Abstract
Natural resistance to infection with mouse cytomegalovirus (MCMV) is controlled by a dominant locus, Cmv1. Cmv1 is linked to the Ly49 family of natural killer receptors on distal chromosome 6. While some studies localized Cmv1 as distal to the Ly49 gene cluster, genetic and functional analysis identified Ly49h as a pivotal factor in resistance to MCMV. The role of these two independent genomic domains in MCMV resistance was evaluated by functional complementation using transgenesis of bacterial artificial chromosomes (BAC) in genetically susceptible mice. Phenotypic and genetic characterization of the transgenic animals traced the resistance gene to a single region spanning the Ly49h gene. The appearance of the Ly49H protein in NK cells of transgenic mice coincided with the emergence of MCMV resistance, and there was a threshold Ly49H protein level associated with full recovery. Finally, transgenic expression of Ly49H in the context of either of the two independent susceptibility alleles, Cmv1 sBALB or Cmv1 sFVB, conferred resistance to MCMV infection. These results demonstrate that Ly49h is necessary and sufficient to confer MCMV resistance, and formally demonstrate allelism between Cmv1 and Ly49h. This panel of transgenic animals provides a unique resource to study possible pleiotropic effect of Cmv1.
Keywords: genetic complementation test, natural immunity, animal model, immunogenetics, natural killer cells
Introduction
Experimental models of infection of mice with mouse cytomegalovirus (MCMV)* have provided an excellent tool for the in vivo dissection of the individual components of the host response responsible for mediating MCMV resistance. Indeed, resistance/susceptibility to MCMV is under multigenic control, with contributions from both H2 and non-H2 genes exerting their effects at different levels. The H2 complex influences susceptibility to the lethal effect of MCMV and viral titers in target organs (1, 2). However, non-H2 linked genes are also important in determining the outcome of MCMV infection during the initial phase of the infection. For example, in inbred mice, infection with a sublethal dose of MCMV presents an early nonimmune phase (0–6 d) characterized by either rapid proliferation of the virus or reduced viral titers in target organs (3). This early phase is controlled by a single dominant locus named Cmv1 (4). Cmv1 presents two allelic forms in mouse strains, either the dominant Cmv1 r (low virus titer, resistant) or the recessive Cmv1 s (high virus titer, susceptible), with Cmv1 r mouse strains being fivefold more resistant to lethal MCMV infection, and exhibiting 3–4 log10 lower level of viral replication in the spleen than Cmv1 s strains (4). The Cmv1 locus is related to NK cells, a set of lymphocytes involved in natural killing of tumor or virally infected cells. Indeed, when NK cells are depleted with an anti-NK1.1 mAb, resistant C57BL/6 (Cmv1 r) mice become susceptible (5). In addition, genetic mapping data indicated that Cmv1 maps to the mouse chromosome 6 and is tightly linked to the NK cell gene complex (NKC; references 4, 6, and 7). The NKC clusters numerous genes and gene families coding for highly related receptors of the C-lectin-type family important for signaling activation or inhibition of NK cell function (8).
Because of the genetic complexity of the NKC and the absence of a reliable in vitro assay to assess susceptibility/resistance to MCMV, at least two groups initiated a positional cloning approach to isolate Cmv1 (7, 9–12). This approach is based strictly on the knowledge of the chromosomal location of a specific gene to achieve its molecular cloning, after chromosome walking toward the gene from the most closely linked genetic markers on that chromosome (13). Through meiotic and physical mapping using classical inbred strains of mice, a minimal Cmv1 interval of 0.35 cM was defined (7, 10). This interval translated into 1.6 Mb of genomic DNA and contained a minimum of 20 transcription units (11). Among them, members of the Ly49 (Klra) gene family of NK cell receptors appeared to be the most attractive candidate genes for Cmv1. However, an independent mapping effort positioned Cmv1 within a 390-kb region distal to the Ly49b (9), which excludes all the members of Ly49 gene family as a candidate for Cmv1. The combination of this interval with a 1.6-Mb genetic interval generated by our group produced a 200-kb overlapping region, suggesting the possibility of modifier genes of MCMV-resistance in the BALB/c or A/J strains used for mapping.
The genetic complexity of this genomic domain was further underscored by haplotype analysis for a set of 25 markers in the vicinity of Cmv1, which revealed the presence of different haplotypes in a panel of 18 inbred strains of mice (14). Interestingly, the majority of strains analyzed (16/18) were MCMV susceptible. Accordingly, the MCMV resistance allele, Cmv1 r, was associated with a haplotype unique to strains of the C57BL background. Susceptible strains could be classified in two groups based on their haplotype at Cmv1, indicating the presence of two unrelated MCMV susceptibility alleles, named Cmv1 sBALB and Cmv1 sFVB, raising the possibility of genetic heterogeneity at Cmv1, such as mutations in alternate Ly49 members in different inbred strains. While the Cmv1 sBALB haplotype is distinct from Cmv1 r at every locus, identical alleles are shared at several loci between Cmv1 sFVB and Cmv1 r, indicating that they share a common ancestral haplotype. Close inspection of these haplotypes pointed to two regions in disequilibrium with susceptibility corresponding to the Ly49d-Ly49a interval and to the genomic region distal to Ly49b (14). A possible interpretation of this result is that tightly linked loci at Cmv1 contribute to MCMV susceptibility or resistance.
Recently, three independent groups reported the Ly49h gene as an important determinant in MCMV resistance (15–17). We took advantage of the recombinant inbred strain BXD-8, derived from Cmv1 r C57BL/6 and Cmv1 s DBA/2 parents (18). This strain is of particular interest, as it is highly susceptible to MCMV infection but harbors a C57BL/6 haplotype at Cmv1 (4). Using a combination of linkage analyses in an informative (BXD-8 × C57BL/6)F2 population, together with genetic complementation in vivo using mouse strains congenic for different Cmv1 segments, physical mapping and expression analysis, we demonstrated that the MCMV susceptibility trait of BXD-8 is associated with a 23-kb deletion encompassing Ly49h (15). At the same time, using a panel of monoclonal antibodies, the laboratory of Yokoyama demonstrated that the Ly49H receptor was absent from BXD-8 mice and, moreover, that treatment with the anti-Ly49H monoclonal antibody 3D10 before MCMV infection abrogated MCMV-resistance in the C57BL/6 mice (16). Welsh and coworkers confirmed and extended these results by demonstrating that depletion of Ly49H+ NK cells specifically affects MCMV response but not that to vaccinia virus or lymphocytic choriomeningitis virus (17). The Ly49H protein is an activating receptor expressed in approximately half of the NK cell population in adult C57BL/6 mice (19). Ly49H signals through the kinase-associated adaptor molecule DAP12 (20) probably upon recognition of the MCMV-encoded molecule m157 (21, 22), a glycoinositol phospholipids (GPI)-linked protein structurally related to nonclassical MHC.
While several lines of experiments clearly demonstrate that absence of Ly49H is associated with MCMV susceptibility, it was important to evaluate the role of the distal 200-kb region during the MCMV response as well as to determine whether expression of Ly49H is sufficient to confer MCMV resistance in order to provide the formal demonstration of Ly49h/Cmv1 identity. To achieve this aim we have used an approach of functional in vivo complementation by genetic transfer of bacterial artificial chromosome (BAC) clones overlapping important regions on the Cmv1 interval that we report here. The results obtained from this study clearly pinpointed the precise location of Cmv1, providing conclusive evidence that Cmv1 and Ly49h are allelic. In addition, these transgenic mice provided an opportunity to investigate several questions regarding the role of Ly49H during MCMV infection such as the threshold of Ly49H+ NK cells for Cmv1, the possible genetic heterogeneity of Cmv1, and the rescue by the transfer of Ly49h on different Cmv1 susceptible backgrounds.
Materials and Methods
Mice.
All mice were initially purchased from The Jackson Laboratory and subsequently maintained at the animal facility at the University of Ottawa in agreement with guidelines and regulations of the Canadian Council of Animal Care.
Generation of BAC Transgenic Mice.
BAC clones 13J11 and 128D23 spanning the corresponding candidate regions within our Cmv1 interval were previously described (11). These BAC clones were obtained from RPCI-23 mouse BAC library originated from DNA of C57BL/6 (Cmv1 r MCMV-resistant) strain (Research Genetics). The BAC DNA was isolated by the alkaline lysis method with gentle handling to prevent the breakage of BAC DNA, linearized by either NotI or XhoI/SalI digestion, and size fractionated on 1% low melting agarose gel using pulsed field gel electrophoresis (PFGE; 6 V/cm, 120 angle, 6 s of switching time for 24 h). The BAC insert DNA was recovered from the gel by β-Agarase I treatment (New England BioLabs, Inc.) without exposure to UV light, as described (23), and purified by dialysis for 4 h against microinjection buffer (10 mM Tris, pH 7.5, 0.1 mM EDTA) by floating on a Millipore VSWP 2500 (pore 0.05 μm) filter disc. The BAC DNA was microinjected into fertilized pronucleus of fertilized eggs derived from FVB/N at 1.5 ng/μl concentration. After injection, the eggs were transferred into the oviduct of 0.5-d pseudo-pregnant foster CD-1 female mice. All procedures for generating BAC transgenic mice were performed at the transgenic facility of the Ottawa Hospital Research Institute or McGill University. Transgenic founders were identified via PCR (see below) and subsequently bred to BXD-8, FVB/N, and BALB/c mice by serial backcrossing.
Genomic DNA Analysis.
To identify the presence of BAC DNA, tail DNA was extracted using alkaline lysis. Briefly, a 2-mm tail biopsy was incubated in 600 μl of a 50 mM NaOH solution at 95°C for 20 min with mixing every 5 min. At the end of the incubation, the DNA solution was vortexed for 30 s and neutralized by adding 50 μl of 1 M Tris (pH 7.0). Aliquots of 2 μl of this solution were used for PCR reactions. A first round of PCR screening was done using the following set of primers: 5′-GCCGCTAATACGACTCACTATAGGG-3′ and 5′-GAAGATTTGCAGGCCAGGAG-3′ (amplified a 163-bp fragment from the T7-end of BAC 13J11 vector-insert junction) and 5′-CCGTCGACATTTAGGTGACAC-3′ and 5′-CTGAGTTCAATTCCTAATTC-3′ (amplified a 187-bp fragment from the SP6-end of BAC 13J11 vector-insert junction); 5′-GCCGCTAATACGACTCACTATAGGG-3′ and 5′-TCCTGCAGAAATCAGAGTTCTGTT-3′ (amplified a 74-bp fragment from the T7-end of BAC 128D23 vector-insert junction) and 5′-CCGTCGACATTTAGGTGACAC-3′ and 5′-GGGGACTGAAAGATAGAAGTG-3′ (amplified a 222-bp fragment from the SP6-end of BAC 128D23 vector-insert junction). To confirm the integrity of BAC DNA in the transgenic mice, several internal polymorphic markers were tested by PCR with previously reported primer pairs (14, 15). In addition, Ly49e and D6Ott115 primer pairs (10) were used to determine the Cmv1 phenotype of mice bred into the BALB/c background. To determine the copy number of transgene on founder animal, 20 ng of tail DNA was semiquantitatively analyzed by PCR at three different cycles and their intensity compared with corresponding sample from homozygous C57BL/6 mice. Oligos specific for Ly49h intron1 were used to amplify B6 copies and the sequence of the oligos were, D6Ott151-F: 5′-GTGCTACCACTGAAAACCATTG-3′ and D6Ott151-R: 5′-CTGTCTCTTGAGTCACCTGCAC-3′.
Detection of Transgene Expression by PCR Analysis.
Transgene expression was detected by semiquantitative RT-PCR. Briefly, total splenic RNA was extracted using Trizol reagent (Life Technologies), and 2 μg of total RNA was reverse transcribed using oligo-dT primers and Superscript II reverse transcriptase according to the manufacturer's protocols (Life Technologies). Gene specific oligonucleotides for the PCR step were, EST335500-F: 5′-GGCCGTGGTCGCAGTGAACT-3′ and EST335500-R: 5′-GCTGGCTTCTCTGGCCACCT-3′; Ly49d-F: 5′-GGCTCAAGGAGACACGGAAG-3′ and Ly49d-R: 5′-TCCCAAAATCTTCCAGATTG-3′; Ly49h-F: 5′-AGCCTCTTAGGGGATACAGAC-3′ and Ly49h-R: 5′-TGTCAAGATAGAT-AGGAGAGG-3′; Ly49i-F: 5′-GATGAATGAGCCGGAGGTC-3′ and Ly49i-R: 5′-TTTCACTGTTCCATCTGTCCT-3′; Gapdh-F: 5′-ACCACAGTCCATGCCATCAC-3′ and Gapdh-R: 5′-TCCACCACCCTGTTGCTGTA-3′.
Production of Anti-Ly49H Antibodies.
Antibodies against Ly49H were generated in rabbits using a bacterial glutathione S transferase (GST)-fusion protein encompassing amino acids residues 1 to 44 which corresponds to the entire cytoplasmic domain of Ly49H. The anti-Ly49H serum was adsorbed on GST-sepharose 4B beads and anti-Ly49H antibodies were immunoaffinity purified on GST/Ly49H fusion protein coupled to sepharose beads as described (24).
Flow Cytometric Analysis.
To isolate splenic leukocytes, spleen was ground between rough surfaces of glass slides and treated with Red Blood Cell Lysing Buffer (Sigma-Aldrich) to remove red blood cells. For isolation of leukocytes from liver, the organ was homogenized with a tissue grinder and the homogenate was passed through a fine metal screens to remove large debris and cell clumps. Then, 25% of hepatic cells were layered over Lympholyte-M (Cedarlane) and centrifuged for 20 min at 1,500 g at room temperature to remove red blood cells. After centrifugation, a lymphocyte layer at the interface was collected using Pasteur pipette. NK cells were enriched from spleen and liver leukocytes by magnetic positive separation using anti-DX5 microbeads following the manufacturer's protocol (MACS®; Miltenyi Biotec). The freshly purified DX5+ NK cells were stained for surface expression of DX5 using PE-conjugated murine anti-DX5 (BD Biosciences). After washing, the cells were fixed, permeabilized and internally stained for cytoplasmic tails of Ly49H receptor. FITC-conjugated anti–rabbit IgG (BD Biosciences) was used to stain polyclonal anti-Ly49H that was previously bound. Cell population was analyzed by flow cytometry on an Epics analyzer (Coulter).
Cmv1 Phenotype.
Mice between 6 and 8 wk of age were challenged with a virulent stock of the MCMV Smith strain (7) originally obtained from the American Type Culture Collection. Mice were injected intraperitoneally with 0.2 ml of PBS containing 5 × 103 PFU of MCMV. At 3 d after infection, the degree of viral replication in the spleen and the liver was determined by plaque assay in mouse embryonic fibroblasts as described (7).
Statistical Analysis.
Significance of the differences observed for viral titers or the size of Ly49H+ NK cell population in reference to control nontransgenic groups or between transgenic lines was assessed using the two-tailed Student's t test.
Results
Characterization of BAC Clones Used in Transgenesis.
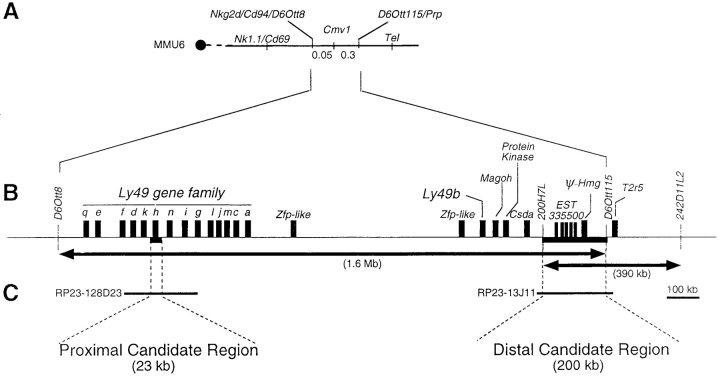
In the absence of an in vitro system for Cmv1, we used in vivo complementation with BAC clones expressed in transgenic mice as a functional assay for the Cmv1 gene. We reasoned that introduction of the dominant Cmv1 r (C57BL/6) allele onto a recessive Cmv1 s background should produce F1 progeny uniformly resistant to MCMV infection. High resolution genetic and physical mapping of the Cmv1 interval identified a minimal genetic interval that is flanked by the markers D6Ott8 and D6Ott115 and which corresponds to a physical distance of ∼1.6 Mb of genomic DNA (Fig. 1, A and B ; references 10 and 11). Within this large interval, two regions were of particular interest for complementation analysis: a distal candidate region of ∼200 kb that corresponds to the common domain for Cmv1 as defined from two independent mapping efforts (9, 10), and a proximal candidate region of ∼23 kb encompassing Ly49h whose absence is associated with susceptibility to cytomegalovirus in BXD-8 mice (Fig. C; references 15 and 16).
Figure 1.
Genomic targets for transgenic analysis of the Cmv1 locus. (A) Composite genetic linkage map of mouse chromosome 6 in the vicinity of Cmv1. The order and distances of the loci was determined by pedigree analysis. The centromere is represented by a black circle. Recombination frequencies in centimorgan (cM) are shown below the chromosome. Nk1.1, Cd69, Cd94, and Nkg2d code for C-lectin type receptors. Prp, prolin-rich protein; Tel, translocation-ets-leukemia (reference 7). (B) Blow-up of the minimal genetic interval between D6Ott8 and D6Ott115 and transcriptional map showing candidate genes for Cmv1. Arrows indicate the minimal Cmv1 interval as defined by Depatie et al. (references 10 and 11) and Brown et al. (reference 9) (C). Localization of BAC clones used for transgenesis (reference 11).
In the process of cloning Cmv1, we have previously completed a physical map in the Cmv1 interval using a series of 73 overlapping BAC clones and screened the BAC clones to provide genomic segments fully encompassing each candidate region (11). Based on these considerations, BAC clones 13J11 and 128D23 were identified for complementation analysis (Fig. 1 C). The BAC 13J11 spans the common Cmv1 interval (200 kb) as shown by the presence of marker 200H7L, a YAC end defining the proximal boundary of Cmv1 in Brown et al.'s map (9), as well as D6Ott115, a marker defining the distal end of the Cmv1 interval in Depatie et al.'s map (10). Genomic DNA sequence analysis indicated the presence of at least three different transcription units in BAC 13J11, including a pseudogene for one of the chromatine proteins of the High Mobility Group (25) and the gene for the taste receptor T2R (26) localized distal to D6Ott115. The third gene, which presents 5 copies, corresponds to EST335500 (GenBank/EMBL/DDBJ accession no. W18806), a novel spleen-specific gene whose expression is absent in the susceptible BALB/c strain (see below). The BAC clone 128D23 is located in the middle of Ly49 gene cluster where it overlaps the 23-kb deletion present in the BXD-8 strain. This BAC clone contains five Ly49 genes, three functional genes (Ly49d, Ly49h, and Ly49i) and two pseudogenes (Ly49k and Ly49n) as deduced from its available genomic DNA sequence (GenBank/EMBL/DDBJ accession no. AC090127).
Production and Analysis of BAC Transgenic Mouse Lines for Clone 13J11.
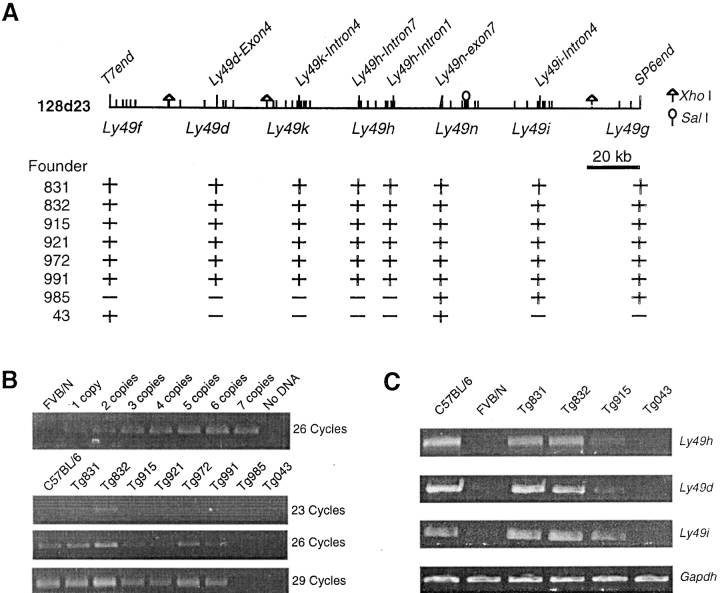
We generated transgenic mice by injection of purified BAC insert DNA into the male pronucleus of FVB/N zygotes. Transgenic mice were identified by PCR amplification of the BAC vector-insert junctions ends and by the presence of internal polymorphic markers as indicated in Fig. 2 A. In summary, 3 out of 20 founder animals were positive for the transgenic markers and carried an intact 210 kb transgenic construct (Fig. 2 A). The F1 progeny of founder animals with BALB/c were analyzed by the PCR approach, indicating transmission of the transgene to ∼50% of their progeny as expected for nonmosaic germ-line transmission.
Figure 2.
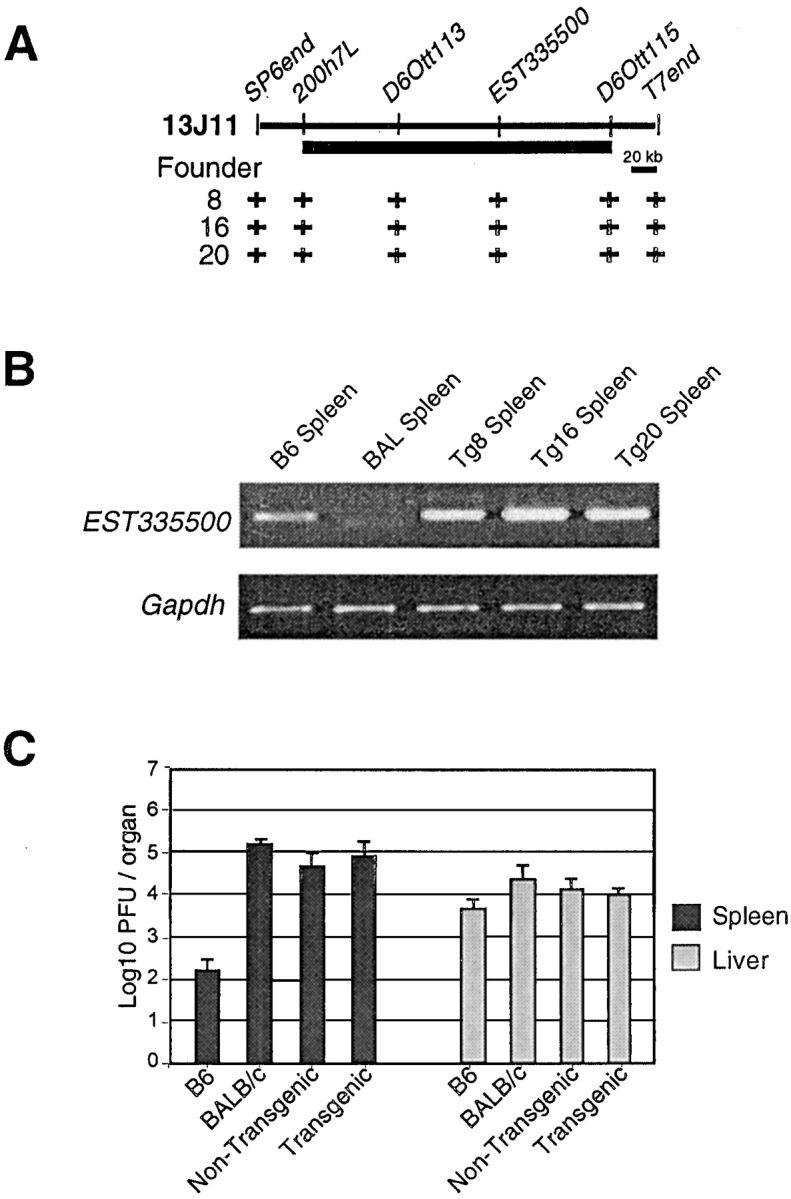
Characterization of BAC clone 13J11 transgenic lines. (A) Physical map of BAC clone 13J11 with the localization of markers used for identification of transgenic lines by PCR. The black bar indicates the physical domain common to two independent mapping efforts for Cmv1 (references 9 and 10). The (+) indicates a positive result in STS content analysis of transgenic founder animals. (B) RT-PCR for EST335500 and Gapdh control RNA from spleens of wild-type strains and transgenic lines mice. (C) Replication of MCMV in the spleen and liver of inbred and transgenic lines. Viral titers in the organs of mice per group were determined by plaque assay 3 d after infection with 5 × 103 PFU of MCMV.
To demonstrate proper expression of genes encoded in BAC 13J11, we took advantage of the fact that only the C57BL/6 allele of EST335500 is expressed in the mouse spleen. As shown in Fig. 2 B, high levels of EST335500 RT-PCR product were obtained in spleen of the three transgenic lines as well as from C57BL/6 RNA. No product was observed in BALB/c samples. A control RT-PCR with Gapdh primers demonstrates similar results for all samples, indicating that the RNA used as starting material was consistent in quality and quantity in all the animals and that genes in the construct were expressed in transgenic animals (Fig. 2 B).
To examine the effect of BAC 13J11 in resistance to MCMV, transgenic founders were backcrossed with BALB/c, a susceptible strain used for mapping of Cmv1 (9, 10). After the acquisition of the context of homozygous Cmv1 sBALB haplotype in the Cmv1 interval, transgenic and nontransgenic siblings were infected with MCMV and viral titers in the spleen were measured 3 d after infection. As shown in Fig. 2 C, no protective effect was observed in transgenic mice, which presented a viral load identical to BALB/c (Cmv1 s) and 102–103 times higher than the resistant C57BL/6 mice. Taken together, these results indicate that neither EST335500 nor any other genes in the 210-kb region covered by 13J11 confer resistance to cytomegalovirus infection, excluding the presence of MCMV-resistance genes within this genomic domain.
Production and Analysis of BAC Transgenic Mouse Lines for Clone 128D23.
The BAC 128D23 transgenic mice were produced by microinjection of FVB/N (Cmv1 s) zygotes, and founder animals were analyzed by PCR with several markers as shown in Fig. 3 A to identify transgenic mice. A summary of the BAC transgenic lines and their characteristics is presented in Table I. Out of 40 progenies, six founders were shown to present an intact 128D23 insert, while two of them missed one of two BAC ends (Fig. 3 A). Interestingly, founder 043 showed discontinuous sequence tagged site (STS) content, indicating possible rearrangement between Ly49f and Ly49n. Transgene copy number in founder animals was determined using semiquantitative PCR with Ly49h-specific primers. The range of transgene copy numbers was 1 copy in line Tg915, 2 copies in line Tg831 and 3–4 copies in line Tg832 (Fig. 3 B). Interestingly, copy number correlated well with expression of functional genes present in BAC 128D23 as shown by RT-PCR analysis using Ly49d, Ly49h, and Ly49i specific primer pairs (Fig. 3 C). To note, primer pairs were also allele specific for B6 transcripts since no product was detected from nontransgenic FVB/N control RNA.
Figure 3.
Characterization of BAC clone 128D23 transgenic lines. (A) Physical map of BAC clone 128D23 with the localization of markers used for identification of transgenic lines by PCR. The localization of genes present in the BAC clone is also indicated. The (+) indicates a positive result in STS content analysis of transgenic founder animals. Restriction sites of XhoI and SalI were shown. (B) Semiquantitative PCR to determine transgene copy number on founders. Ly49h-specific primers were used to amplify 1–7 copies of BAC DNA. Results were compared with those obtained from transgenic lines in 26 cycles of amplification. (C) RT-PCR for genes contained in BAC clone 128D23 and Gapdh control RNA from spleens of wild-type mice and transgenic lines. Ly49h, Ly49d, and Ly49i oligonucleotide primer pairs were designed to amplify specifically the C57BL/6 alleles (see Materials and Methods).
Table I.
Summary of Genetic and Phenotypic Analysis of Transgenic Mice Expressing Ly49H
| Transgenic line | Integration of BAC DNA |
Ly49h
copy no. |
Ly49h RNA expression |
Protein expression of Ly49Ha |
MFI of Ly49H+ cellsb |
Viral titer on spleenc |
|---|---|---|---|---|---|---|
| 212 kb full BAC 128D23 | ||||||
| Tg832 | Intact | 3–4 | +++ | 45.3 ± 3% | 5.5 ± 0.4 | 2.1 ± 0.4 |
| Tg915 | Intact | 1 | + | 28 ± 2% | 3.6 ± 0.3 | 3.1 ± 0.4 |
| Tg985 | Partial | 0 | No | 6 ± 1% | N/D | 4.8 ± 0.3 |
| Tg043 | Partial | 0 | No | 3 ± 1% | N/D | 4.7 ± 0.4 |
| 79 kb SalI-XhoI fragment of BAC 128D23 | ||||||
| Tg796 | Intact | N/D | No | N/D | N/D | 4.5 ± 0.2 |
| Tg814 | Intact | N/D | ++ | 35 ± 2% | 4.6 ± 0.3 | 2.3 ± 0.2 |
Data analyzed on four mice in each group on FVB/N background, except for Tg814, of which three mice were analyzed. Similar data were obtained for transgenic mice in the context of the BXD-8 or BALB/c.
Indicates the percentage of Ly49H+ population among DX5+ cells in the spleen (mean ± SD).
Indicates median of fluorescence intensity of Ly49H+ cells (mean ± SD).
Indicates log10 PFU/organ (mean ± SD). For controls, viral titers for resistant C57BL/6 and susceptible FVB/N were 2.0 ± 0.3 and 4.5 ± 0.2, respectively.
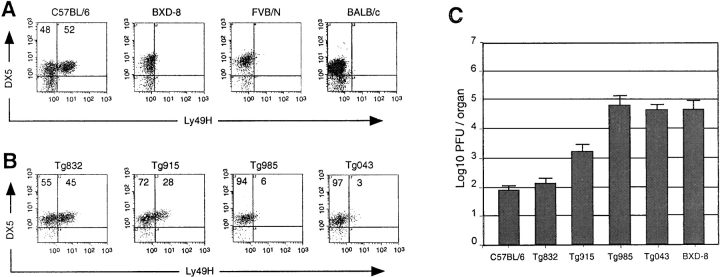
To monitor expression of Ly49H in different mouse strains, we produced a polyclonal antibody against the cytoplasmic tail of the receptor. This antisera stained 52% of DX5+ NK cells isolated from C57BL/6 mouse spleen consistent with previous results using an antibody directed against the extracellular domain of Ly49H (19; Fig. 4 A). No signal was observed in either FVB/N, BALB/c, or BXD-8 indicating that this anti-Ly49H polyclonal antibody does not cross-react with other members of the Ly49 family. In particular, we have shown that, with the exception of Ly49h, the BXD-8 strain expresses the full C57BL/6 Ly49 repertoire, strongly supporting the specificity of the antibody (15).
Figure 4.
Acquisition of MCMV-resistance in BAC clone 128D23 transgenic lines. (A) Enriched NK cell preparations from spleens of wild-type strains were stained with the mAb DX5 and the rabbit polyclonal antibody against the cytoplasmic domain of the Ly49H peptide reported here. The numbers in the density plots indicate the percentage of DX5+ lymphocytes either Ly49H− or Ly49H+. (B) Enriched NK cell preparations from spleens of BAC 128D23 transgenic lines were stained with the monoclonal antibody DX5 and the rabbit polyclonal antibody against the cytoplasmic domain of the Ly49H. The numbers in the density plots indicate the percentage of DX5+ lymphocytes either Ly49H− or Ly49H+. (C) Replication of MCMV in the spleen of wild-type and transgenic lines. Viral titers in the spleen of five mice per group were determined by plaque assay 3 d after infection with 5 × 103 PFU of MCMV. Statistically significant differences were observed between transgenic (Tg832, Tg915) and nontransgenic mice at P values of < 0.0005.
Interestingly, the RNA expression of Ly49h strongly correlated with protein expression as determined by the size of Ly49H+ NK cell population and the level of its expression on single cell. The size of the Ly49H+ NK cell population varied in transgenic lines with 45 and 28% of Ly49H+ NK cells in lines Tg832 and Tg915, respectively (P < 0.005; Fig. 4 B). Moreover, the median of fluorescence intensity (MFI) of the Ly49H+ NK cells was significantly different with a MFI of 5.5 ± 0.4 for line Tg832 and MFI of 3.6 ± 0.3 for line Tg915 (P < 0.005). These differences correlated with RNA expression of Ly49h and may depend on either the number of Ly49h copy or the site of integration of the constructs. As expected, no significant expression was observed in lines Tg985 and Tg043, which lack Ly49h coding sequences.
Ly49H Confers Resistance to MCMV Infection.
The phenotypic consequences of BAC128D23 expression in resistance to infection with MCMV was evaluated in the progeny of a cross between transgenic animals and the susceptible mouse strain BXD-8. This strain expresses the full Ly49 C57BL/6 repertoire with the exception of Ly49h (15). As shown in Fig. 4 C, transgenic expression of the 212-kb BAC 128D23 clone conferred resistance to MCMV infection in BXD-8 mice. Among the four BAC 128D23 lines included in our analysis, line Tg832 is of particular interest because it presented splenic viral titers comparable to those of resistant C57BL/6 mice (102 PFU/spleen), whereas line Tg915 presented an intermediate phenotype (103-104 PFU/spleen; Fig. 4 C). As expected, lines Tg985 and Tg043 lacking Ly49h allowed uncontrolled viral replication at the same level as those of susceptible strains (104–105 PFU/spleen).
To reduce the rescuing interval further, we used the same functional assay to test a 79-kb XhoI and SalI fragment of BAC128D23 encompassing a single functional gene, Ly49h. Out of two transgenic lines carrying the complete construct (Fig. 5 A), only one line Tg814, was shown to express Ly49H in ∼35% of DX5+ NK cells (Fig. 5 B). Phenotypic analysis of line Tg814 demonstrated that transgenic animals acquired MCMV resistance upon introduction of the 79-kb transgene (Fig. 5 C). It is important to note, however, that at this early time point the effect of Cmv1 (or the transgene) is observed primarily in the spleen (see below; reference 4). The proper expression of Ly49H in line Tg814 indicates that Cmv1 gene and its requisite regulatory sequences are contained within the genomic construct. These results showed that MCMV susceptibility can be reversed by proper expression of a single functional gene, Ly49h, thereby providing the formal demonstration of allelism between Ly49h and Cmv1. In addition, partial recovery observed in line Tg915 shows a gene dosage effect indicating a threshold value in providing resistance above 28% of Ly49H+ cells.
Figure 5.
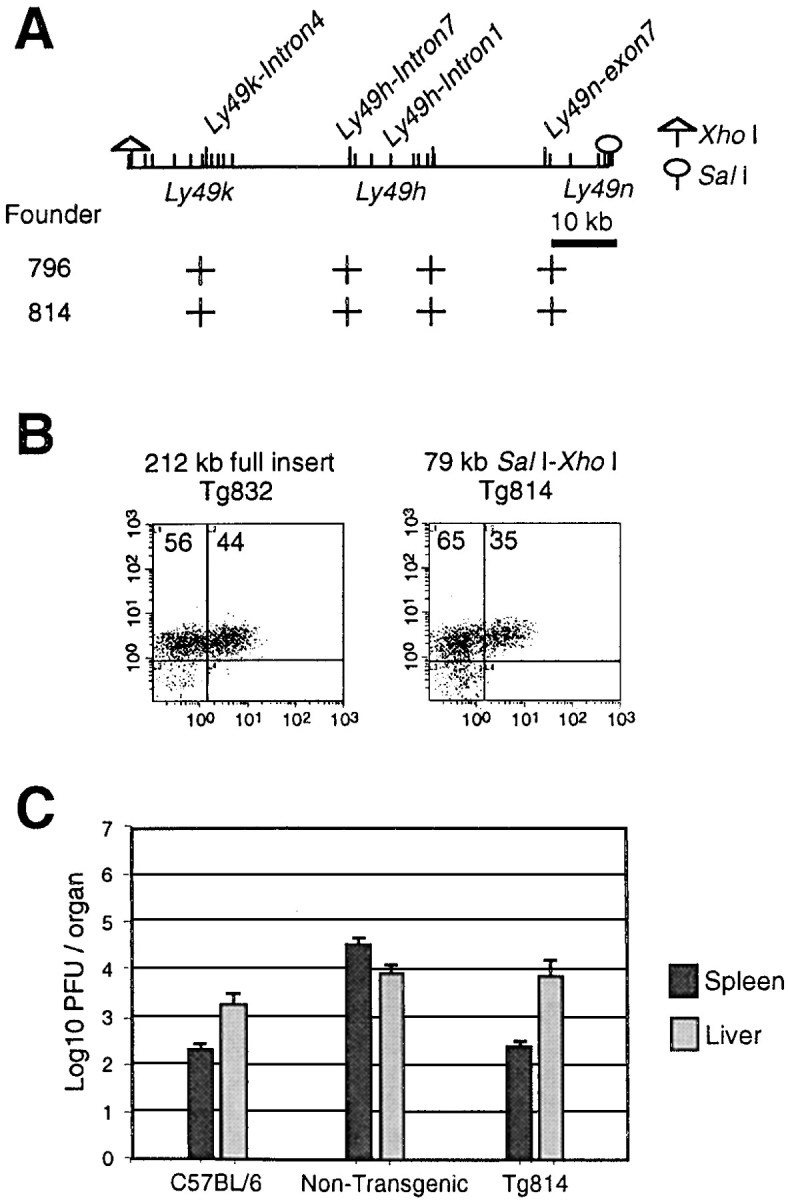
A 79-kb genomic construct containing Ly49H confers resistance to MCMV infection. (A) Physical map of a 79-kb fragment with the localization of markers used for identification of transgenic lines by PCR. The localization of genes present in the BAC clone is also indicated. Restriction sites of XhoI and SalI used to generate the 79-kb fragment were shown. The (+) indicates a positive result in STS content analysis of transgenic founder animals. (B) Enriched NK cell preparations from spleens of lines Tg832 and Tg814 were stained with the mAb DX5 and the rabbit polyclonal antibody against the cytoplasmic domain of the Ly49H. The numbers in the density plots indicate the percentage of DX5+ lymphocytes either Ly49H− or Ly49H+. (C) Replication of MCMV in the spleen and liver of wild-type and transgenic lines. Viral titers in the spleen of five mice per group were determined by plaque assay 3 d after infection with 5 × 103 PFU of MCMV.
Ly49H Confers Resistance to MCMV Infection Independently of the Genetic Background.
With the exception of the 23-kb deletion encompassing Ly49h, the susceptible BXD-8 and the resistant C57BL/6 strains are identical at the Cmv1 locus (15). In contrast, classical inbred strains present two independent susceptible haplotypes. In particular, susceptible BALB/c and A/J (used for genetic mapping) and FVB/N (used for transgenesis) present unrelated haplotypes for all loci analyzed in the vicinity of Cmv1 (14). These results indicated two independent origins for the MCMV susceptibility Cmv1 s allele and suggested the possibility of genetic heterogeneity at Cmv1, such as mutations in alternate Ly49h members in different inbred strains. In addition, a BALB/c modifier gene of MCMV resistance has been proposed to be present in the distal candidate region (9). Moreover the susceptibility to MCMV has been shown to be controlled by various cytokines and chemokines such as IFN-α, β, IL-12, TNF, and macrophage inflammatory protein 1α (MIP-1α; references 27–29) whose expression is very diverse in different genetic backgrounds (30, 31). Thus, the rescue by the transfer of Ly49h might be influenced by the variable level of expression of these molecules.
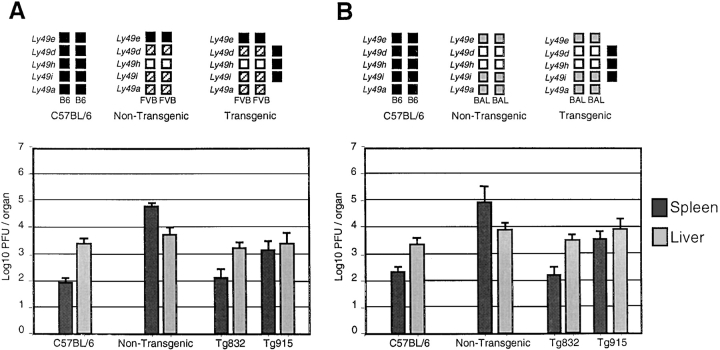
To evaluate a possible effect of the genetic background in MCMV susceptibility, the response of transgenic mice to MCMV infection was investigated in the context of a homozygous Cmv1 sBALB and Cmv1 sFVB background. Five animals from lines Tg832 and Tg915 were analyzed as described above. As shown in Fig. 6, A and B , both lines supported similar levels of MCMV replication to those obtained in the BXD-8 background, supporting a critical role for Ly49H in MCMV resistance independent of the genetic background.
Figure 6.
Ly49H confers resistance to MCMV infection independently of the genetic background. Replication of MCMV in wild-type and transgenic Tg832 and Tg915 mice in the context of homozygous FVB/N (Cmv1 sFVB; A) and BALB/c (Cmv1 sBALB; B) was shown. Haplotypes of Ly49 family for Cmv1 r (C57BL/6), Cmv1 sFVB and Cmv1 sBALB are shown; black boxes, B6 alleles; hatched boxes, FVB alleles; gray boxes, BALB/c alleles; white boxes indicate the absence of alleles. Viral titers in the spleen and liver of five mice per group were determined by plaque assay 3 d after infection with 5 × 103 PFU of MCMV.
Regulation of Trans-Ly49H Expression in Transgenic Mice.
The identification of Ly49H as the critical resistance factor in host response to MCMV predicted that cells expressing Ly49H should display evidence of specific activation during MCMV infection in vivo. Soon after MCMV infection, there is a general activation of NK cells to produce IFN-γ and proliferate in the spleen (32). At later time points, there is a specific expansion of Ly49H+ cells in C57BL/6 mice (32), while expression of MIP-1α promotes recruitment of NK cells and formation of inflammatory foci in the infected liver (29).
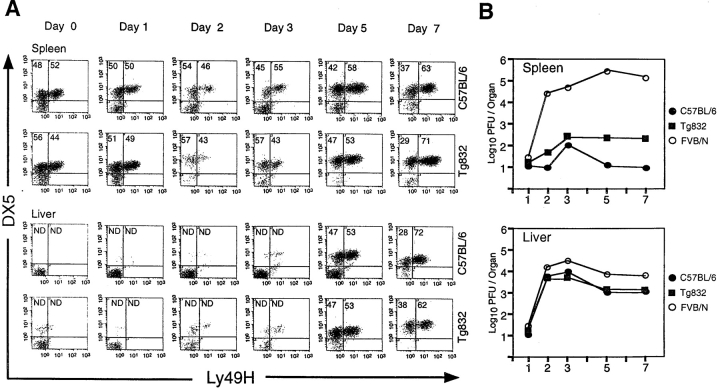
To investigate whether control sequences for clonal expansion are present in the transgenic construct, we followed Ly49H expression by FACS® analysis of DX5+ NK cells isolated from spleen and liver over a 1-wk period after infection. The kinetics of the Ly49H+ NK cell expansion was identical for wild-type C57BL/6 mice and the line Tg832 (Fig. 7 A). The percentage of splenic and liver NK cells expressing Ly49H gradually increased during infection (Fig. 7 A). By day 7, the proportion of NK cells in spleen and liver expressing Ly49H was ∼70%, compared with only 50% in spleens of mock-infected mice. Importantly, the increased percentage of Ly49H+ cells in the liver was accompanied by an enhanced control of viral titers in these organs, supporting a direct role for this receptor in MCMV resistance in the liver (Fig. 7 B). Taken together, these results indicate a proliferation of Ly49H+ NK cells during infection and demonstrate that regulation of the Ly49h transgene is indistinguishable from that of its endogenous counterpart in C57BL/6 mice, possibly due to specific activation through the Ly49H receptor itself.
Figure 7.
Kinetics of trans-Ly49H expression. (A) Ly49H expression was followed during a 1-wk period in the transgenic line Tg832 and C57BL/6 by staining with the mAb DX5 and a rabbit polyclonal antibody against the cytoplasmic domain of the Ly49H. The numbers in the density plots indicate the percentage of DX5+ lymphocytes either Ly49H− or Ly49H+. Enriched NK cell preparations were obtained from spleen and liver tissues after 0, 1, 2, 3, 5, and 7 d after infection. The proportion of NK cells in liver until 3 d after infection was not determined owing to the insignificant number of DX5+ cells on samples. (B) A fraction of the organ homogenates used in panel A were used to determine the course of MCMV infection at the indicated time points in transgenic Tg832 mice and wild-type C57BL/6 (Cmv1 r) and FVB/N (Cmv1 s) controls. Viral titers were determined by plaque after infection with 5 × 103 PFU of MCMV.
Discussion
Ly49h was identified as a strong candidate for Cmv1 based on the following observations: (a) it mapped within the minimal genetic and physical intervals delineating the Cmv1 region (11); (b) a 23-kb deletion encompassing Ly49h in the context of a C57BL/6 (Cmv1 r) is associated with susceptibility (15); (c) expression of Ly49h is restricted to Cmv1 r mice (15, 16); (d) Ly49h encodes for a C-type lectin activating receptor specifically expressed on NK cells (19); (e) depletion of Ly49H+ NK cell populations abrogates MCMV-resistance (16, 17); (f) Ly49H recognizes an MCMV-encoded molecule, m157, a GPI-membrane bound protein that is expressed early during infection (21, 22).
While these data provide compelling evidence of the role of Ly49h in MCMV infection, important questions remain to be clarified concerning the association between Ly49h and Cmv1: (a) an independent genetic mapping effort identified a region distal to the Ly49 cluster as associated with the MCMV-susceptibility phenotype (9), this region, referred to as the distal candidate region, was also detected by haplotype analysis in genealogically and phylogenetically unrelated inbred strains of mice (14); (b) the minimal Cmv1 physical interval was shown to contain other genes that were potential candidates for an infection resistance locus. For example Csda, that encodes a putative repressor for human GM-CSF, a key molecule for macrophage maturation (33), or the spleen-specific gene, EST335500, described here, are both localized in the vicinity of the distal candidate region; (c) functional analysis with monoclonal antibodies could be misleading because the possibility of cross-reactivity with a yet to be identified member of the Ly49 family; (d) the presence of two independent origins for the Cmv1 s alleles, Cmv1 sFVB or Cmv1 sBALB, suggested that MCMV susceptibility could be associated with either independent mutations on the same gene or mutations in tightly linked genes within the Cmv1 interval (14).
To address these questions, we performed experiments in which we transferred cloned copies of two important regions within the minimal Cmv1 interval onto a susceptible (Cmv1 sFVB or Cmv1 sBALB) genetic background and analyzed the effects of this transfer on resistance to infection. In the present study, we have shown that transfer of DNA constructs that overlap the distal candidate region did not affect resistance, whereas transfer of genomic constructs overlapping Ly49h caused gain-of-function on an otherwise susceptible background. Moreover, using a 79-kb genomic fragment containing no functional gene other than Ly49h, the phenotypic rescue obtained in these experiments was complete, unambiguous, and quantitative. Combined with the previous positional cloning studies and the antibody depletion experiments, the transgenic transfer of Ly49h presented here unequivocally establishes that Ly49h and Cmv1 are allelic.
Our results did not detect either a gain-of-function after genetic transfer of the distal construct BAC 13J11 or a loss-of-function in Ly49h-transgenic animals in the BALB/c context. A possible effect of genes distal to the Ly49 cluster was previously suggested by genetic analysis using crosses between the MCMV-resistant C57BL/6 and the MCMV-susceptible BALB/c or A/J mouse strains (9). The gene EST335500 presented here was of particular interest because it is flanked by markers defining the distal region involved in MCMV resistance and it is specifically expressed in the spleen, which is a main site of control of MCVM replication. Moreover, EST335500 transcript is present in tissues of Cmv1r C57BL/6 mice, while it is absent in those of Cmv1s BALB/c or A/J mice, indicating a possible role in MCMV resistance. However, lack of complementation with 13J11 transgenic mice indicates that polymorphisms in this genomic domain, including those identified in EST335500, do not affect the MCMV response, as measured by splenic or liver viral titers. However, we cannot rule-out at this time the possibility of a MCMV strain–specific effect as we have used the original Smith strain (34), while others have used the strain K181 (9, 12, 35). K181 is considered to be a more virulent strain, as at a similar infectious dose it produces more plaques and cytopathic effects than the Smith strain, which might allow the detection of more subtle effects of genomic regions distinct from Ly49h in antiviral response (36). The transgenic models reported here will allow us to test this possibility.
The use of genomic DNA in transgenesis has the advantage of allowing the function and regulation of genes to be studied in conditions approaching their natural context. Indeed, this is the first study reporting successful production of mice that are transgenic for an activating NK receptor whereas several transgenic mice expressing inhibitory Ly49s were previously generated (37–40). Ly49 gene expression is known to be regulated in an isoform-specific fashion, by a process that restricts the number of Ly49 genes expressed per single NK cell and generates clonal variability (19, 41, 42). Furthermore, Ly49H protein is expressed on the surface of 56% of NK1.1+CD3− splenic NK cells (19), the cell population known to be engaged by Cmv1. Using a polyclonal antibody specific for the cytoplasmic tail of Ly49H we observed the appearance of the Ly49H protein in transgenic NK cells and restoration of resistance to infection, suggesting that the regulatory sequences for proper expression of the gene are contained within the genomic fragment used for transgenesis. Moreover, trans-Ly49H expression seems to be NK cell specific since no expression was observed in other lymphocyte lineages, such as T or B cells in flow cytometric analysis of total splenocytes. So far, little is known about cis- and trans-elements that regulate activating Ly49 receptors. A comparative analysis of the promoter regions of mouse Ly49 genes indicated the presence of two consensus binding sites for the T cell factor 1 (TCF-1), a member of the high mobility group protein family (43), immediately upstream of a predicted TATA box (44). Using TCF-1−/− mice, Held and coworkers demonstrated that TCF-1 was essential for acquisition but not maintenance of expression of Ly49 receptors, such as Ly49A and Ly49D, as the size of the Ly49A and D populations were greatly reduced in the knockout mice in contrast to the level of receptor expression in individual cells (45). Our results indicate that sequences within the 79-kb genomic construct are sufficient to direct proper acquisition and maintenance of Ly49H expression by a mechanism distinct to that involving TCF-1. This mechanism, however, would also act at the transcriptional level, as Ly49H protein levels, determined by both the size of Ly49H+ population and MFI per individual cell, parallels Ly49h RNA expression levels in the various transgenic lines produced here.
Our observation that the transgenic line Tg915 presented a Ly49H+ cell population of reduced size and splenic viral titers intermediate to those present in Cmv1 r or Cmv1 s mouse strains revealed a previously unforeseen gene dosage effect for Ly49H function and defined a threshold level of Ly49H+ NK cells (28%) necessary for full resistance to MCMV infection. This result indicates that line Tg915 carries a hypomorphic Cmv1 allele, which is an allele with reduced expression compared with Cmv1 r. This leads one to predict that transgenic overexpression of Cmv1/Ly49h in more than 50% of NK cells should have the hypermorphic effect of increased resistance - that is lower splenic viral titers and shorter span of the acute infectious phase - in comparison with C57BL/6. However, we have not observed higher levels of Ly49h transcript in any of the transgenic animals obtained, suggesting that upper levels of transcript and/or protein expression are tightly regulated. This might reflect the fact that expression of Ly49 genes is developmentally controlled (42). It is thought that NK cells acquire Ly49 receptors until they generate sufficient inhibitory signals to balance their activation signals (42, 46). Thus, overexpression of activating receptors could be deleterious for mouse development by preventing a process that is crucial to ensure the induction of NK cell self-tolerance.
Complementation was observed in two independent genetic backgrounds carrying either the susceptibility allele Cmv1 sBALB or Cvm1 sFVB. We have shown that a group of susceptible strains including FVB/N and 129 carry a similar haplotype at Cmv1 that is related to that of C57BL/6, in opposition to susceptible strains such as BALB/c that do not share alleles at any of the 13 loci studied (14). This is particularly noteworthy at the level of the sequence and functional diversity for genes of the Ly49 family. The role of inhibitory NK cell receptors in the response to MCMV is underscored by the recent characterization of MCMV mechanisms to escape NK cell recognition. MCMV is able to subvert the NK cell recognition system by deploying MHC class I homologues that confer resistance to both the innate and adaptive immune systems. Interestingly, those mechanisms may differ in different inbred strains of mice. The first of these viral encoded class I homologues to be identified (m144) was found to confer resistance to NK cell mediated attack in BALB/c mice (47) and is thus likely to represent a ligand for an inhibitory NK receptor in this strain. In a recent report, Arase et al. established that Ly49H was being activated by a second viral-encoded class I homologue known as m157 (21). Interestingly, m157 also engages the inhibitory receptor Ly49I from 129 mice but it does not bind to NK cells from BALB/c mice (21). While the presence of Ly49I in FVB/N mice remains to be confirmed, initial cloning experiments (data not shown) and haplotype analysis (14) indicate that FVB/N and 129 share a similar Ly49 repertoire. Thus, results presented here indicate that engagement of Ly49H is sufficient to override inhibitory signals prompted by either m144 or m157.
Selective recognition of the pathogen is a key step for an effective innate immune response. The major targets of innate immune recognition are pathogen-associated molecular patterns (PAMPs; reference 48). PAMPs represent conserved molecular structures produced only by microbes and essential for their survival. Peptidoglycan and lipopolysaccharide are good examples of bacterial PAMPs. Although recognition of viral double-stranded RNA is an example of PAMP recognition, viruses pose a particular problem for specific recognition, as all the components of the virus are synthesized and assembled in the host cell. So far, two alternative strategies were shown to be used by NK cells to detect unhealthy cells: recognition of the absence of self in the case of MHC class I recognition by NK cell inhibitory receptors (49) and recognition of altered self, in the case of stress molecule recognition (ULBPs, H-60, RAE-1; reference 50). The demonstration that specific resistance to MCMV is attributed to Ly49H protein expression independently of other MCMV-mediated effects clearly demonstrates that NK cells are also able to mediate specific recognition and destruction of infected cells through individual surface receptors. Important next questions are whether Ly49H is a component of the innate immune system that is ‘hard-wired’ to detect MCMV (21, 22) or whether it is less restricted in that it recognizes other yet to be identified ligands. In addition to the Cmv1 gene, the murine NKC region has been linked to loci contributing to susceptibility to cutaneous leishmaniasis (51), ectromelia virus (52), and herpes simplex I virus (53). These susceptibility traits all involve NK cell–mediated immunity, and the C57BL/6 mouse presents the active, “resistant” allele. Transgenic mice reported here will be instrumental to test the proposal that such functions are strictly dependent on activating members of the Ly49 family and will support a direct role of these molecules in recognition of foreign antigens.
Acknowledgments
We are grateful to Michael Wilson and Ben Koop (University of Victoria, Canada) for DNA sequence analysis of BAC 13J11. We wish to thank Svetlana Zykova, Janice Penney, Susan Gauthier, and Eduardo Diez for technical help and logistics with mouse transgenics.
Our laboratories are supported by grants from the Canadian Institutes of Health Research (CIHR). Seung-Hwan Lee is supported by a CIHR doctoral scholarship and Silvia M. Vidal is a Junior Scientist of CIHR.
Footnotes
Abbreviations used in this paper: BAC, bacterial artificial chromosome; GST, glutathione S transferase; MCMV, mouse CMV; NKC, NK cell gene complex; PAMP, pathogen associated molecular pattern; STS, sequence tagged site; TCF-1, T cell factor 1.
References
- 1.Chalmer, J.E., J.S. Mackenzie, and N.F. Stanley. 1977. Resistance to murine cytomegalovirus linked to the major histocompatibility complex of the mouse. J. Gen. Virol. 37:107–114. [DOI] [PubMed] [Google Scholar]
- 2.Grundy, J.E., J.S. Mackenzie, and N.F. Stanley. 1981. Influence of H-2 and non-H-2 genes on resistance to murine cytomegalovirus infection. Infect. Immun. 32:277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allan, J.E., and G.R. Shellam. 1984. Genetic control of murine cytomegalovirus infection: virus titres in resistant and susceptible strains of mice. Arch. Virol. 81:139–150. [DOI] [PubMed] [Google Scholar]
- 4.Scalzo, A.A., N.A. Fitzgerald, A. Simmons, A.B. La Vista, and G.R. Shellam. 1990. Cmv-1, a genetic locus that controls murine cytomegalovirus replication in the spleen. J. Exp. Med. 171:1469–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scalzo, A.A., N.A. Fitzgerald, C.R. Wallace, A.E. Gibbons, Y.C. Smart, R.C. Burton, and G.R. Shellam. 1992. The effect of the Cmv-1 resistance gene, which is linked to the natural killer cell gene complex, is mediated by natural killer cells. J. Immunol. 149:581–589. [PubMed] [Google Scholar]
- 6.Brown, M.G., S. Fulmek, K. Matsumoto, R. Cho, P.A. Lyons, E.R. Levy, A.A. Scalzo, and W.M. Yokoyama. 1997. A 2-Mb YAC contig and physical map of the natural killer gene complex on mouse chromosome 6. Genomics. 42:16–25. [DOI] [PubMed] [Google Scholar]
- 7.Depatie, C., E. Muise, P. Lepage, P. Gros, and S.M. Vidal. 1997. High-resolution linkage map in the proximity of the host resistance locus Cmv1. Genomics. 39:154–163. [DOI] [PubMed] [Google Scholar]
- 8.Brown, M.G., A.A. Scalzo, K. Matsumoto, and W.M. Yokoyama. 1997. The natural killer gene complex: a genetic basis for understanding natural killer cell function and innate immunity. Immunol. Rev. 155:53–65. [DOI] [PubMed] [Google Scholar]
- 9.Brown, M.G., J. Zhang, Y. Du, J. Stoll, W.M. Yokoyama, and A.A. Scalzo. 1999. Localization on a physical map of the NKC-linked Cmv1 locus between Ly49b and the Prp gene cluster on mouse chromosome 6. J. Immunol. 163:1991–1999. [PubMed] [Google Scholar]
- 10.Depatie, C., A. Chalifour, C. Pare, S.H. Lee, S.M. Vidal, and S. Lemieux. 1999. Assessment of Cmv1 candidates by genetic mapping and in vivo antibody depletion of NK cell subsets. Int. Immunol. 11:1541–1551. [DOI] [PubMed] [Google Scholar]
- 11.Depatie, C., S.H. Lee, A. Stafford, P. Avner, A. Belouchi, P. Gros, and S.M. Vidal. 2000. Sequence-ready BAC contig, physical, and transcriptional map of a 2-Mb region overlapping the mouse chromosome 6 host-resistance locus Cmv1. Genomics. 66:161–174. [DOI] [PubMed] [Google Scholar]
- 12.Scalzo, A.A., M.G. Brown, D.T. Chu, J.W. Heusel, W.M. Yokoyama, and C.A. Forbes. 1999. Development of intra-natural killer complex (NKC) recombinant and congenic mouse strains for mapping and functional analysis of NK cell regulatory loci. Immunogenetics. 49:238–241. [DOI] [PubMed] [Google Scholar]
- 13.Boehm, T. 1998. Positional cloning and gene identification. Methods. 14:152–158. [DOI] [PubMed] [Google Scholar]
- 14.Lee, S.H., J. Gitas, A. Zafer, P. Lepage, T.J. Hudson, A. Belouchi, and S.M. Vidal. 2001. Haplotype mapping indicates two independent origins for the Cmv1s susceptibility allele to cytomegalovirus infection and refines its localization within the Ly49 cluster. Immunogenetics. 53:501–505. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S.H., S. Girard, D. Macina, M. Busa, A. Zafer, A. Belouchi, P. Gros, and S.M. Vidal. 2001. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat. Genet. 28:42–45. [DOI] [PubMed] [Google Scholar]
- 16.Brown, M.G., A.O. Dokun, J.W. Heusel, H.R. Smith, D.L. Beckman, E.A. Blattenberger, C.E. Dubbelde, L.R. Stone, A.A. Scalzo, and W.M. Yokoyama. 2001. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 292:934–937. [DOI] [PubMed] [Google Scholar]
- 17.Daniels, K.A., G. Devora, W.C. Lai, C.L. O'Donnell, M. Bennett, and R.M. Welsh. 2001. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J. Exp. Med. 194:29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor, B.A., C. Wnek, B.S. Kotlus, N. Roemer, T. MacTaggart, and S.J. Phillips. 1999. Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps. Mamm. Genome. 10:335–348. [DOI] [PubMed] [Google Scholar]
- 19.Smith, H.R., H.H. Chuang, L.L. Wang, M. Salcedo, J.W. Heusel, and W.M. Yokoyama. 2000. Nonstochastic coexpression of activation receptors on murine natural killer cells. J. Exp. Med. 191:1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosselin, P., L.H. Mason, J. Willette-Brown, J.R. Ortaldo, D.W. McVicar, and S.K. Anderson. 1999. Induction of DAP12 phosphorylation, calcium mobilization, and cytokine secretion by Ly49H. J. Leukoc. Biol. 66:165–171. [DOI] [PubMed] [Google Scholar]
- 21.Arase, H., E.S. Mocarski, A.E. Campbell, A.B. Hill, and L.L. Lanier. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 296:1323–1326. [DOI] [PubMed] [Google Scholar]
- 22.Smith, H.R., J.W. Heusel, I.K. Mehta, S. Kim, B.G. Dorner, O.V. Naidenko, K. Iizuka, H. Furukawa, D.L. Beckman, J.T. Pingel, et al. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. USA. 99:8826–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huxley, C. 1998. Exploring gene function: use of yeast artificial chromosome transgenesis. Methods. 14:199–210. [DOI] [PubMed] [Google Scholar]
- 24.Beattie, R.E., S.G. Volsen, D. Smith, A.L. McCormack, S.E. Gillard, J.P. Burnett, S.B. Ellis, A. Gillespie, M.M. Harpold, and W. Smith. 1997. Preparation and purification of antibodies specific to human neuronal voltage-dependent calcium channel subunits. Brain Res. Brain Res. Protoc. 1:307–319. [DOI] [PubMed] [Google Scholar]
- 25.Gariboldi, M., L. De Gregorio, S. Ferrari, G. Manenti, M.A. Pierotti, M.E. Bianchi, and T.A. Dragani. 1995. Mapping of the Hmg1 gene and of seven related sequences in the mouse. Mamm. Genome. 6:581–585. [DOI] [PubMed] [Google Scholar]
- 26.Adler, E., M.A. Hoon, K.L. Mueller, J. Chandrashekar, N.J. Ryba, and C.S. Zuker. 2000. A novel family of mammalian taste receptors. Cell. 100:693–702. [DOI] [PubMed] [Google Scholar]
- 27.Orange, J.S., and C.A. Biron. 1996. Characterization of early IL-12, IFN-alphabeta, and TNF effects on antiviral state and NK cell responses during murine cytomegalovirus infection. J. Immunol. 156:4746–4756. [PubMed] [Google Scholar]
- 28.Dalod, M., T.P. Salazar-Mather, L. Malmgaard, C. Lewis, C. Asselin-Paturel, F. Briere, G. Trinchieri, and C.A. Biron. 2002. Interferon alpha/beta and interleukin 12 responses to viral infections: pathways regulating dendritic cell cytokine expression in vivo. J. Exp. Med. 195:517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salazar-Mather, T.P., J.S. Orange, and C.A. Biron. 1998. Early murine cytomegalovirus (MCMV) infection induces liver natural killer (NK) cell inflammation and protection through macrophage inflammatory protein 1alpha (MIP-1alpha)-dependent pathways. J. Exp. Med. 187:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He, X., H. Yoshida, Y. Minamishima, and K. Nomoto. 1995. Analysis of the role of CD4+ T-cells during murine cytomegalovirus infection in different strains of mice. Virus Res. 36:233–245. [DOI] [PubMed] [Google Scholar]
- 31.Charles, P.C., K.S. Weber, B. Cipriani, and C.F. Brosnan. 1999. Cytokine, chemokine and chemokine receptor mRNA expression in different strains of normal mice: implications for establishment of a Th1/Th2 bias. J. Neuroimmunol. 100:64–73. [DOI] [PubMed] [Google Scholar]
- 32.Dokun, A.O., S. Kim, H.R. Smith, H.S. Kang, D.T. Chu, and W.M. Yokoyama. 2001. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol. 2:951–956. [DOI] [PubMed] [Google Scholar]
- 33.Coles, L.S., P. Diamond, F. Occhiodoro, M.A. Vadas, and M.F. Shannon. 1996. Cold shock domain proteins repress transcription from the GM-CSF promoter. Nucleic Acids Res. 24:2311–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selgrade, M.K., J.G. Nedrud, A.M. Collier, and D.E. Gardner. 1981. Effects of cell source, mouse strain, and immunosuppressive treatment on production of virulent and attenuated murine cytomegalovirus. Infect. Immun. 33:840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forbes, C.A., M.G. Brown, R. Cho, G.R. Shellam, W.M. Yokoyama, and A.A. Scalzo. 1997. The Cmv1 host resistance locus is closely linked to the Ly49 multigene family within the natural killer cell gene complex on mouse chromosome 6. Genomics. 41:406–413. [DOI] [PubMed] [Google Scholar]
- 36.Hudson, J.B., D.G. Walker, and M. Altamirano. 1988. Analysis in vitro of two biologically distinct strains of murine cytomegalovirus. Arch. Virol. 102:289–295. [DOI] [PubMed] [Google Scholar]
- 37.Lowin-Kropf, B., and W. Held. 2000. Positive impact of inhibitory Ly49 receptor-MHC class I interaction on NK cell development. J. Immunol. 165:91–95. [DOI] [PubMed] [Google Scholar]
- 38.Oberg, L., M. Eriksson, L. Fahlen, and C.L. Sentman. 2000. Expression of Ly49A on T cells alters the threshold for T cell responses. Eur. J. Immunol. 30:2849–2856. [DOI] [PubMed] [Google Scholar]
- 39.Liu, J., M.A. Morris, P. Nguyen, T.C. George, E. Koulich, W.C. Lai, J.D. Schatzle, V. Kumar, and M. Bennett. 2000. Ly49I NK cell receptor transgene inhibition of rejection of H2b mouse bone marrow transplants. J. Immunol. 164:1793–1799. [DOI] [PubMed] [Google Scholar]
- 40.Hanke, T., and D.H. Raulet. 2001. Cumulative inhibition of NK cells and T cells resulting from engagement of multiple inhibitory Ly49 receptors. J. Immunol. 166:3002–3007. [DOI] [PubMed] [Google Scholar]
- 41.Kubota, A., S. Kubota, S. Lohwasser, D.L. Mager, and F. Takei. 1999. Diversity of NK cell receptor repertoire in adult and neonatal mice. J. Immunol. 163:212–216. [PubMed] [Google Scholar]
- 42.Raulet, D.H., R.E. Vance, and C.W. McMahon. 2001. Regulation of the natural killer cell receptor repertoire. Annu. Rev. Immunol. 19:291–330. [DOI] [PubMed] [Google Scholar]
- 43.Cadigan, K.M., and R. Nusse. 1997. Wnt signaling: a common theme in animal development. Genes Dev. 11:3286–3305. [DOI] [PubMed] [Google Scholar]
- 44.Wilhelm, B.T., K.L. McQueen, J.D. Freeman, F. Takei, and D.L. Mager. 2001. Comparative analysis of the promoter regions and transcriptional start sites of mouse Ly49 genes. Immunogenetics. 53:215–224. [DOI] [PubMed] [Google Scholar]
- 45.Held, W., B. Kunz, B. Lowin-Kropf, M. van de Wettering, and H. Clevers. 1999. Clonal acquisition of the Ly49A NK cell receptor is dependent on the trans-acting factor TCF-1. Immunity. 11:433-442. [DOI] [PubMed] [Google Scholar]
- 46.Lowin-Kropf, B., B. Kunz, P. Schneider, and W. Held. 2002. A role for the src family kinase Fyn in NK cell activation and the formation of the repertoire of Ly49 receptors. Eur. J. Immunol. 32:773–782. [DOI] [PubMed] [Google Scholar]
- 47.Farrell, H.E., H. Vally, D.M. Lynch, P. Fleming, G.R. Shellam, A.A. Scalzo, and N.J. Davis-Poynter. 1997. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature. 386:510–514. [DOI] [PubMed] [Google Scholar]
- 48.Janeway, C.A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197–216. [DOI] [PubMed] [Google Scholar]
- 49.Karre, K. 2002. NK cells, MHC class I molecules and the missing self. Scand. J. Immunol. 55:221–228. [DOI] [PubMed] [Google Scholar]
- 50.Lanier, L.L. 2001. On guard–activating NK cell receptors. Nat. Immunol. 2:23–27. [DOI] [PubMed] [Google Scholar]
- 51.Beebe, A.M., S. Mauze, N.J. Schork, and R.L. Coffman. 1997. Serial backcross mapping of multiple loci associated with resistance to Leishmania major in mice. Immunity. 6:551–557. [DOI] [PubMed] [Google Scholar]
- 52.Delano, M.L., and D.G. Brownstein. 1995. Innate resistance to lethal mousepox is genetically linked to the NK gene complex on chromosome 6 and correlates with early restriction of virus replication by cells with an NK phenotype. J. Virol. 69:5875–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pereira, R.A., A. Scalzo, and A. Simmons. 2001. Cutting edge: a NK complex-linked locus governs acute versus latent herpes simplex virus infection of neurons. J. Immunol. 166:5869–5873. [DOI] [PubMed] [Google Scholar]