Abstract
CD4 T cells regulate immune responses that cause chronic graft rejection and graft versus host disease but their target antigens remain virtually unknown. We developed a new method to identify CD4 T cell–stimulating antigens. LacZ-inducible CD4 T cells were used as a probe to detect their cognate peptide/MHC II ligand generated in dendritic cells fed with Escherichia coli expressing a library of target cell genes. The murine H46 locus on chromosome 7 was thus found to encode the interleukin 4–induced IL4i1 gene. The IL4i1 precursor contains the HAFVEAIPELQGHV peptide which is presented by Ab major histocompatibility complex class II molecule via an endogenous pathway in professional antigen presenting cells. Both allelic peptides bind Ab and a single alanine to methionine substitution at p2 defines nonself. These results reveal novel features of H loci that regulate CD4 T cell responses as well as provide a general strategy for identifying elusive antigens that elicit CD4 T cell responses to tumors or self-tissues in autoimmunity.
Keywords: transplantation, antigens, antigen processing, CD4 T cells, GVHD
Introduction
The histocompatibility H loci are polymorphic genes that determine the outcome of transplanted tissue grafts as well as graft versus host disease reactions in both mouse and man (1, 2). Despite the complete match of MHC molecules between the donor and host strains natural polymorphisms within the H loci result in the presentation of novel peptides by the MHC to T cells (3). For example, different H loci located on the male Y chromosome encode peptides that are presented by both MHC I and MHC II molecules and elicit potent CD8 and CD4 responses respectively (4, 5). At least 60 H loci, located largely on autosomal chromosomes are known to exist in mice and a similar number is likely to be present in the human as well (2). Determining the molecular identity of H loci is critical not only because of their clinical relevance but also for understanding the fundamental mechanisms that make the H antigens immunogenic within the context of highly complex mammalian cells.
In recent years several H loci have been identified at the molecular level as the source of naturally processed peptides presented by MHC I molecules to CD8 T cells (6, 7, and references therein). In contrast, despite the appreciation of their critical role in regulating immune responses to tissue grafts and tumors (8, 9), the H loci that encode antigenic peptides presented by MHC II molecules to CD4 T cells remain virtually unknown at the molecular level. For instance it is not known whether the genetically defined H loci that regulate CD4 T cell responses do so because they encode the antigenic peptides or because they directly or indirectly regulate the MHC II antigen processing pathway as seen for one of the CD8 T cell defined human H antigen (10). It is also unclear whether professional APCs acquire the H antigens from exogenous sources as are most antigens presented by MHC II molecules or whether processing of H peptide/MHC II utilizes precursors synthesized endogenously within the professional APCs (11). The recently discovered Y chromosome encoded Dby H antigen was presented by MHC II when professional dendritic cells (DCs)*acquired it from other cells expressing the Dby gene (5). Whether this is a general paradigm for all CD4 T cell–defined H loci is unclear. Finally, to what extent the donor H antigens differ from self to elicit potent CD4 T cell responses remains to be determined.
The relative success in the identification of CD8 T cell–stimulating H loci is due to the availability of robust methods for expression cloning the antigenic precursor genes or for biochemical purification and identification of the processed antigenic peptides (12–14). By contrast, the identification of CD4 T cell–stimulating antigens has remained an extraordinarily difficult task. Unlike the homogenous peptides presented by MHC class I, the peptides presented by MHC class II are heterogeneous with varying N- and COOH termini flanking the core antigenic peptide (11, 15–17). Peptides eluted from MHC II therefore do not elute in a single peak when fractionated by HPLC and are thus difficult to purify. The peptide purification problem is further magnified by the generally low sensitivity of CD4 T cells to exogenous antigens, in the nano to micromolar concentration range. In comparison, CD8 T cells respond to exogenous peptides at femto to picomolar concentrations which makes it simpler to follow the purification of endogenously processed peptides from the target cells.
The expression cloning strategies for identifying CD4 T cell antigens are also challenging. The MHC I presentation pathway operates in almost all cell types, and it is usually sufficient to introduce an antigen encoding cDNA to obtain expression of the peptide/MHC I ligand (18). By contrast, the MHC class II processing pathway operates primarily in professional APCs (11). These APCs are specialized to present antigenic peptide/MHC II complexes from exogenous proteins but endogenously synthesized proteins are usually excluded. Thus, even if antigenic precursors were expressed in the professional APCs, their presentation as peptide/MHC II complexes may not occur. We and others circumvented this problem by targeting antigenic precursors directly into the MHC II processing compartment by expressing them as invariant chain fusion proteins, which significantly improved the efficiency with which peptide/MHC II complexes were generated (19, 20). Nevertheless, the limitations in transfection efficiency have so far precluded the use of this method in professional APCs. The invariant chain/fusion protein strategy has so far been successful only in specially engineered, readily transfectable cell lines allowing the identification of several CD4 T cell–stimulating tumor antigens (21, 22). In an elegant method that combined the expression of target antigen in transfectable cell lines with the use of DCs as professional APCs, the Simpson group recently identified the Y-chromosome encoded Dby gene as the source of peptides presented by the Ab and Ek MHC class II molecules to CD4 T cells (5). This method is presently limited to single candidate genes and has thus far not permitted the screening of complex cDNA libraries.
Here we developed a new method for identifying CD4 T cell stimulating antigens in mammalian cells containing thousands of potential precursors. The p/MHC II ligands were generated in professional APCs fed with pools of recombinant Escherichia coli expressing the target antigens. We took advantage of the high phagocytic and antigen presentation capacity of bone marrow–derived immature DCs to generate peptide/MHC class II complexes from proteins expressed in recombinant E. coli (23). The peptide/MHC II complexes were probed with the exquisitely sensitive lacZ-inducible CD4 T cells to identify the bacteria expressing the recombinant antigen. We used this method to identify the elusive autosomal CD4 T cell–stimulating H loci. We report that the polymorphic H46 locus first discovered more than a decade ago (24), encodes the IL-4–induced IL4i1 gene that is located on chromosome 7. Furthermore, we have defined the antigenic peptide within the IL4i1 gene presented by the Ab MHC class II to CD4 T cells, the antigenic relationship between the donor and host strains as well as the antigen processing pathways used by the APCs.
Materials and Methods
Mice, Cells, and Peptides.
Inbred mice were obtained from The Jackson Laboratory or bred within the OLAC facility at UC Berkeley. All procedures were performed in compliance with the institutional ACUC guidelines. Procedures for immunization, generation and maintenance of T cell lines and β-galactosidase (lacZ)-inducible T cell hybrids have been described (24, 25). Briefly, the H46a-specific CD4 T cell line, 2PB-TH3 was generated by immunizing the congenic B10.129-H46 bH47b (21M) with C57BL/10J (H46 aH47a) splenocytes and was provided by Dr. Derry Roopenian (The Jackson Laboratory, Bar Harbor, ME). The T cells were used to generate the lacZ-inducible CD4 T cell hybrid TH3Z by fusing them with the lacZ-inducible fusion partner, BWZ.36 (25).
Bone marrow cells obtained from the thigh bone of C57BL/6J (B6) or 129 P3/J (129/J mice) were cultured with RPMI 1640 supplemented with 5% fetal bovine serum (Hyclone), 10 ng/ml GM-CSF (Biosource International), 2 mM glutamine, 50 μM 2-mercaptoethanol, 200 U/ml penicillin, 200 μg/ml streptomycin, and 20 μg/ml gentamicin. After 5 d the characteristic immature DCs in the CD11c+, CD86−, and Ab+ low cultures were greater than 65% by FACS® analysis and are referred to as bone marrow–derived immature DCs (BMDCs) and used as APCs. Peritoneal macrophages were elicited by an intraperitoneal injection of aged thioglycolate (Difco). The mice were killed 4 d later, the macrophages harvested by peritoneal lavage with PBS, plated out in dishes in complete RPMI medium, and allowed to adhere at 37°C. After 2 h the nonadherent cells were washed away and the remaining cells were cultured overnight in complete RPMI medium supplemented with 100 U/ml IFN-γ (Genzyme). B cells were obtained from the spleen and purified by MACS® system (Auburn, CA) with CD45R/B220-FITC conjugated mAb (BD Biosciences). Purified B cells were cultured with 1 μg/ml IL-4 (Biosource International) for 6 h. The peptides with the sequences shown in Fig. 5 and Fig. 6 were synthesized, purified by HPLC, and confirmed by mass spectrometry.
Figure 5.
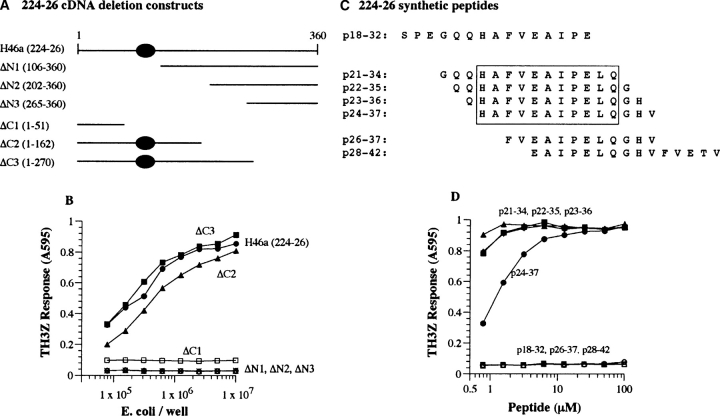
Identification of the TH3Z-stimulating H46 a antigenic epitope within clone 224–26. (A) Schematic representation of the deletion constructs used to identify the antigenic activity encoded within clone 224–26. ΔN1−3 represent deletions that truncate the NH2 terminus of the potential polypeptide while ΔC1−3 cause COOH-terminal deletions. The specific nucleotides included in the constructs are shown in parentheses and the region with antigenic activity is indicated by a shaded oval. (B) The TH3Z T cell lacZ response to the N- and COOH-terminal deletion constructs expressed in E. coli that were fed to 129/J BMDC. (C) Overlapping synthetic peptides used for fine mapping the TH3Z T cell epitope. The numbering refers to the predicted protein sequence of clone 224–26 shown in Fig. 4 A. The core sequence required for antigenic activity is boxed. (D) 129/J BMDCs were incubated for 60 min with the indicated concentration of the synthetic peptides shown in C. The TH3Z T cells were added and their lacZ response measured after an overnight culture.
Figure 6.
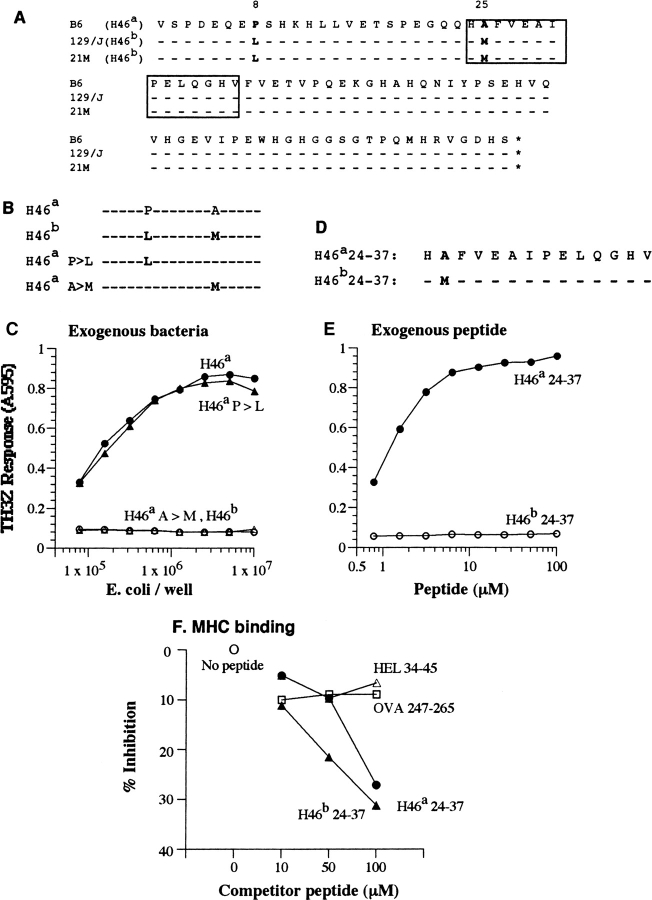
Non-self at the H46 locus is defined by a single amino acid polymorphism. (A) The deduced amino acid sequences of H46 cDNA from the B6 (H46a), the congenic 21M (H46 b), and the parental 129/J (H46 b) mouse strains. The cDNA fragments corresponding to the B6-derived clone 224–26 were obtained by RT-PCR. The H46a antigenic peptide is boxed. (B) Constructs containing the H46a and H46b wild-type as well as single point mutations of each of the two polymorphic residues H46a P>L and H46a A>M subcloned into the bacterial expression vector pLEX. (C) TH3Z lacZ response to 129/J BMDCs fed with bacteria expressing the constructs shown in B. (D) Amino acid sequences of the synthetic peptide representing the H46a and H46b homologues. Bold letters indicate the single amino acid polymorphism between the B6 and 129 strains. (E) TH3Z lacZ response to the H46a and H46b peptides presented by 129/J-derived BMDCs. (F) The relative Ab MHC II binding property of H46a and H46b peptides compared with the Ak MHC II binding peptides HEL34–45 and OVA247–265. The data show the varying concentrations of indicated peptides cause competitive inhibition of the binding of the Eα peptide to the Ab MHC II on Ab-L cells. The Eα/Ab complex was detected with the Yae mAb by flow cytometry as described in Materials and Methods.
T Cell Activation Assays.
LacZ-inducible T cell hybrids were cocultured overnight with APCs from mice, or cells that had ingested transformed bacteria, were transfected with cDNAs, or pulsed with exogenous peptides. In some experiments the APCs were preincubated with anti-H2-Db (B22.249) or anti-I-Ab (M5/114.5.2) mAb for 1 h at 4°C before coculture with T cells. The results shown are the lacZ activity measured as the absorbance at 595 nm (with 635 nm as the reference wavelength) of the chromogenic product released after cleavage of the substrate chlorophenol red β-pyrranoside (25).
cDNA Library and Recombinant Constructs.
PolyA+ mRNA from day 5 BMDCs was used for the construction of the cDNA library using the cDNA synthesis kit from Invitrogen. The cDNA fragments were inserted unidirectionally into the pLEX prokaryotic expression between the EcoRI/NotI sites and used to transform GI724 competent bacteria to yield ∼107 cfu with an average insert size of 0.5–3 kbp. The 5′ forward and 3′ reverse PCR primers for the DNA constructs used here are shown below. All primers included EcoRI and XbaI restriction sites except those used for amplification of IL4i1 gene which contained EcoRI and NotI sites to allow cloning into appropriate expression vectors (Table I).
Table I.
Oligonucleotide Primers
| Construct | Forward (5′–3′) | Reverse (5′–3′) | Figure |
|---|---|---|---|
| ΔN1 | CCGGAATTCCATCACGTGTTCGTGGAGACT | TGTAAAACGACGGCCAGTGC | 5, A and B |
| ΔN2 | CCGGAATTCCATGAGTGGCATGGTCATGGG | TGTAAAACGACGGCCAGTGC | 5, A and B |
| ΔN3 | CCGGAATTCCATTCGCAAAGAGGAAGTGAG | TGTAAAACGACGGCCAGTGC | 5, A and B |
| ΔC1 | GGTGACGCTCTTAAAAATTAAGCC | GACTCTAGACGTTTCCACCAACAAGTG | 5, A and B |
| ΔC2 | GGTGACGCTCTTAAAAATTAAGCC | GACTCTAGAATATATATTCTGGTGGGC | 5, A and B |
| ΔC3 | GGTGACGCTCTTAAAAATTAAGCC | GACTCTAGATTGCGATTAGGAGTGGTC | 5, A and B |
| H46b | CCGGAATTCCATGTCAGCCCAGATGAACAG | GACTCTAGATTGCGATTAGGAGTGGTC | 6, A–C |
| H46b
P>L |
CAGATGAACAGGAGCTCTCTCACAAACAC | GTGTTTGTGAGAGAGCTCCTGTTCATCTG | 6, B and C |
| H46b
A>M |
GAGGGGCAGCAACACATGTTTGTGGAGGCCA | TGGCCTCCACAAACATGTGTTGCTGCCCCTC | 6, B and C |
| IL4i1 | CCGGAATTCATGGCTGGGCTGGCCCTG | ATAGTTTAGCGGCCGCTTGCGATTAGGAGTGGT | 7, B, C, and D |
First strand cDNA from polyA+ mRNA from day 5 BMDC from B6, 129/J, or 21M mice was used to obtain the full-length IL4i1 homologues. PCR was performed using the high fidelity Pfu Turbo polymerase system (Stratagene). The PCR products were purified by agarose gel, sequenced directly with gene-specific primers, and subcloned into pLEX, pcDNA3, or the MSCV IRES GFP retroviral vector which includes the IRES-green fluorescent protein (GFP) to allow detection of transduced cells. The point mutants H46 a P8L and A22M were prepared using the Stratagene QuickChange Site-Directed Mutagenesis Kit.
Expression Cloning.
Bacterial transformants were grown and induced to express the cDNA encoded proteins according to instructions (PL Expression System; Invitrogen). Briefly, 2 × 104 transformants were plated out on drug selection agar plates and incubated overnight at 30°C. The colonies were harvested and resuspended in the culture medium. The transformant density was determined by OD600 (OD 1.0 = 2 × 109 cells/ml) and the bacteria were plated out in 96-well round bottomed plates at 10–20 cfu/well in 100 μl medium. After 24–36 h at 30°C when the cultures had reached a 0.5 OD600, 5 μl of each well was replica plated into a separate 96-well plate containing 100 μl medium/well and cultured for 1–2 h at 30°C to attain log growth. Induction medium containing 10 μg/ml L-tryptophan was then added and the incubation continued for 3 h at 37°C. The number of transformants was again estimated by OD600 and bacteria were transferred to 96-well plate to obtain a final concentration of 5 × 106 cfu/well and were used as the antigen source. The BMDCs from 129/J mice were resuspended in RPMI medium without fetal bovine serum and antibiotics and 105 cells/well were incubated with the induced bacteria for 1 h at 37°C to allow phagocytosis. These plates were centrifuged for 2 min at 1,500 rpm, the supernatant was removed, and 105 TH3Z hybridoma cells in medium containing 100 μg/ml gentamicin (to eliminate residual bacteria) were added to each well. T cell activation was measured as their lacZ response described above.
Ab Competitive Peptide Binding Assay.
The ability of peptide analogs to bind Ab was determined by incubating Ab-L cell transfectants with 10 μM Eα56–73 peptide (ASFEAQGALANIAVDKA) together with varying concentrations of the indicated competitor peptides. After overnight incubation, the cells were stained with Yae mAb specific for the Eα/Ab complex and PE-conjugated goat anti–mouse Ig and analyzed by flow cytometry. The mean fluorescence intensity values were used to calculate the percent inhibition. Both the Eα peptide and the Yae mAb supernatant were gifts of A. Rudensky (University of Washington).
Transfections and Retroviral Transduction.
Simian COS7 (COS) cells were electroporated in 270 mM sucrose, 7 mM sodium phosphate at pH 7.4, and 1 mM MgCl2 with either full-length IL4i1 genes from the B6, 129/J or the 224–26 fragment subcloned in the pcDNA3 vector using the BTX800 electroporator. The electrode had a 1.9-mm gap-prong and was set to deliver 5 99 ms pulses at 700 V. 2 d later the cells were titrated and cocultured with the TH3Z T cells. Antigen-expressing retrovirus supernatants were prepared using 293T as packaging cells. The cells were transiently transfected with B6 or 129/J derived full-length IL4i1 gene in the MIG vector together with pGP-KV plasmid encoded Moloney murine leukemia virus structural genes, gag-pol (26), and VSV-G plasmids (27) by Superfect transfection as described (QIAGEN). After 24 h the supernatants were filtered through a 0.45 μ filter and 4 μg/ml polybrene was added before infection. Bone marrow cultures of 129/J mice plated out in a 24-well plate at 106 cells/well were infected with the retroviral supernatants after 2 d. The plate was centrifuged at 2,500 rpm for 2 h and the medium was replaced (28). After 2 d in culture ∼8–10% CD11c+ cells were found to express the vector encoded GFP marker and used as APCs.
Results and Discussion
CD4+ TH3Z Recognize a Polymorphic Antigen Presented by the Ab MHC II Molecule.
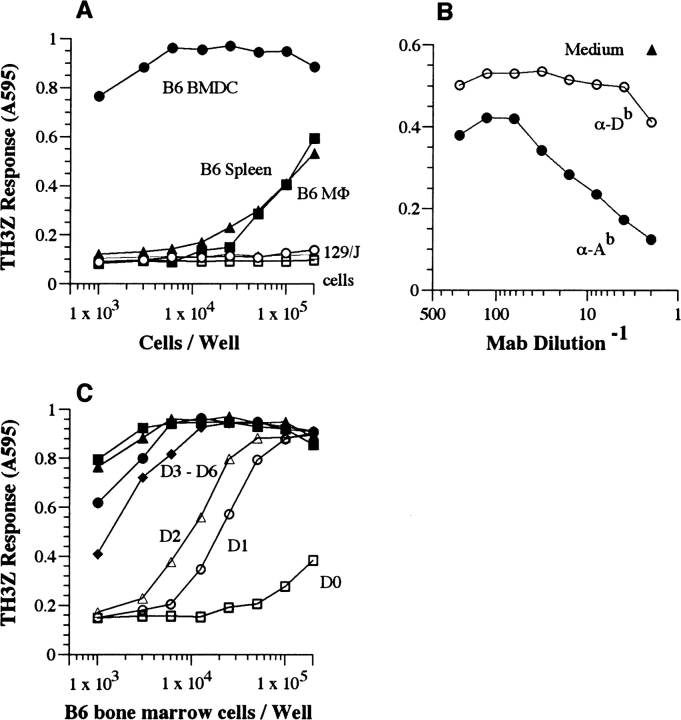
As the first step toward the identification of CD4 T cell antigens, we generated a lacZ inducible CD4+ T cell hybridoma TH3Z (25). We used the CD4 T cell clone 2PB-TH3 that was specific for the mouse H46 antigen based upon its reactivity to a panel of congenic recombinant mice (24). This genetic analysis of the H46 locus had indicated that it was located within or was regulated by a gene within the H4 complex on chromosome 7 of C57BL/6 (B6) mice. In contrast to the H47 locus, located in the same H4 complex that elicited a CD8 T cell response, the closely linked H46 locus elicited a CD4 T cell response when 129/J-derived congenic B10.129/J-H46 b,H47b (21M) mice were immunized with B10 or B6 spleen cells (6, 24). Accordingly, the lacZ activity was stimulated only when the TH3Z T cell hybridoma was cultured with donor B6-derived splenocytes, macrophages, or bone marrow cultures enriched for dendritic cells (BMDCs), but not with the same cell types from the MHC-matched 129/J strain (Fig. 1 A). Furthermore, the TH3Z response required expression of the Ab MHC II molecule because mAb specific for Ab, but not Db MHC class I molecules inhibited its response to the APC (Fig. 1 B). The TH3Z hybridoma was therefore specific for a polymorphic antigen that was presented by the Ab MHC II molecule in the B6 background.
Figure 1.
TH3Z hybridoma recognizes a polymorphic H antigen which is highly expressed in BMDCs and is presented by the Ab MHC II molecule. (A) The indicated number of cells derived from C57BL/6 (B6) or MHC-matched 129/J mice were cocultured with the lacZ-inducible TH3Z hybridoma. (B) 105/well B6 spleen cells were cultured with TH3Z T cells in the presence of the indicated mAb or medium alone. (C) Bone marrow cells were cultured for the indicated days and varying number of cells were used as APCs for stimulating TH3Z T cells. The lacZ response of the TH3Z hybridoma after an overnight incubation was determined after lysis of the cultures and addition of the lacZ substrate CPRG. The absorbance of the cleaved chlorophenol red product at 595 nm is shown for each culture.
Interestingly, BMDCs derived from B6 mice were by far the most potent APCs in comparison to splenocytes or peritoneal macrophages. To identify the optimal source of the antigen we tested the APC function of B6 bone marrow cultures after different days in culture. On a per cell basis the TH3Z stimulating activity in bone marrow cells was low on day 0, but increased dramatically during the next 2 d, reached its peak on day 3, and remained at this level for up to 6 d of culture (Fig. 1 C). The expression of the TH3Z stimulating activity correlated well with a concomitant increase in the CD11c+ cells in the bone marrow cultures strongly suggesting that the antigen gene was expressed in immature DCs (unpublished data). We therefore chose day 5 BMDCs as the optimal source of mRNA to screen for the TH3Z stimulating antigen gene.
Generation of CD4 T Cell–stimulating Peptide/MHC Class II Ligands from Precursors Expressed in Recombinant Bacteria.
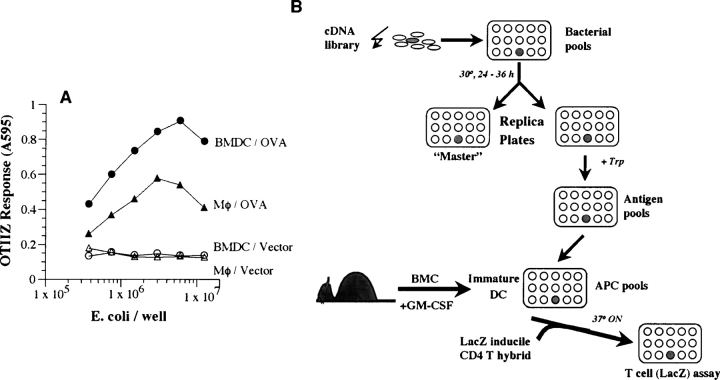
We had previously shown that recombinant E. coli expressing heterologous proteins can serve as an antigen source for generating peptide/MHC class II complexes in activated macrophages (29, 30). The observed efficiency was adequate for screening small prokaryotic genomes such as that of Listeria monocytogenes for antigens recognized by lacZ inducible CD4 T cells. To determine if the efficiency of introducing exogenous proteins into the MHC class II pathway could be further improved we compared peritoneal macrophages and BMDCs as APCs for processing exogenous ovalbumin to OTIIZ T cells. Both macrophages and immature BMDCs generated the OVA peptide/Ab complex from recombinant E. coli expressing OVA (Fig. 2 A). The BMDCs were, however, consistently superior APCs in their requirement for lower number of E. coli to obtain a detectable T cell response as well as in stimulating a higher magnitude of the OTIIZ response. Similar results were also observed with the OVA/Ak and HEL/Ak-specific T cell hybridomas (unpublished data). The approximately threefold increase in efficiency encouraged us to undertake the task of screening cDNA libraries from mammalian cells that contain tens of thousands of different proteins as potential precursors for the antigenic peptide.
Figure 2.
BMDCs are superior to peritoneal macrophages in the presentation of antigen expressed in E. coli. (A) Varying numbers of E. coli expressing OVA or vector alone were fed to 105 peritoneal macrophages (MΦ) or BMDCs. After a brief incubation, OVA/Ab-specific OTIIZ T cells were added and their lacZ response measured after an overnight incubation. (B) A schematic representation of the strategy for expression cloning CD4 T cell–stimulating antigens. A cDNA library from appropriate donor tissue is prepared in a prokaryotic, tryptophan inducible, expression vector. The transformed E. coli are plated in small pools in a 96-well plate. Expression of the cDNA encoded proteins is obtained in a replica plate of the bacterial cultures by inducing with tryptophan (Trp). The bacterial pools containing the putative antigen are fed to host day 5 BMDCs which phagocytose the bacteria, process and present the antigen/MHC II complexes. The presence of these CD4 T cell ligands is detected by the lacZ response of the CD4 T cell hybrid. The individual bacterial clone is then identified by subcloning the bacteria in the replicate well from the “Master” plate and repeating the screen. See Materials and Methods for details.
Based upon these results we devised a strategy schematically outlined in Fig. 2 B to search for CD4 T cell stimulating antigens using recombinant E. coli as the antigen source and BMDCs as recipient APCs. We planned first to construct a cDNA library in a prokaryotic expression vector which would allow expression of the antigenic precursor proteins in transformed bacteria. We chose to use the commercially available pLEX vector because it allows regulated transcriptional control in the presence of tryptophan (Trp) thereby reducing the chance that potentially toxic proteins would be excluded from the bacterial pools. Second, we planned to screen the potential precursor pools by feeding the bacteria to BMDCs and to probe them with the TH3Z cells for the presence of their cognate peptide/Ab ligand. If precursor pools were detected individual bacteria harboring the antigenic cDNA could be identified by cloning and rescreening those in the original well of the master plate.
Isolation of the TH3Z-stimulating Antigen Gene.
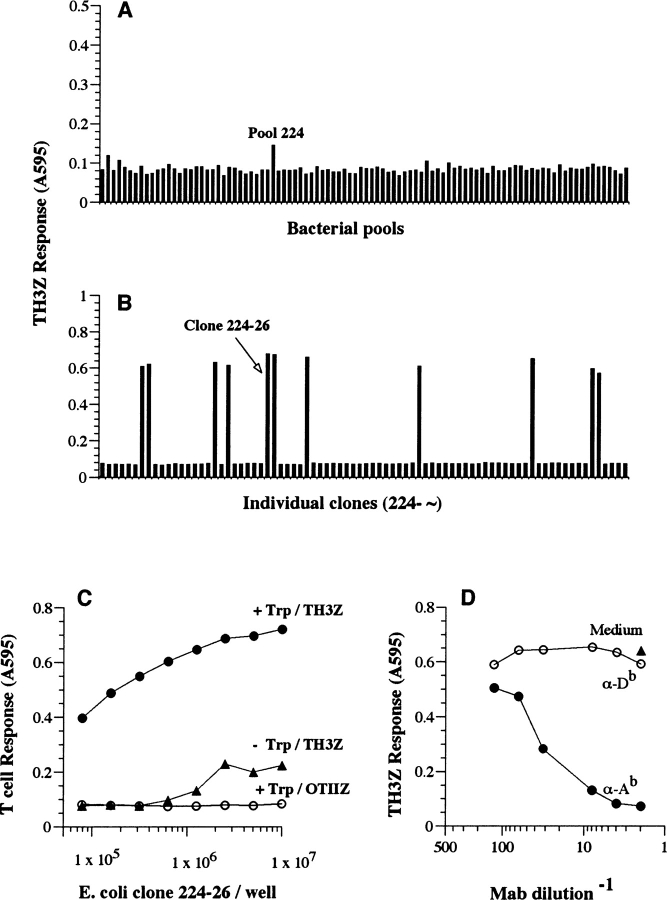
A cDNA library was prepared from polyA+ mRNA isolated from day 5 B6 BMDCs because they were by far the most potent stimulators of TH3Z T cell response (Fig. 1, A and C). The cDNA library was screened in pools of ∼10 cfu/well using immature BMDCs from the host 129/J strain as APCs because they did not express the cognate antigen (Fig. 1 A). The TH3Z response to pool #224, one of the approximately 1,000 different pools screened, showed a response that was above that of the other pools (Fig. 3 A). Pool 224 was fractionated into individual colonies several of which strongly stimulated the TH3Z response in the rescreen (Fig. 3 B). One of these bacterial colonies was designated 224–26 and selected for further analysis. A second clone 296–76 was also identified in the screen which after sequence analysis was found to be identical to clone 224–26.
Figure 3.
Identification of the TH3Z cognate antigen by expression cloning. (A) A prokaryotic cDNA expression library from day 5 BMDCs derived from B6 mice was screened in 129/J-derived BMDCs using the lacZ-inducible TH3Z T cells as described in Fig. 2 B. The lacZ response of TH3Z hybridoma to the bacterial pools in one of the 96-well plates screened is shown indicating the higher than background response to pool 224. (B) TH3Z response to individual bacterial colonies isolated from pool 224. Clone 224–26 was selected for further analysis. (C and D) The TH3Z response to clone 224–26 is antigen-specific and Ab MHC class II restricted. Clone 224–26 bacteria were cultured in medium alone (−Trp) or in medium containing the transcriptional inducer tryptophan (+Trp). Varying number of bacteria were fed to 105 129/J BMDCs/well. 105 TH3Z or the OVA/Ab-specific OTIIZ T cells were added and their lacZ response was assayed after overnight culture. (D) The right panel shows the TH3Z response to 129/J BMDC that were fed 106 clone 224–26 bacteria/well in medium alone or in the presence of mAb specific for the Ab MHC II or the Db MHC I molecules.
The TH3Z response to 224–26 was antigen-specific and restricted by the Ab MHC II molecule. The 224–26 bacteria failed to stimulate the OVA/Ab-specific OTIIZ T cells (Fig. 3 C). The TH3Z response to 224–26 bacteria was enhanced over 100-fold when the bacteria were cultured with tryptophan that activated expression of the insert in the pLEX vector. Furthermore, TH3Z response to 224–26 bacteria was completely inhibited in the presence of anti-Ab MHC II mAb while anti-Db MHC I specific mAb had no effect (Fig. 3 D). We conclude that clone 224–26 allowed the expression of the peptide/Ab complex in the recipient 129/J APC that was recognized by the TH3Z T cells most likely because clone 224–26 encoded the antigenic peptide.
The Murine H46a Locus Encodes the IL4i1 Gene on Chromosome 7.
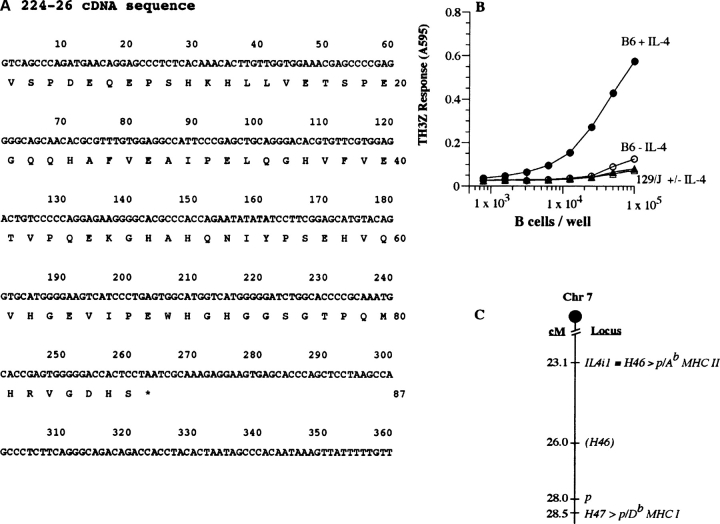
The nucleotide sequence of 224–26 cDNA was determined (Fig. 4 A). A search of the sequence databases at the National Center for Biotechnology Information showed that this sequence was identical to nucleotides 1,680–2,059 encoding the 87 COOH-terminal amino acids of the IL-4–induced IL4i1 gene (accession no. NM_010215). The IL4i1 gene was originally identified as an IL-4–induced transcript in B cells from BALB/c strain (31). In agreement with the identity of the IL4i1 gene in the B6 background analyzed here the Ab expressing BALB.B spleen cells were also capable of stimulating the TH3Z response (unpublished data). Furthermore, the TH3Z response to purified B cells was observed only after the B cells had been cultured in IL-4 (Fig. 4 B). This result was consistent with the notion that the IL-4–stimulated transcription of the IL4i1 in B cells led to the generation of the peptide/Ab complex.
Figure 4.
TH3Z T cell stimulating H46 locus encodes the IL-4–induced IL4i1 gene located on chromosome 7. (A) Nucleotide sequence and the predicted translation of the insert in plasmid DNA of bacterial clone 224–26. The antigenic peptide experimentally identified below is boxed. (B) IL-4–stimulated B cells derived from B6 mice stimulate TH3Z T cells. Purified B cells from donor B6 or 129/J strains were cultured in medium alone or with IL-4 for 6 h. Varying number of these cells were used as APC for TH3Z T cells. Data show the lacZ response of TH3Z cells after overnight culture. (C) The H46 locus is defined by the IL4i1 gene that is presented as a peptide/Ab MHC II complex. The H46 locus is closely linked to the H47 locus that encodes a peptide presented by the Db MHC I molecule and the pink-eyed dilution locus p on murine chromosome 7. The map positions indicated in centi-morgan (cM) are based upon the mouse genome database (http://www.informatics.jax.org/.) and exclude other genes located in this region.
An analysis of the predicted 630 amino acid protein sequence of the full-length IL4i1 gene revealed a number of potential N-linked glycosylation sites, tyrosine phosphorylation sites as well as a flavin cofactor binding domain, suggesting that it is a member of the flavin monoamine oxidase family of enzymes (http://www.ebi.ac.uk/interpro/). The analysis also showed that it contains a hydrophobic ER translocation signal sequence at its NH2 terminus and is therefore predicted to be translocated into the ER. It has been suggested that the IL4i1 gene may be involved in systemic lupus erythematosus because of its genomic location (31, 32). However, the normal function of this protein and its intracellular location have not been experimentally established. In addition, the impact of the natural polymorphisms (see below) in the IL4i1 gene on its function remains to be determined.
The H46 locus had previously been genetically mapped to the H4 complex on chromosome 7 (24). To determine whether the 224–26 cDNA encoding the antigenic precursor mapped to the same chromosomal location or was regulated by another gene located on chromosome 7, we searched the mouse genome database (http://www.informatics.jax.org/.) Most significantly, the IL4i1 gene was located on chromosome 7 within 3 centi-morgan (cM) of the previously suggested location of the H46 locus. Based upon the current map of chromosome 7 both CD4 and CD8 T cell defined H46 and H47 polymorphic loci are located within 5.4 cM and close to the pink-eyed dilute locus p (Fig. 4 C). These data are in complete agreement with the previous genetic analysis and confirm that the H46 locus is physically located on chromosome 7. Note that the close linkage seen here between the CD8 (H47) and CD4 (H46) T cell defined autosomal loci was also seen earlier for the Y-chromosome encoded CD8 (Smcy, Uty) and CD4 (Dby) T cell defined loci (5). Thus, the discovery of the tight linkage between CD4 and CD8 T cell defined loci on the sex as well as autosomal chromosomes is in complete agreement with the original Roopenian hypothesis that the CD8 and CD4 T cell–stimulating H loci must cosegregate (33).
Identification of the Antigenic Peptide Encoded by 224–26 cDNA.
To understand the molecular basis for the antigenicity of the H46 locus we defined the putative antigenic peptide within the 224–26 encoded polypeptide. We generated a series of deletions at the 5′ and 3′ ends of the 224–26 cDNA (Fig. 5 A). The deleted fragments were recloned in the original pLEX vector and recombinant E. coli expressing the truncated polypeptides were tested for antigenic activity after feeding BMDC from 129/J mice (Fig. 5 B). All deletions of 106 or more nucleotides from the 5′ end (ΔN1) or 309 nucleotides from the 3′ end (ΔC1) caused complete loss of antigenic activity. Based upon the full antigenic activity of the ΔC2 and the loss of antigenic activity of ΔN1 and ΔC1 constructs we inferred that the antigenic activity within the 224–26 cDNA was dependent upon the presence of nucleotides 52–106 encoding aa18–36 (Fig. 4 A).
To directly establish the validity of this assignment as well as to determine whether the antigenic activity was contained within the peptide itself or was somehow induced in the recipient APC by the 224–26 polypeptide we tested a panel of overlapping synthetic peptides using 129/J BMDC as APC (Fig. 5, C and D). Peptide 24–37, but not peptides 18–32 or 28–42 stimulated a strong TH3Z response at submicromolar concentrations. The antigenic activity was therefore contained within the 14 amino acids of peptide 24–37. Additional analogs of peptide 24–37 were prepared to define the N- and COOH-terminal boundaries of the antigenic epitope. The antigenic activity of all the analogs with up to three additional amino acids at the NH2 terminus and three fewer amino acids at the COOH terminus (p21–34) was comparable to that of p24–37. From these data we infer that the 11-mer sequence HAFVEAIPELQ (aa 24–34) within the 224–26 cDNA defines the core antigenic epitope that is presented by the Ab MHC II molecule.
Consensus sequence motifs for peptides presented by MHC II have been difficult to define. Nevertheless systematic mutagenesis of single amino acids has suggested that the Ab bound peptides share an aromatic tyrosine or phenylalanine residue at the i anchor position, an uncharged amino acid at the i+5 position, with no positively charged residues at the i+8 position (34). The H46a peptide defined by the sequence HAFVEAIPELQ shares each of these features as well. Assuming that the phenylaline (F) occupies the i position, the i+5 and i+8 residues are uncharged proline (P) and a nonpositively charged glutamine (Q) residues. However, systematic amino acid substitutions are required to confirm the importance of these positions to the Ab binding characteristics of the H46a peptide.
A Single Amino Acid Polymorphism Defines the Antigenicity of H46 Alleles.
To determine the relationship between the donor and host strains that define the polymorphism at the H46 locus we first analyzed BMDC mRNA in the B6, the congenic B10.129/J-H46 b-H47b (21M) and the parental 129/J strains by Northern blot. Transcripts hybridizing with the 224–26 probe were expressed in each strain (unpublished data). To determine if there were any sequence polymorphisms we generated by RT-PCR the 21M and 129/J-derived fragments corresponding to the 224–26 cDNA from B6 mice (Fig. 6 A). The nucleotide sequences of these fragments were identical to that expressed in the B6 strain except for three nucleotide substitutions C23>T, G73>A, and C74>T. These changes caused the Pro8>Leu and Ala25>Met substitutions in the predicted amino acid sequence of the H46 b allele in the parental 129/J as well as its congenic derivative 21M strains.
Both amino acid substitutions were located in the proximity of the H46a antigenic peptide. The P8>L substitution was located 18 residues upstream while the A25>M substitution was contained within the antigenic peptide. To determine which one or both of these substitutions affected the antigenicity of the H46 a (P-A) versus the H46 b allele (L-M), we prepared mutant constructs in the pLEX bacterial expression vector that would yield precursors with only a single (P>L or A>M) substitution (Fig. 6 B). Recombinant E. coli expressing the H46 a, H46 b as well as the mutant constructs were fed to 129/J BMDCs and tested for their ability to stimulate the TH3Z T cells (Fig. 6 C). Clearly the B6-derived H46 a and the mutant H46 a P>L precursors were active in stimulating the TH3Z response while the allelic 129/J-derived H46 b and the H46 a A>M precursors were completely inactive at all concentrations tested. Thus, the antigenicity of the H46 a allele was dependent upon the single A>M substitution within the antigenic peptide and was not affected by the upstream P>L substitution.
To directly establish that the difference in antigenicity was due to the A>M substitution we also synthesized the 14 amino acid H46 b (residues 24–37) homologue of the H46 a antigenic peptide (Fig. 6 D). In an exogenous presentation assay only the H46a peptide was active, and the H46b peptide with a single A>M substitution was completely inactive even at 100-fold higher concentration (Fig. 6 E). Thus, the difference in antigenicity of the two H46 alleles could be directly attributed to the single alanine to methionine amino acid substitution within the core antigenic sequence.
The lack of TH3Z response to the H46 b peptide could have been due to inability of this peptide to bind the Ab MHC II molecule. We compared the two H46 analogs for their ability to compete with the Eα peptide for binding to Ab MHC on the surface of Ab expressing L cells. Differences in the level of the Eα/Ab complex were monitored by flow cytometry using the Yae mAb (35). In contrast to Ak binding HEL34–45 and OVA247–265 peptides which competed poorly, both H46a and H46b peptides were comparable in their ability to compete with the Eα peptide for binding Ab (Fig. 6 F). Thus, the failure of TH3Z T cells to recognize the H46b peptide is likely due to changes in the conformation of the peptide and/or the Ab surface caused by the single Ala>Met substitution which may interfere with TCR recognition, rather than a simple failure to bind Ab MHC. It would be interesting to determine whether the H46b peptide, which is predicted to be presented by DCs in the 129/J strain, is immunogenic in the H46 a B6 strain.
The H46a Peptide/Ab Complex Is Generated via an Endogenous Processing Pathway.
MHC II molecules acquire their peptides primarily from exogenous sources but can also present some endogenously synthesized proteins (11). The IL4i1 gene was originally identified as a transcript induced in IL-4–treated B cells, but is now recognized to be constitutively expressed in many tissues (reference 32; and http://www.ncbi.nlm.nih.gov/UniGene/). The intracellular location of the IL4i1 protein is not known, and the presence of an ER translocation signal raised the possibility that IL4i1 precursor may be secreted by other cells and acquired by professional APC as an exogenous antigen. Alternatively, because DCs appear to constitutively express the IL4i1 gene as judged by Northern blots and direct APC function (Fig. 1, A and C), it is possible that the H46/Ab epitope is generated via an endogenous processing pathway.
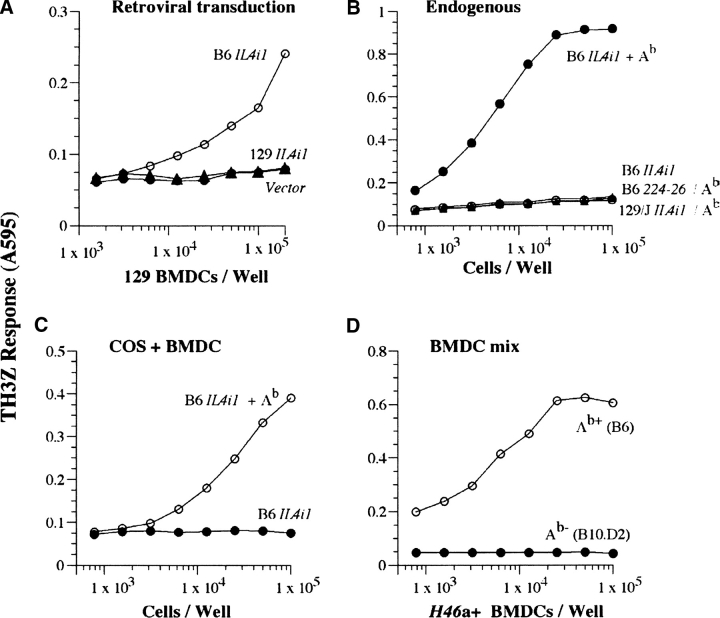
To distinguish between these two possibilities, we first isolated the full-length cDNAs for the IL4i1 gene from the B6 (H46 a) and 129/J (H46 b) strains. Similar to the COOH-terminal fragments represented by the 224–26 cDNA the full-length cDNA could also serve as antigenic precursors for the TH3Z epitope whether they were first expressed in E. coli and fed to 129/J BMDC (unpublished data) or expressed directly in the 129/J DCs via retroviral transduction (Fig. 7 A). Likewise transient transfection of the B6 but not 129/J-derived full-length IL4i1 cDNA together with Abα and Abβ cDNAs in COS cells resulted in the expression of the TH3Z stimulating epitope and was completely dependent upon the presence of the Ab MHC II (Fig. 7 B). Interestingly, the cytoplasmic 224–26 fragment was insufficient for generating the TH3Z epitope in COS cells, suggesting that other features of the full-length protein are required for its entry into and/or processing within the MHC II processing compartments. These features remain to be identified.
Figure 7.
The H46a/Ab complex is generated from the full-length IL4i1 gene via an endogenous presentation pathway. (A) 129/J BMDCs were transduced with retroviruses expressing either vector alone or the B6 or 129/J-derived full-length IL4i1 cDNAs as described in Materials and Methods. 2 d later the transduced cells were titrated and used as APC for TH3Z T cells. About 8–10% cells expressed the retrovirus as indicated by the presence of CD11c+GFP+ cells. (B) COS cells were transiently transfected with the indicated cDNA constructs representing the Ab MHC II subunits Ab α and Ab β together with the B6 or 129/J derived full-length IL4i1 genes or the 224–26 fragment. After 48 h, indicated number of transfected cells were cocultured with TH3Z T cells and their lacZ response was measured after overnight incubation. (C and D) The H46a/Ab epitope is not acquired by BMDC from other cells expressing the precursor gene. COS cells were transfected with the B6-derived full-length IL4i1 cDNA with or without the Ab subunits. 2 d later indicated numbers of transfected COS cells (C) or BMDCs from B6 (H46 a+, Ab+) or B10.D2 (H46 a+, Ab-). (D) were added to 129/J BMDC (H46 b, Ab+). The lacZ response of TH3Z T cells was obtained only when the H46 a precursor and the Ab MHC II is expressed in the same cells.
The processing of the full-length precursor occurred via an endogenous processing pathway. A mixture of 129/J BMDCs and COS cells transiently transfected only with B6-derived IL4i1 cDNA failed to generate the TH3Z ligand while COS cells expressing both the IL4i1 precursor and Ab stimulated TH3Z cells strongly (Fig. 7 D). Similarly, mixture of BMDCs from the H46 a expressing B10.D2, that lack the restricting Ab MHC II and 129/J (H46 b, H2b) derived BMDCs also failed to generate the TH3Z stimulating ligand (Fig. 7 E). In contrast, mixture of H46a and Ab expressing BMDCs from B6 and 129/J mice were strongly stimulatory. Thus, whether the IL4i1 precursor was expressed in COS cells or BMDCs, it could not be used as an exogenous source by host 129/J-derived professional APCs. We therefore conclude that the generation of the H46a/Ab epitope occurred via an endogenous MHC class II processing pathway within the APCs. How the IL4i1 is targeted to the MHC II processing pathway remains to be determined.
In conclusion, we have established a generally applicable method for the identification of CD4 T cell stimulating antigens. This method allowed us to screen large cDNA libraries from the target tissue in professional APCs that were generated from the host strain. Note that the ability to use professional APCs as recipients for the screen is a significant advantage because it obviates the necessity of generating designer recipient cell lines expressing the appropriate MHC II molecule, invariant chain, and DM (21). In fact, not only are the cDNAs of the appropriate MHC II subunits unnecessary even the knowledge of which particular MHC II presents the antigenic peptide is not essential. This could be very important in human applications where multiple heterodimers are formed between the MHC II α and β subunits and it is often difficult to unambiguously identify the MHC II molecules involved. We envisage that this method will now permit identification of CD4 T cell antigens involved in autoimmunity as well as tumors.
The identification of the polymorphic H46 locus and its antigenic peptide presented by the Ab MHC II molecule shows that even a subtle, single amino acid difference between self and nonself can lead to potent CD4 T cell responses (24). The foreign H46a peptide differed only in a single conserved methionine to alanine substitution in the self H46b peptide. Single amino acid substitution have often been observed among CD8 T cell stimulating H peptides and their self-homologues (2, 6, 36), but all the known H epitopes recognized by CD4 T cells have generally differed in several amino acids from their self counterparts. For example, the Y-chromosome encoded Dby gene yields different peptides presented by the Ab, Ek, and HLA-DQ5 MHC II molecules (5, 37). Each of these peptides differ in at least 3 amino acid residues from their counterparts in the X-chromosome encoded Dbx gene, and unlike the H46a precursor, they can be presented by the DCs when expressed in other cells. The distinction between the presentation of H46a versus Dby peptides via endogenous and exogenous processing pathways raises an interesting and now testable hypothesis. Large differences between the tolerated self and foreign nonself may not be required when the processed peptides are generated endogenously within the professional APCs: a distinct advantage of being at the right place all the time.
Acknowledgments
We are grateful to Derry Roopenian, The Jackson Laboratory, for providing the H46a-specific CD4 T cell line. Marcella Fasso, UC Berkeley, William Heath, Walter and Eliza Hall Institute, John Monaco, University of Cincinnati, and Alexander Rudensky, University of Washington for kindly providing us with key reagents and mice. We thank Fred Gonzales, Tom Serwold, Susan Schwab, and Melanie Foster for help and comments.
This research was supported by grants to N. Shastri from the National Institutes of Health.
H. Sahara's present address is Marine Biomedical Institute, Sapporo Medical University School of Medicine, Rishirifuji-cho, Hokkaido 097-0101, Japan.
Footnotes
Abbreviations used in this paper: BMDC, bone marrow–derived immature DC; DC, dendritic cell.
References
- 1.Goulmy, E., R. Schipper, J. Pool, E. Blokland, J.H.F. Falkenburg, J. Vossen, A. Gratwohl, G.B. Vogelsang, H.C. Van Houwelingen, and J.J. Van Rood. 1996. Mismatches of minor histocompatibility antigens between HLA-identical donors and recipients and the development of graft-versus-host disease after bone marrow transplantation. N. Engl. J. Med. 334:281–285. [DOI] [PubMed] [Google Scholar]
- 2.Roopenian, D.C., and E. Simpson. 2000. Minor histocompatibility antigens: from the laboratory to the clinic. Landes Bioscience, Georgetown, TX.
- 3.Wallny, H.J., and H.G. Rammensee. 1990. Identification of classical minor histocompatibility antigen as cell-derived peptide. Nature. 343:275–278. [DOI] [PubMed] [Google Scholar]
- 4.Simpson, E., D. Scott, and P. Chandler. 1997. The male-specific histocompatibility antigen, H-Y: A history of transplantation, immune response genes, sex determination and expression cloning. Annu. Rev. Immunol. 15:39–61. [DOI] [PubMed] [Google Scholar]
- 5.Scott, D., C. Addey, P. Ellis, E. James, M. Mitchell, S. Jurcevic, and E. Simpson. 2000. Dendritic cells permit identification of genes encoding MHC class II-restricted epitopes of transplantation antigens. Immunity. 12:711–720. [DOI] [PubMed] [Google Scholar]
- 6.Mendoza, L., G. Villaflor, P. Eden, D.C. Roopenian, and N. Shastri. 2001. Distinguishing self from nonself. Immunogenicity of the murine H47 locus is determined by a single amino acid substitution in an unusual peptide. J. Immunol. 166:4438–4445. [DOI] [PubMed] [Google Scholar]
- 7.Pierce, R.A., E.D. Field, T. Mutis, T.N. Golovina, C. Von Kap-Herr, M. Wilke, J. Pool, J. Shabanowitz, M.J. Pettenati, L.C. Eisenlohr, et al. 2001. The HA-2 minor histocompatibility antigen is derived from a diallelic gene encoding a novel human class I myosin protein. J. Immunol. 167:3223–3230. [DOI] [PubMed] [Google Scholar]
- 8.Hung, K., R. Hayashi, A. Lafond-Walker, C. Lowenstein, D. Pardoll, and H. Levitsky. 1998. The central role of CD4(+) T cells in the antitumor immune response. J. Exp. Med. 188:2357–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zelenika, D., E. Adams, A. Mellor, E. Simpson, P. Chandler, B. Stockinger, H. Waldmann, and S.P. Cobbold. 1998. Rejection of H-Y disparate skin grafts by monospecific CD4+ Th1 and Th2 cells: no requirement for CD8+ T cells or B cells. J. Immunol. 161:1868–1874. [PubMed] [Google Scholar]
- 10.Brickner, A.G., E.H. Warren, J.A. Caldwell, Y. Akatsuka, T.N. Golovina, A.L. Zarling, J. Shabanowitz, L.C. Eisenlohr, D.F. Hunt, V.H. Engelhard, and S.R. Riddell. 2001. The immunogenicity of a new human minor histocompatibility antigen results from differential antigen processing. J. Exp. Med. 193:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unanue, E.R. 2002. Perspective on antigen processing and presentation. Immunol. Rev. 185:86–102. [DOI] [PubMed] [Google Scholar]
- 12.Scott, D.M., I.E. Ehrmann, P.S. Ellis, C.E. Bishop, A.I. Agulnik, E. Simpson, and M.J. Mitchell. 1995. Identification of a mouse male-specific transplantation antigen, H-Y. Nature. 376:695–698. [DOI] [PubMed] [Google Scholar]
- 13.Wang, W., L.R. Meadows, J.M.M. den Haan, N.E. Sherman, Y. Chen, E. Blokland, J. Shabanowitz, A.I. Agulnik, R.C. Hendrickson, C.E. Bishop, et al. 1995. Human H-Y: A male-specific histocompatibility antigen derived from the SMCY protein. Science. 269:1588–1590. [DOI] [PubMed] [Google Scholar]
- 14.Shastri, N. 1996. Needles in haystacks. Identifying specific peptide antigens for T-cells. Curr. Opin. Immunol. 8:271–277. [DOI] [PubMed] [Google Scholar]
- 15.Rudensky, A.Y., P. Preston-Hurlburt, S.-C. Hong, A. Barlow, and C.A.J. Janeway. 1991. Sequence analysis of peptides bound to MHC class II molecules. Nature. 353:622–627. [DOI] [PubMed] [Google Scholar]
- 16.Engelhard, V.H. 1994. Structure of peptides associated with class I and class II molecules. Annu. Rev. Immunol. 12:181–207. [DOI] [PubMed] [Google Scholar]
- 17.Rammensee, H.G., J. Bachmann, and S. Stevanovic. 1997. MHC ligands and peptide motifs. Landes Bioscience, Austin, TX. 457 pp.
- 18.Shastri, N., S. Schwab, and T. Serwold. 2002. Producing nature's gene-chips. The generation of peptides for display by MHC class I molecules. Annu. Rev. Immunol. 20:463–493. [DOI] [PubMed] [Google Scholar]
- 19.Sanderson, S., K. Frauwirth, and N. Shastri. 1995. Expression of endogenous peptide-major histocompatibility complex class II complexes derived from invariant chain-antigen fusion proteins. Proc. Natl. Acad. Sci. USA. 92:7217–7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodmer, H., S. Viville, C. Benoist, and D. Mathis. 1994. Diversity of endogenous epitopes bound to MHC class II molecules limited by invariant chain. Science. 263:1284–1286. [DOI] [PubMed] [Google Scholar]
- 21.Wang, R.F., X. Wang, A.C. Atwood, S.L. Topalian, and S.A. Rosenberg. 1999. Cloning genes encoding MHC class II-restricted antigens: mutated CDC27 as a tumor antigen. Science 284:1351–1354. [DOI] [PubMed] [Google Scholar]
- 22.Wang, H.Y., J. Zhou, K. Zhu, A.I. Riker, F.M. Marincola, and R.F. Wang. 2002. Identification of a mutated fibronectin as a tumor antigen recognized by CD4+ T cells: its role in extracellular matrix formation and tumor metastasis. J. Exp. Med. 195:1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellman, I., and R.M. Steinman. 2001. Dendritic cells: Specialized and regulated antigen processing machines. Cell. 106:255–258. [DOI] [PubMed] [Google Scholar]
- 24.Davis, A.P., and D.C. Roopenian. 1990. Complexity at the mouse minor histocompatibility locus H-4. Immunogenetics. 31:7–12. [DOI] [PubMed] [Google Scholar]
- 25.Sanderson, S., and N. Shastri. 1994. LacZ inducible peptide/MHC specific T-hybrids. Int. Immunol. 6:369–376. [DOI] [PubMed] [Google Scholar]
- 26.Takahara, Y., K. Hamada, and D.E. Housman. 1992. A new retrovirus packaging cell for gene transfer constructed from amplified long terminal repeat-free chimeric proviral genes. J. Virol. 66:3725–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns, J.C., T. Friedmann, W. Driever, M. Burrascano, and J.K. Yee. 1993. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA. 90:8033–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahnson, A.B., J.T. Dunigan, B.E. Baysal, T. Mohney, R.W. Atchison, M.T. Nimgaonkar, E.D. Ball, and J.A. Barranger. 1995. Centrifugal enhancement of retroviral mediated gene transfer. J. Virol. Methods. 54:131–143. [DOI] [PubMed] [Google Scholar]
- 29.Sanderson, S., D.J. Campbell, and N. Shastri. 1995. Identification of a CD4+ T cell-stimulating antigen of pathogenic bacteria by expression cloning. J. Exp. Med. 182:1751–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell, D., and N. Shastri. 1998. Bacterial surface proteins recognized by CD4+ T cells during murine infection with Listeria monocytogenes. J. Immunol. 161:2339–2347. [PubMed] [Google Scholar]
- 31.Chu, C.C., and W.E. Paul. 1997. Fig1, an interleukin 4-induced mouse B cell gene isolated by cDNA representational difference analysis. Proc. Natl. Acad. Sci. USA. 94:2507–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chavan, S.S., W. Tian, K. Hsueh, D. Jawaheer, P.K. Gregersen, and C.C. Chu. 2002. Characterization of the human homolog of the IL-4 induced gene-1 (Fig1). Biochim. Biophys. Acta. 1576:70–80. [DOI] [PubMed] [Google Scholar]
- 33.Roopenian, D.C. 1992. What are minor histocompatibility loci? A new look at an old question. Immunol. Today. 13:7–10. [DOI] [PubMed] [Google Scholar]
- 34.Wall, K.A., J.Y. Hu, P. Currier, S. Southwood, A. Sette, and A.J. Infante. 1994. A disease-related epitope of Torpedo acetylcholine receptor. Residues involved in I-Ab binding, self-nonself discrimination, and TCR antagonism. J. Immunol. 152:4526–4536. [PubMed] [Google Scholar]
- 35.Rudensky, A., S. Rath, P. Preston-Hurlburt, D.B. Murphy, and C.A. Janeway, Jr. 1991. On the complexity of self. Nature. 353:660–662. [DOI] [PubMed] [Google Scholar]
- 36.Mendoza, L., P. Paz, A.R. Zuberi, G. Christianson, D.C. Roopenian, and N. Shastri. 1997. Minors held by majors. The H13 minor histocompatibility locus defined as a peptide/MHC class I complex. Immunity. 7:461–472. [DOI] [PubMed] [Google Scholar]
- 37.Vogt, M.H., J.W. van den Muijsenberg, E. Goulmy, E. Spierings, P. Kluck, M.G. Kester, R.A. van Soest, J.W. Drijfhout, R. Willemze, and J.H. Falkenburg. 2002. The DBY gene codes for an HLA-DQ5-restricted human male-specific minor histocompatibility antigen involved in graft-versus-host disease. Blood. 99:3027–3032. [DOI] [PubMed] [Google Scholar]