Abstract
Genome-scale in silico metabolic networks of Escherichia coli have been reconstructed. By using a constraint-based in silico model of a reconstructed network, the range of phenotypes exhibited by E. coli under different growth conditions can be computed, and optimal growth phenotypes can be predicted. We hypothesized that the end point of adaptive evolution of E. coli could be accurately described a priori by our in silico model since adaptive evolution should lead to an optimal phenotype. Adaptive evolution of E. coli during prolonged exponential growth was performed with M9 minimal medium supplemented with 2 g of α-ketoglutarate per liter, 2 g of lactate per liter, or 2 g of pyruvate per liter at both 30 and 37°C, which produced seven distinct strains. The growth rates, substrate uptake rates, oxygen uptake rates, by-product secretion patterns, and growth rates on alternative substrates were measured for each strain as a function of evolutionary time. Three major conclusions were drawn from the experimental results. First, adaptive evolution leads to a phenotype characterized by maximized growth rates that may not correspond to the highest biomass yield. Second, metabolic phenotypes resulting from adaptive evolution can be described and predicted computationally. Third, adaptive evolution on a single substrate leads to changes in growth characteristics on other substrates that could signify parallel or opposing growth objectives. Together, the results show that genome-scale in silico metabolic models can describe the end point of adaptive evolution a priori and can be used to gain insight into the adaptive evolutionary process for E. coli.
Biological systems are fundamentally complex, and thus a systems approach is necessary to account for the diversity of interactions that can occur among the myriad of molecular components that comprise living cells (1, 5, 14). The use of genome-scale metabolic reconstructions of an organism may prove to be a valuable tool in attempts to account for biological complexity and to elucidate the genotype-phenotype relationship. The annotation of full microbial genome sequences (2, 7) has enabled reconstruction of whole-cell metabolic networks (5, 15, 19, 29). By using these reconstructed networks, detailed analyses of specific biological functions and system properties have been performed (11, 12, 21, 25, 27, 31). In addition, numerous different in silico approaches have been developed and are available to analyze the properties of metabolic networks (11, 16, 24, 28, 34, 36). While the rationales underlying the various methods are becoming widely accepted, there still has been limited prospective experimental verification of genome-scale in silico models with regard to their abilities to interpret and predict complex biological processes, such as adaptive evolution.
In several studies the workers have productively combined computational and experimental approaches (4, 17, 32, 33). In these studies, the in silico models were constructed and used to analyze specific metabolic subsystems accounting for a relatively small number of metabolic reactions. More recently, a constraint-based in silico model of a genome-scale metabolic reconstruction of Escherichia coli has been used to describe the metabolic phenotypes for growth on several substrates and the end point of adaptive evolution for growth on glycerol (8, 13).
In this study we elaborated on the conclusion reached previously that adaptive evolution drives E. coli to a predicted optimal growth phenotype (13), and we evaluated the overall predictive capabilities of the genome-scale in silico model for adaptive evolution of E. coli by examining adaptive evolution on three different substrates, lactate, pyruvate, and α-ketoglutarate. Additional phenotypic characterization of the evolved strains generated in this study also allowed us to better define some of the underlying biological changes that occurred in these strains during laboratory adaptive evolution.
MATERIALS AND METHODS
Computational methods.
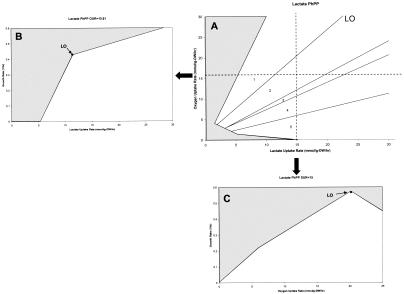
We a developed a genome-scale in silico model of E. coli K-12 including 906 genes and 1,327 reactions based upon the constraint-based approach utilizing genomic annotation, biochemical stoichiometry, physiological data, and thermodynamics extending the model described by Edwards and Palsson (9). For this study, phenotype phase plane (PhPP) analysis (Fig. 1) was applied to our model, whose details have been described previously (10, 26). Briefly, the PhPP is a representation of the constrained solution space (allowable phenotypes for growth in a given environment) for a given organism and can be used to visualize optimal metabolic phenotypes of the organism. In two dimensions, the substrate uptake rate (SUR) and oxygen uptake rate (OUR) comprise the two axes (x and y, respectively). The cellular growth rate (GR) or any other objective of interest can be added as a dependent, computed variable and can be plotted over the PhPP to obtain a three-dimensional surface.
FIG. 1.
Lactate PhPP. The gray regions are regions of metabolic phenotypes that are not feasible. (A) PhPP plot of OUR versus lactate uptake rate. (B) Plot of GR versus lactate uptake rate with a fixed OUR of 15.91 mmol/g (dry weight)/h. (C) Plot of GR versus OUR with a fixed lactate uptake rate of 15 mmol/g (dry weight)/h. DW, dry weight.
The PhPP can be used to represent all the optimal phenotypes that an organism can exhibit for anaerobic and aerobic growth on single carbon substrates. In the PhPP, a computationally determined line of optimality (LO) is found that represents the predicted optimal phenotype for maximizing biomass yield. Growth on the LO thus represents the best possible fully aerobic conversion of the carbon substrate into biomass. Other lines on the PhPP delineate regions of similar metabolic phenotypes, as shown in Fig. 1A (regions 1 to 5). Higher GRs are sometimes possible due to partial anaerobic metabolism of the substrate in addition to the amount that can be fully aerobically metabolized (Fig. 1B). Prior to any experimental work, a PhPP and the LO can be calculated for a particular substrate, and the computational prediction can be compared with subsequent experimental results. Thus, a strain can then be subjected to adaptive evolution, and the changes in growth phenotype properties can be traced in the PhPP (13).
In addition to generating PhPPs to predict and interpret the outcome of the adaptive evolution process, the in silico model was used to calculate and analyze theoretical GRs, uptake rates, and metabolic flux distributions for the results obtained experimentally. Two different parameters, optimal yield and optimal growth, were calculated for the end point phenotype of each evolved strain. By constraining only the OUR, the computational model was allowed to determine optimal values for the SUR and GR to obtain the optimal yield. For the optimal growth calculation, both uptake rates (OUR and SUR) were constrained to the experimental values at the end point of evolution, resulting in a computed GR. For every optimal growth phenotype calculated, a predicted metabolic flux distribution was also determined, which allowed comparisons to be made between growth on the evolutionary carbon source and growth on alternative carbon sources.
Experimental methods.
E. coli K-12 wild-type strain MG1655, obtained from the American Type Culture Collection (Rockville, Md.), was used for all experiments. We started with fresh cultures, and E. coli was grown on lactate, pyruvate, or α-ketoglutarate at 30 and 37°C and subsequently examined to measure growth and uptake rates, analyzed for by-product secretion, and monitored for growth on alternative substrates. The two temperatures selected for each type of culture conditions represent the optimal (37°C) and suboptimal (30°C) temperatures for growth of E. coli. Seven distinct evolved strains were generated in the following conditions: growth on lactate at 37°C (strain L1), growth on lactate at 30°C (strains L2 and L3), growth on pyruvate at 37°C (strain P1), growth on pyruvate at 30°C (strain P2), growth on α-ketoglutarate at 37°C (strain A1), and growth on α-ketoglutarate at 30°C (strain A2).
Evolution of E. coli was conducted in 250 ml of M9 minimal medium supplemented with 2 g of a carbon source per liter in Erlenmeyer flasks by using magnetic stir bars for aeration (13). The cells were grown overnight and allowed to reach the mid-exponential growth phase (A600, ≤ 0.5) before they were diluted by passage in fresh medium. The level of dilution at each passage was adjusted daily to account for changes in the GR. Typically, following dilution the A600 was ≤2.4 × 10−6. This process of batch growth and serial passage was conducted for 45 to 75 days for the various cultures until a stable GR was achieved. This serial passage maintained a state of prolonged exponential growth so that each culture never entered the stationary phase. Throughout the course of evolution, samples of each evolved strain were flash frozen by using liquid nitrogen and stored in a freezer at −80°C.
The evolving cultures were phenotypically characterized at regular time intervals over the course of adaptive evolution to quantitatively determine the phenotypic changes during the adaptive evolutionary process. For each time point examined, precultures were grown overnight and used to inoculate fresh medium for a batch culture. The GR, SUR, and OUR were measured throughout exponential growth for each culture (13). A biomass correlation was also determined for each time point, and medium samples were taken throughout exponential growth and into the stationary phase to monitor metabolic by-product secretion. The GRs were determined by measuring the optical densities of the cultures with a spectrophotometer (A600 and A420). The SUR was determined by monitoring the depletion of the carbon source in filtered medium samples over time by using UV detection by high-performance liquid chromatography (HPLC) or an enzymatic assay. The OUR was determined by measuring dissolved oxygen depletion with a respirometer by using a polarographic dissolved oxygen probe. The biomass correlation was determined by measuring the optical density of a culture and filtering a set volume of the culture onto a preweighed filter which was weighed after it was dried to a constant weight. The by-product secretion patterns were determined by UV detection by HPLC, and samples were taken regularly throughout batch culture testing. All phenotype testing was performed more than once for each time point of evolution tested.
Growth on alternative carbon sources was evaluated by using the Bioscreen C system (Thermo Labsystems, Franklin, Mass.). This system measured the optical densities of up to 200 cultures (by using two 100-well plates) for each experiment in a temperature-controlled environment. For each experiment conducted with the Bioscreen C system, precultures were grown overnight and allowed to reach the mid-exponential growth phase (A600, ≤ 0.5). Portions (2.5 to 15 μl) of these cultures were used to inoculate the multiwell Bioscreen C plates containing 300 μl of medium, which yielded initial A600 of between 0.06 and 0.07. Each experimental run included a blank well containing medium as a negative control and a well with wild-type cells as a positive control. Growth on 10 different carbon sources (acetate, α-ketoglutarate, citrate, glucose, glycerol, lactate, malate, pyruvate, ribose, and succinate) was tested. The plates were incubated and monitored with the Bioscreen C system for 24 h; measurements were taken every 15 min, and there was continuous shaking between measurements. Each evolved strain was tested with the Bioscreen C system at the temperature at which it was evolved. GRs were averaged from replicate cultures and normalized to the wild-type GR.
RESULTS
Phenotype assessment.
Seven distinct evolved strains were generated during this study by using E. coli K-12 wild-type strain MG1655 as the parent. The results obtained from phenotype characterizations of these strains are described below.
(i) Lactate.
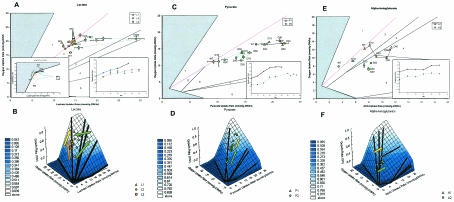
When E. coli K-12 wild-type strain MG1655 was grown on 2 g of lactate per liter, we found that at 30°C it operated in region 2 of the lactate PhPP (Fig. 2A and B), which is a region characterized by partial anaerobic growth and acetate secretion. At 37°C, the wild-type cells functioned in region 1 of the lactate PhPP, a region exhibiting metabolic futile cycles that leads to suboptimal GRs (9). One culture was evolved and examined on lactate at 37°C (strain L1), and two cultures were evolved and examined on lactate at 30°C (strains L2 and L3). At 37°C, the L1 culture was evolved for 45 days, or approximately 870 generations, and it exhibited an 80% increase in GR, from 0.40 to 0.72 h−1. We found that by day 20 of evolution the culture was operating along the LO. The cells continued to operate along the LO from days 20 to 30 of evolution, exhibiting increases in GR, SUR, and OUR. By day 45, the cells had drifted slightly off the LO into region 2 to achieve a higher GR.
FIG. 2.
Adaptive evolution on lactate, pyruvate, and α-ketoglutarate. The data points represent uptake and GR values obtained throughout the course of evolution. The error bars indicate standard deviations for tests of replicate cultures. (A) PhPP plot of OUR versus SUR for evolution on lactate at 37°C (strain L1) and 30°C (strains L2 and L3). The inset on bottom left is a plot of SUR versus GR for a set OUR of 20 mmol/g (dry weight)/h, showing the three lactate evolved strains. The inset on bottom right shows GR changes during evolution. (B) Three-dimensional representation of the GR over the PhPP for all lactate strains. (C) PhPP plot of OUR versus SUR for evolution on pyruvate at 37°C (strain P1) and 30°C (strain P2). The inset shows the GR changes during evolution. (D) Three-dimensional representation of the GR over the PhPP (OUR, SUR, and GR) for pyruvate strains. (E) PhPP plot of OUR versus SUR for evolution on α-ketoglutarate at 37°C (strain A1) and 30°C (strain A2). The inset shows the GR changes during evolution. (F) Three-dimensional representation of the GR over the PhPP (OUR, SUR, and GR) for α-ketoglutarate strains. WT, wild type.
The two cultures evolved on lactate at 30°C (strains L2 and L3) had two distinct evolutionary trajectories, but they exhibited similar growth phenotypes at the end of evolution on day 60 (approximately 950 generations), as shown in Fig. 2A and B. L2 showed a 147% increase in GR from 0.23 to 0.56 h−1, and L3 showed a 132% increase in GR from 0.23 to 0.53 h−1. At the start of evolution, the L2 strain showed large increases in SUR and only a slight increase in OUR. The increased SUR moved the cells away from the LO by day 20 of evolution. After day 20, the SUR of L2 decreased, bringing it to operation on the LO (Fig. 2A, inset). In contrast to the evolutionary trajectory of strain L2, strain L3 showed an increase in OUR at the beginning of evolution with almost no change in SUR. By day 10 of evolution, strain L3 operated close to the LO and remained along the LO throughout the remainder of evolution. L2 and L3 had almost identical growth phenotypes at the end of evolution.
The by-product secretion patterns measured by UV detection with an HPLC for strains L2 and L3 also qualitatively reflected the observed phenotypic differences between the two strains. At day 20, when the two strains were most divergent phenotypically, the by-product secretion patterns were distinctly different, with strain L3 secreting an unidentified by-product that strain L2 did not secrete. After the end of evolution, when the two strains were phenotypically similar, the by-product secretion patterns were also similar (data not shown).
(ii) Pyruvate.
Evolution of E. coli K-12 wild-type strain MG1655 on pyruvate was examined at both 37 and 30°C, as shown in Fig. 2C and D. At both temperatures, the wild-type strain operated in region 2, which is a partially anaerobic growth region characterized by acetate secretion. Cultures were evolved and tested at both 37°C (strain P1) and 30°C (strain P2); P1 was evolved for 60 days or approximately 1,200 generations, and P2 was evolved for 75 days or approximately 1,000 generations. P1 showed a 69% increase in GR from 0.42 to 0.70 h−1, and P2 showed a 115% increase in GR from 0.23 to 0.49 h−1. The P1 strain showed an increase in GR by increasing the SUR without a significant increase in the OUR (possibly due to oxygen limitation) and thus moved away from the LO to a region of faster growth. The P2 strain evolved parallel to the LO by increasing both OUR and SUR for the first 40 days of evolution, moving from fully aerobic growth with the maximal biomass yield to partially anaerobic growth with a higher GR. After day 40, the P2 strain drifted away from the LO into region 2.
(iii) α-Ketoglutarate.
When E. coli K-12 wild-type strain MG1655 was tested on 2 g of α-ketoglutarate per liter, it operated close to the predicted LO when it was tested at both 37°C (strain A1) and 30°C (strain A2), as shown in Fig. 2E and F. The evolved strains of E. coli were characterized phenotypically after every 10 days of evolution to monitor changes in the strains, and each strain was allowed to evolve for a total of 50 days, or approximately 625 generations for A1 and 440 generations for A2. The GR changed from 0.21 to 0.31 h−1 at 30°C (48% increase) and from 0.32 to 0.45 h−1 at 37°C (41% increase), and there were small increases in OUR but large increases in SUR. Adaptive evolution on α-ketoglutarate resulted in changes that moved the cells away from the predicted LO to a region of faster growth with partially anaerobic metabolism, similar to adaptive evolution on pyruvate.
Computations based on in silico model.
At the end of adaptive evolution, the end point phenotype of each strain was compared to computationally derived optimal phenotypes. Two different calculations were made for each strain, optimal yield and optimal growth.
(i) Optimal yield.
The optimal yield for each strain was calculated by using the experimentally measured OUR as a set parameter in the in silico model, thus allowing the model to calculate the predicted optimal GR and corresponding SUR, as illustrated in Fig. 1C. The results are shown in Table 1. For the two strains whose end points operated on the LO, strains L2 and L3, there is good agreement between the experimental values and the computationally determined optimal GR and SUR. All of the other strains (L1, P1, P2, A1, and A2) showed large deviations between the experimental results and the calculated optimal phenotype where the computed GR and SUR were lower than the experimental results.
TABLE 1.
Comparison between the phenotype for the experimental end point of evolution and the computationally calculated optimal phenotype for each evolved straina
| Strain | Experimental end point
|
Calculated optimal yield
|
Calculated optimal GR (h−1) | |||
|---|---|---|---|---|---|---|
| GR (h−1) | SUR (mmol/g [dry wt]/h) | OUR (mmol/g [dry wt]/h) | GR (h−1) | SUR (mmol/g [dry wt]/h) | ||
| L1 | 0.72 | 17.30 ± 0.24 | 20.60 ± 0.74 | 0.57 | 15.17 | 0.60 |
| L2 | 0.56 | 14.88 ± 0.10 | 19.88 ± 0.04 | 0.55 | 14.60 | 0.55 |
| L3 | 0.53 | 14.50 ± 0.40 | 20.25 ± 0.15 | 0.56 | 14.90 | 0.55 |
| P1 | 0.70 | 21.80 ± 0.50 | 16.20 ± 0.40 | 0.44 | 14.19 | 0.52 |
| P2 | 0.49 | 14.70 ± 0.21 | 12.50 ± 0.50 | 0.32 | 10.64 | 0.36 |
| A1 | 0.45 | 11.15 ± 0.15 | 12.00 ± 0.10 | 0.36 | 6.88 | 0.43 |
| A2 | 0.31 | 8.32 ± 0.07 | 8.71 ± 0.03 | 0.24 | 4.73 | 0.30 |
The optimal yield was calculated by setting the OUR. The optimal growth was calculated by setting both the OUR and SUR. The experimental errors are the standard deviations for tests of replicate cultures.
(ii) Optimal growth.
The theoretical optimal growth for the end point of each strain was calculated for each evolved strain by setting both the SUR and OUR to the experimentally measured values and allowing the model to calculate the GR (Table 1). There was good agreement (the differences were within 5%) between the computational and experimental results for strains L2, L3, A1, and A2. Strain L1 experimentally grew approximately 20% faster than the computational prediction (Fig. 2A, inset). Both of the strains evolved on pyruvate, P1 and P2, experimentally grew 35% faster than the computational prediction, suggesting that the in silico model is incomplete in its representation of pyruvate metabolism.
Growth on alternative substrates.
To further assess the characteristics of each of the evolved strains, the GR of each strain was measured on 10 carbon substrates as a function of evolutionary time. The results were expressed as a ratio of the measured GR of the evolved strain to the measured GR of the wild-type strain for each substrate, where unity indicated that the GRs of the evolved strain and the wild-type strain were identical. The graphic results shown in Fig. 3 are limited to tests on 5 of the 10 substrates tested for clarity, and the inset summarizes all of the results obtained. The GRs obtained by using the Bioscreen C system were consistently lower than the GRs measured in stirred 250-ml batch cultures due to the partially anaerobic growth conditions of the Bioscreen C system.
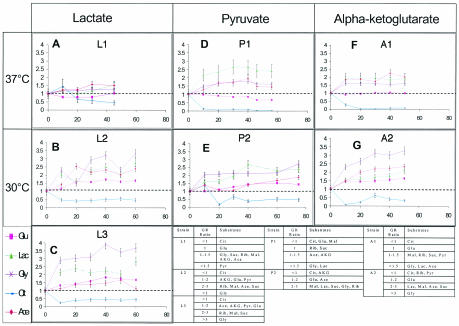
FIG. 3.
Assessment of GRs on alternative carbon substrates. GR changes on alternate substrates for each evolved strain are shown for growth on glucose (Glu), lactate (Lac), glycerol (Gly), citrate (Cit), and acetate (Ace). The normalized growth (ratio of the measured GR to the wild-type GR) is shown as a function of evolutionary time (in days). The dashed lines indicate unity, where the observed GR was identical to the wild-type GR. The results for all alternative carbon substrates tested are also shown. Suc, sucrose; Rib, ribose; Mal, maltose; AKG, α-ketoglutarate; Pyr, pyruvate.
(i) Lactate.
The strains evolved on lactate at different temperatures showed markedly different growth characteristics when they were tested on alternative substrates, as shown in Fig. 3A to C. The L1 strain evolved at 37°C exhibited most of the GR changes within the first 10 days of evolution. Growth on citrate was decreased at the end point of evolution, and the growth on glucose was no different from that of the wild-type strain. Growth on all other substrates was increased, with growth on pyruvate showing almost a 100% increase in the GR compared with the growth of the wild-type strain.
The two strains evolved at 30°C, L2 and L3, produced interesting results. There were noticeable differences in the GR patterns over the course of evolution between these two strains, with large differences occurring on day 20 of evolution, when the two strains were most different phenotypically (Fig. 2A and B). Strain L2 showed a large increase in the GR on acetate and decreases in growth on glycerol and ribose, whereas strain L3 showed no changes for growth on acetate or glycerol and an increase for growth on ribose. By the end of evolution, the two strains showed almost identical GRs on all alternative substrates except acetate. Strain L2 exhibited an ability to grow much faster on acetate (138% increase compared with the wild type) than the L3 strain (13% increase compared with the wild type).
(ii) Pyruvate.
The two strains evolved on pyruvate showed different growth characteristics when they were grown on alternative substrates, as shown in Fig. 3D and E. The P1 strain experienced most of its GR changes within the first 10 days of evolution for growth on alternative substrates. At the end point the P1 strain showed a decreased GR on glucose and malate, a loss of growth on citrate, and no change for growth on ribose and succinate. Growth on acetate, α-ketoglutarate, glycerol, and lactate was improved by evolution on pyruvate, with the GR on lactate increasing more than 100%. The P2 strain showed GR changes on alternative substrates throughout all 75 days of evolution. Growth on citrate and α-ketoglutarate was decreased for the end point of the P2 strain. Growth on all of the other substrates increased, with the GRs on glycerol and ribose increasing more than 150% compared with the wild-type GR.
(iii) α-Ketoglutarate.
Differences between the two strains evolved on α-ketoglutarate were demonstrated by determining GRs on alternative substrates, as shown in Fig. 3F and G. Most GR changes on the alternative substrates for the A1 stain occurred within the first 10 days of evolution. At the end point strain A1 showed a decreased ability to grow on citrate and no change in growth on glucose compared with the wild-type strain. The GRs on malate, ribose, succinate, pyruvate, glycerol, lactate, and acetate all increased compared with the GRs of the wild-type strain, with growth on glycerol, lactate, and acetate improving more than 50%. In contrast to the A1 strain, the A2 strain exhibited GR changes on alternative substrates throughout the course of evolution. At the end point strain A2 showed a decreased GR on citrate, ribose, and pyruvate. Growth on all other substrates increased, with the largest GR increase occurring on glycerol (more than a 200% increase in the GR compared with the GR of the wild-type strain).
Taken together, the GRs of each evolved strain on alternative carbon substrates showed several general trends. Evolution at 37°C stimulated most changes to occur within the first 10 days of evolution, whereas changes occurred throughout evolution for cultures grown at 30°C. Larger increases in GRs were observed for cultures evolved at 30°C than for cultures evolved at 37°C, and evolution at 30°C always induced some change in growth on alternative substrates (either an increase or a decrease in the GR), but evolution at 37°C showed no change in GRs on some of the alternative substrates (glucose, ribose, and succinate). In addition, it was observed that growth on citrate always decreased regardless of what primary substrate E. coli was evolved on, and the increases in the GRs on glycerol and lactate consistently were among the largest increases in GRs for all of the evolved strains.
DISCUSSION
In this study, a genome-scale in silico model of E. coli was used to predict the phenotype at the end point of adaptive evolution on lactate, pyruvate, and α-ketoglutarate, and seven strains of E. coli were evolved and characterized phenotypically. We found that (i) during laboratory adaptive evolution in serially passed batch cultures cells evolve to a region of the highest GR, (ii) the outcome of laboratory adaptive evolution can be predicted and described by using a genome-scale in silico model, and (iii) parallel or opposing physiological growth objectives can be identified by using assessment of the GRs on alternative carbon sources.
The finding that E. coli evolves to a region of greater growth (away from the LO) was based upon the results obtained for all seven evolved strains. Deviation from the LO could have occurred because of inadequacies in the model or because of the selection pressure imposed by the experimental system. While the three strains evolved on lactate moved towards the predicted LO (with the L1 strain eventually moving off the LO), the strains evolved on α-ketoglutarate and pyruvate evolved away from the LO. For five of the seven evolved strains (L1, A1, A2, P1, and P2), the observed experimental growth was faster than the predicted growth based on the optimal biomass yield. Thus, the cells consumed the carbon substrate faster than it could be fully aerobically metabolized and secreted the excess carbon as by-products that could potentially be reconsumed. This phenomenon was observed experimentally during batch culture testing with the secretion and reconsumption of acetate (data not shown) and has been observed previously (18, 35).
The computational results showed that in all cases except strains L2 and L3, the experimental SUR was higher than the calculated SUR that resulted in the maximum biomass yield (Table 1). Thus, strains L1, P1, P2, A1, and A2 overconsumed substrate, resulting in an excess of carbon which had to be secreted as by-products. The secretion of by-products was correctly predicted computationally for the uptake rates measured for these five strains. Taken together, these results indicate that adaptive evolution does not always drive cells to operate on the LO, which represents perfect aerobic metabolism, and that in a batch culture the imposed selection pressure drives cells to the highest GR.
It should be noted that for a subset of growth conditions the region of fastest growth coincides with the LO that defines the maximum biomass yield. In such cases (for example, growth on acetate [8]), GR and biomass yield are simultaneously optimized through adaptive evolution.
The parallel strains evolved on lactate, L2 and L3, produced interesting results due to their convergence toward a common end point of evolution. Strains L2 and L3 were evolved side by side from the same parental strain; however, they took vastly divergent evolutionary paths that began and ended at similar points. Repeated phenotype testing of the two strains along with the by-product secretion patterns and growth characteristics on alternative substrates verified the divergence of the two strains during evolution and the convergence at the end of evolution. Together with the parallel evolution completed on glycerol (13), these results suggest that there are global optimum phenotypes that can be attained through adaptive evolution independent of the starting point or the path taken. There may be multiple ways of reaching the same equivalent end point of evolution.
While the two strains evolved on lactate (L2 and L3) exhibited almost identical growth phenotypes, they clearly were not identical in their underlying metabolic functionalities, as shown in their by-product secretion patterns and growth characteristics on alternative substrates. Cells may be able to utilize their metabolic networks differently to achieve the same external phenotype. The differences between these two strains became apparent when they were grown on acetate minimal medium, on which L2 grew more than twice as fast as L3. Thus, adaptive evolution may create silent phenotypes (3, 22, 30) within a strain, some of which can be probed by characterization of growth on alternative substrates.
Adaptive evolution led to some changes in growth on alternative substrates that were common to all strains. For all of the evolved strains obtained, a decrease in growth on citrate and significant increases in growth on glycerol and lactate were observed. These results suggest that different cellular growth objectives may inhibit or facilitate each other such that growth on one substrate leads to either an increase or a decrease in functionality on another substrate. As many of the changes were observed to occur early in each strain, the changes may have been associated with the initial adaptation to the growth environment, which could later be fixed into the genome through long-term evolution and mutation. Adaptive evolution may lead to an overall increase in functionality of the organism and not just metabolic refinement for the single evolution condition.
We have started to identify mutations that occur during adaptive evolution (23). Once completed, this information not only should allow us to gain a better understanding of the evolutionary process but also should allow us to characterize the genotype-phenotype relationship in the evolved strains.
The development of a genome-scale in silico model of E. coli and testing of this model by using adaptive evolution suggest that E. coli evolves towards a computationally predicted optimal growth phenotype on acetate, succinate, glucose, malate, and glycerol (8, 13). Despite the success of the genome-scale in silico model to predict cellular phenotypes in these cases, the full applicability of this model to diverse biological conditions need to be evaluated to determine what limitations there are to its accuracy and what improvements can be made to the current model. One area of refinement that needs to be made in the model was revealed by the calculations for optimal growth for the two strains evolved on pyruvate, P1 and P2, as shown in Table 1. In both cases, the calculated GR was 35% lower than the experimental value. To investigate this discrepancy, growth on pyruvate was computationally compared to growth on lactate since these two substrates are separated by only one metabolic reaction (mediated by lactate dehydrogenase), and there was good agreement between the computational and experimental results for lactate. By computationally removing reactions and running simulations, we found that the erroneous calculations for growth on pyruvate were not attributable to the lactate dehydrogenase reaction or energetic considerations connected to NADH. Thus, there may be metabolic processes related to pyruvate metabolism that are not accounted for in the in silico model. This model can be used as a basis for probing metabolic questions and systematically reconciling discrepancies (6, 20, 25).
With a genome-scale model and suitable experimental methods in place, the research described here may be one of the first steps in the growing field of combining computational and experimental methods. Thus, we may be at the doorstep of using computational models to prospectively design strains and experiments; this could help reduce the amount of experimental work needed to achieve a goal, but we must also be careful to fully evaluate the limitations of any computational model and its applicability to the complexities of real-life biology.
Acknowledgments
This work was supported by NIH grants GM57089 and GM62791 and by NSF grant MCB 9873384.
We thank Rafael Ibarra, Jennie Reed, Susan Toyama, Eric Knight, Henry Kang, Monica Mo, Markus Covert, and Grace Lim for their technical assistance and critical input.
REFERENCES
- 1.Bailey, J. E. 2001. Complex biology with no parameters. Nat. Biotechnol. 19:503-504. [DOI] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Bouche, N., and D. Bouchez. 2001. Arabidopsis gene knockout: phenotypes wanted. Curr. Opin. Plant Biol. 4:111-117. [DOI] [PubMed] [Google Scholar]
- 4.Carlson, R., D. Fell, and F. Srienc. 2002. Metabolic pathway analysis of a recombinant yeast for rational strain development. Biotechnol. Bioeng. 79:121-134. [DOI] [PubMed] [Google Scholar]
- 5.Covert, M.W., C. H. Schilling, I. Famili, J. S. Edwards. I. I. Goryanin, E. Selkov, and B. O. Palsson. 2001. Metabolic modeling of microbial strains in silico. Trends Biochem. Sci. 26:179-186. [DOI] [PubMed] [Google Scholar]
- 6.Covert, M. W., B. O. Palsson, and C. H. Schilling. 2002. A more-palatable Helicobacter pylori: iterative model building through the peer review process. ASM News 68:529-530. [Google Scholar]
- 7.Drell, D. 2002. The Department of Energy Microbial Cell Project: a 180° paradigm shift for biology. Omics A J. Integr. Biol. 6:3-9. [DOI] [PubMed] [Google Scholar]
- 8.Edwards, J. S., R. U. Ibarra, and B. O. Palsson. 2001. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat. Biotechnol. 19:125-130. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, J. S., and B. O. Palsson. 2000. The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc. Natl. Acad. Sci. USA 97:5528-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, J. S., R. Ramakrishna, and B. O. Palsson. 2002. Characterizing the metabolic phenotype: a phenotype phase plane analysis. Biotechnol. Bioeng. 77:27-36. [DOI] [PubMed] [Google Scholar]
- 11.Fell, D. 1996. Understanding the control of metabolism. Frontiers in metabolism, vol. 2. Portland Press, London, UK.
- 12.Gombert, A. K., and J. Nielsen. 2000. Mathematical modelling of metabolism. Curr. Opin. Biotechnol. 11:180-186. [DOI] [PubMed] [Google Scholar]
- 13.Ibarra, R. U., J. S. Edwards, and B. O. Palsson. 2002. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420:186-189. [DOI] [PubMed] [Google Scholar]
- 14.Ideker, T., and L. Hood. 2001. A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2:343-372. [DOI] [PubMed] [Google Scholar]
- 15.Karp, P. D., M. Riley, M. Saier, I. T. Paulsen, J. Collado-Vides, S. M. Paley, A. Pellegrini-Toole, C. Bonavides, and S. Gama-Castro. 2002. The EcoCyc Database. Nucleic Acids Res. 30:56-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao, J. C., and J. Delgado. 1993. Advances in metabolic control analysis. Biotechnol. Prog. 9:221-233. [Google Scholar]
- 17.Liao, J. C., S. Y. Hou, and Y. P. Chao. 1996. Pathway analysis, engineering and physiological considerations for redirecting central metabolism. Biotechnol. Bioeng. 52:129-140. [DOI] [PubMed] [Google Scholar]
- 18.Mahadevan, R., J. S. Edwards, and F. J. Doyle. 2002. Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys. J. 83:1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Overbeek, R., N. Larsen, G. D. Pusch, M. D'Souza, E. Selkov, Jr., N. Kyrpides, M. Fonstein, N. Maltsev, and E. Selkov. 2000. WIT: integrated system for high-throughput genome sequence analysis and metabolic reconstruction. Nucleic Acids Res. 28:123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palsson, B. O. 2000. The challenges of in silico biology. Nat. Biotechnol. 18:1147-1150. [DOI] [PubMed] [Google Scholar]
- 21.Price, N. D., J. A. Papin, and B. O. Palsson. 2002. Determination of redundancy and systems properties of the metabolic network of Helicobacter pylori using genome-scale extreme pathway analysis. Genome Res. 12:760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raamsdonk, L. M., B. Teusink, D. Broadhurst, N. Zhang, A. Hayes, M. C. Walsh, J. A. Berden, K. M. Brindle, D. B. Kell, J. J. Rowland, H. V. Westerhoff, K. van Dam, and S. G. Oliver. 2001. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 19:45-50. [DOI] [PubMed] [Google Scholar]
- 23.Raghunathan, A., and B. O. Palsson. 2003. Scalable method to determine mutations that occur during adaptive evolution of Escherichia coli. Biotechnol. Lett. 25:435-441. [DOI] [PubMed] [Google Scholar]
- 24.Savinell, J. M., and B. O. Palsson. 1992. Network analysis of intermediary metabolism using linear optimization. I. Development of mathematical formalism. J. Theor. Biol. 154:421-454. [DOI] [PubMed] [Google Scholar]
- 25.Schilling, C. H., M. W. Covert, I. Famili, G. M. Church, J. S. Edwards, and B. O. Palsson. 2002. Genome-scale metabolic model of Helicobacter pylori 26695. J. Bacteriol. 184:4582-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schilling, C. H., J. S. Edwards, D. Letscher, and B. O. Palsson. 2000. Combining pathway analysis with flux balance analysis for the comprehensive study of metabolic systems. Biotechnol. Bioeng.. 71:286-306. [PubMed] [Google Scholar]
- 27.Schuster, S., D. A. Fell, and T. Dandekar. 2000. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat. Biotechnol. 18:326-332. [DOI] [PubMed] [Google Scholar]
- 28.Schuster, S., and C. Hilgetag. 1994. On elementary flux modes in biochemical reaction systems at steady state. J. Biol. Systems 2:165-182. [Google Scholar]
- 29.Selkov, E., N. Maltsev, G. J. Olsen, R. Overbeek, and W. B. Whitman. 1997. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene 197:GC11-GC26. [DOI] [PubMed] [Google Scholar]
- 30.Thorneycroft, D., S. M. Sherson, and S. M. Smith. 2001. Using gene knockouts to investigate plant metabolism. J. Exp. Bot. 52:1593-1601. [PubMed] [Google Scholar]
- 31.Tomita, M., K. Hashimoto, K. Takahashi, T. S. Shimizu, Y. Matsuzaki, F. Miyoshi, K. Saito, S. Tanida, K. Yugi, J. C. Venter, and C. A. Hutchison 3rd. 1999. E-CELL: software environment for whole-cell simulation. Bioinformatics 15:72-84. [DOI] [PubMed] [Google Scholar]
- 32.Vallino, J., and G. Stephanopoulos. 1993. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Biotechnol. Bioeng. 41:633-646. [DOI] [PubMed] [Google Scholar]
- 33.Van Dien, S. J., and M. E. Lidstrom. 2002. Stoichiometric model for evaluating the metabolic capabilities of the facultative methylotroph Methylobacterium extorquens AM1, with application to reconstruction of C(3) and C(4) metabolism. Biotechnol. Bioeng. 78:296-312. [DOI] [PubMed] [Google Scholar]
- 34.Varma, A., and B. O. Palsson. 1994. Metabolic flux balancing: basic concepts, scientific and practical use. Bio/Technology 12:994-998. [Google Scholar]
- 35.Varma, A., and B. O. Palsson. 1994. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl. Environ. Microbiol. 60:3724-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voit, E. S. 2000. Computational analysis of biochemical systems. Cambridge University Press, Cambridge, UK.