Med. 188:897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Correia-Neves, M., C. Waltzinger, D. Mathis, and C. Benoist. 2001. The shaping of the T cell repertoire. Immunity. 14:21–32. [DOI] [PubMed] [Google Scholar]
- 59.Santori, F.R., W.C. Kieper, S.M. Brown, Y. Lu, T.A. Neubert, K.L. Johnson, S. Naylor, S. Vukmanovic, K.A. Hogquist, and S.C. Jameson. 2002. Rare, structurally homologous self-peptides promote thymocyte positive selection. Immunity. 17:131–142. [DOI] [PubMed] [Google Scholar]
- 60.Casanova, J.L., and J.L. Maryanski. 1993. Antigen-selected T-cell receptor diversity and self-nonself homology. Immunol. Today. 14:391–394. [DOI] [PubMed] [Google Scholar]
- 61.Petrie, H.T., F. Livak, D.G. Schatz, A. Strasser, I.N. Crispe, and K. Shortman. 1993. Multiple rearrangements in T cell receptor alpha chain genes maximize the production of useful thymocytes. J. Exp. Med. 178:615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fink, P.J. 2000. If at first you don't succeed. Nat. Immunol. 1:271–272. [DOI] [PubMed] [Google Scholar]
- 63.Wang, F., C.Y. Huang, and O. Kanagawa. 1998. Rapid deletion of rearranged T cell antigen receptor (TCR) Valpha-Jalpha segment by secondary rearrangement in the thymus: role of continuous rearrangement of TCR alpha chain gene and positive selection in the T cell repertoire formation. Proc. Natl. Acad. Sci. USA. 95:11834–11839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang, C., and O. Kanagawa. 2001. Ordered and coordinated rearrangement of the TCR alpha locus: role of secondary rearrangement in thymic selection. J. Immunol. 166:2597–2601. [DOI] [PubMed] [Google Scholar]
- 65.McGargill, M.A., J.M. Derbinski, and K.A. Hogquist. 2000. Receptor editing in developing T cells. Nat. Immunol. 1:336–341. [DOI] [PubMed] [Google Scholar]