Abstract
Rearrangement of antigen receptor genes generates a vast array of antigen receptors on lymphocytes. The establishment of allelic exclusion in immunoglobulin genes requires differential treatment of the two sequence identical alleles. In the case of the κ immunoglobulin locus, changes in chromatin structure, methylation, and replication timing of the two alleles are all potentially involved in regulating rearrangement. Additionally, germline transcription of the κ locus which precedes rearrangement has been proposed to reflect an opening of the chromatin structure rendering it available for rearrangement. As the initial restriction of rearrangement to one allele is critical to the establishment of allelic exclusion, a key question is whether or not germline transcription at the κ locus is monoallelic or biallelic. We have used a sensitive reverse transcription-polymerase chain reaction (RT-PCR) assay and an RNA–fluorescence in situ hybridization (FISH) to show that germline transcription of the κ locus is biallelic in wild-type immature B cells and in recombination activating gene (RAG)−/−, μ+ B cells. Therefore, germline transcription is unlikely to dictate which allele will be rearranged first and rather reflects a general opening on both alleles that must be accompanied by a mechanism allowing one of the two alleles to be rearranged first.
Keywords: immunoglobulin, germline transcription, allele exclusion
Introduction
The rearrangement of antigen receptor genes presents a remarkably elegant solution to the problem of how to generate an extremely large number of different specificities for each type of antigen receptor deployed by the immune system (1, 2). RAG-1 and RAG-2 together play an essential role in the rearrangement events by recognizing recombination signal sequences (RSSs)* (3–5). Stringent regulation of rearrangement is important in allowing the different types of lymphocytes to function properly. For example, rearrangements in B cells produce the immunoglobulin type of antigen receptor found on B cells whereas rearrangement in T cells leads to surface expression of T cell receptors. This distinct regulation is accomplished despite conservation in RSSs and the fact that RAG-1 and RAG-2 are involved in both types of rearrangements. Indeed, rearrangements of, for example, T cell receptor genes in a B cell are rarely observed.
Allelic exclusion is a fascinating aspect of the regulation of antigen-specific receptors, first observed by Pernis and colleagues (6). Their observation predated the identification of the antigen receptor genes and the discovery of rearrangement as a mechanism for generation of diversity. Allelic exclusion, in its original definition refers to the expression of either the maternally derived or the paternally derived allele of each antigen receptor gene in each individual cell. This monoallelic expression is random, with parity in the number of cells expressing the maternal allele and the number of cells expressing the paternal allele. Monoallelic expression, or allelic exclusion is critical to the functioning of the immune system as it allows each lymphocyte to elaborate an antigen receptor of a single type. Chaos in immune system regulation might ensue if, for example, a B cell expressed both an antibody that responded to a pathogen and a second antibody that would cause damage to a certain host tissue.
How is allelic exclusion established in the face of the sequence identity of the two alleles of a given antigen receptor gene? Any proposed mechanism must explain the observation that in many lymphocytes rearrangement has occurred nonproductively on one allele (first) and productively on the second allele (7). Furthermore, some cells have a functional rearrangement on one allele and the second allele is in the germline configuration. A feedback mechanism plays a role in keeping the second allele from rearranging as demonstrated in transgenic mouse models (8, 9). This feedback mechanism is certainly important in maintaining allelic exclusion. However, in order for feedback inhibition to establish allelic exclusion the process of rearrangement would have to be very inefficient, allowing time for a productively rearranged allele to have its protein made and for the appropriate signal transduction to occur to silence the other allele. Alternatively, a mechanism which allows one allele to be preferentially accessible for rearrangement could accomplish the establishment of allelic exclusion; such a mechanism would then collaborate with the feedback inhibition mediated by a functionally rearranged allele to maintain allelic exclusion. In the absence of a functional rearrangement on the first allele, the second allele would ultimately become available for rearrangement. We have recently shown that asynchronous replication of the two alleles of a given antigen receptor gene represents an epigenetic difference that is established before B cell differentiation (10). This epigenetic difference is associated with the initial choice of the allele that will first undergo demethylation and subsequent rearrangement.
Germline transcription at a number of antigen receptor gene loci has been observed to begin before rearrangement. Germline transcription is not thought to produce functional proteins and the promoter and initiation sites are often lost in subsequent rearrangements. Rather, germline transcription is thought to be involved in regulating the rearrangement process. Germline transcription of the κ immunoglobulin locus is initiated after the production of a functional immunoglobulin heavy chain (11). The onset of κ germline transcription just before rearrangement has led to the proposal that germline transcription directs or reflects a change in the chromatin structure that is a prelude to rearrangement. In support of this idea, it has been observed that perturbations of enhancer elements required for rearrangement also effect germline transcription before rearrangement (7). In the case of the germline transcription at the κ locus, it has been demonstrated that LPS treatment of Abelson virus-transformed pre-B cells induces germline transcription and induces rearrangement (12).
Whether germline transcription plays a role in the selection of one of the two alleles for initial rearrangement events remains an open question. If germline transcription plays a role in the establishment of allelic exclusion, the germline transcript would have to be monoallelic. To address this issue, we have employed a sensitive single cell RT-PCR analysis to analyze germline transcription in pre-B cells. We find biallelic germline transcription in these RT-PCR analyses and in RNA–fluorescence in situ hybridization (FISH) analyses. Therefore, germline transcription reflects or dictates a general opening on both alleles that must be accompanied by a distinct mechanism allowing one of the two alleles to be rearranged first.
Materials and Methods
Single Cell RT-PCR from RAG−/−μ+ Mice.
Bone marrow cells from the rag−/−μ+ mice containing a Mus spretus and a Mus musculus polymorphism were stained with a rat anti–mouse B220 monoclonal antibody (RA3–6B2; BD Biosciences; FITC conjugated). Individual B220+ cells were FACS® sorted directly into 12.5 μl RT buffer (1× RT buffer [Promega], 20 U Rnasin [Promega], 500 ng oligo dT (GIBCO BRL) and 100 ng RT primer RMB1: 5′-CACTCATTCCTGTTGAAGCTCTTG-3′) in individual PCR tubes. The tubes were incubated at 65°C for 1 min, then at 22°C for 3 min, and then placed on ice. The reverse transcriptase from Promega (1× RT buffer [Promega], 20 U Rnasin [Promega], 150 U AMV reverse transcriptase) was added to a final volume of 25 μl. The tubes were incubated at 22°C for 3 min followed by the reverse transcription reaction at 42°C for 50 min. The RT reaction was split into eight PCR tubes and used as a template for the PCR reactions. The PCR was performed under standard conditions using GIBCO BRL Taq (1× PCR buffer from GIBCO BRL, 1.5 mM MgCl2, 200 μM each of all 4 dNTPs 25 ng each of primer).
PCR for the 1.1 kb Transcript.
The first round PCR was performed in 30 μl reactions using primers RMB1 and RMF1 (5′-GTGAAGTGAAATGGCTGTAGCCTAATG-3′). Amplification was carried as follows: 4 min at 94°C; 35 cycles of 35 s at 94°C, 40 s at 55°C, and 1 min at 72°C; and, finally, 4 min at 72°C. Second-round PCR conditions were identical to the first round except the PCR were performed in 25 μl reactions and 2 μl of the first round was added as a template. The PCR primers were KG.1 (5′-CCTTTCTTCAGGGACAAGTGGG-3′) and KG.5 (5′-TGTCGTTCATACTCGTCCTTGGTC). A third round of PCR was performed with 2 μl of the second round amplification product with the primers KG.2 (5′-AAGTGGGA- ATGGACATAAGGAGC-3′) and KG.3 (5′TGTAGGTGCTGTCTTTGCTGTCC).
PCR for the 0.8 kb Transcript.
The first round PCR was performed in 30 μl reactions using primers RMB1 and KG.10 (5′- CAGTGAGGAGGGTTTTTGTACAGCCAGACAG-3′). Amplification was carried as follows: 4 min at 94°C; 35 cycles of 35 s at 94°C, 40 s at 55°C, and 1 min at 72°C; and, finally, 4 min at 72°C. Second-round PCR conditions were identical to the first round except the PCR were performed in 25 μl reactions and 2 μl of the first round was added as a template. The PCR primers were KG.7 (5′-GCTGGAAATCAAACGGGCTG-3′) and KG.5 (5′-TGTCGTTCATACTCGTCCTTGGTC). A third round of PCR was performed with 2 μl of the second round amplification product with the primers KG.7 and KG.3.
BCC' Fraction Bone Marrow from WT Mice.
The BCC' fraction cells were isolated using the expression of cell surface markers as identified by reference 13. Cell suspensions from bone marrow were prepared by flushing four femurs of F1 progeny of a cross between Mus musculus and Mus spretus and washing with PBA (PBS + 0.2% BSA). A million cells were resuspended in 200 μl PBA. The cells were incubated with 4 μl each of the following three antibodies; rat anti–mouse B220 monoclonal antibody (RA3–6B2; BD Biosciences; APC conjugated), rat anti–mouse CD43 monoclonal antibody (S7; BD Biosciences; FITC conjugated), and rat anti–mouse HSA monoclonal antibody (MI/69; BD Biosciences; PE conjugated). Cells positive for all the above cell surface markers were FACS® sorted into PCR tubes either as single cells or 50 cells/tube. Isotype control experiments (for each of the above antibodies in combination with the other two antibodies) were performed to ensure the specificity of staining. 50 cells or single cells were analyzed by RT-PCR for the 0.8 kb transcript as described above.
RNA-FISH was performed essentially as described previously (14, 15). A plasmid containing sequences between the J region and the C region of the κ immunoglobulin locus (pSPIg8) was labeled with Cy3 hybridized to fixed (4% formaldehyde, 5% acetic acid in phosphate-buffered saline), dehydrated RAG-1−/− Abelson-transformed pre-B cells which had been stimulated with LPS. After washing, slides were mounted in vectashield with DAPI and viewed with a Nikon E600 fluorescence microscope.
Results
An RT-PCR Assay for Analyzing Allele Specificity of κ Germline Transcription in Single Cells.
A single cell, allele-discriminating assay is required to test whether there is allele-specific transcription of the κ germline transcript. We therefore developed a single cell RT-PCR assay to distinguish the two alleles of the κ germline transcript in individual pre-B cells. The assay is specifically designed to assess the allele specificity even if the level of germline transcript present in each cell is quite low. The two alleles are distinguished using a polymorphism between the maternal and paternal alleles; the animals analyzed are heterozygotes carrying one Mus musculus allele and one Mus spretus allele. This approach allows us to examine a given gene and distinguish amongst four possibilities with respect to allelic expression: (a) transcription in individual cells is monoallelic, (b) transcription is biallelic, (c) a mix of monoallelic and biallelic cells is present, and (d) the sensitivity of the assay is too low to distinguish monoallelic and biallelic expression.
One immediate concern in a single cell RT-PCR analysis is the potential for the assay to incorrectly indicate monoallelic expression. For example, if one were to PCR amplify from only a single (cDNA) template molecule, it is predetermined that one will amplify only one allele. Thus, if the starting cell in the example were in fact expressing transcripts from both alleles, the low efficiency of conversion of mRNA into cDNA would have caused the RT-PCR experiment to provide the erroneous conclusion that transcription is monoallelic. To avoid this potential problem, our assay has at its heart the following principle: a single molecule (in this case a single cDNA) can only be in one place at a time (in this case in one PCR tube). Therefore, by splitting up a RT reaction from a single cell into eight separate PCRs, we can be sure that each ‘positive’ PCR represents at least one (if not more than one) individual template(s). Each cDNA template corresponds to an individual mRNA molecule present in the analyzed cell. While this assay may or may not be more sensitive at detecting expression of a given gene, it is more sensitive at distinguishing between monoallelic and biallelic expression.
Another concern in a single cell RT-PCR analysis is that the assay can incorrectly indicate biallelic expression if more than one cell is analyzed at a time rather than a single cell. To assure that we were indeed analyzing single cells, we used a FACS® to place individual F1 B cells directly into PCR tubes. Control experiments demonstrated that the FACS® reliably places only a single cell in each tube (reference 16; and data not shown). A reconstruction control experiment was performed in which we artificially create a situation with pure “monoallelic” expression. This is accomplished by mixing together equal numbers of Mus spretus B cells and Mus musculus B cells. This mixture of cells was then run through the full single cell RT-PCR assay including FACS® sorting of single cells into individual tubes, followed by RT, and then eight separate PCRs. We analyzed the highly expressed β2 microglobulin gene to allow this control experiment to demonstrate the power of the assay to call monoallelic expression when it is present. Consistently, we observe expression from only one type of allele (Mus musculus or Mus spretus) in each cell (Table I). We analyzed a total of 28 cells, 22 of which yielded from one to eight PCR products per cell. All of these analyses confirm the expected result that this mixture of Mus spretus B cells and Mus musculus B cells should reveal “monoallelic” expression. This control experiment demonstrates that our single cell RT-PCR assay can detect monoallelic expression when it is present (see also Rhoades et al. [reference 13]). Thus, all aspects of the assay including sorting, RT, PCR, and allele discrimination were tested in these control experiments. These controls are important to mention given the extraordinary amplification that PCR affords.
Table I.
Analysis of β2 Microglobulin Allelic Expression in a Mixture of Spretus and Musculus B Lymphocytes
| Positive PCRs per cell |
No. of cells | Percent monoallelic |
|
|---|---|---|---|
| 8 | 2 | 100% | 2M |
| 7 | 1 | 100% | 1M |
| 6 | 1 | 100% | 1S |
| 5 | 5 | 100% | 2M, 3S |
| 4 | 2 | 100% | 2M |
| 3 | 5 | 100% | 3M, 2S |
| 2 | 1 | 100% | 1M |
| 1 | 5 | 100% | 4M, 1S |
| 0 | 6 |
The analysis was performed on 28 cells. M, musculus; S, spretus. As expected, this control reconstruction experiment shows that when we artificially create pure “monoallelic” expression, the single cell RTPCR assay can indicate monoallelic expression consistently.
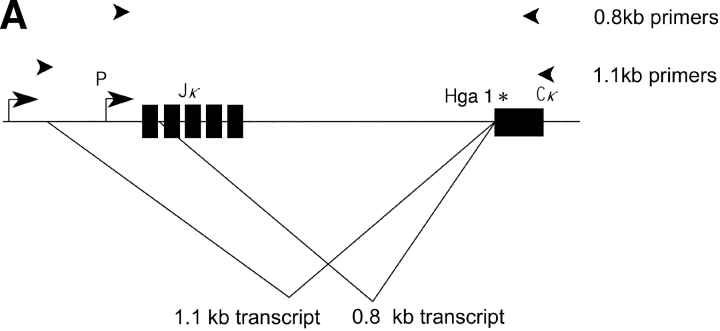
Germline transcripts in the vicinity of the κ J/C region are present in two forms: a short and long form 0.8 and 1.1 kb, respectively (11). As outlined in Fig. 1 A, these two transcripts have different initiation sites and 5′ splice donors, and share a splice acceptor which is the same splice acceptor used to generate the postrearrangement RNA once rearrangement has occurred.
Figure 1.
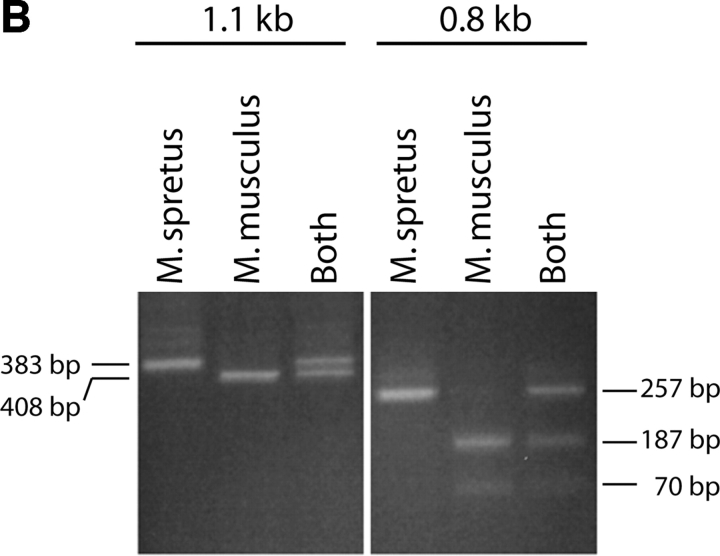
(A) The κ germline transcripts initiate from two promoters yielding a 8.4 and a 4.7 kb products which are further spliced to 1.1 and a 0.8 kb transcripts, respectively. The arrowheads indicate PCR primers used to amplify the 0.8 or the 1.1 kb transcript. Restriction site polymorphism (Hga I) between the Mus musculus and Mus spretus allele is indicated by the asterisk. (B) The 1.1 kb or the 0.8 kb transcript was subjected to RT-PCR with Mus musculus RNA, Mus spretus RNA or a mix of RNA from both Mus musculus and Mus spretus. The PCR products were then digested with the restriction endonuclease Hga I.
To analyze the 0.8 and 1.1 kb germline transcripts (in separate experiments), we have identified a polymorphism between the Mus spretus and the Mus musculus alleles in a portion of the sequence that is present in both the 0.8 and 1.1 kb germline transcripts. This polymorphism allows the two alleles of both types of transcript to be distinguished by restriction endonuclease digestion (Fig. 1 B). As we wished to analyze the 0.8 and 1.1 kb germline transcripts respectively (in separate experiments), we designed PCR primers that flank the polymorphic site and also span introns in each germline transcript. The fact that in each case the primers are from sequences flanking an intron assures that the PCR products we detect came from template cDNAs representing spliced mRNAs. We designed an RT primer that efficiently generates cDNA from either the short or the long germline transcript and allows PCR amplification of the region of the germline transcript cDNA containing the restriction enzyme-detectable polymorphism. HgaI digests only the Mus musculus allele yielding a 383 and a 25 bp fragment from the 408 bp PCR product representing the 1.1 kb transcript. The same polymorphic site in the 257 bp PCR product representing the 0.8 kb transcript yields fragments of 187 and 70 bp. The Mus spretus allele is not digested. Control experiments demonstrate that the RT and PCR primers amplify both alleles equally and that the germline transcription from a bulk population of bone marrow cells derives from both alleles.
κ Germline Transcription in RAG-1−/−/μ+ B Cells.
We first examined germline transcription in individual bone marrow cells from RAG-1−/−/μ+ mice (17) by RT-PCR. In these RAG-1−/−/μ+ mice, early B cells in the bone marrow can advance to the pre-B stage of development because of the functional heavy chain expressed from the μ transgene. Then, they are unable to progress further in B cell development, because they are unable to rearrange the light chain genes secondary to the deletion of the RAG-1 gene. These animals therefore provide a large number of cells at the appropriate stage of B cell development to examine κ germline transcription.
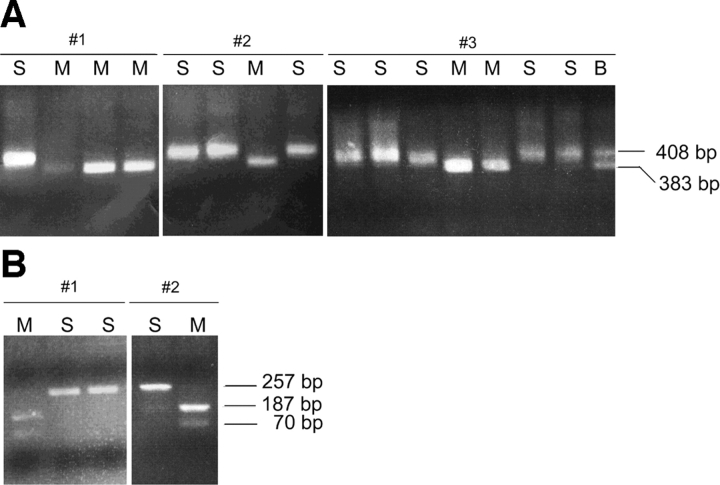
Single bone marrow cells from RAG-1−/−/μ+ mice expressing the pan B cell marker B220 were placed in individual tubes by FACS®. An RT reaction was set up in the same tubes and the completed RT reaction was split into eight tubes for independent PCR amplifications. PCR products derived from the 1.1 kb transcript were digested with HgaI (which digests only the Mus musculus allele) and then visualized on an agarose gel to determine the parental origin of κ germline transcripts. Representative analyses are shown in Fig. 2 A. If one examines the parental origins of the transcripts within individual cells, one can clearly observe biallelic expression (Fig. 2 A). 23 cells were informative for assessing whether transcription is monoallelic or biallelic. In cell #1 for example, four of the eight PCRs from the single cell–derived RT reaction show a product and both alleles are observed: the lane on the left is uncut (Mus spretus allele) and the other three are digested. It is important to note that in many of these single cell analyses, one or more of the eight individual PCRs can reveal only one allele while other PCRs from the same RT reaction may show the other allele or in some cases both alleles are present in the same PCR. This is due to the low number of cDNA templates that are available for PCR reflecting the expected low level of expression of the germline transcript. The low level of expression was also found for the germline transcript in WT cells (see below). These results highlight the importance of using multiple PCR reactions to analyze each single cell RT reaction.
Figure 2.
(A) Single-cell RT-PCR analysis of 1.1 kb κ germline transcript in BM cells from Rag−/− μ+ mice. RT-PCR products were digested with Hga1 restriction endonuclease which recognizes only the Mus musculus allele. M is the Mus musculus allele, S is the Mus spretus allele, and B indicates the presence of both the Mus musculus and Mus spretus allele. The number of PCR products obtained ranged from one to eight out of eight. Cell # 1 and #2 are examples with four PCR products. Cell # 3 is an example with 8 PCR products. (B) Single-cell RT-PCR analysis of 0.8 kb κ germline transcript in BM cells from Rag−/− μ+/+ mice. RT-PCR products were digested with HgaI restriction endonuclease which recognizes only the Mus musculus allele. M is the Mus musculus allele and S is the Mus spretus allele. The number of PCR products obtained ranged from one to eight out of eight. Cell # 1 is an example with three PCR products. Cell #2 is an example with two PCR products.
Examination of all the data for RAG-1−/−/μ+ bone marrow cells demonstrates that the 1.1 kb germline transcript is biallelically expressed in individual cells (Table II). While some cells show only one allele amplified by RT-PCR, the frequency of occurrence of these cells is no greater than would be expected in a mathematical reconstruction of the experiment. The 23 cells (out of 63 analyzed) that had two or more of the eight PCR reactions positive were used for analysis. For cells that only yield two PCR products, one would expect half of these to appear ‘monoallelic’ in the assay. This is similar to the expectation that if one flips a ‘true’ coin twice, 50% of the time one will get either two heads or two tails. Indeed, examination of cells for which there was a product in two out of eight PCRs reveals that half (7 of 14) of them show biallelic expression, in line with the statistical prediction. The cells that revealed a product in more than two of eight PCR samples also had a distribution of the alleles consistent with biallelic expression. Therefore, these data demonstrated that individual RAG-1−/−/μ1 B cells express both alleles and that the levels of the two alleles appear to be equal.
Table II.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|
| Rag−/−μ+ 1.1 kb | ||||||||
| Monoallelic | 9 | 7 | 2 | |||||
| Biallelic | 7 | 2 | 4 | 1 | ||||
| Rag−/−μ+ 0.8 kb | ||||||||
| Monoallelic | 3 | 1 | 1 | |||||
| Biallelic | 1 | 1 | 2 | 1 | 1 | 1 | 1 | |
| BCC', 0.8 kb | ||||||||
| Monoallelic | 4 | 2 | ||||||
| Biallelic | 2 | 2 | 5 | 5 | 2 | 1 | 1 | 1 |
The numbers of cells with monoallelic or biallelic PCR products are listed as a function of the number of PCR products per cell (1–8).
We performed a similar analysis of the 0.8 kb κ germline transcript yielding similar results as the analysis of the 1.1 kb germline transcript. Primers were designed to allow specific amplification of the 0.8 kb transcript. The 257 bp product is digested with HgaI yielding 187 and 70 bp bands for the Mus musculus allele and a single 257 bp band for the Mus spretus allele. Representative single cells are shown in Fig. 2 B. The 10 cells that revealed a product in two or more of eight PCR samples (out of 21 cells analyzed) had a distribution of the alleles reflecting biallelic expression (Table II). Note that some single cells reveal five or more out of eight PCRs with a product while other cells reveal fewer than five out of eight PCRs with a product. This result is similar to the result observed for the 1.1 kb germline transcript in RAG-1−/−/μ1 bone marrow cells.
κ Germline Transcription in WT Immature B Cells.
Since RAG-1−/−/μ1 bone marrow cells are cells that are artificially arrested in their development (due to the absence of RAG-1), we wished to confirm our observation of biallelic κ germline transcription with an analysis of WT immature B cells. We therefore performed a similar analysis on WT immature B cells from a Mus spretus and Mus musculus F1 mouse. We used the FACS® criteria of Hardy et al. (13) to isolate immature B cells before κ rearrangement (BCC' fraction).
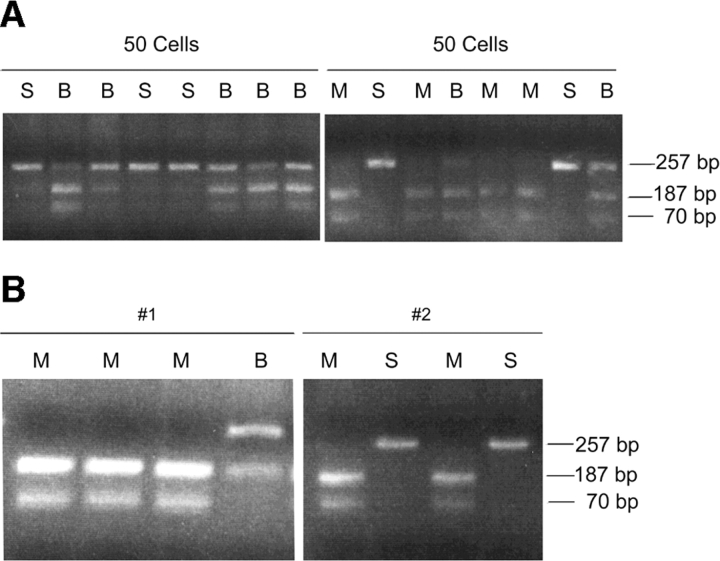
To assess the level of expression of the germline transcript in the population of BCC' fraction pre-B cells, we first sorted 50 cells per tube and analyzed each group of 50 cells by RT-PCR. The RT was divided into 8 tubes for PCR as was done with the single cell analyses. We analyzed the 0.8 kb germline transcript and in each case, when 50 cells are placed in a single tube for the RT reaction, we observe a PCR product in all 8 tubes. When these PCR products are subjected to restriction digestion we are able to observe both the Mus musculus and Mus spretus alleles (Fig. 3 A). The observation that some of the PCR products reveal only one of the two alleles indicates that as expected, the level of germline transcript expression in these BCC' fraction cells is not very high. We then proceeded to analyze single BCC' fraction cells (Fig. 3 B).
Figure 3.
(A) RT-PCR analysis of 0.8 kb κ germline transcript in BCC' fraction of BM preB cells from WT mice. 50 cells were FACS® sorted into a single tube and subjected to a RT reaction. This RT reaction was divided to eight PCR tubes. The RT-PCR products were digested with Hga1 restriction endonuclease which recognizes only the Mus musculus allele. M is the Mus musculus allele, S is the Mus spretus allele, and B indicates the presence of both the Mus musculus and Mus spretus allele. (B) Single-cell RT-PCR analysis of 0.8 kb κ germline transcript in BCC' fraction of BM preB cells from WT mice. RT-PCR products were digested with Hga1 restriction endonuclease which recognizes only the Mus musculus allele. M is the Mus musculus allele, S is the Mus spretus allele, and B indicates the presence of both the Mus musculus and Mus spretus allele. The number of PCR products obtained ranged from one to eight out of eight. Cell # 1 and #2 are examples with four PCR products.
20 single FACS®-sorted BCC' fraction cells analyzed (out of 77) revealed a PCR product representing the 0.8 kb transcript in at least two of eight tubes and were thus informative for assessing monoallelic vs. biallelic transcription. As we observed in the experiments using RAG-1−/−/μ1 B cells, WT BCC' fraction cells that revealed a product in more than two of eight PCR samples had a distribution of the alleles consistent with biallelic expression (Table II). Again, in line with the theoretical prediction, two of the four cells that had exactly two of eight PCRs with a product revealed biallelic expression. For the cells with from three to eight PCRs positive (out of eight possible) all of them revealed biallelic expression. These data provide a clear demonstration of biallelic expression in individual BCC' fraction cells.
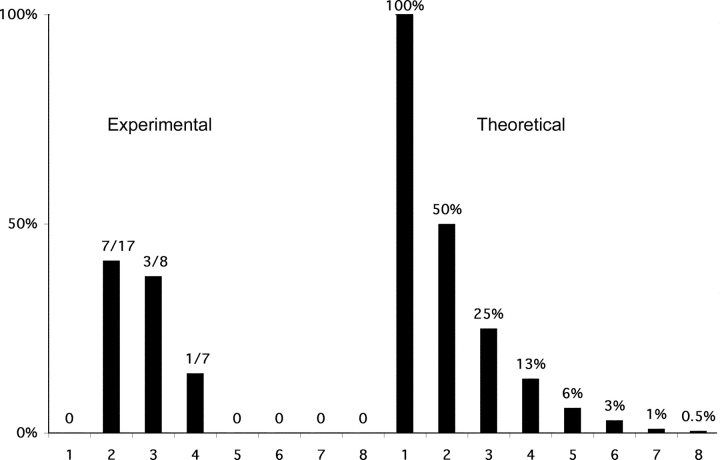
Fig. 4 shows a comparison of the observed data with the theoretical expectation for an analysis of a biallelically expressed gene that is expressed at a low level. 100% of the cells with one product are expected to show ‘monoallelic expression’ in such an assay even though expression is in reality biallelic. Similarly, half of the cells with two of eight PCRs positive would be expected to reveal ‘monoallelic expression’ and one quarter of the cells with three of eight PCRs positive would be expected to reveal ‘monolallelic expression’. A similar calculation for four to eight of eight PCR reactions revealing a product is also shown. We also present a compiled data we have gathered for germline transcription in the left hand panel. Our data are similar to the theoretical data for an unskewed biallelically transcribed gene. This graphical representation of the data illustrates that the germline transcript from the κ immunoglobulin locus is biallelically transcribed.
Figure 4.
Graph of κ germline transcription in pre-B cells. Left panel: the percentage of cells appearing to be monoallelic is plotted as a function of the number of positive PCR reactions observed per cell. 16 cells revealed one PCR product/cell, 17 cells revealed two, 8 cells revealed three, 7 cells revealed four, 2 cells revealed five, six and seven PCR product/cell, and 3 cells revealed eight PCR products/cell. Note that grouping either the cells with 2 or 3 products per cell or the cells with 4 or more products per cell gives a fraction of the cells appearing monoallelic which is slightly below and statistically indistinguishable from the theoretical prediction for a biallelically expressed gene. Right panel: the theoretical expectation for an analysis of a monoallelically expressed gene. The formula 2/2n (where n is the number of PCR products observed) approximates the fractions for low values of n. The percentages presented leave out a minor correction in the formula for rare instances in which a given signal actually represents more than one cDNA product. The line across at the 100% level indicates what would be theoretically expected if monoallelic expression were absolute.
RNA-FISH Analysis of κ Germline Transcription.
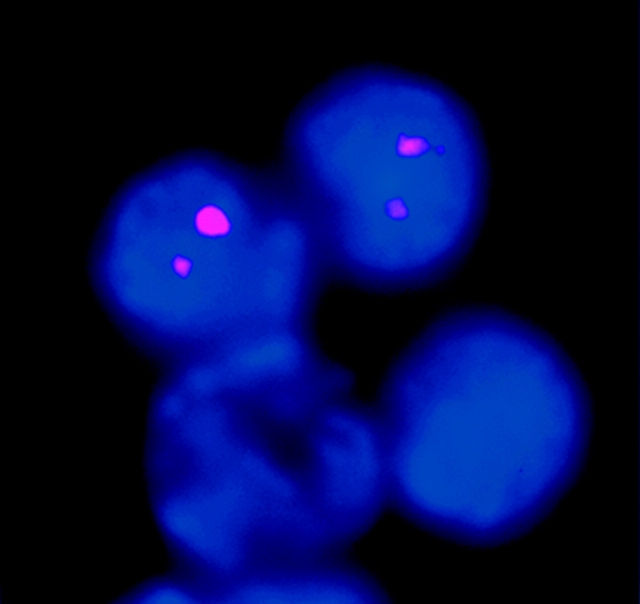
We have also confirmed biallelic κ germline transcription using a non-PCR–based approach. We performed RNA-FISH analysis on a population of RAG-1−/− Abelson-transformed pre-B cells. These cells activate germline transcription when stimulated with LPS. A probe for the κ germline transcript revealed that 40% of these cells express the κ germline transcript. 91% (91/100) of the cells expressing the κ germline transcript revealed two hybridization signals (Fig. 5) . The small number of cells with only one signal could either represent limitations in detection, or could represent a small subset of cells that have monoallelic expression. Control experiments included analyses of fibroblast cells which as expected revealed no κ germline transcription. We also analyzed the variable region for the possibility of κ-variable region germline transcription. Consistent with reports in the literature (18), we observed no germline transcripts emanating from the κ-variable region. These RNA-FISH observations are consistent with the single cell RT-PCR analyses in indicating that κ germline transcription in the J-C region is biallelic most if not all of the time.
Figure 5.
RNA-FISH confirms biallelic κ germline transcription. A Cy3 labeled probe (pSPIg8) for the κ germline transcript revealed biallelic transcription in 91% of the RAG-1−/− Abelson-transformed pre-B cells in which any κ germline transcript was detected. DAPI allows visualization of DNA in nuclei. Two of the four cells in the field presented are expressing the κ germline transcript and both of them have two RNA-FISH signals.
Discussion
When a developing B cell undergoes κ immunoglobulin gene rearrangement, how are the two alleles differentially treated such that at first rearrangement occurs on only one of the two alleles? This represents a particularly intriguing problem for the obvious reason that the two alleles can have identical sequences. Either epigenetic differences must be established between the two alleles, or alternatively the mechanism which allows only one allele to be chosen does not require a difference between the two alleles. Prior observations that germline transcription precedes rearrangement suggested the possibility that since rearrangement occurs on one of two alleles, germline transcription might also be from only one allele. We have used a sensitive, single cell RT-PCR assay to assess germline transcription of the two alleles in both RAG-1−/−/μ+ cells and in WT pre-B cells. We find that in fact, germline transcription is biallelic. We have also confirmed these data with RNA-FISH experiments. It is important to note that in previous analyses of IL-2 transcription, we were able to observe both monoallelic cells and biallelic cells. Thus, our data suggest that κ germline transcription remains biallelic in all cells until rearrangement renders one of the alleles incompetent for germline transcription. Germline transcription persists even in mature B cells (unpublished data). Our finding of biallelic κ germline transcription suggests that while the onset of germline transcription is clearly a marker of the general availability of the κ gene locus for rearrangement, it does not dictate which allele will be preferentially rearranged first.
Germline Transcription.
Germline transcription has been found to occur before rearrangement of a number of antigen receptor families. Sources of germline transcription have been found near constant and J regions, emanating from promoters that can be eliminated by subsequent rearrangement events (19). Additionally, V region germline transcripts have been observed in which the promoter used is the same promoter used subsequent to rearrangement. However, it is important to note that the critical post rearrangement enhancer elements are contributed by the constant regions. While these variable region germline transcripts have been shown to be differentially regulated, it is not clear whether differential transcription of different V regions plays a role in the selection of a V region for rearrangement. Prior studies of germline transcription have assessed germline transcripts without regard for the two different alleles.
Germline transcription also plays a role in the rearrangements allowing immunoglobulin heavy chain class switching (20). Class switching proceeds through mechanisms distinct from the RSS mediated VDJ recombination. Class switching is regulated by the signals that a given B cell receives and can be skewed toward different C regions. This skewing is preceded by germline transcription in the regions where rearrangement will take place. Thus, in the case of class switching, the germline transcript presages where rearrangement will take place.
Possible Mechanisms Suggested by Biallelic Germline Transcription.
The biallelic nature of the germline transcription at the κ locus suggests that germline transcription itself, is not involved in the choice of the allele that will be rearranged first. Germline transcription may either be a cause or an effect of general opening of the chromatin on both alleles. If germline transcription is opening up (or reflects the opening up) of both κ alleles, how then is rearrangement limited (at least initially) to one allele? Note that it is formally possible that κ germline transcription switches from biallelic to monoallelic immediately before rearrangement. Other epigenetic differences between the two alleles may play critical roles in the establishment of allelic exclusion. Differences in methylation are could provide such an epigenetic mark. We have previously identified sites in the vicinity of the J region that are monoallelically demethylated immediately before rearrangement (21). The fact that the observed monoallelic demethylation only immediately precedes rearrangement means it is not suited to be the mark that differentiates the two alleles. It is possible that there are other differentially methylated sites that could provide a mark perhaps even earlier than the onset of B cell differentiation.
Other epigenetic modifications might also underlie a difference between the two alleles. There could be differences in chromatin structure such as in histone subunit usage, or acetylation status. Another intriguing possible mark involves the rendering of the two alleles asynchronously replicating. Replicative asynchrony is associated with monoallelically expressed genes such as imprinted genes in which transcription and the asynchrony of replication are both in one direction with respect to the parental legacy of the two alleles (22). Randomly monoallelically transcribed genes also show replicative asynchrony. This has been known for a long time for the X chromosome in female cells (23). More recently, olfactory receptors and interleukin genes have been shown to have both monoallelic transcription and replicative asynchrony (16, 24–27). We have recently shown that the antigen receptor genes are asynchronously replicated early in development and this may provide a primary basis for the establishment of allelic exclusion (10).
Other Gene Families Where Monoallelic Expression Plays a Role in Gene Regulation.
In addition to antigen receptors on B and T lymphocytes, monoallelic transcription has been observed for two other gene families in the immune system. The Ly49 family of natural killer cell receptors has been demonstrated to be monoallelically expressed in certain cells (28). Additionally, we and others have analyzed interleukin genes and found evidence for monoallelic transcription (16, 24, 26, 27). Outside of the immune system, both the olfactory receptor genes (29) and at least one of the two classes of pheromone receptor genes (30, 31) are strictly monoallelically expressed. Besides monoallelic expression, all of these genes are also characterized by their expression patterns in which individual cells express one (or a subset) of a family of genes. It will be interesting to further explore similarities in the regulation of these various gene families.
Acknowledgments
We thank G. Paradis for FACS® analyses, Peimin Qi for technical assistance, P. Sharp and members of the laboratory for discussions, J. Chen, F. Ebrahimi, P. Sklar for comments on the manuscript, and H. Higgins for preparation of the manuscript.
This work was supported by grants from the National Institutes of Health (A. Chess, Y. Bergman, H. Cedar) and A. Chess is a Rita Allen Foundation Scholar.
Footnotes
Abbreviation used in this paper: RSS, recombination signal sequence.
References
- 1.Tonegawa, S. 1983. Somatic generation of antibody diversity. Nature. 302:575–581. [DOI] [PubMed] [Google Scholar]
- 2.Rajewsky, K. 1996. Clonal selection and learning in the antibody system. Nature. 381:751–758. [DOI] [PubMed] [Google Scholar]
- 3.Schatz, D.G., M.A. Oettinger, and D. Baltimore. 1989. The V(D)J recombination activating gene, RAG-1. Cell. 59:1035–1048. [DOI] [PubMed] [Google Scholar]
- 4.Oettinger, M.A., D.G. Schatz, C. Gorka, and D. Baltimore. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 248:1517–1523. [DOI] [PubMed] [Google Scholar]
- 5.Gellert, M. 1996. A new view of V(D)J recombination. Genes Cells. 1:269–275. [DOI] [PubMed] [Google Scholar]
- 6.Pernis, B., G. Chiappino, A.S. Kelus, and P.G. Gell. 1965. Cellular localization of immunoglobulins with different allotypic specificities in rabbit lymphoid tissues. J. Exp. Med. 122:853–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sleckman, B.P., J.R. Gorman, and F.W. Alt. 1996. Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu. Rev. Immunol. 14:459–481. [DOI] [PubMed] [Google Scholar]
- 8.Ritchie, K.A., R.L. Brinster, and U. Storb. 1984. Allelic exclusion and control of endogenous immunoglobulin gene rearrangement in kappa transgenic mice. Nature. 312:517–520. [DOI] [PubMed] [Google Scholar]
- 9.Nussenzweig, M.C., A.C. Shaw, E. Sinn, J. Campos-Torres, and P. Leder. 1988. Allelic exclusion in transgenic mice carrying mutant human IgM genes. J. Exp. Med. 167:1969–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mostoslavsky, R., N. Singh, T. Tenzen, M. Goldmit, C. Gabay, S. Elizur, P. Qi, B.E. Reubinoff, A. Chess, H. Cedar, and Y. Bergman. 2001. Asynchronous replication and allelic exclusion in the immune system. Nature. 414:221–225. [DOI] [PubMed] [Google Scholar]
- 11.Martin, D.J., and B.G. van Ness. 1990. Initiation and processing of two kappa immunoglobulin germ line transcripts in mouse B cells. Mol. Cell. Biol. 10:1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlissel, M.S., and D. Baltimore. 1989. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 58:1001–1007. [DOI] [PubMed] [Google Scholar]
- 13.Hardy, R.R., C.E. Carmack, S.A. Shinton, J.D. Kemp, and K. Hayakawa. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173:1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Corput, M.P., and F.G. Grosveld. 2001. Fluorescence in situ hybridization analysis of transcript dynamics in cells. Methods. 25:111–118. [DOI] [PubMed] [Google Scholar]
- 15.Wijgerde, M., F. Grosveld, and P. Fraser. 1995. Transcription complex stability and chromatin dynamics in vivo. Nature. 377:209–213. [DOI] [PubMed] [Google Scholar]
- 16.Rhoades, K.L., N. Singh, I. Simon, B. Glidden, H. Cedar, and A. Chess. 2000. Allele-specific expression patterns of interleukin-2 and Pax-5 revealed by a sensitive single-cell RT-PCR analysis. Curr. Biol. 10:789–792. [DOI] [PubMed] [Google Scholar]
- 17.Spanopoulou, E., C.A. Roman, L.M. Corcoran, M.S. Schlissel, D.P. Silver, D. Nemazee, M.C. Nussenzweig, S.A. Shinton, R.R. Hardy, and D. Baltimore. 1994. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 8:1030–1042. [DOI] [PubMed] [Google Scholar]
- 18.Mather, E.L., and R.P. Perry. 1981. Transcriptional regulation of immunoglobulin V genes. Nucleic Acids Res. 9:6855–6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willerford, D.M., W. Swat, and F.W. Alt. 1996. Developmental regulation of V(D)J recombination and lymphocyte differentiation. Curr. Opin. Genet. Dev. 6:603–609. [DOI] [PubMed] [Google Scholar]
- 20.Lutzker, S., P. Rothman, R. Pollock, R. Coffman, and F.W. Alt. 1988. Mitogen- and IL-4-regulated expression of germ-line Ig gamma 2b transcripts: evidence for directed heavy chain class switching. Cell. 53:177–184. [DOI] [PubMed] [Google Scholar]
- 21.Mostoslavsky, R., N. Singh, A. Kirillov, R. Pelanda, H. Cedar, A. Chess, and Y. Bergman. 1998. Kappa chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 12:1801–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitsberg, D., S. Selig, M. Brandeis, I. Simon, I. Keshet, D.J. Driscoll, R.D. Nicholls, and H. Cedar. 1993. Allele-specific replication timing of imprinted gene regions. Nature. 364:459–463. [DOI] [PubMed] [Google Scholar]
- 23.Lyon, M.F. 1986. X chromosomes and dosage compensation. Nature. 320:313. [DOI] [PubMed] [Google Scholar]
- 24.Hollander, G.A., S. Zuklys, C. Morel, E. Mizoguchi, K. Mobisson, S. Simpson, C. Terhorst, W. Wishart, D.E. Golan, A.K. Bhan, and S.J. Burakoff. 1998. Monoallelic expression of the interleukin-2 locus. Science. 279:2118–2121. [DOI] [PubMed] [Google Scholar]
- 25.Naramura, M., R.J. Hu, and H. Gu. 1998. Mice with a fluorescent marker for interleukin 2 gene activation. Immunity. 9:209–2016. [DOI] [PubMed] [Google Scholar]
- 26.Bix, M., and R.M. Locksley. 1998. Independent and epigenetic regulation of the interleukin-4 alleles in CD4+ T cells. Science. 281:1352–1354. [DOI] [PubMed] [Google Scholar]
- 27.Riviere, I., M.J. Sunshine, and D.R. Littman. 1998. Regulation of IL-4 expression by activation of individual alleles. Immunity. 9:217–228. [DOI] [PubMed] [Google Scholar]
- 28.Held, W., J. Roland, and D.H. Raulet. 1995. Allelic exclusion of Ly49-family genes encoding class I MHC-specific receptors on NK cells. Nature. 376:355–358. [DOI] [PubMed] [Google Scholar]
- 29.Chess, A., I. Simon, H. Cedar, and R. Axel. 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell. 78:823–834. [DOI] [PubMed] [Google Scholar]
- 30.Belluscio, L., G. Koentges, R. Axel, and C. Dulac. 1999. A map of pheromone receptor activation in the mammalian brain. Cell. 97:209–220. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez, I., P. Feinstein, and P. Mombaerts. 1999. Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell. 97:199–208. [DOI] [PubMed] [Google Scholar]