Abstract
The gram-positive soil bacterium Bacillus subtilis often faces increases in the salinity in its natural habitats. A transcriptional profiling approach was utilized to investigate both the initial reaction to a sudden increase in salinity elicited by the addition of 0.4 M NaCl and the cellular adaptation reactions to prolonged growth at high salinity (1.2 M NaCl). Following salt shock, a sigB mutant displayed immediate and transient induction and repression of 75 and 51 genes, respectively. Continuous propagation of this strain in the presence of 1.2 M NaCl triggered the induction of 123 genes and led to the repression of 101 genes. In summary, our studies revealed (i) an immediate and transient induction of the SigW regulon following salt shock, (ii) a role of the DegS/DegU two-component system in sensing high salinity, (iii) a high-salinity-mediated iron limitation, and (iv) a repression of chemotaxis and motility genes by high salinity, causing severe impairment of the swarming capability of B. subtilis cells. Initial adaptation to salt shock and continuous growth at high salinity share only a limited set of induced and repressed genes. This finding strongly suggests that these two phases of adaptation require distinctively different physiological adaptation reactions by the B. subtilis cell. The large portion of genes with unassigned functions among the high-salinity-induced or -repressed genes demonstrates that major aspects of the cellular adaptation of B. subtilis to high salinity are unexplored so far.
The soil bacterium Bacillus subtilis is particularly subject to changes in the supply of water and to the concomitant alterations in salinity and osmolality resulting from frequent drought and flooding of its habitat (11, 43, 70). This threatens the cell with dehydration under hypertonic conditions or with rupture under hypotonic conditions. Like many other bacteria (9, 12), B. subtilis avoids these devastating alternatives by actively modulating its ion and organic solute pool to retain a suitable level of cytoplasmic water and turgor (11).
Following a sudden increase in salinity, cells maintain turgor within physiologically acceptable boundaries by first increasing their potassium (K+) content and then replacing part of the accumulated K+ with compatible solutes in the second phase of osmoadaptation (67, 68). Two Ktr-type K+ transporters (KtrAB and KtrCD) are critically involved in providing the B. subtilis cell with sufficient K+, both during its initial adaptation and during prolonged exposure to high salinity (29). Proline serves as the primary endogenously synthesized compatible solute for B. subtilis (67), and during growth at high salinity, large quantities are produced via a dedicated osmostress-responsive synthesis pathway that depends on the ProHJ and ProA enzymes (3; J. Brill and E. Bremer, unpublished data). In addition, B. subtilis can efficiently scavenge a wide variety of preformed compatible solutes from environmental sources (11) by means of five osmoregulated transport systems (OpuA to OpuE) (31-33, 66). Furthermore, it can synthesize the osmoprotectant glycine betaine via the GbsA and GbsB enzymes from exogenously provided choline that is taken up by the cell via the OpuB and OpuC ABC transporters (7, 32). The intracellular accumulation of compatible solutes offsets the detrimental effects of high salinity on cell physiology and permits growth of B. subtilis over a wide range of environmental osmolalities (6, 38). When the osmolality drops suddenly (9), B. subtilis expels these accumulated compatible solutes via mechanosensitive channels (T. Hoffmann, C. D. Boiangiu, and E. Bremer, unpublished data) to counteract the influx of water into the cell and the concomitant increase in turgor.
Under conditions where the salt stress is so strong that growth is no longer permitted, a nonspecific and preemptive general stress response system is engaged to ensure the survival of B. subtilis (26, 52). High salinity is among the environmental cues that cause the activation of the central regulator (SigB) of this regulon (10, 63) and lead to the transient induction of more than 150 SigB-dependent genes (27, 51, 53). Loss of SigB causes sensitivity of the cells to growth-preventing salt stress (64). Osmoprotection by compatible solutes and the general stress response are linked, because the structural genes for the proline uptake system OpuE (60, 66) and the glycine betaine transporter OpuD (31; F. Spiegelhalter and E. Bremer, unpublished data) are partially dependent on SigB for their expression.
Transcriptional profiling studies have also indicated induction of the SigW regulon following salt shock (51), but the functional contribution of this regulon to cellular adaptation to high salinity has not yet been elucidated. Furthermore, mutants lacking SigM are sensitive to high salt concentrations (30), but this might be an indirect phenotype related to the major cell wall defects exhibited by such mutants.
High salinity exerts pleiotropic effects on the physiology of B. subtilis. Increases in salinity affect the phospholipid composition of the cytoplasmic membrane (39) and the properties of the cell wall (40). In addition, the production of levansucrase (SacB), alkaline protease (AprE), and the cell wall-associated protein WapA is regulated in a DegS/DegU-dependent manner at high salinity (17, 37). Furthermore, under such growth conditions, one observes changes in the supercoiling of reporter plasmids (2, 35) and the transient induction of the ftsH gene, which encodes an ATP-dependent, membrane-associated protease (18). Finally, sporulation is severely impaired by high salinity (37, 57), due to an early block in the sporulation process (57).
A recent proteome analysis of salt-adapted B. subtilis cells revealed yet another facet of the cellular response to high salinity (28). Such cells experience a severe iron limitation that leads to the induction of genes encoding the iron siderophore bacillibactin (42) and putative iron uptake systems (28). This proteome analysis showed a surprisingly small number of proteins (18 spots) that displayed significantly different intensities in cells grown at high versus low salinity.
As exemplified by the analysis of the SigB-dependent general stress response, transcriptional profiling studies (27, 51, 53) provide a more complete view of the cellular response to a particular stress than do proteome studies, which cover primarily soluble proteins (14). To gain a comprehensive overview of the cellular response of B. subtilis to high salinity, we performed a genome-wide comparative transcriptional profiling of exponentially growing, fully salt adapted, and salt-shocked cells.
MATERIALS AND METHODS
Bacterial strains, media and growth conditions.
The B. subtilis sigB mutant BLOB22 (trpC2 pheA1 sfp0 sigBΔ2::cat) (66), a derivative of the wild-type strain JH642 (BGSC1A96; a kind gift from J. Hoch, La Jolla, Calif.), was routinely grown with vigorous agitation in minimal medium containing 0.5% glucose (wt/vol) as the carbon source and the amino acids l-tryptophan (20 mg/liter) and l-phenylalanine (18 mg/liter). For experiments that required a defined iron concentration, the modified minimal medium (MM) described by Chen et al. (16) was used and iron was supplied to the cells from a freshly prepared 20 mM stock solution of FeCl3 (Merck; Darmstadt, Germany) to a final concentration of either 5 or 250 μM. To raise the salinity of MM, NaCl from a 5 M stock solution was added to produce a final concentration of 1.2 M. The osmolality of the growth medium was determined with a Vapour pressure osmometer (model 5500; Wescor). The growth of the bacterial cultures was monitored spectrophotometrically at a wavelength of 578 nm (optical density at 578 nm [OD578]). For continuous growth experiments, precultures of strain BLOB22 were inoculated from exponentially growing overnight cultures propagated in MM with NaCl concentrations of 0 or 1.2 M to a final OD578 of 0.1. These precultures were then allowed to grow to an OD578 of 1 to 2 and were subsequently used to inoculate 70 ml of MM to an OD578 of 0.1 in a 500-ml Erlenmeyer flask. This final culture was then propagated at 37°C and 220 rpm in a shaking water bath until the cultures had reached an OD578 of approximately 1. Cells were harvested by mixing 15 ml of culture with an equal volume of frozen killing buffer (20 mM NaN3, 20 mM Tris-HCl [pH 7.5], 5 mM MgCl2) and subsequently centrifuging for 10 min at 6,000 × g and 4°C. The cell pellets were stored at −80°C until further use for total-RNA preparation.
For salt shock experiments, cultures were grown in Spizizen′s minimal medium (SMM) supplemented with glucose, l-tryptophan, and l-phenylalanine as described above and an additional solution of trace elements (24). An exponentially growing overnight culture (OD578 = 0.3) was diluted into 700 ml of SMM (in a 5-liter Erlenmeyer flask) to an OD578 of 0.025 and then incubated at 37°C in a shaking water bath set at 220 rpm. After this culture reached an OD578 of 0.25, NaCl was added from a 4 M stock solution prepared in SMM to a final concentration of 0.4 M. Samples (15 ml) for RNA preparations were harvested as described above either 10 min prior to NaCl addition or at different time points after salt shock.
Cell lysis and RNA isolation.
RNA was isolated after mechanical disruption of the cells in a Micro-Dismembrator (B. Braun Biotec Int., Melsungen, Germany) as described by Hauser et al. (25) with the modifications introduced by Petersohn et al. (51).
Preparation of labeled cDNA, array hybridization, and DNA macroarray regeneration.
Prior to cDNA labeling, the overall integrity of the total-RNA preparation was analyzed using a capillary electrophoresis system (Bioanalyser 2100; Agilent Technologies, Waldbronn, Germany). For cDNA synthesis, 2 μg of total RNA was mixed with 4 μl of a commercial primer mix (Sigma-Genosys Ltd., The Woodlands, Tex.) and 3 μl of 5× hybridization buffer (50 mM Tris [pH 7.9], 0.2 mM EDTA, 1.25 M KCl) and was adjusted to a total volume of 15 μl with nuclease-free water. The primer mix consisted of 4,107 specific oligonucleotide primers complementary to the 3′ ends of all B. subtilis mRNAs (Sigma-Genosys Ltd.). Subsequently, the sample was heated to 95°C for 2 min and then cooled to 42°C. After primer annealing for 1 h, reverse transcription was performed with SuperScript II reverse transcriptase (Invitrogen Life Technologies GmbH, Karlsruhe, Germany) in the presence of 50 μCi of [α-33P]dCTP (Amersham Biosciences, Freiburg, Germany) in a buffer supplied by Invitrogen Life Technologies GmbH in a total volume of 30 μl for 1 h. Enzymes were inactivated by heating the reaction mixture to 70°C for 15 min, and after addition of 1 μl of 10% sodium dodecyl sulfate (SDS), 1 μl of 0.5 M EDTA(pH 8.0), and 3 μl of 3 M NaOH, the remaining RNA was hydrolyzed by incubation at 65°C for 30 min. The cDNA solution was neutralized with 10 μl of 1 M Tris-HCl (pH 8.0) and 3 μl of 2 N HCl. After addition of 5 μl of 3 M sodium acetate (pH 5.2), 5 μl of carrier tRNA (Sigma, Steinheim, Germany), and 60 μl of isopropanol, the labeled cDNA was precipitated at −20°C for 1 h. The cDNA pellet was then washed twice with 70% ethanol and resolved in 60 μl of nuclease-free water. Labeling efficiency was determined with a liquid scintillation counter.
This study was performed with commercially available Panorama B. subtilis DNA-macroarrays from Sigma Genosys Ltd. that carry duplicate spots of PCR products representing 4,107 B. subtilis genes. cDNA denaturation, probe hybridization, and washing of the filters were performed as described by Petersohn et al. (51) with the following modifications. Prehybridization was carried out in 5 ml of hybridization solution (5× Denhardt solution, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 2% sodium dodecyl sulfate [SDS], 100 μg of denatured herring sperm DNA [Sigma]/ml) for 2 h at 65°C. Hybridization was performed for 20 h at 65°C in 5 ml of hybridization solution containing the labeled cDNA probe that had been heated to 98°C for 2 min. After hybridization, arrays were washed twice with 200 ml of 2× SSC and 0.1% (wt/vol) SDS for 5 min at room temperature and twice with 200 ml of 0.2× SSC and 0.1% (wt/vol) SDS for 20 min at 65°C. Arrays were then exposed to storage phosphor screens (Molecular Dynamics, Sunnyvale, Calif.) for 2 to 4 days and were subsequently scanned with a Storm 840/860 PhosphorImager (Molecular Dynamics) at a resolution of 50 μm and a color depth of 16 bits. Bound cDNA was stripped off the DNA-macroarray membranes by three washing cycles involving a short (1-min) washing step with 250 ml of boiling buffer (5 mM sodium phosphate [pH 7.5]-0.1% SDS) and an incubation in 250 ml of fresh buffer at 95°C for 20 min.
Data analysis.
Data analysis followed a three-step procedure. First, ArrayVision software (version 6.1; Imaging Research, St. Catherine's, Ontario, Canada) was used for the quantification of the hybridization signals after direct import of the phosphorimager files. The analysis yielded the artifact-removed volumes and background values, calculated from the median of lines surrounding each group of eight spots on the array. These data were then used in a second step in Microsoft Excel: calculating for every spot on the array a quality score that reflected the ratio between the signal intensity and the background intensity (see supplemental material for details [http://www.medizin.uni-greifswald.de/funkgenom/supplemental_material]). This quality score was utilized to identify the hybridization signals close to the detection limit, thereby avoiding artificially high induction ratios for those genes. Data normalization and data analysis were done in a third step with GeneSpring (version 5.02; Silicon Genetics, Redwood City, Calif.). Gene expression for a particular comparison of conditions was considered to be changed when three criteria were fulfilled: (i) expression of the gene had to exceed the background signal level by a threshold determined as described in the supplemental material, (ii) changes in expression of the gene had to be statistically significant as defined in a statistical group comparison of the values for the selected conditions with a nonparametric test (Wilcoxon-Mann-Whitney-test) and a Benjamini and Hochberg false discovery rate correction, with a P value cutoff of 0.005 as defined in the GeneSpring software package, and (iii) the change in expression had to exceed a factor of 3. Ratios were calculated with averages of the parallel samples described below.
For all studies of gene expression in cells grown in MM (with either 5 μM FeCl3 and 0 M NaCl, 0 μM FeCl3 and 0 M NaCl, 5 μM FeCl3 and 1.2 M NaCl, or 250 μM FeCl3 and 1.2 M NaCl), mRNA was prepared from three independent cultivations and then used for independent cDNA synthesis and DNA array hybridizations. The Panorama B. subtilis DNA macroarrays from Sigma Genosys Ltd. contained duplicated DNA samples for each of the 4,107 B. subtilis genes; therefore, the processing of three independent samples for each growth condition yielded six data points for the calculation of signal intensities for each gene.
For the salt shock adaptation experiments, control values immediately prior to the shift to high salinity were calculated for three independently processed exponentially growing cultures in SMM. The time course of the salt shock adaptation was studied by analyzing samples derived from the same culture at appropriate time points after addition of NaCl to a final concentration of 0.4 M.
World Wide Web access.
The complete data set for all growth conditions investigated is available online (http://www.medizin.uni-greifswald.de/funkgenom/supplemental_material).
Northern blot analysis.
Aliquots of the total RNA prepared for the DNA macroarray experiments were used for Northern blot analysis of the expression profiles of the opuBC, yocC, feuA, and wapA genes. Digoxigenin-labeled antisense RNA probes were generated by in vitro transcription using a StripEZ-kit (Ambion, Inc., Woodward, TX, USA) and gene-specific PCR products as templates. In each of the PCRs with chromosomal DNA prepared from the B. subtilis strain JH642, one of the DNA primers carried the sequence of the T7 promoter. The PCR fragment was subsequently used for in vitro RNA synthesis with commercially available T7 RNA polymerase (Ambion, Inc., Woodward, Tex.), yielding hybridization probes of the following sizes internal to the structural genes: for opuBC, 572 nucleotides (nt); for feuA, 423 nt; for wapA, 463 nt; and for yocC, 385 nt. Denaturing RNA electrophoresis on agarose gels, RNA transfer by diffusion onto a nylon membrane (NY13N; Schleicher & Schuell, Dassel, Germany), hybridization to gene-specific probes, and signal detection were performed as described by Holtmann et al. (29).
Cell motility assay.
Motility assays of the B. subtilis wild-type strain 168 were performed with cells grown either in SMM alone or in SMM with 1.2 M NaCl. Bacteria were propagated until they had reached an OD578 of 1.0 to 1.5. These cultures were then diluted to an OD578 of 0.2 in the original growth medium, and 5-μl aliquots of these cell suspensions were subsequently spotted in the middle of an agar plate prepared with 1% tryptone and 0.25% agar (low osmolality) or onto an agar plate prepared with 1% tryptone, 0.25% agar, and 1.2 M NaCl (high osmolality). The plates were incubated at 37°C overnight and were inspected for swarming of the B. subtilis 168 strain.
RESULTS AND DISCUSSION
Experimental strategy.
The response of B. subtilis to high salinity can be divided into two phases: an initial reaction to a sudden rise in salinity and the subsequent cellular adaptation to prolonged growth under high-salinity conditions. Consequently, we performed two separate sets of experiments by monitoring both the changes in the transcriptional profile on a time-resolved scale following a salt shock with 0.4 M NaCl and the persisting changes in the gene expression pattern in cells that were continuously cultured at high salinity (1.2 M NaCl). All experiments were conducted with synthetic media, since complex media frequently contain compatible solutes (e.g., glycine betaine) (20) which are known to down-regulate the expression of salt-responsive genes in B. subtilis (60).
Because many of the studies investigating the effects of high salinity on the cellular physiology of B. subtilis have been performed with the wild-type strain JH642, we chose this genetic background to complement the previous functionally driven studies (11) with a genome-wide transcriptional profiling approach.
Salt shock is known to transiently induce the large SigB-dependent general stress regulon (10, 65). The structure of this regulon (at least 150 genes) has been rather well defined through both proteome (5, 14) and transcriptome (27, 51, 53) studies. Therefore, we used a JH642-derived sigB mutant (strain BLOB22) (66) for our transcriptional profiling of salt-stressed B. subtilis cells to avoid the complex and already well-studied changes in gene expression associated with the activation of SigB following a sudden rise in salinity (51).
Salt-induced changes in gene expression in cells continuously growing at high salinity.
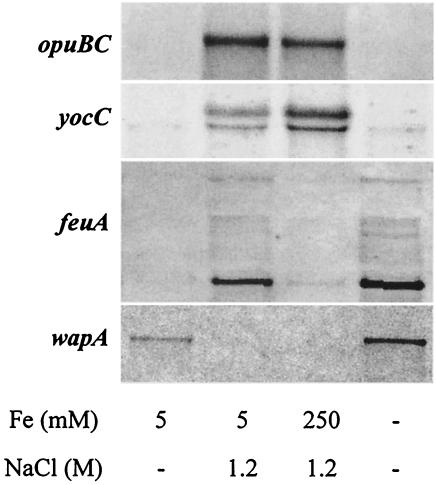
To monitor differences in the transcriptional profile between cells continuously growing at low versus high salinity, we propagated triplicate cultures of strain BLOB22 (sigBΔ2::cat) in MM alone (260 mosmol/kg of water) and in MM with 1.2 M NaCl (2,490 mosmol/kg of water). RNA prepared from exponentially growing cultures (OD578 = 1) was used for the preparation of radiolabeled cDNA, which was then hybridized to commercially available Panorama B. subtilis DNA macroarrays (Sigma Genosys Ltd.) that represent 4,107 protein-encoding genes from B. subtilis. The expression pattern in each of the cultures grown in parallel was highly reproducible and varied by less than a factor of 2 (Pearson correlation coefficient, 0.9968 [data not shown]). In contrast, the expression patterns of samples grown at low versus high salinity differed significantly (Pearson correlation coefficient, 0.9014). A group of 123 genes displayed at least a threefold induction in cultures grown at high salinity, and 101 genes displayed at least threefold repression under high-salinity growth conditions (Tables 1 and 2, respectively). The expression patterns for selected genes induced (yocC, opuBC, and feuA) or repressed (wapA) under high-salinity growth conditions were verified by Northern blot analysis (Fig. 1). For all four genes tested, Northern blot analysis fully confirmed the expression profile found in the DNA macroarray analysis (Fig. 2 and 3; Tables 1 and 2).
TABLE 1.
High-salinity-induced genes
| Genea | Functionb | Signal strength ratioc for cells growing in MM with:
|
|
|---|---|---|---|
| 1.2 M NaCl, 5 μM Fe | 1.2 M NaCl, 250 μM Fe | ||
| bofA | Inhibition of the pro-sigma-K processing machinery | 3.2 | 1.2 |
| yabE | Unknown; similar to cell wall-binding protein | 5.4 | 5.7 |
| ksgA | Dimethyladenosine transferase | 4.4 | 4.8 |
| opuAA | Glycine betaine ABC transporter (ATP-binding protein) | 4.3 | 4.8 |
| opuAB | Glycine betaine ABC transporter (permease) | 3.2 | 3.7 |
| opuAC | Glycine betaine ABC transporter (glycine betaine-binding protein) | 4.7 | 3.9 |
| ydcE | Unknown; similar to unknown proteins | 3.1 | 3.0 |
| ydcK | Unknown; similar to unknown proteins | 3.2 | 3.4 |
| ydcO | Unknown | 3.1 | 5.2 |
| yddJ | Unknown | 13.8 | 9.8 |
| yddT | Unknown | 3.3 | 1.9 |
| groES | Class I heat shock protein (chaperonin) | 3.5 | 3.7 |
| yflJ | Unknown | 3.4 | 3.1 |
| yfjQ | Unknown; similar to divalent cation transport protein | 3.2 | 2.3 |
| yfjC | Unknown | 4.1 | 5.0 |
| yfjB | Unknown | 6.7 | 10.2 |
| yfjA | Unknown | 5.4 | 7.2 |
| yhaA | Unknown; similar to aminoacylase | 3.2 | 4.4 |
| yhjM | Unknown; similar to transcriptional regulator (LacI family) | 7.0 | 6.5 |
| yitM | Unknown; similar to unknown proteins from B. subtilis | 4.0 | 2.6 |
| yjcE | Unknown | 3.3 | 2.4 |
| yjhA | Unknown | 4.2 | 4.5 |
| yjiA | Unknown | 4.1 | 3.1 |
| ykaA | Unknown | 4.9 | 2.5 |
| yklA | Unknown; similar to unknown proteins | 3.4 | 4.6 |
| ykrI | Unknown | 3.7 | 4.8 |
| ykwD | Unknown; similar to unknown proteins from B. subtilis | 3.4 | 3.4 |
| ykuH | Unknown | 6.8 | 5.7 |
| ykzF | Unknown | 3.1 | 2.7 |
| ykuL | Unknown; similar to unknown proteins | 4.7 | 5.1 |
| ylbA | Unknown; similar to unknown proteins | 4.1 | 3.9 |
| ylbB | Unknown; similar to IMP dehydrogenase | 4.4 | 3.4 |
| ylbC | Unknown; similar to unknown proteins from B. subtilis | 3.8 | 3.3 |
| lspA | Signal peptidase II | 3.4 | 3.6 |
| pyrR | Transcriptional attenuation of the pyrimidine operon/uracil phosphoribosyltransferase activity | 3.5 | 2.1 |
| ylqD | Unknown; similar to unknown proteins | 3.7 | 3.2 |
| frr | Ribosome recycling factor | 3.0 | 2.1 |
| pksC | Involved in polyketide synthesis | 3.3 | 3.0 |
| sspN | Small acid-soluble spore protein (minor) | 3.3 | 1.4 |
| proJ | Glutamate 5-kinase | 16.2 | 17.7 |
| proH | Pyrroline-5-carboxylate reductase | 22.5 | 24.7 |
| yoaG | Unknown; similar to unknown proteins | 3.8 | 2.6 |
| yoaJ | Unknown; similar to extracellular endoglucanase precursor | 3.3 | 1.4 |
| penP | Beta-lactamase | 3.3 | 3.9 |
| yocA | Unknown; similar to transposon-related protein | 3.1 | 3.3 |
| yocC | Unknown; similar to unknown proteins | 7.0 | 7.3 |
| yocH | Unknown; similar to cell wall-binding protein | 12.3 | 13.8 |
| dhaS | Aldehyde dehydrogenase | 3.0 | 3.8 |
| yotH | Unknown | 3.4 | 3.5 |
| yomL | Unknown | 3.0 | 1.6 |
| bdbA | Thiol-disulfide oxidoreductase | 3.5 | 3.4 |
| sunT | Sublancin 168 lantibiotic transporter | 4.1 | 4.6 |
| bsaA | Putative glutathione peroxidase | 4.3 | 4.2 |
| sspL | Small acid-soluble spore protein (minor) | 3.9 | 2.0 |
| ypsB | Unknown; similar to unknown proteins | 3.7 | 3.2 |
| ypqE | Unknown; similar to phosphotransferase system enzyme II | 3.1 | 2.7 |
| ypjD | Unknown; similar to unknown proteins | 3.2 | 2.7 |
| qcrC | Menaquinol:cytochrome c oxidoreductase (cytochrome b/c subunit) | 3.4 | 5.9 |
| yphF | Unknown; similar to unknown proteins | 3.1 | 2.7 |
| ypfD | Unknown; similar to ribosomal protein S1 homolog | 3.2 | 2.5 |
| ypdA | Unknown; similar to thioredoxin reductase | 4.0 | 3.7 |
| spoIIAB | Anti-sigma factor/serine kinase | 3.1 | 3.3 |
| yqjX | Unknown | 3.1 | 2.1 |
| yqiI | Unknown; similar to N-acetylmuramoyl-l-alanine amidase | 6.8 | 4.4 |
| yqiH | Unknown; similar to lipoprotein | 14.9 | 9.8 |
| mntR | Transcriptional regulator of manganese uptake | 4.2 | 3.2 |
| gcvT | Probable aminomethyltransferase | 3.1 | 2.2 |
| yqxJ | Unknown | 12.8 | 13.0 |
| yqxI | Unknown | 8.7 | 10.9 |
| yqaO | Unknown; similar to phage-related protein | 4.3 | 3.0 |
| yrdR | Unknown; similar to unknown proteins | 3.7 | 2.5 |
| yraG | Unknown; similar to spore coat protein | 3.9 | 2.0 |
| yrvC | Unknown; similar to unknown proteins | 4.7 | 5.1 |
| ysfC | Unknown; similar to glycolate oxidase subunit | 3.7 | 1.8 |
| citZ | Citrate synthase II (major) | 4.3 | 6.3 |
| ytxG | Unknown; similar to general stress protein | 3.2 | 3.4 |
| yteV | Unknown | 3.0 | 1.7 |
| yukC | Unknown; similar to unknown proteins | 6.0 | 5.3 |
| yukD | Unknown; similar to unknown proteins | 3.7 | 2.3 |
| yuxI | Unknown | 3.8 | 3.9 |
| yukJ | Unknown | 3.5 | 2.8 |
| lipA | Probable lipoic acid synthetase | 4.4 | 3.8 |
| yusF | Unknown | 3.0 | 2.5 |
| opuBC | Choline ABC transporter (choline-binding protein) | 4.8 | 5.2 |
| opuBA | Choline ABC transporter (ATP-binding protein) | 7.9 | 9.3 |
| yvaW | Unknown; similar to unknown proteins from B. subtilis | 9.0 | 9.4 |
| yvzA | Unknown | 6.1 | 5.4 |
| yvcB | Unknown | 4.0 | 3.8 |
| yvcA | Unknown | 12.7 | 10.1 |
| degU | Two-component response regulator involved in degradative enzyme and competence regulation | 3.9 | 3.6 |
| ywqJ | Unknown; similar to unknown proteins from B. subtilis | 4.0 | 3.2 |
| ywqI | Unknown; similar to unknown proteins from B. subtilis | 4.3 | 4.0 |
| ywqH | Unknown | 4.4 | 4.1 |
| ywfH | Unknown; similar to 3-oxoacyl-acyl-carrier protein reductase | 3.0 | 2.7 |
| ywdA | Unknown | 4.2 | 4.2 |
| yxiD | Unknown; similar to unknown proteins | 3.0 | 2.6 |
| yydJ | Unknown | 4.1 | 3.9 |
| yydI | Unknown; similar to ABC transporter (ATP-binding protein) | 6.2 | 5.3 |
| yydH | Unknown | 5.6 | 5.0 |
| yydG | Unknown | 6.2 | 7.2 |
| rapG | Response regulator aspartate phosphatase | 3.0 | 2.6 |
| phrG | Phosphatase (RapG) regulator | 10.7 | 9.4 |
Sorted according to the order in the B. subtilis genome. Members of the DegS/DegU regulon are boldfaced.
Derived from the SubtiList database.
Ratio between the signal strength in cells propagated under the conditions indicated and the signal strength in cells continuously growing in synthetic medium with 5 μM FeCl3 and no extra NaCl added.
TABLE 2.
High-salinity-repressed genes
| Genea | Functionb | Signal strength ratioc for cells growing in MM with:
|
|
|---|---|---|---|
| 1.2 M NaCl, 5 μM Fe | 1.2 M NaCl, 250 μM Fe | ||
| ybdO | Unknown | 4.6 | 3.3 |
| ybfG | Unknown; similar to unknown proteins | 3.6 | 2.5 |
| ycbJ | Unknown; similar to macrolide 2′-phosphotransferase | 5.6 | 4.5 |
| ycbU | Unknown; similar to NifS protein homolog | 4.9 | 3.2 |
| ldh | l-Lactate dehydrogenase | 26.0 | 0.6 |
| lctP | l-Lactate permease | 4.2 | 0.7 |
| ydaD | Unknown; similar to alcohol dehydrogenase | 3.5 | 2.0 |
| ydcR | Unknown; similar to transposon protein | 5.1 | 1.5 |
| ydfR | Unknown; similar to unknown proteins | 3.2 | 2.1 |
| yfmT | Unknown; similar to benzaldehyde dehydrogenase | 8.6 | 4.5 |
| yfmS | Unknown; similar to methyl-accepting chemotaxis protein | 11.4 | 12.7 |
| pel | Pectate lyase | 4.3 | 5.0 |
| yfiQ | Unknown; similar to surface adhesion protein | 3.4 | 2.1 |
| katA | Vegetative catalase 1 | 4.4 | 21.3 |
| glpD | Glycerol-3-phosphate dehydrogenase | 3.0 | 3.2 |
| lytF | Gamma-d-glutamate-meso-diaminopimelate muropeptidase (major autolysin) | 11.3 | 6.5 |
| yhdO | Unknown; similar to 1-acylglycerol-3-phosphate O-acyltransferase | 3.8 | 3.4 |
| yhxC | Unknown; similar to alcohol dehydrogenase | 3.3 | 1.7 |
| yjfB | Unknown | 3.0 | 3.1 |
| motB | Motility protein (flagellar motor rotation) | 22.3 | 9.6 |
| motA | Motility protein (flagellar motor rotation) | 26.4 | 9.7 |
| mcpC | Methyl-accepting chemotaxis protein | 4.1 | 4.2 |
| abh | Transcriptional regulator of transition state genes (AbrB-like) | 11.5 | 16.8 |
| ylqB | Unknown | 10.0 | 4.0 |
| flgB | Flagellar basal-body rod protein | 3.6 | 3.3 |
| flgC | Flagellar basal-body rod protein | 3.2 | 2.7 |
| fliF | Flagellar basal-body M-ring protein | 5.9 | 4.5 |
| fliG | Flagellar motor switch protein | 6.4 | 5.3 |
| fliH | Flagellar assembly protein | 5.9 | 4.6 |
| fliI | Flagellum-specific ATP synthase | 4.7 | 3.8 |
| fliJ | Flagellar protein required for formation of basal body | 5.1 | 4.5 |
| ylxF | Unknown; similar to unknown proteins | 25.7 | 15.5 |
| ylxG | Unknown; similar to flagellar hook assembly protein | 6.8 | 7.1 |
| flgE | Flagellar hook protein | 7.1 | 7.5 |
| fliL | Flagellar protein required for flagellar formation | 7.3 | 6.7 |
| fliM | Flagellar motor switch protein | 9.0 | 10.4 |
| fliY | Flagellar motor switch protein | 5.4 | 4.0 |
| cheY | Two-component response regulator involved in modulation of flagellar switch bias | 4.5 | 4.2 |
| fliZ | Flagellar protein required for flagellar formation | 6.1 | 5.7 |
| fliP | Flagellar protein required for flagellar formation | 9.3 | 9.9 |
| fliR | Flagellar protein required for flagellar formation | 3.4 | 4.4 |
| flhB | Flagellum-associated protein | 5.9 | 7.5 |
| flhA | Flagellum-associated protein | 6.0 | 4.9 |
| flhF | Flagellum-associated protein | 7.6 | 5.2 |
| ylxH | Unknown; similar to flagellar biosynthesis switch protein | 5.2 | 5.0 |
| cheC | Inhibition of CheR-mediated methylation of MCPs | 5.6 | 7.2 |
| sigD | RNA polymerase flagella, motility, chemotaxis, and autolysis sigma factor | 3.4 | 4.5 |
| ylxL | Unknown; similar to unknown proteins | 3.3 | 4.2 |
| yoeB | Unknown | 40.9 | 28.9 |
| yolA | Unknown | 7.5 | 6.1 |
| trpF | Phosphoribosyl anthranilate isomerase | 4.8 | 14.5 |
| trpC | Indole-3-glycerol phosphate synthase | 4.3 | 7.7 |
| sigX | RNA polymerase ECF-type sigma factor | 3.7 | 2.9 |
| tasA | Translocation-dependent antimicrobial spore component | 28.9 | 41.9 |
| sipW | Signal peptidase 1 | 5.5 | 5.8 |
| hemC | Porphobilinogen deaminase | 3.1 | 2.1 |
| yteR | Unknown; similar to unknown proteins | 3.2 | 1.9 |
| tlpB | Methyl-accepting chemotaxis protein | 3.4 | 2.6 |
| mcpA | Methyl-accepting chemotaxis protein | 11.3 | 8.9 |
| mcpB | Methyl-accepting chemotaxis protein | 3.6 | 3.0 |
| maeN | Na+/malate symporter | 3.2 | 1.6 |
| guaC | GMP reductase | 3.4 | 3.0 |
| yusP | Unknown; similar to multidrug efflux transporter | 5.2 | 9.9 |
| mrgA | Metalloregulation DNA-binding stress protein | 3.2 | 4.7 |
| yvfE | Unknown; similar to spore coat polysaccharide biosynthesis | 19.1 | 10.6 |
| yvfC | Unknown; similar to UDP-galactose phosphate transferase | 4.7 | 3.4 |
| yveS | Unknown; similar to unknown proteins | 3.5 | 2.5 |
| yveO | Unknown; similar to glycosyltransferase | 3.1 | 3.1 |
| yveK | Unknown; similar to capsular polysaccharide biosynthesis | 4.2 | 3.2 |
| yvzB | Unknown; similar to flagellin | 14.2 | 11.5 |
| yvjB | Unknown; similar to carboxy-terminal processing protease | 3.8 | 3.3 |
| fliT | Flagellar protein | 3.8 | 4.3 |
| fliS | Flagellar protein | 8.2 | 7.1 |
| fliD | Flagellar hook-associated protein 2 (HAP2) | 14.5 | 9.5 |
| hag | Flagellin protein | 21.2 | 10.3 |
| flgK | Flagellar hook-associated protein 1 (HAP1) | 3.1 | 3.5 |
| yvyG | Unknown; similar to flagellar protein | 3.7 | 4.2 |
| ywtD | Unknown; similar to murein hydrolase | 8.8 | 5.0 |
| flhP | Flagellar hook-basal body protein | 4.6 | 4.2 |
| flhO | Flagellar basal-body rod protein | 7.9 | 5.2 |
| thrZ | Threonyl-tRNA synthetase (minor) | 3.6 | 3.6 |
| ywbG | Unknown; similar to unknown proteins | 3.1 | 2.0 |
| epr | Minor extracellular serine protease | 6.9 | 4.8 |
| dltA | d-Alanyl-d-alanine carrier protein ligase | 5.8 | 6.6 |
| dltB | d-Alanine transfer from Dcp to undecaprenol phosphate | 3.8 | 5.8 |
| dltE | Involved in lipoteichoic acid biosynthesis | 5.4 | 5.7 |
| cydD | ABC transporter required for expression of cytochrome bd (ATP-binding protein) | 3.2 | 0.4 |
| cydC | ABC transporter required for expression of cytochrome bd (ATP-binding protein) | 3.1 | 0.6 |
| yxkC | Unknown | 8.4 | 4.7 |
| yxjH | Unknown; similar to unknown proteins from B. subtilis | 3.4 | 3.7 |
| yxjG | Unknown; similar to unknown proteins from B. subtilis | 3.1 | 3.8 |
| deaD | ATP-dependent RNA helicase | 3.7 | 3.2 |
| yxiM | Unknown; similar to rhamnogalacturonan acetylesterase | 3.6 | 3.1 |
| yxzC | Unknown | 5.6 | 4.2 |
| yxxG | Unknown | 9.4 | 9.9 |
| wapA | Cell wall-associated protein precursor | 7.7 | 6.9 |
| hutI | Imidazolone-5-propionate hydrolase | 3.0 | 1.8 |
| iolH | myo-Inositol catabolism | 3.1 | 3.3 |
| idh | myo-Inositol 2-dehydrogenase | 3.5 | 2.3 |
| yxaL | Unknown; similar to serine/threonine protein kinase | 3.3 | 0.9 |
| yxaD | Unknown; similar to transcriptional regulator (MarR family) | 4.4 | 4.3 |
Sorted according to the order in the B. subtilis genome. Members of the DegS/DegU regulon are boldfaced.
Functions are derived from the SubtiList database.
Ratio between the signal strength in cells propagated under the conditions indicated and the signal strength in cells continuously growing in synthetic medium with 5 μM FeCl3 and no extra NaCl added.
FIG. 1.
Influence of high salinity and iron limitation on the expression pattern of the opuBC, yocC, feuA, and wapA genes. The sigB mutant strain BLOB22 was continuously grown either in synthetic medium with 5 μM FeCl3 and no extra NaCl added (control condition), in a medium without any extra NaCl or FeCl3 (iron limitation), or in a high-salinity synthetic medium with 1.2 M NaCl and either 5 μM FeCl3 (high-salinity growth) or 250 μM FeCl3 (high-salinity growth with excess iron). Total RNA was prepared from cultures of each of the four different conditions as described in Materials and Methods. RNA samples were electrophoretically resolved on denaturing agarose gels and transferred to nylon membranes. RNA blots were then hybridized with specific, digoxigenin-labeled probes internal to the structural genes of feuA, opuBC, wapA, and yocC. Transcript sizes were estimated based on the relative positions of appropriate size markers.
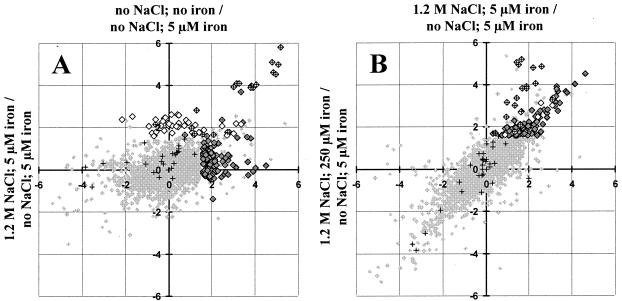
FIG. 2.
Influence of high salinity and iron limitation on the transcriptional pattern of the B. subtilis sigB mutant strain BLOB22. Log2 values of the ratios between the normalized signal strength for the individual conditions and the signal strength from control cultures continuously grown in synthetic medium with 5 μM FeCl3 and no extra NaCl added are plotted. (A) Iron limitation (no NaCl, no iron) (y axis) compared to high-salinity growth (1.2 M NaCl, 5 μM iron) (x axis). (B) High-salinity growth (1.2 M NaCl, 5 μM iron) (y axis) compared to high-salinity growth in the presence of excess iron (1.2 M NaCl, 250 μM iron) (x axis). The intensity ratios of all 4,107 genes represented on the Panorama B. subtilis DNA macroarrays from Sigma Genosys Ltd. are represented by small, light shaded diamonds in the background. These data are not filtered to remove spurious induction ratios; thus, light shaded background symbols displaying seemingly strong regulation are not statistically significant. The following groups of genes are emphasized with specific symbols: members of the Fur regulon as described by Fuangthong et al. (22) (crosses), iron limitation-induced genes (large open diamonds), and high-salinity-induced genes (large, dark shaded diamonds). Genes belonging to the Fur regulon and displaying significant induction by iron limitation or high salinity are represented by diamonds with superimposed crosses.
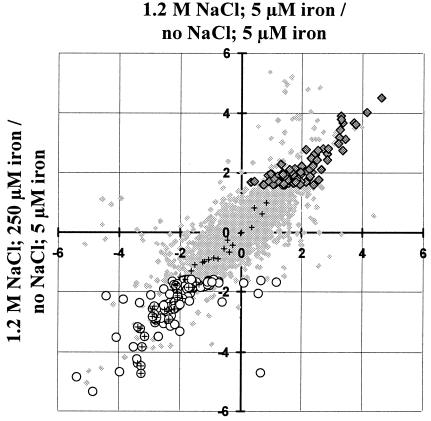
FIG. 3.
High-salinity-regulated transcription in B. subtilis. Shown are log2 values of the ratios between the normalized signal strength in cells grown at high salinity and the signal strength from control cultures continuously grown in synthetic medium with 5 μM FeCl3 and no extra NaCl added. High-salinity growth (1.2 M NaCl, 5 μM iron) (y axis) is compared to high-salinity growth in the presence of excess iron (1.2 M NaCl, 250 μM iron) (x axis). The intensity ratios of all 4,107 genes represented on the Panorama B. subtilis DNA macroarrays from Sigma Genosys Ltd. are represented by small, light shaded diamonds in the background. These data are not filtered to remove spurious induction ratios; thus, light shaded background symbols displaying seemingly strong regulation are not statistically significant. The following groups of genes are emphasized with specific symbols: genes whose products are predicted to be involved in chemotaxis or motility (crosses) (1), true high-salinity-induced genes (large, dark shaded diamonds) (see Table 1), and genes repressed by high salinity (open circles) (see Table 2). Genes with a potential function in chemotaxis or motility that display significant repression by high salinity are represented by circles with superimposed crosses.
High-salt-mediated iron limitation.
High-salinity-grown B. subtilis cultures experience severe iron limitation (28) leading to the induction of genes that are subjected to regulation by the Fur repressor (22, 28). This effect is particularly pronounced in strains derived from JH642 due to the presence of a mutation (sfp0) (23) that prevents or strongly reduces the phosphopantethylation of DhbB and DhbF. Consequently, such strains fail to effectively convert 2,3-dihydroxybenzoate (DHB) into the high-affinity iron siderophore bacillibactin via a modular peptide synthetase (42).
To distinguish which of the 123 genes expressed at higher levels in high-salinity-grown cells are truly induced by this stimulus from those whose induction is primarily a consequence of iron limitation, we used two additional cultivation conditions for our transcriptional profiling experiments. First, cells were propagated in MM with no added iron to identify the genes induced as a result of severe iron limitation. A group of 59 genes displayed at least threefold induction in cultures propagated in MM without added iron. Many of those genes were also induced in high-salinity-grown cultures (Fig. 2A), thus corroborating findings from the proteome study of high-salinity-induced iron limitation in B. subtilis (28). A representative example of this class of genes is feuA, and its expression pattern was verified by Northern blot analysis (Fig. 1). FeuA is the extracellular binding protein of the ABC transporter FeuABC, which is putatively involved in iron siderophore uptake.
A comparison of our results with those obtained in a genome-wide transcriptional profiling study of Fur-regulated B. subtilis genes by Fuangthong et al. (22) assigned 21 of the 59 iron limitation-induced genes to the Fur regulon (Fig. 2A; see also supplemental material, Table 3). Of these 21 genes, 15 displayed at least threefold induction in high-salinity-grown cells (see supplemental material, Table 3); many more members of the Fur regulon (22) were induced by high salt but did not pass the threefold induction criterion adopted in our study for salinity-mediated gene induction (Fig. 2A). The products of most of these genes are functionally associated with iron acquisition through either synthesis of bacillibactin or iron uptake (see supplemental material, Tables 3 and 4). Not all of the iron limitation-induced genes could be assigned to the Fur regulon, and this might reflect pleiotropic effects caused by the poor growth of severely iron limited B. subtilis cultures. These culture conditions trigger partial induction of competence, because we recognized 22 members of the competence-associated ComK regulon (4) among the genes that were induced by severe iron limitation (see supplemental material, Table 5).
To specifically investigate the contribution of iron availability to gene expression under high-salinity conditions, bacteria were also cultivated in MM with 1.2 M NaCl containing 250 μM FeCl3, since excess iron should reduce the expression of Fur-controlled genes (13). Furthermore, it has also been shown that such an excess of iron reduces the level of genes that are induced under high salinity but primarily respond to low iron availability in B. subtilis (28). Inclusion of a large amount of iron (250 μM) in the growth medium reduced the high-salinity-mediated induction for 21 genes.
By taking into account those genes that were induced by iron limitation and those genes whose high-salinity-induced expression was reduced by excess iron, we were able to show that 21 of the 123 salt-induced genes mentioned above (Fig. 2B; see also supplemental material, Table 4) actually represent genes that respond primarily to iron availability rather than to increases in salinity. Of those 21 genes, 15 were members of the Fur regulon (22). The remaining six genes (Fig. 2B), encoding proteins of unknown function, displayed a different induction pattern: they were induced by either high salinity or iron limitation, but their high-salinity-mediated induction could be reduced only marginally by excess iron.
Addition of excess iron (250 μM) to cells propagated in the presence of 1.2 M NaCl strongly increases the growth of the B. subtilis strain JH642 (28). This enhancement of growth is substantial, since it is similar to that caused by the potent osmoprotectant glycine betaine (28). Besides the repressive effect of high iron on high salinity gene induction, we also noted a group of 53 genes whose transcription was induced by excess iron. Many of their deduced gene products are connected with energy conservation (e.g., components of the respiratory chain) and heme-biosynthesis (see supplementary material, Table 6).
Taking all these data into account, we consider 102 B. subtilis genes to be truly salt induced (by at least a factor of 3) in cultures continuously propagated under high-salinity growth conditions (Fig. 3). These genes, their physiological functions as predicted in the current SubtiList database (http://genolist.pasteur.fr/SubtiList/) (45), and their induction ratios are listed in Table 1. The remaining 21 of the 123 originally discovered high-salinity-induced genes actually represent iron limitation-responsive genes and are not further considered here.
Physiological functions associated with salt-induced genes.
The uptake and synthesis of compatible solutes are important facets of the cellular defense of B. subtilis against high salinity (11). The proHJ genes, which are centrally involved in the osmoregulatory synthesis of the compatible solute proline (Brill and Bremer, unpublished), displayed the strongest induction ratios (22-fold for proH; 16-fold for proJ) of all 102 high-salinity-induced genes (Table 1). The proHJ locus is transcribed as an operon, and its disruption causes a strong growth defect under hypertonic conditions (Brill and Bremer, unpublished). Of the five compatible solute uptake systems known to operate in B. subtilis (11), we detected salt stress-mediated induction of the opuA and opuB operons, while the opuC operon and the opuD gene displayed some salt induction but did not pass the threefold induction criterion adopted in our transcriptional profiling study. Salt or stress induction of opuE has not been detected in this or previous transcriptional profiling studies (51) employing the commercially available Panorama B. subtilis DNA macroarrays from Sigma Genosys Ltd. However, it is known from detailed Northern blot and gene fusion analyses that the transcription of opuE is induced by high salinity (60). Transcription of the gbsAB genes, which are necessary for glycine betaine synthesis from the precursor choline (7), was not found to be inducible in cells grown at high salinity; this is fully consistent with the Northern blot analysis of this operon (G. Nau-Wagner and E. Bremer, unpublished data).
Of the 36 histidine kinases and 35 response regulators of two-component regulatory systems detected in B. subtilis (21, 34, 36), only degSU displayed a higher expression level in high- versus low-salt growth conditions (1.8-fold for degS; 3.9-fold for degU). The sensory kinase DegS and the response regulator DegU are involved in a complex network that mediates the regulation of transition state-specific processes by contributing to the regulation of degradative enzyme synthesis and the development of natural competence for DNA uptake (46). Furthermore, this two-component regulatory system has been implicated in sensing salt stress (37). The DegS/DegU regulon has recently been characterized in transcriptome studies that used either a hyperactive DegU allele (degU32Hy) (41) or artificial induction of the response regulator gene degU under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter on a multicopy plasmid (49). A comparison of the 102 high-salinity-induced genes of B. subtilis (Table 1) with the DegS/DegU regulon as defined by Mäder et al. (41) or Ogura et al. (49) assigned 20 of the high-salinity-induced genes to the DegS/DegU regulon (Table 1). Of these 20 genes, only degU (response regulator), frr (ribosome recycling factor), lipA (a probable lipoic acid synthetase), lspA (signal peptidase II), and yoaJ (a probable endoglucanase) encode products with functions assigned in databases.
The transcriptional control of a significant portion of hypertonicity-induced genes (20 out of 102) by DegS/DegU indicates that this two-component regulatory system plays an important role in regulating gene expression in cells that are continuously growing under high-salinity conditions. The physiological importance of the DegS/DegU system for the cellular adaptation of B. subtilis to high salinity became even more apparent when we analyzed the genes whose transcription is repressed by high salinity (see below).
Lopez et al. (39, 40) have provided evidence that the properties of the B. subtilis cell wall change when cells are cultured in a rich medium (Luria-Bertani medium) containing 1.5 M NaCl, and it has been reported that expression of the structural gene for the cell wall-associated protein WapA is strongly decreased in the presence of 0.7 M disodium succinate (17) (for Northern blot analysis of wapA, see Fig. 1). Among the 102 high-salinity-induced genes, we found 2 (yabE and yocH) that encode putative cell wall binding proteins and 1 (yqiL) that encodes a putative N-acetylmuramoyl-l-alanine amidase, likely to be involved in peptidoglycan biosynthesis (Table 1). Furthermore, expression of the genes for four transport proteins (yfjQ, ypqE, yydI, and sunT) displayed induction by hypertonicity (Table 1). Interestingly, the transcription of lspA, the structural gene for the sole prolipoprotein signal peptidase (type II) of B. subtilis (61), was increased 3.4-fold (Table 1), while that of the type-I signal peptidase SipW, which has only a minor role in protein secretion (61), was 5.5-fold repressed (Table 2).
The genes induced by high salinity also encode additional regulatory proteins (Table 1). These include RapG, a member of the response regulator aspartate phosphatase gene family, and the phosphatase regulatory peptide PhrG (50), but the physiological functions of these two proteins have not yet been elucidated. We also found substantial induction (sevenfold) of the yhjM gene, which encodes a transcriptional regulator of the LacI family, but it is currently unknown which genes are under the control of this regulatory protein. Finally, mntR, which encodes a transcriptional regulator of the manganese uptake systems MntABCD and MntH (55), was found to be induced (fourfold) in cells grown at high versus low salinity. MntR had a dramatic effect in B. subtilis cells grown with 1.2 M NaCl and an excess of iron (250 μM), where expression of the mntABCD operon and the mntH gene were more than 20- and 10-fold repressed, respectively.
For 53 of the 102 B. subtilis genes induced at high salinity (Table 1), database searches revealed no significant homology with proteins of known function. Such a large portion of proteins with thus far undefined functions indicates that a significant part of the cellular and physiological adaptation reactions of B. subtilis to high-salinity growth conditions is still unexplored.
High-salt-mediated gene repression.
The presence of 1.2 M salt in the growth medium not only induced gene expression but also caused at least threefold repression of 101 B. subtilis genes (Fig. 3 and Table 2). Strikingly, 38 of these 101 salt-repressed genes are predicted to be involved in chemotaxis and cell motility (Table 2 and Fig. 3). A first indication of the repressing effect of high salinity on cell motility was obtained in a recent proteome analysis, where it was found that the cellular level of Hag, the structural protein of the flagellum, was drastically reduced in cells cultured in the presence of 1.2 M NaCl (28). In excellent agreement with the proteome study, transcription of the hag gene was 21-fold reduced in cells grown at high salinity (Table 2). Extending the proteome study, we now show that salt repression is not confined to the hag gene: 36 of the 56 known chemotaxis and motility genes (1) are repressed by this environmental insult as well (Table 2). High-salinity-mediated repression of chemotaxis and motility genes was not significantly influenced by the iron concentration in the growth medium (Table 2).
In B. subtilis, motility and chemotaxis genes have been grouped into two classes that constitute a hierarchy of gene expression (1). All genes belonging to the first class are localized in a single 26-kb fla/che operon, expression of which is critically dependent on a SigA-dependent promoter located upstream of the first gene of the operon. Genes constituting the second class are scattered in several operons on the B. subtilis chromosome and require the alternative sigma factor SigD for expression. High-salinity-mediated repression targets both classes of chemotaxis and motility genes. The transcriptional profiling experiments offer several hints why such a large number of B. subtilis genes associated with chemotaxis and cell motility are so strongly repressed under high-saline environmental conditions. The GTP-binding protein CodY, which functions as a nutrition-responsive repressor in B. subtilis (56), appears to repress flagellin gene expression in response to the availability of amino acids (44). CodY has been shown to bind specifically to DNA fragments containing the promoters for both the fla/che operon and the hag gene (1, 44), and we found that expression of codY increased by a factor of approximately 2 in B. subtilis cells propagated at high salinity (see supplemental material). CodY-mediated reduction of fla/che operon expression might also partially account for the threefold reduction in sigD transcription observed at high versus low salinity (Table 2). An additional level of control of chemotaxis and motility gene expression might be exerted by the two-component regulatory system DegS/DegU, since both SigD activity and sigD expression are subject to control by the DegS/DegU system (46, 48, 62). Inspection of the list of 101 high-salinity-repressed genes (Table 2) revealed that 50 of them actually belong to the currently defined DegS/DegU regulon (41, 49) (Table 2), further supporting a physiologically relevant function of this two-component regulatory system in cellular adaptation to high salinity.
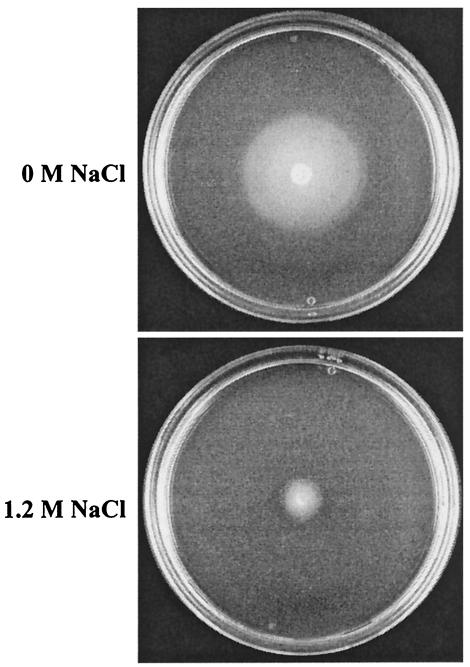
Consequently, the combined effects of the DegS/DegU-mediated decrease in cellular gene levels, the activity of the sigma factor SigD, and the increased synthesis of the repressor CodY might together accomplish the drastic reduction in chemotaxis and cell motility gene expression observed in B. subtilis under hypertonic growth conditions. Since such a large portion (38 of 101) of the salt-repressed genes is functionally associated with chemotaxis and cell motility, we experimentally tested the effect of high salinity on cell motility. The motility of B. subtilis was strongly impaired when cells were grown on swarming plates containing 1.2 M NaCl (Fig. 4), corroborating the findings from the proteome (28) and transcriptional profiling (Fig. 3 and Table 2) studies.
FIG. 4.
Influence of high salinity on motility of B. subtilis. The B. subtilis wild-type strain 168 was inoculated onto swarming plates without or with 1.2 M NaCl. After growth at 37°C overnight, plates were inspected for swarming capability.
The high-saline-repressed genes also included 11 genes (wapA, dltABE, lytF, ywtD, yfiQ, yveKNO, and yvfC) that encode either proteins associated with the cell wall or enzymes involved in peptidoglycan, lipoteichoic acid, or capsular polysaccharide synthesis, indicating that major changes in the cell envelope take place when B. subtilis is cultivated under hypertonic conditions. We also noted significant repression at high salinity of the genes encoding the alternative transcription factor SigX, the transition state regulator Abh, and the MarR-type family transcriptional regulator YxaD (Table 2). As with the hypertonicity-induced genes, a significant portion (18 out of 101) of the high-salinity-repressed genes encode proteins whose functions are thus far undefined (Table 2).
Cellular response to salt shock.
In natural settings, B. subtilis might also encounter sudden changes in salinity. We therefore performed time-resolved genome-wide transcriptional profiling of cells that were grown in a minimal medium whose salinity was suddenly increased by addition of 0.4 M NaCl. This rather moderate increase in salinity was specifically chosen to avoid the strong growth retardation that is associated with a severe salt shock. We monitored the transcriptional pattern of the B. subtilis strain BLOB22 (sigB) up to 6 h after the salt shock, when cells started to enter the stationary phase (Fig. 5A).
FIG. 5.
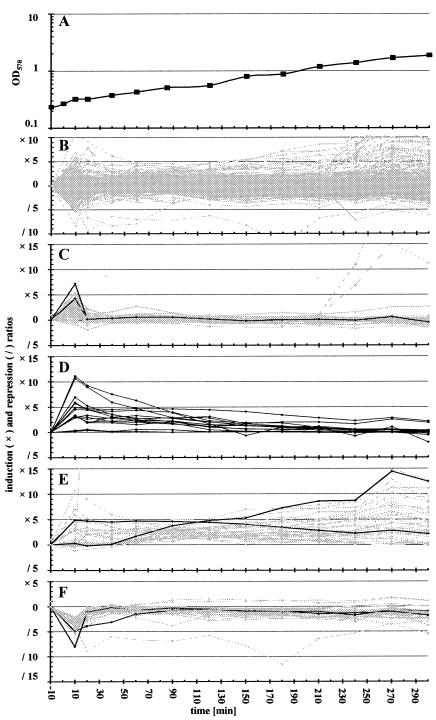
Influence of salt shock on the transcriptional profile of B. subtilis. The sigB mutant strain BLOB22 was grown in synthetic medium (SMM) with 5 μM FeCl3. During exponential growth cells were exposed to salt shock (final NaCl concentration, 0.4 M), and total RNA was prepared from samples collected 10 min prior to and at various time points after the imposition of salt stress. Radioactively labeled cDNA prepared with these RNA samples as templates was hybridized to Panorama B. subtilis DNA macroarrays from Sigma Genosys Ltd., and signal intensities were determined as described in Materials and Methods. The figure displays the induction and repression ratios between the normalized signal strength in salt-shocked cells and the signal strength from control cultures 10 min prior to the addition of NaCl. (A) Bacterial growth in SMM. (B) Expression pattern of all B. subtilis genes displaying significant expression on the Panorama DNA macroarrays under the conditions investigated. (C) Influence of salt shock on the expression of the SigW regulon (15, 51, 69). Shaded lines, members of the SigW regulon; solid lines, transcription patterns of yuaG and yvlB. (D) Induction of genes and operons involved in the uptake (opuA, opuB, opuC, opuD, and opuE) and synthesis (proHJ) of compatible solutes. (E) Shaded lines, genes with prolonged induction; solid lines, two representative examples (comEA and proH). (F) Genes repressed by salt shock. Shaded lines, the 51 genes showing significant repression immediately after addition of 0.4 M NaCl to cultures. Solid lines, two genes (argJ and lysC) encoding products involved in amino acid biosynthesis.
Most of the B. subtilis genes did not display any significant variation in their expression level following the imposition of salt stress. Inspection of the response pattern of genes that showed a significant change in expression revealed two distinct classes (Fig. 5B): (i) genes that responded rapidly with an increase or decrease in expression and then returned within a very short time (20 min) to their preshock expression level and (ii) genes whose expression responded more slowly to the salt shock but did not rapidly return to the preshock level.
Even at the first time point (10 min) investigated after the imposition of salt stress, the transcription of 75 genes was increased (see supplemental material, Table 7), and that of 51 genes was decreased (see supplemental material, Table 8), by a factor of at least 3. Of the 75 salt shock-induced genes, 31 could be assigned to the SigW regulon (15, 51, 69). Many of the remaining members of the SigW regulon also responded to salt shock but did not pass the threefold cutoff level adopted in this study. Induction of the whole SigW regulon was almost completely shut off 20 min after salt shock (Fig. 5C). Among the rapidly induced genes were those encoding transporters (OpuA, OpuB, and OpuC) for osmoprotectants and enzymes mediating the synthesis of the compatible solute proline (ProHJ). The response of these genes to salt shock differed from the pattern described above in that their expression remained elevated throughout the time course of the experiment (Fig. 5D and E). This continued elevated expression is consistent with the physiological function of these genes, since growing salt-shocked cells need to replenish their compatible solute pools (11). The list of genes discovered to be induced immediately following salt shock is also in good agreement with the data from a recent transcriptional profiling study reported by Petersohn et al. (51). Of the 64 genes assigned by those authors to the group of salt-specific but SigB-independent stress genes, 45 were confirmed in this study.
A distinct group of 41 genes exhibited longer-lasting induction following the imposition of moderate salt stress (Fig. 5E). The products of these genes are either involved in the uptake or synthesis of compatible solutes (see above) or largely represent proteins involved in the development of competence (see supplemental material, Table 9) (19). Induction of these genes was somewhat unexpected. Although induction is observed while the B. subtilis cells are still growing, we cannot exclude the possibility that it is related to higher cell density and/or the upcoming stationary phase (Fig. 5A).
The repression of 51 genes immediately after the addition of 0.4 M salt (Fig. 5F) most likely reflects a reduced need for intermediates in metabolism caused by the reduction in growth. This notion is supported by the observation that 29 of these genes encode enzymes involved in amino acid or vitamin biosynthesis and an additional 4 participate in ATP generation (see supplemental material, Table 8). Repression of the majority of the 51 genes is rapidly relieved, presumably because the moderate shock exerted by the addition of 0.4 M NaCl caused only short growth retardation (Fig. 5A). Due to the moderate strength of the salt shock, we did not observe genes displaying a lasting significant repression of their transcription.
Conclusions.
In natural settings, high salinity is an important abiotic factor that determines the growth and survival of B. subtilis. As a bacterium that inhabits the upper layers of soil, B. subtilis is exposed to sustained drought-mediated increases in salt concentration and is also washed off into the sea. Consequently, bacilli are frequently found in coastal waters and estuarine saline sediments (8, 54). While the moderate salt shock employed in this study provoked only a short, transient change in the expression pattern, continuous propagation of the cells in a medium with 1.2 M NaCl had profound effects on their transcriptional profile. Approximately 5% of the 4,107 protein-encoding genes differed significantly in their expression (at least threefold) between high- and low-salinity-grown cells. Fifty percent of the 102 hypertonicity-induced genes have not yet been assigned biochemical functions, and determining their role in the adaptation to high-salinity surroundings remains a challenge for future studies. The proportion of genes with unassigned functions is much smaller within the group of 101 hypertonicity-repressed genes, since genes associated with chemotaxis and motility are prevalent within this group. The observation that cell motility is severely impaired at high salinity is certainly interesting; however, its physiological relevance in natural settings remains unexplained at present.
Only a small portion of the genes that are immediately induced or repressed by salt shock also displayed significant differences in cells continuously cultivated at low or high salinity. This observation indicates that salt shock and continuous growth at high salinity require quite different adaptation reactions by B. subtilis cells.
The DegS/DegU two-component regulatory system has previously been implicated in the genetic control of salt-mediated induction (sacB) and repression (aprE and wapA) of gene expression (17, 37) and in the development of osmostress resistance (58, 59). Substantial fractions of the groups of genes induced (20 of 102 genes [Table 1]) and repressed (50 of 101 genes [Table 2]) in cells continuously grown under high-salinity conditions seem to belong to the DegS/DegU regulon (41, 49), suggesting an important physiological function for the DegS sensory kinase and the DegU response regulator in sensing salt stress and transmitting this environmental signal to the transcriptional apparatus of the cell. The cytosolic location of DegS (47) implies that this sensor kinase perceives a intracellular signal elicited by growth of the cells in high-salinity media. Future studies must unravel the molecular mechanism of this high-salt perception by DegS and the physiological role of DegS/DegU-controlled genes in the adaptation of B. subtilis cells to high salinity.
Acknowledgments
We thank V. Koogle for help in editing the manuscript.
Financial support for this study was provided by the Deutsche Forschungsgemeinschaft through the SFB-395, the Max-Planck-Institute for terrestrial Microbiology (Marburg, Germany), the European Union (contract IAC4-CT-2000-30041), the Bundesministerium für Bildung und Forschung through the “Genomnetzwerk Göttingen,” and the Fonds der Chemischen Industrie (to E.B. and U.V.).
REFERENCES
- 1.Aizawa, S. I., I. B. Zhulin, L. M. Marquez-Magana, and G. W. Ordal. 2002. Chemotaxis and motility, p. 437-452. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. From genes to cells. ASM Press, Washington, D.C.
- 2.Alice, A. F., and C. Sanchez-Rivas. 1997. DNA supercoiling and osmoresistance in Bacillus subtilis 168. Curr. Microbiol. 35:309-315. [DOI] [PubMed] [Google Scholar]
- 3.Belitsky, B. R., J. Brill, E. Bremer, and A. L. Sonenshein. 2001. Multiple genes for the last step of proline biosynthesis in Bacillus subtilis. J. Bacteriol. 183:4389-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. Cui, A. Sloma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. [DOI] [PubMed] [Google Scholar]
- 5.Bernhardt, J., U. Völker, A. Völker, H. Antelmann, R. Schmid, H. Mach, and M. Hecker. 1997. Specific and general stress proteins in Bacillus subtilis—a two-dimensional protein electrophoresis study. Microbiology 143:999-1017. [DOI] [PubMed] [Google Scholar]
- 6.Boch, J., B. Kempf, and E. Bremer. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 176:5364-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boch, J., B. Kempf, R. Schmid, and E. Bremer. 1996. Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: characterization of the gbsAB genes. J. Bacteriol. 178:5121-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonde, G. J. 1981. Bacillus from marine habitats, allocation to phena established by numerical techniques, p. 181-215. In R. C. W. Berkeley and M. Goodfellow (ed.), The aerobic endospore-forming bacteria, classification and identification. Academic Press, London, England.
- 9.Booth, I. R., and P. Louis. 1999. Managing hypoosmotic stress: aquaporins and mechanosensitive channels in Escherichia coli. Curr. Opin. Microbiol. 2:166-169. [DOI] [PubMed] [Google Scholar]
- 10.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bremer, E. 2002. Adaptation to changing osmolarity, p. 385-391. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. From genes to cells. ASM Press, Washington, D.C.
- 12.Bremer, E., and R. Krämer. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes in bacteria, p. 79-97. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 13.Bsat, N., and J. D. Helmann. 1999. Interaction of Bacillus subtilis Fur (ferric uptake repressor) with the dhb operator in vitro and in vivo. J. Bacteriol. 181:4299-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Büttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mader, C. Eymann, H. Antelmann, A. Völker, U. Völker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908-2935. [DOI] [PubMed] [Google Scholar]
- 15.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. [DOI] [PubMed] [Google Scholar]
- 16.Chen, L., L. P. James, and J. D. Helmann. 1993. Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions. J. Bacteriol. 175:5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dartois, V., M. Debarbouille, F. Kunst, and G. Rapoport. 1998. Characterization of a novel member of the DegS-DegU regulon affected by salt stress in Bacillus subtilis. J. Bacteriol. 180:1855-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deuerling, E., B. Paeslack, and W. Schumann. 1995. The ftsH gene of Bacillus subtilis is transiently induced after osmotic and temperature upshift. J. Bacteriol. 177:4105-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubnau, D., and K. Turgay. 2000. Regulation of competence in Bacillus subtilis and its relation to stress, p. 249-260. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 20.Dulaney, E. L., D. D. Dulaney, and E. L. Rickes. 1968. Factors in yeast extract which relieve growth inhibition of bacteria in defined medium of high osmolarity. Dev. Ind. Microbiol. 163:260-269. [Google Scholar]
- 21.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuangthong, M., A. F. Herbig, N. Bsat, and J. D. Helmann. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grossman, T. H., M. Tuckman, S. Ellestad, and M. S. Osburne. 1993. Isolation and characterization of Bacillus subtilis genes involved in siderophore biosynthesis: relationship between B. subtilis sfpo and Escherichia coli entD genes. J. Bacteriol. 175:6203-6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harwood, C. R., and A. R. Archibald. 1990. Growth, maintenance and general techniques, p. 1-26. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom.
- 25.Hauser, N. C., M. Vingron, M. Scheideler, B. Krems, K. Hellmuth, K. D. Entian, and J. D. Hoheisel. 1998. Transcriptional profiling on all open reading frames of Saccharomyces cerevisiae. Yeast 14:1209-1221. [DOI] [PubMed] [Google Scholar]
- 26.Hecker, M., and U. Völker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 27.Helmann, J. D., M. F. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann, T., A. Schütz, M. Brosius, A. Völker, U. Völker, and E. Bremer. 2002. High-salinity-induced iron limitation in Bacillus subtilis. J. Bacteriol. 184:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holtmann, G., E. P. Bakker, N. Uozumi, and E. Bremer. 2003. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in the adaptation to hypertonicity. J. Bacteriol. 185:1289-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horsburgh, M. J., and A. Moir. 1999. σM, an ECF RNA polymerase sigma factor of Bacillus subtilis 168, is essential for growth and survival in high concentrations of salt. Mol. Microbiol. 32:41-50. [DOI] [PubMed] [Google Scholar]
- 31.Kappes, R. M., B. Kempf, and E. Bremer. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kappes, R. M., B. Kempf, S. Kneip, J. Boch, J. Gade, J. Meier-Wagner, and E. Bremer. 1999. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32:203-216. [DOI] [PubMed] [Google Scholar]
- 33.Kempf, B., and E. Bremer. 1995. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J. Biol. Chem. 270:16701-16713. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi, K., M. Ogura, H. Yamaguchi, K. Yoshida, N. Ogasawara, T. Tanaka, and Y. Fujita. 2001. Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J. Bacteriol. 183:7365-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krispin, O., and R. Allmansberger. 1995. Changes in DNA supertwist as a response of Bacillus subtilis towards different kinds of stress. FEMS Microbiol. Lett. 134:129-135. [DOI] [PubMed] [Google Scholar]
- 36.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, and A. Danchin. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 37.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, Y., and J. N. Hansen. 1995. Characterization of a chimeric proU operon in a subtilin-producing mutant of Bacillus subtilis 168. J. Bacteriol. 177:6874-6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez, C. S., H. Heras, H. Garda, S. Ruzal, C. Sanchez-Rivas, and E. Rivas. 2000. Biochemical and biophysical studies of Bacillus subtilis envelopes under hyperosmotic stress. Int. J. Food Microbiol. 55:137-142. [DOI] [PubMed] [Google Scholar]
- 40.Lopez, C. S., H. Heras, S. M. Ruzal, C. Sanchezrivas, and E. A. Rivas. 1998. Variations of the envelope composition of Bacillus subtilis during growth in hyperosmotic medium. Curr. Microbiol. 36:55-61. [DOI] [PubMed] [Google Scholar]
- 41.Mäder, U., H. Antelmann, T. Buder, M. K. Dahl, M. Hecker, and G. Homuth. 2002. Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol. Genet. Genomics 268:455-467. [DOI] [PubMed] [Google Scholar]
- 42.May, J. J., T. M. Wendrich, and M. A. Marahiel. 2001. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276:7209-7217. [DOI] [PubMed] [Google Scholar]
- 43.Miller, K. J., and J. M. Wood. 1996. Osmoadaptation by rhizosphere bacteria. Annu. Rev. Microbiol. 50:101-136. [DOI] [PubMed] [Google Scholar]
- 44.Mirel, D. B., W. F. Estacio, M. Mathieu, E. Olmsted, J. Ramirez, and L. M. Marquez-Magana. 2000. Environmental regulation of Bacillus subtilis σD-dependent gene expression. J. Bacteriol. 182:3055-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moszer, I., L. M. Jones, S. Moreira, C. Fabry, and A. Danchin. 2002. SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res. 30:62-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Msadek, T., F. Kunst, and G. Rapoport. 1995. A signal transduction network in Bacillus subtilis includes the DegS/DegU and ComP/ComA two-component systems, p. 447-471. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 47.Msadek, T., F. Kunst, and G. Rapoport. 1993. Two-component regulatory systems, p. 729-745. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, D.C.
- 48.Ogura, M., and T. Tanaka. 1996. Transcription of Bacillus subtilis degR is σD dependent and suppressed by multicopy proB through σD. J. Bacteriol. 178:216-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogura, M., H. Yamaguchi, K. Yoshida, Y. Fujita, and T. Tanaka. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29:3804-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perego, M., and J. A. Hoch. 2002. Two-component system, phosphorelays, and regulation of their activities by phosphatases, p. 473-481. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. From genes to cells. ASM Press, Washington, D.C.
- 51.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price, C. W. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria, p. 179-197. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 53.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 54.Priest, F. G. 1993. Systematics and ecology of bacilli, p. 3-16. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, D.C.
- 55.Que, Q., and J. D. Helmann. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35:1454-1468. [DOI] [PubMed] [Google Scholar]
- 56.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruzal, S. M., C. Lopez, E. Rivas, and C. Sanchez-Rivas. 1998. Osmotic strength blocks sporulation at stage II by impeding activation of early sigma factors in Bacillus subtilis. Curr. Microbiol. 36:75-79. [DOI] [PubMed] [Google Scholar]
- 58.Ruzal, S. M., and C. Sanchez-Rivas. 1998. In Bacillus subtilis DegU-P is a positive regulator of the osmotic response. Curr. Microbiol. 37:368-372. [DOI] [PubMed] [Google Scholar]
- 59.Ruzal, S. M., and C. Sanchez-Rivas. 1994. Physiological and genetic characterization of the osmotic stress response in Bacillus subtilis. Can. J. Microbiol. 40:140-144. [DOI] [PubMed] [Google Scholar]
- 60.Spiegelhalter, F., and E. Bremer. 1998. Osmoregulation of the opuE proline transport gene from Bacillus subtilis—contributions of the σA- and σB-dependent stress-responsive promoters. Mol. Microbiol. 29:285-296. [DOI] [PubMed] [Google Scholar]
- 61.Tjalsma, H., A. Bolhuis, J. D. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tokunaga, T., M. H. Rashid, A. Kuroda, and J. Sekiguchi. 1994. Effect of degS-degU mutations on the expression of sigD, encoding an alternative sigma factor, and autolysin operon of Bacillus subtilis. J. Bacteriol. 176:5177-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Völker, U., S. Engelmann, B. Maul, S. Riethdorf, A. Völker, R. Schmid, H. Mach, and M. Hecker. 1994. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 140:741-752. [DOI] [PubMed] [Google Scholar]
- 64.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Völker, U., A. Völker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.von Blohn, C., B. Kempf, R. M. Kappes, and E. Bremer. 1997. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25:175-187. [DOI] [PubMed] [Google Scholar]
- 67.Whatmore, A. M., J. A. Chudek, and R. H. Reed. 1990. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136:2527-2535. [DOI] [PubMed] [Google Scholar]
- 68.Whatmore, A. M., and R. H. Reed. 1990. Determination of turgor pressure in Bacillus subtilis: a possible role for K+ in turgor regulation. J. Gen. Microbiol. 136:2521-2526. [DOI] [PubMed] [Google Scholar]
- 69.Wiegert, T., G. Homuth, S. Versteeg, and W. Schumann. 2001. Alkaline shock induces the Bacillus subtilis σW regulon. Mol. Microbiol. 41:59-71. [DOI] [PubMed] [Google Scholar]
- 70.Wipat, A., and C. R. Harwood. 1999. The Bacillus subtilis genome sequence: the molecular blueprint of a soil bacterium. FEMS Microbiol. Ecol. 28:1-9. [Google Scholar]